Abstract
The profile of T-cell receptor beta-chain variable (TRBV) genes usually skews in subjects with virus infection or cancer. The gene melting spectral pattern (GMSP) can be used to determine the profile of the TRBV gene family. To explore the portrait of the TRBV family in peripheral blood lymphocytes from subjects who have recovered from acute hepatitis B virus infection (AHI), peripheral blood mononuclear cells (PBMCs) were separated and further sorted into CD4+ and CD8+ T-cell subsets. The molecular features of the TRBV complementary determining region 3 (CDR3) motifs were determined using GMSP analysis. When a GMSP profile showed a single peak, the monoclonally expanded TRBV gene was cloned and sequenced. Skewed expansions of multiple TRBV genes were observed among the CD4+ and CD8+ T-cell subsets and the PBMCs. The frequency of monoclonally expanded TRBV genes in the CD8+ T-cell subset was significantly higher than that of the CD4+ T-cell subset and the PBMCs. Compared to other members of the TRBV gene family, TRBV11, BV15 and BV20 were predominantly expressed in the repertoire of peripheral blood lymphocytes in recovered AHI subjects. The relatively conserved amino acid motifs of TRBV5.1 and BV20 CDR3 were also detected in the CD4+ and CD8+ T-cell subsets. These results demonstrate the presence of multiple biased TRBV families in recovered AHI subjects. TRBV11, BV15 and BV20, especially from the CD8+ T-cell subset, may be relevant to the pathogenesis of subjects with AHI. The preferentially selected TRBV5.1 and BV20 with the relatively conserved CDR3 motif may be potential targets for personalized treatments of chronic HBV infection.
Keywords: acute hepatitis B virus infection, gene melting spectral pattern, gene therapy, molecular profile, T-cell receptor beta-chain variable
Introduction
Hepatitis B virus (HBV) infection remains a global public health problem. According to a 2010 report from the World Health Organization, approximately two billion people were infected with HBV worldwide, of which 350 million had chronic HBV infection. Of those infected, approximately one million died each year from liver disease related to HBV infection.1 Chronic HBV infection has been positively correlated with morbidity from a spectrum of liver diseases, including chronic hepatitis B, liver cirrhosis, liver failure and hepatocellular carcinoma.2
The age of subjects is the most important factor affecting the outcome of HBV infections. Ninety percent of subjects infected with HBV perinatally or 25%–30% of infants will develop chronic HBV infection. However, only 5%–10% of individuals infected with HBV after 5 years of age will develop chronic HBV infection, and 90% of infected people will recover after presenting with acute HBV infection without any clinical symptoms.3,4 In the peripheral blood of subjects who recovered from AHI (recovered AHI subjects), polyclonal and polyspecific cytotoxic T lymphocytes and T helper cells develop against antigenic peptides of HBV, and protection with hepatitis B surface antibodies (HBsAbs) subsequently emerges.
In general, clonal or oligoclonal T cells in the peripheral circulation of patients with viral infection appear to be antigen-specific T cells. The composition and function of specific T cells can be determined by the T-cell receptor (TCR) profile in their membranes.5 There are several reports that address the immunological characteristics of recovered acute hepatitis B virus infection (AHI) subjects.6,7 However, the molecular characteristics of TCR gene families in the peripheral blood from recovered AHI subjects have not been well characterized. In previous studies, we have developed and used gene melting spectral pattern (GMSP) to monitor T-cell receptor beta-chain variable (TRBV) gene distribution and monoclonality.8,9 In the current study, this technique was used to detect the distribution of TRBV complementary determining region 3 (CDR3) gene families expressed in peripheral blood mononuclear cells (PBMCs) and T-cell subsets separated and isolated from recovered AHI subjects. The TRBV families were sequenced and analyzed when the shapes of their GMGP profiles presented single peaks. This study will help clarify the molecular portrait of TRBVs in peripheral blood from recovered AHI subjects and the role of cellular immunity in the pathogenesis of AHI as well as assist in the diagnosis and provision of individualized treatment for chronic HBV infection.
Materials and methods
Subjects
Between February and June 2012, 38 subjects who had recovered from AHI were enrolled in our study. The subjects were selected in the health examination center of the First Affiliated Hospital, College of Medicine, Zhejiang University. The diagnoses for the recovered AHI subjects were categorized based on normal biochemical markers of liver function: no previous history of hepatitis, positive for the HBsAb and/or hepatitis B core antibody without any clinical symptoms, the absence of the hepatitis B surface antigen (HBsAg) and HBV DNA levels below the detection limit. Moreover, the chosen subjects had not previously been vaccinated with the hepatitis B vaccine. Our diagnostic criteria for recovered AHI subjects complied with the proposal of the prevention and treatment for viral hepatitis (2000, Xi'an),10 and the criteria for AHI diagnosis have also been described in other reports.11
Co-infection with hepatitis A, C, D or E, the Epstein–Barr virus or overt cytomegalovirus was ruled out according to negative serologic test results. Confounding etiologies of liver disease, human immunodeficiency virus infection, and other infectious diseases were not found in any subject. Patients diagnosed with autoimmune diseases, blood diseases or other malignancies were also excluded. The 27 healthy controls were negative for all HBV serological markers and displayed no clinical or laboratory evidence of other infectious diseases or immunological disorders. Additional characteristics of enrolled subjects at the time of the study are shown in Table 1.
Table 1. Demographic and clinical characteristics of the enrolled subjectsa.
| Characteristics | Recovered AHI | HDs |
|---|---|---|
| No. subject | 38 | 27 |
| Gender (male/female) | 23/15 | 19/8 |
| Mean age (years) | 30.6±7.8 | 31.9±6.9 |
| ALT level (IU/l) | 26.61±6.39 | <40 |
| Total bilirubin level (µmol/l) | 13.32±4.07 | <21 |
| HBV DNA | Negative | Negative |
| HBeAb-positive | 10 (26.3%) | Negative |
Abbreviations: AHI, acute hepatitis B virus infection; ALT, alanine aminotransferase; HBeAb, hepatitis B e antibody; HBV, hepatitis B virus; HDs, healthy donors; Negative, below the detection limit; Recovered AHI, subjects who have recovered from acute hepatitis B infection.
All subjects are of Chinese Han ethnicity.
Data are expressed as the mean±s.d. (standard deviation), unless otherwise indicated.
Normal values: ALT ≤40 IU/l; total bilirubin ≤21 µmol/l.
The study was approved by the local research ethics committee (Medical Ethics Committee of the First Affiliated Hospital, Zhejiang University, Hangzhou, China) in accordance with the 2008 Declaration of Helsinki, and all subjects provided written informed consent.
Biochemical, serological and virological assays
Biochemical indicators of liver function were determined using an automatic biochemical analyzer (HITACHI 7600, Japan) in the clinical laboratory center of our unit. Serum samples were tested for HBsAg, HBsAb, hepatitis B e antigen and antibodies to hepatitis B e antigen and HBcAg using enzyme immunoassays (Abbott Laboratories, Chicago, IL, USA) according to the manufacturer's instructions. Serum HBV DNA was quantitatively detected using real-time polymerase chain reaction (PCR) with commercial kits as described previously.12 The minimal detection standard for PCR contained 500 copies/ml. HBV genotypes were determined with sequence detection via PCR. All products were directly sequenced with an HBV Genotype Real Time PCR Kit (ZJ Bio-Tech, Shanghai, China) and run on MegaBACE 500 according to the manufacturer's instructions, as has been previously described in detail.12
PBMC separation and T-cell subset sorting
PBMCs were prepared from whole blood treated with 10 ml of fresh EDTA–K2 anticoagulate by a Ficoll-Paque density gradient separation (CEDARLANE, Burlington, CANADA). Because we could not recover enough PBMCs useful for sorting T cells into subsets, PBMC samples were taken from 30 of 38 recovered AHI subjects and 25 of 27 healthy controls for further sorting. The positive selection for CD4+ and CD8+ T cells was performed using anti-CD4 and anti-CD8 monoclonal antibodies with magnetic activated cell sorting according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). Each T-cell subset purity was shown to be greater than 90% (data not shown), as determined with flow cytometry using mouse anti-human antibodies CD4-FITC and CD8-PE (BD Biosciences, San Jose, CA, USA).
RNA extraction and cDNA synthesis
Total RNA was extracted from PBMCs and CD4+ and CD8+ T-cell populations using an SV Total RNA Isolation System (Promega, Madison, WI, USA) according to the manufacturer's instructions. RNA was quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA), and RNA integrity was confirmed by the observation of 18S and 28S rRNA bands in 2% agarose gel electrophoresis. cDNA was synthesized immediately from RNA using Oligo dT18 primers and a RevertAid First Strand cDNA Synthesis Kit (MBI, Fermentas, EU) according to the manufacturer's instructions.
GMSP analysis of the TRBV gene family
Real-time PCR reactions were performed with 0.5 µl of cDNA sample in an amplification mixture of 25 µl for each TRBV gene family. Following a PCR-product melting curve analysis, the peak shape pattern of the melting curve for the 24 TRBV gene families was determined by plotting the negative of the first derivative of the reduction in the fluorescence signal (−dF/dT) as a function of temperature (Tm) and the GMSPs of the TRBV families were determined, as has been described in more detail previously.8 Gene segments from TCR beta chain constant 1 (TRBC1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were added to each reaction as internal reference controls for monoclonal expansion, and a PCR reaction without template was used as a blank control. The primers were designed for the TRBV family as previously reported,13 and the primers used in this study are provided in Supplementary Table 1).
Skewed TRBV gene family identification
Skewed TRBV gene families were identified by GMSP profile using Opticon Monitor 3.0 software attached to a MJ Opticon 2 DNA engine (Bio-Rad, Hercules, CA, USA) and then divided into two categories: (i) ‘oligoclonal expansion’, which appeared as a main peak associated with other small peaks and a small peak with a height less than fifth-eighths the height of the main peak, and (ii) the ‘monoclonal’ category, which appeared as one main peak and, in some instances, a short small peak with a height less than three-eighths the height of the main peak. If the height of any secondary peak was more than fifth-eighths the height of the main peak, the TRBV gene family was categorized as a multiclonal expansion (not skewed), which is a development of our previously published reports.14
Relative quantification of each TRBV gene family
To quantify the relative expression of each TRBV gene in recovered AHI subjects,
the relative expression of a single TRBV family in PBMCs was evaluated. The
expression of each TRBV gene was calculated based on the signal strength in a
real-time PCR reaction and expressed as the ratio of the copy number of TRBV
relative to GAPDH (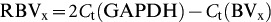 ), where Ct refers to the threshold cycle. The relative percentage (%) of
TRBVx gene expression was calculated according to the following
formula, which was modified from previous reports:15,16
), where Ct refers to the threshold cycle. The relative percentage (%) of
TRBVx gene expression was calculated according to the following
formula, which was modified from previous reports:15,16
Cloning and sequencing of monoclonal TRBV gene families
The GMSP profile of a TRBV gene family showing a single peak was selected for cloning and sequencing to determine the degree of homogeneity within the CDR3 region. This process is described in our previous report.17 The resulting TRBV gene sequences were translated into the corresponding amino-acid sequence using Chromas software (version 2.22; Technelysium, QLD, Australia), and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to define the CDR3 and BJ segments in the TRBV families.
HLA genotype determination
Genomic DNA was extracted from EDTA-treated peripheral whole blood, and the major histocompatibility complex (MHC) class I and class II haplotypes of enrolled subjects were determined by PCR and sequence-specific oligonucleotide-probe hybridization, as has been described in more detail in previous reports.16,18
Statistical analysis
All data were analyzed using SPSS software version 16.0 (SPSS Inc., Chicago, IL, USA). Differences in skewed TRBV frequencies between PBMC and CD4+ T-cell and CD8+ T-cell populations from recovered AHI subjects were analyzed using the Kruskal–Wallis H test or the Mann–Whitney non-parametric U test. Differences between two TRBV families were examined using a χ2 test or Student's t-test with P<0.05 considered to be statistically significant.
Results
Biased TRBV families within T-cell repertoires in recovered AHI subjects
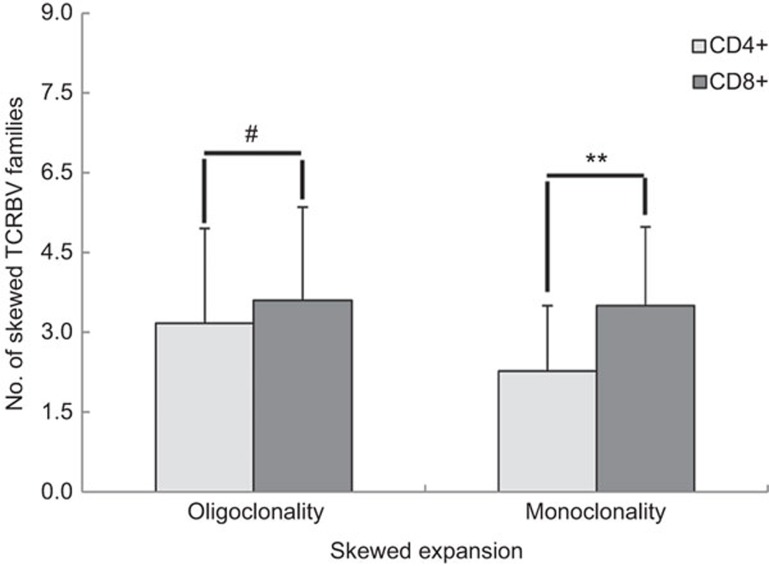
Biased TRBV families were compared between the two sorted cell populations (30 samples) and the 38 PBMC samples isolated from recovered AHI subjects. Among the three cell populations analyzed, the average number of biased oligoclonal and monoclonal TRBV families in the CD8+ T-cell subset was higher than in the CD4+ T-cell subset or in PBMCs (P<0.01). There was no significant difference between the latter two groups (P>0.05) (Table 2). In addition, there was a higher number of monoclonally expanded TRBV patterns in the CD8+ fraction compared to the CD4+ fraction (Figures 1 and 2 and Table 3). However, the number of oligoclonal TRBV families expressed in the CD4+ T-cell and CD8+ T-cell populations of recovered AHI subjects was not significantly different (P>0.05) (Figure 2 and Table 3).
Table 2. Frequencies of skewed TRBV in CD4+ and CD8+ T cells and PBMCs from subjects who have recovered from AHIa.
| TRBV families | CD4+ cells Incidence (%) b | CD8+ cells Incidence (%) b | PBMCs Incidence (%) b |
|---|---|---|---|
| 1 | 4 | 6 | 5 |
| 2 | 5 | 7 | 6 |
| 3 | 4 | 6 | 9 |
| 4 | 5 | 7 | 8 |
| 5.1 | 8 | 12 (40.0) | 14 (36.8) |
| 5.2 | 6 | 5 | 9 |
| 6 | 4 | 6 | 5 |
| 7 | 6 | 8 | 6 |
| 8 | 7 | 7 | 7 |
| 9 | 5 | 5 | 3 |
| 10 | 6 | 6 | 5 |
| 11 | 9 (32.1) | 16 (53.3) | 19 (50.0) |
| 12 | 9 (32.1) | 8 | 11 |
| 13.1 | 7 | 10 (33.3) | 10 |
| 13.2 | 5 | 6 | 8 |
| 14 | 6 | 7 | 7 |
| 15 | 12 (42.9) | 13 (43.3) | 18 (47.4) |
| 16 | 3 | 6 | 5 |
| 17 | 4 | 7 | 9 |
| 18 | 8 | 8 | 10 |
| 19 | 5 | 7 | 5 |
| 20 | 9 (32.1) | 14 (46.7) | 14 (36.8) |
| 21 | 7 | 6 | 4 |
| 22 | 6 | 7 | 6 |
| 23 | 6 | 8 | 7 |
| 24 | 7 | 15 (50.0) | 8 |
| Total no. of skewed BV (average ratio for a case) | 163 (5.82)c,d | 213 (7.10)c | 218 (5.74)c,d |
| No. of patients examined with normal patterne (ratio, %) | 2 (6.67)f | 0 (0.00)f | 0 (0.00)f |
| No. of patients examined | 30 | 30 | 38 |
Abbreviations: AHI, acute hepatitis B virus infection; GMSP, gene melting spectral pattern; PBMC, peripheral blood mononuclear cell; TRBV, T-cell receptor beta-chain variable.
The number of samples with specific skewed (oligoclonal or monoclonal) TRBV families is summarized for the subjects who have recovered from AHI.
The number (percentage) of samples showing skewed clonal expansion among the total detected samples. Samples with normal GMSP are excluded from the percentage calculation.
The average skewed expansion ratio of the TRBV families was higher in CD8+ T-cell population than in the other two cell populations (P<0.01 by χ2 test).
There was no significant difference between the two groups (P>0.05 by χ2 test). Samples with normal GMSPs are excluded from the average ratio calculation.
Normal patterns of GMSPs indicates that there are no monoclonal or oligoclonal patterns in any TRBV gene family in a subject.
The incidence of the normal GMSP was significantly higher in CD4+ T-cell populations than that in the other two cell populations (P<0.01 by χ2 test).
Figure 1.
The number of subjects with a monoclonally expanded TRBV family. The number of subjects with a monoclonal TRBV family is summarized using peripheral CD4+ or CD8+ T cells from 30 subjects who have recovered from AHI. AHI, acute hepatitis B virus infection; TRBV, T-cell receptor beta-chain variable.
Figure 2.
Comparing the number of skewed (oligoclonal or monoclonal) TRBV families in the CD4+ and CD8+ T-cell subsets from subjects who have recovered from AHI. Values are expressed as the mean±s.d. (standard deviation). #P>0.05; **P<0.01. AHI, acute hepatitis B virus infection; TRBV, T-cell receptor beta-chain variable.
Table 3. GMSP assay-generated profiles of skewed (oligoclonal or monoclonal) TRBV families in CD4+ and CD8+ T cells in subjects who have recovered from AHI.
| Subjects no. | CD4+ cells | CD8+ cells | ||
|---|---|---|---|---|
| TRBV oligoclonality | TRBV monoclonality | TRBV oligoclonality | TRBV monoclonality | |
| S1 | 8/9/22 | 14/17 | 5.1/7/14/15/20 | 4/8/11/12/13.1 |
| S2 | 3/7/15/18/20/24 | 11/12 | None | 1/15/20/21/22/24 |
| S3 | 15/16 | 10/13.1/23 | 11/12/15/17/19 | 2/5.1/6/7 |
| S4 | 9/22 | 7/19 | 13.2/22/24 | 3/4/10/15/18/20 |
| S5 | 3/5.1/11/21/23 | 2/7/15 | 5.1/9/13.1/14/17/21 | 11/19 |
| S6 | 5.2/9/24 | 4/8/11/13.1 | 9 | 4/13.1/17/20/22 |
| S7 | None | None | 6/18 | 5.1/5.2/7/8/19 |
| S8 | 3/10/11/12/19/21 | 15/24 | 19/23 | 11/14/20/24 |
| S9 | 12/20/22 | 18 | 3/13.2/15/20/22/24 | 9 |
| S10 | 1/4/11/14/24 | 8/20 | 1/5.2 | 13.2/18 |
| S11 | 6/14/24 | 4/5.1/12/20 | 5.1/19/23 | 10/12/13.1 |
| S12 | 8/13.2 | 1/9/24 | 4/8/10/18 | 2/11/15/23/24 |
| S13 | 2/5.1/13.1/17/21 | 13.2/19 | 2/20/24 | 7/11/17/19 |
| S14 | 7/10/13.1 | 5.2/23 | 4/8/11/12/13.1/15 | 5.1/16 |
| S15 | 5.2/6/14/15/16 | 7 | 3/21/23/24 | 13.2/20 |
| S16 | 10/17/23 | 18 | 7/11/13.1/16/24 | 5.1/20 |
| S17 | 3/5.1/11/13.1/20/22 | 4/12/14 | 1/5.1/10/14/17/20/22 | 12/23 |
| S18 | 15/18 | 6/8/11 | 3/5.2/19/24 | 8/11/13.1/15 |
| S19 | 2/10/16/19 | 12/13.2 | 2/11/13.2/20 | 24 |
| S20 | 15 | 5.1/13.1/20/23 | 5.1/12/15 | 1/11 |
| S21 | 2/5.2 | 8 | 1 | 6/16/18/22 |
| S22 | 11/18 | 5.2/24 | 7/10/15/20/23/24 | 2/5.1/13.2/21 |
| S23 | 7/12/21 | 1/10/13.1/15/18 | 3/6/11 | 9/15/17/18 |
| S24 | None | None | 7/16/18 | 1/5.2/11/13.1/14/23 |
| S25 | 4/5.2/8/13.2/20 | 14/15/19 | 2/5.1/11/15/21 | 6/24 |
| S26 | 6/15/22 | 1/5.1/18 | 10/14/21 | 2/5.1/7/20 |
| S27 | 2/12/23 | 9/15/21 | 4/13.1/16 | 12/24 |
| S28 | 5.1/11/17/18/20/21 | None | 8/12/18/23/24 | 6/11/13.1/16 |
| S29 | 15 | 12/13.2 | 17 | 4/8/15/22 |
| S30 | 21 | 5.1/20/22 | 5.2/9/11 | 3/14/20/24 |
| Total no. of skewed TRBV families (no. of subjects) | 95 (30)a | 68 (30)b | 108 (30)a | 105 (30)b |
Abbreviations: AHI, acute hepatitis B virus infection; GMSP, gene melting spectral pattern; PBMC, peripheral blood mononuclear cell; TRBV, T-cell receptor beta-chain variable.
The ratio of oligoclonal-expanded TRBV families was not significantly different between the two groups (P>0.05 by χ2 test).
The monoclonal ratio was lower in the CD4+ cells than that in the CD8+ cells (P<0.01 by χ2 test).
None, no skewed TRBV family was detected.
Comparison of TRBV frequency between recovered AHI subjects and healthy donors (HDs)
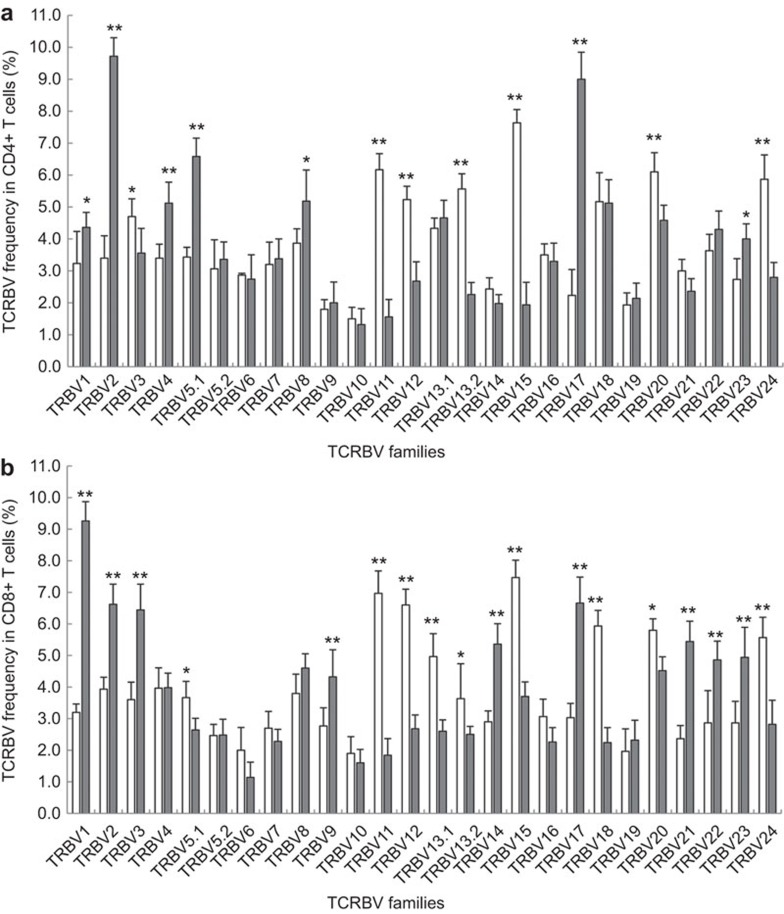
The relative percentage of a TRBV family was used to determine relative TRBV use in the CD4+ T-cell and CD8+ T-cell subsets. In CD4+ T cells, we observed seven TRBV gene families with increased expression (BV3, BV11, BV12, BV13.2, BV15, BV20 and BV24) and seven with reduced expression (BV1, BV2, BV4, BV5.1, BV8, BV17 and BV23) (Figure 3a). Moreover, in CD8+ T cells, nine TRBV families had increased expression (BV5.1, BV11, BV12, BV13.1, BV13.2, BV15, BV18, BV20 and BV24), and nine TRBV families (BV1, BV2, BV3, BV9, BV14, BV17, BV21, BV22 and BV23) showed a reduction in relative expression (Figure 3b).
Figure 3.
Relative frequencies of each TRBV gene family in the CD4+ and CD8+ T cell subsets from recovered AHI subjects and HDs. PBMCs from recovered AHI subjects (open bars) and HDs (filled bars) were sorted into CD4+ (A) and CD8+ (B) T-cell subsets. Data represent the relative percentages (%) of each TRBV gene family (1–24). The data are presented as the mean and standard deviation (s.d.) of the relative expression of each TRBV from 30 recovered AHI subjects and 20 HDs. *P<0.05; **P<0.01. AHI, acute hepatitis B virus infection; HD, healthy donor; PBMC, peripheral blood mononuclear cell; recovered AHI subjects, subjects who have recovered from AHI; TRBV, T-cell receptor beta-chain variable.
Monoclonal TRBV profile in CD4+ and CD8+ T-cell subsets and PBMCs
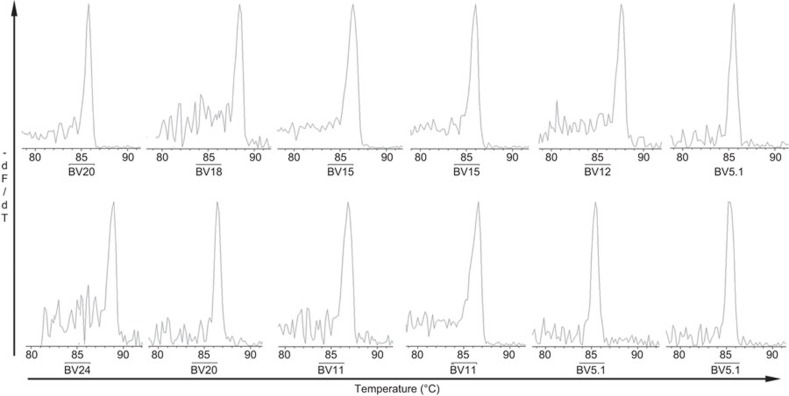
We identified the monoclonal expansion of TRBV families among CD4+ T cells, CD8+ T cells and PBMC populations based on our judgment criteria. Although a single-peak GMSP for a TRBV family may have been detected in any one of the TRBV families derived from the three cell populations (CD4+ and CD8+ T-cell subsets and PBMCs), a monoclonal (single peak) TRBV pattern was often detected in the following TRBV families: BV5.1, BV11, BV13.1, BV15, BV20 and BV24 (Table 3). Representative GMSP profiles of monoclonal TRBV expressed in the CD4+ and CD8+ T-cell subsets and in PBMCs are shown in Figures 4 and 5, respectively. Although monoclonality was identified in different TRBV families within the CD4+ and CD8+ T-cell subsets, we cannot discriminate which monoclonal TRBV families were derived from the CD4+ or CD8+ T-cell subsets by their GMSP profile shape alone, which was always expressed as a single peak in the GMSP whether the melting temperature was the same.
Figure 4.
A representative GMSP with a single-peak (monoclonal expansion) for TRBV families in the CD4+ and CD8+ T-cell subsets in recovered AHI subjects. The TRBV families shown in the top graphs correspond to S10 CD4+ (BV20), S9 CD4+ (BV18), S5 CD4+ (BV15), S8 CD4+ (BV15), S11 CD4+ (BV12) and S20 CD4+ (BV5.1), and the TRBVs shown in the bottom graphs correspond to S2 CD8+ (BV24), S4 CD8+ (BV20), S5 CD8+ (BV11), S8 CD8+ (BV11), S12 CD8+ (BV5.1) and S19 CD8+ (BV5.1). The corresponding amino-acid sequences are shown in Table 4. The melting temperature is on the x-axis of each plot. The negative first derivative of the reduction in fluorescence as a function of temperature (−dF/dT) is shown on the y-axis. AHI, acute hepatitis B virus infection; GMSP, gene melting spectral pattern; TRBV, T-cell receptor beta-chain variable.
Figure 5.
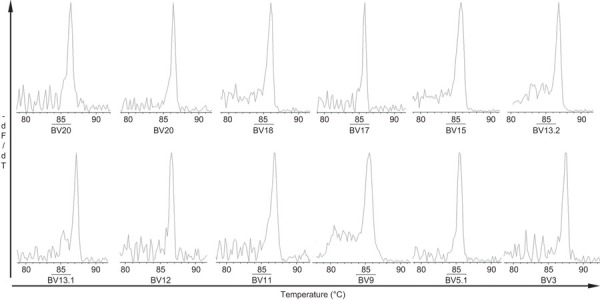
A representative GMSP with a single peak (monoclonal expansion) of the TRBV families in PBMCs from recovered AHI subjects. The TRBV families shown in the top graphs correspond to S1 (BV20), S15 (BV20), S6 (BV18), S3 (BV17), S1 (BV15) and S9 (BV13.2), and the TRBVs shown in the bottom graphs correspond to S12 (BV13.1), S5 (BV12), S2 (BV11), S16 (BV9), S32 (BV5.1) and S15 (BV3). The corresponding amino-acid sequences are shown in Table 5. The melting temperature is on the x-axis of each plot. The negative first derivative of the reduction in fluorescence as a function of temperature (−dF/dT) is shown on the y-axis. AHI, acute hepatitis B virus infection; GMSP, gene melting spectral pattern; PBMC, peripheral blood mononuclear cell; TRBV, T-cell receptor beta-chain variable.
Conservative CDR3 sequence of monoclonal TRBV in recovered AHI subjects
Representative amino acid sequences of TRBV CDR3 from the CD4+ and CD8+ T-cell subsets and PBMCs from recovered AHI subjects are shown in Tables 4 and 5. We observed that the monoclonality of the same or different TRBV CDR3s among different recovered AHI subjects was relatively well conserved, including BV15 (XXXX-GRTNEQ with BJ2.1) and BV12 (XXX-DSYEQ with BJ2.7) in the CD4+ T-cell subset. We also found CDR3 conservation in TRBV families from different patients in CD8+ T cells; for example, X-GEL (BJ2.2) and LDYS-XXX-I (BJ1.3) were found in CDR3s from the TRBV11 and BV5.1 families, respectively. In addition, TGTG-XXXX (BJ1.6) was found in the CD4+ and CD8+ T-cell subsets and PBMCs from BV20. Moreover, conserved CDR3 motifs from BV15 (XXXX-GRTNEQ with BJ2.1) and BV5.1 (LDYS-XXX-I with BJ1.3) were also found in the PBMC population from recovered AHI subjects, and the ‘TEA’ (BJ1.1) motif was observed in both TRBV18 and BV17 families. In addition, the relatively conserved CDR3 of TRBV11 was expressed as X-GEL (BJ2.2) or VYNEQ (BJ2.1), respectively, in the PBMC population.
Table 4. Representative amino-acid sequences of monoclonal TRBV families in CD4+ or CD8+ T cells from subjects who have recovered from AHI.
| Subjects | Vbeta | CDR3 | BJ | Ratio | ||
|---|---|---|---|---|---|---|
| S10 (CD4+)a | BV20 | SGFYLCAWS | TGTGKYAL | HFGNGTRLTVTED | 1.6 | 20/23 |
| S9 (CD4+) | BV18 | SAAYFCASS | RTGDTEA | FFGQGTRLTVVED | 1.1 | 18/23 |
| S5 (CD4+) | BV15 | TALYFCATS | RFGYGRTNEQ | FFGPGTRLTVLED | 2.1 | 22/23 |
| S8 (CD4+) | BV15 | TALYFCATS | DPASGRTNEQ | FFGPGTRLTVLED | 2.1 | 20/23 |
| S23 (CD4+) | BV15 | TALYFCATS | DLVGGRTNEQ | FFGPGTRLTVLED | 2.1 | 21/25 |
| S11 (CD4+) | BV12 | TSVYFCAIR | HTADSYEQ | YFGPGTRLTVTED | 2.7 | 19/23 |
| S17 (CD4+) | BV12 | TSVYFCAIR | KQGDSYEQ | YFGPGTRLTVTED | 2.7 | 18/21 |
| S20 (CD4+) | BV5.1 | SALYLCASS | LDYSGNTI | YFGEGSWLTVVED | 1.3 | 21/22 |
| S2 (CD8+) | BV24 | AMYLCATS | ARGTGDQETQ | YFGPGTRLLVLED | 2.5 | 16/23 |
| S4 (CD8+) | BV20 | SGFYLCAWS | TGTGHSPL | HFGNGTRLTVTED | 1.6 | 21/22 |
| S5 (CD8+) | BV11 | TSQYLCASS | FGEL | FFGEGSRLTVLED | 2.2 | 21/23 |
| S8 (CD8+) | BV11 | TSQYLCASS | AGEL | FFGEGSRLTVLED | 2.2 | 22/23 |
| S13 (CD8+) | BV11 | TSQYLCASS | LGEL | FFGEGSRLTVLED | 2.2 | 20/22 |
| S18 (CD8+) | BV11 | TSQYLCASS | PGEL | FFGEGSRLTVLED | 2.2 | 23/23 |
| S22 (CD8+) | BV5.1 | SALYLCASS | LDYSVMSI | YFGEGSWLTVVED | 1.3 | 22/23 |
| S26 (CD8+) | BV5.1 | SALYLCASS | LDYSGNTI | YFGEGRILQED | 1.3 | 21/22 |
Abbreviations: AHI, acute hepatitis B virus infection; BJ, T-cell receptor beta joint region; CDR3, complementary determining regions; PBMC, peripheral blood mononuclear cell; TRBV, T-cell receptor beta-chain variable; Vbeta, T-cell receptor beta-chain variable region.
When the GMSP of a TRBV gene family displayed a single peak (Figure 4), the PCR product of the corresponding TRBV gene family was re-amplified, and the PCR product was sequenced after cloning. Only the amino acid sequences of the CDR3 max ratio are shown. The identical amino acids in the CDR3 sequences are underlined.
The T-cell subset used for analysis is in brackets.
Table 5. Representative amino acid sequences of monoclonal TRBV gene families in PBMCs from subjects who have recovered from AHI.
| Subjects | Vbeta | CDR3 | BJ | Ratio | ||
|---|---|---|---|---|---|---|
| S1 | BV20 | SGFYLCAWS | TGTGHTPV | HFGNGTRLTVTED | 1.6 | 15/20 |
| S15 | BV20 | SGFYLCAWS | TGTGHSPL | HFGNGTRLTVTED | 1.6 | 18/21 |
| S27 | BV20 | SGFYLCAWS | VQNEQ | FFGPGTRLTVLED | 2.1 | 17/20 |
| S6 | BV18 | SAAYFCASS | RTGDTEA | FFGQGTRLTVVED | 1.1 | 23/23 |
| S3 | BV17 | TAFYLCASS | RQLTEA | FFGQGTRLTVVED | 1.1 | 19/23 |
| S1 | BV15 | TALYFCATS | RFAYGRTNEQ | FFGPGTRLTALED | 2.1 | 24/27 |
| S9 | BV13.2 | TSVYFCASS | YSTDEQ | YFGPGTRLTVTED | 2.7 | 21/24 |
| S12 | BV13.1 | TSVYFCAST | PGYSYEQ | YFGPGTRLTVTED | 2.7 | 20/23 |
| S5 | BV12 | TSVYFCAIR | KQGDSYEQ | YFGPGTRLTVTED | 2.7 | 19/23 |
| S9 | BV12 | TSVYFCASC | PGGSGGTGEL | FFGEGSRLTVLED | 2.2 | 20/23 |
| S2 | BV11 | TSQYLCASS | PGEL | FFGEGSRLTVLED | 2.2 | 21/23 |
| S15 | BV11 | TSQYLCASS | AGEL | FFGEGSRLTVLED | 2.2 | 20/23 |
| S21 | BV11 | TSQYLCATG | VYNEQ | FFGPGTRLTVLED | 2.1 | 20/23 |
| S16 | BV9 | SAVYFCASS | PWNTGEL | FFGEGSRLTVLED | 2.2 | 18/23 |
| S32 | BV5.1 | SALYLCASS | LDYSLYTI | YFGEGSWLTVVED | 1.3 | 20/23 |
| S15 | BV3 | TSMYLCASS | LGRETQ | YFGPGTRLLVPED | 2.5 | 17/21 |
| S32 | BV3 | TSMYLCASS | FLLRGMFFYEQ | YFGPGTRLTVTED | 2.7 | 18/23 |
Abbreviations: AHI, acute hepatitis B virus infection; BJ, T-cell receptor beta joint region; CDR3, complementary determining regions; PBMC, peripheral blood mononuclear cell; TRBV, T-cell receptor beta-chain variable; Vbeta, T-cell receptor beta-chain variable region.
When the GMSP of a TRBV gene family displayed a single peak (Figure 5), the PCR product of the corresponding TRBV gene family was re-amplified, and the PCR product was sequenced after cloning. Only the amino-acid sequences of the CDR3 max ratios are shown. The identical sequences are underlined.
Discussion
HBV infection is an important public health problem with a serious impact on human health. The different clinical stages of chronic HBV infection are determined by the status of the immune response in a host. The T-cell response to HBV is strong in a host with a self-limited acute HBV infection.6 In addition, the status and function of T cells is reflected in their TRBV profile, and a TCR has three complementary determining regions (CDR1, CDR2 and CDR3). CDR3 primarily recognizes antigen-derived peptides bound to a MHC molecule, an interaction that has been observed through crystal structure analysis.19,20 Therefore, the analysis of a CDR3 profile can reveal changes in the T-cell population stimulated by a specific antigen,21,22 the amount of a particular T-cell clone and the functional status of T cells.23,24
T lymphocytes are primarily divided into the CD4+ and CD8+ subsets. In peripheral blood from HDs, the TCRs of more than 95% of T cells are composed of alpha and beta chain heterodimers.25 The biased clonal expansion of T cells has been identified in the peripheral circulation after a host has been infected with a virus.5,7 In this study, we analyzed the extent of oligoclonally or monoclonally skewed, expanded TRBV families in the CD4+ and CD8+ T-cell subsets and in PBMCs from recovered AHI subjects and found that the number of skewed TRBV in the CD8+ fraction was significantly higher than in the CD4+ T-cell subset and in PBMCs. These observations are consistent with previous reports that indicate that cellular immunity plays a vital role in controlling HBV infection of a host and that clonal proliferation of T cells for HBV specific antigens are mainly found in CD8+ T cells rather than in CD4+ T cells.26,27 Moreover, monoclonal TCR transcripts could be detected exclusively in CD8+ but not in CD4+ T cells from subjects who had been immunized with a recombinant hepatitis B surface vaccine.28
The analysis of TRBV usage in an individual can help in the evaluation of the immune response in a variety of conditions over the course of the disease.29 Many studies have determined that the usage frequency of antigen-specific TRBV families in subjects with either virus infection or cancer can be different.30,31 In the current study, the increased expression of several TRBV families was detected in CD4+ and CD8+ T-cell populations, which is consistent with observations that the T-cell immune response to HBV antigens involves multiple TRBV families.32 In addition, these results are consistent with the statement that T-cell immune responses are polyclonal and polyspecific in recovered AHI subjects and involve a number of special TRBV families. Similar results were also found in subjects vaccinated with recombinant HBsAg.33 These observations may be due to several factors, including the complexity and fluidity of HBV epitopes34 and the existence of subjects with different HLA phenotypes. In addition, these phenomena may be associated with the structural and spatial configuration of the TCR CDR3, such that different CDR3 sequences may permit the recognition of the same epitope if their molecular spacings are structurally similar.35
Currently, anti-HBV treatment with nucleoside analogs is used to reduce liver inflammation and to prevent the development of cirrhosis and liver cancer. The curative effect of these treatments is closely associated with the restoration of T-cell functionality in the host, particularly the activity of HBV-specific cytotoxic T lymphocyte.36 In addition, antiviral treatment is often accompanied by side effects and drug resistance. Therefore, there is a pressing need to develop new immunotherapeutic interventions for chronic HBV infection that limit the high costs and risks of toxicity and viral resistance.37
Methods that modify T cells with antigen-specific TCR genes to treat specific cancers and infectious diseases have attracted significant interest in recent years. In 2006, data from a clinical trial demonstrated that T cells modified with an antigen-specific TCR gene could be used to treat melanoma.38 It has also been shown that T cells modified with a virus-specific TCR gene are cytotoxic to HBV-infected hepatocytes and a liver cell line expressing an HBV-related antigen.39 Vizcardo et al.40 transformed antigen-specific T cells into multifunctional T cells and then caused the cells to redifferentiate into T cells with the original antigen-specificity, thereby expanding the potential range of applications for specific T cells.40 Nishimura et al.41 reported that CD8+ T cells isolated from human immunodeficiency virus-1-infected patients could be reprogrammed to generate pluripotent cells41 that could be differentiated into original antigen-specific T cells with high proliferative activity and telomere length. These reports also showed that T cells modified with an antigen-specific TCR gene have excellent prospects for application in clinical practice.42
In this report, we found that the TRBV families TRBV5.1, BV11, BV13.1, BV15, BV20 and BV24 were more frequently expanded monoclonally than other TRBV family members in recovered AHI subjects, regardless of the presence of CDR3 in the CD4+ or CD8+ T-cell subsets, further demonstrating that the T-cell immune response was polyclonal and polyspecific in the recovered AHI subjects studied.43 This phenomenon was also discovered in the peripheral blood of patients with chronic hepatitis B, although the prevalent usage of TCR was lower than that of the recovered AHI subjects44,45 and may be associated with the different immune response ability between them.
In addition, we analyzed the HLA types of subjects with prominent TRBV families to determine whether the relatively conserved CDR3 sequence was associated with a common HLA phenotype (Supplementary Table 2). These results indicated that the identity of the conserved CDR3 sequences in recovered AHI subjects were independent of the similarity in their HLA alleles. Moreover, the recognition of antigen by specialized T cells is controlled by MHC-restricted T cells. Our results of relatively conserved CDR3 motifs in the independent HLA phenotypes of recovered AHI subjects may be associated with the CDR reaction to the complex of peptide-MHC with plasticity and cross reactivity.46,47 In future studies, we plan to demonstrate the association of other HLA genotypes in combination with skewed TRBV families with HBV subgenotypes in more cases, in which we could discover the relationship between the HLA phenotypes and the relatively conserved TRBV families.
Additionally, many HBV T-cell epitopes were identified. In this study, we found several prevalent TRBV families that may be specific for HBV-related antigens; however, it is hard to determine to which peptides from HBV these T cells (TRBV) react because more than one quintillion TCR combinations can be generated by somatic recombination, in theory.48 Nevertheless, the determined prevalence of TRBV families can provide a key clue for establishing a TRBV profile as being specific for a specific HBV peptide.
In the production of antibodies in a host infected with an infectious disease, cells from the CD4+ T-cell subset (Th) play a vital role in helping B cells produce antigen-specific antibodies.49 We found that several skewed TRBV families (BV11, BV12, BV15 and BV20) had relatively higher usage and expression than other TRBV families in the CD4+ T-cell subset, indicating that these TRBV families were associated with the HBsAb produced in the recovered AHI subjects. Furthermore, TRBV15 from the CD4+ subset had the highest rate of use and relatively higher expression in recovered AHI subjects, suggesting that the TRBV family was most likely associated with the emergence and maintenance of HBsAb. In addition, the six TRBV families BV5.1, BV11, BV13.1, BV15, BV20 and BV24 have a higher rate of use and relative expression of the 24 TRBV families in recovered AHI subjects. This was particularly true for the TRBV families derived from the CD8+ T-cell subset, which is consistent with previous reports.26 These results suggested that multiple TRBV families, primarily derived from CD8+ T cells, were involved in the response against HBV antigens in recovered AHI subjects; however, these observations should be further confirmed with more cases in future studies.
In summary, the molecular characteristics of TRBV in subjects recovered from AHI are biased and involve multiple TRBV families. The TRBV11, BV12, BV15 and BV20 families in the CD4+ T-cell subset may have contributed more to the emergence and maintenance of HBsAb in recovered AHI subjects, and TRBV11, BV15 and BV20, all of which were preferentially detected in the T-cell subsets and in PBMCs and may have been relevant to the pathogenesis and positive outcome of subjects with AHI. Furthermore, the prevalent use of TRBV5.1 and BV20, which have relative conservative amino acid sequences, would help with the diagnosis of chronic HBV infection and the provision of individualized treatment with specific T cells modified with antigen-specific TCR genes.50
Acknowledgments
This work was supported by the Zhejiang Medical Health Fund of China (2012RCA020), the National High-technology R&D Program of China (2013CB531406), the National Science and Technology Major Project (2012ZX10002-006) and the Program for Zhejiang Leading Team of S&T Innovation (2011R09041-02). The authors thank Ms Ping Ye and Ms Haifeng Lu from the Department of Infectious Diseases at the First Affiliated Hospital, College of Medicine, Zhejiang University for their help in collecting samples for this study.
Footnotes
Supplementary Information accompanies the paper on Cellular & Molecular Immunology’s website. (http://www.nature.com/cmi).
The authors declare no conflict of interest.
Supplementary Information
References
- Wiersma ST, McMahon B, Pawlotsky JM, Thio CL, Thursz M, Lim SG et al. Treatment of chronic hepatitis B virus infection in resource-constrained settings: expert panel consensus. Liver Int 2011; 31: 755–761. [DOI] [PubMed] [Google Scholar]
- McMahon BJ. The natural history of chronic hepatitis B virus infection. Hepatology 2009; 49: S45–S55. [DOI] [PubMed] [Google Scholar]
- Su WJ, Liu CC, Liu DP, Chen SF, Huang JJ, Chan TC et al. Effect of age on the incidence of acute hepatitis B after 25 years of a universal newborn hepatitis B immunization program in Taiwan. J Infect Dis 2012; 205: 757–762. [DOI] [PubMed] [Google Scholar]
- Dandri M, Locarnini S. New insight in the pathobiology of hepatitis B virus infection. Gut 2012; 61 Suppl 1: i6–i17. [DOI] [PubMed] [Google Scholar]
- Balamurugan A, Ng HL, Yang OO. Rapid T cell receptor delineation reveals clonal expansion limitation of the magnitude of the HIV-1-specific CD8+ T cell response. J Immunol 2010; 185: 5935–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalova E, Laccabue D, Boni C, Watanabe T, Tan A, Zong HZ et al. Increased levels of arginase in patients with acute hepatitis B suppress antiviral T cells. Gastroenterology 2012; 143: 78–87. [DOI] [PubMed] [Google Scholar]
- Petrova GV, Naumova EN, Gorski J. The polyclonal CD8 T cell response to influenza M158-66 generates a fully connected network of cross-reactive clonotypes to structurally related peptides: a paradigm for memory repertoire coverage of novel epitopes or escape mutants. J Immunol 2011; 186: 6390–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JZ, Li MW, Wang JG, Lu HF, Yao XS, He JQ et al. Rapid detection of clonal expansion of T-cell receptor-beta gene in patients with HBV using the real-time PCR with DNA melting curve analysis. Hepatol Res 2010; 40: 407–414. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ma R, Luo R, Sun Y, He X, Sun W et al. Primary exploration of CDR3 spectratyping and molecular features of TCR beta chain in the peripheral blood and tissue of patients with colorectal carcinoma. Cancer Epidemiol 2010; 34: 733–740. [DOI] [PubMed] [Google Scholar]
- Chinese Society of Infectious Diseases and Parasitic Diseases CSoH, Chinese Medical Association. Management scheme of diagnostic and therapy criteria of viral hepatitis. Zhong hua Gan Zang Bing Za Zhi 2000; 8: 324–329. Chinese. [Google Scholar]
- Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology 2007; 45: 507–539. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li X, Ye B, Yang X, Wu W, Chen B et al. Effect of telbivudine therapy on the cellular immune response in chronic hepatitis B. Antiviral Res 2011; 91: 23–31. [DOI] [PubMed] [Google Scholar]
- Yao XS, Zhang GW, Ma L, Wen Q, Hou JL, Meng MJ et al. Analysis of the CDR3 length of TCR alphabeta T cells in the peripheral blood of patients with chronic hepatitis B. Hepatol Res 2006; 35: 10–18. [DOI] [PubMed] [Google Scholar]
- Yang J, Yi P, Wei L, Xu Z, Chen Y, Tang L et al. Phenotypes and clinical significance of circulating CD4+CD25+ regulatory T cells (Tregs) in patients with acute-on-chronic liver failure (ACLF). J Transl Med 2012; 10: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreither S, Fusi A, Busse A, Nagorsen D, Schrama D, Becker J et al. Relative quantification of TCR Vbeta-chain families by real time PCR for identification of clonal T-cell populations. J Transl Med 2008; 6: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nie H, Li N, Zang YC, Zhang D, Feng G et al. Skewed T-cell receptor BV14 and BV16 expression and shared CDR3 sequence and common sequence motifs in synovial T cells of rheumatoid arthritis. Genes Immun 2005; 6: 248–261. [DOI] [PubMed] [Google Scholar]
- Yang J, Xu K, Zheng J, Wei L, Fan J, Li L. Limited T cell receptor beta variable repertoire responses to ESAT-6 and CFP-10 in subjects infected with Mycobacterium tuberculosis. Tuberculosis (Edinb) 2013; 93: 529–537. [DOI] [PubMed] [Google Scholar]
- Schmidt AH, Baier D, Solloch UV, Stahr A, Cereb N, Wassmuth R et al. Estimation of high-resolution HLA-A, -B, -C, -DRB1 allele and haplotype frequencies based on 8862 German stem cell donors and implications for strategic donor registry planning. Hum Immunol 2009; 70: 895–902. [DOI] [PubMed] [Google Scholar]
- Marrack P, Kappler J. Positive selection of thymocytes bearing alpha beta T cell receptors. Curr Opin Immunol 1997; 9: 250–255. [DOI] [PubMed] [Google Scholar]
- Miqueu P, Guillet M, Degauque N, Dore JC, Soulillou JP, Brouard S. Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol Immunol 2007; 44: 1057–1064. [DOI] [PubMed] [Google Scholar]
- Okajima M, Wada T, Nishida M, Yokoyama T, Nakayama Y, Hashida Y et al. Analysis of T cell receptor Vbeta diversity in peripheral CD4 and CD8 T lymphocytes in patients with autoimmune thyroid diseases. Clin Exp Immunol 2009; 155: 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du JW, Gu JY, Liu J, Cen XN, Zhang Y, Ou Y et al. TCR spectratyping revealed T lymphocytes associated with graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Leuk Lymphoma 2007; 48: 1618–1627. [DOI] [PubMed] [Google Scholar]
- Sethi DK, Schubert DA, Anders AK, Heroux A, Bonsor DA, Thomas CP et al. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J Exp Med 2011; 208: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimgruber A, Ferber M, Irving M, Hussain-Kahn H, Wieckowski S, Derre L et al. TCRep 3D: an automated in silico approach to study the structural properties of TCR repertoires. PLoS ONE 2011; 6: e26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T, Gleimer M, Garbe AI, von Boehmer H. alphabeta versus gammadelta fate choice: counting the T-cell lineages at the branch point. Immunol Rev 2010; 238: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Hoare M, Davies N, Lopes AR, Dunn C, Kennedy PT et al. Functional skewing of the global CD8 T cell population in chronic hepatitis B virus infection. J Exp Med 2008; 205: 2111–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 2012; 338: 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn H, Neukirch C, Freitag K, Necker A, Hitzler W, Seliger B et al. Longitudinal analysis of the T-cell receptor (TCR)-VA and -VB repertoire in CD8+ T cells from individuals immunized with recombinant hepatitis B surface antigen. Clin Exp Immunol 2002; 129: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo KS, Park JY, Terman DS, Bohach GA. A quantitative real time PCR method to analyze T cell receptor Vbeta subgroup expansion by staphylococcal superantigens. J Transl Med 2010; 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NJ, Bando JK, Schwartz RH. Subsets of nonclonal neighboring CD4+ T cells specifically regulate the frequency of individual antigen-reactive T cells. Immunity 2012; 37: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol 2011; 89: 375–387. [DOI] [PubMed] [Google Scholar]
- Huang CF, Lin SS, Ho YC, Chen FL, Yang CC. The immune response induced by hepatitis B virus principal antigens. Cell Mol Immunol 2006; 3: 97–106. [PubMed] [Google Scholar]
- Yuh K, Sugyo S, Nakamura K, Shijo H, Emi K, Harada K et al. Analysis of human T-cell antigen receptor variable beta gene usage following vaccination with recombinant HBsAg. Dig Dis Sci 1998; 43: 880–886. [DOI] [PubMed] [Google Scholar]
- Desmond CP, Bartholomeusz A, Gaudieri S, Revill PA, Lewin SR. A systematic review of T-cell epitopes in hepatitis B virus: identification, genotypic variation and relevance to antiviral therapeutics. Antivir Ther 2008; 13: 161–175. [PubMed] [Google Scholar]
- Matalon E, Faingold O, Eisenstein M, Shai Y, Goldfarb D. The topology, in model membranes, of the core peptide derived from the T-cell receptor transmembrane domain. Chembiochem 2013; 14: 1867–1875. [DOI] [PubMed] [Google Scholar]
- Martinet J, Leroy V, Dufeu-Duchesne T, Larrat S, Richard MJ, Zoulim F et al. Plasmacytoid dendritic cells induce efficient stimulation of antiviral immunity in the context of chronic hepatitis B virus infection. Hepatology 2012; 56: 1706–1718. [DOI] [PubMed] [Google Scholar]
- Locarnini S. Transmission of antiviral drug resistant hepatitis B virus: implications for public health and patient management. J Gastroenterol Hepatol 2010; 25: 649–651. [DOI] [PubMed] [Google Scholar]
- Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 2006; 314: 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring AJ, Xue SA, Ho ZZ, Teoh D, Ruedl C, Chia A et al. Engineering virus-specific T cells that target HBV infected hepatocytes and hepatocellular carcinoma cell lines. J Hepatol 2011; 55: 103–110. [DOI] [PubMed] [Google Scholar]
- Vizcardo R, Masuda K, Yamada D, Ikawa T, Shimizu K, Fujii S et al. Regeneration of human tumor antigen-specific T cells from iPSCs derived from mature CD8+ T cells. Cell Stem Cell 2013; 12: 31–36. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kaneko S, Kawana-Tachikawa A, Tajima Y, Goto H, Zhu D et al. Generation of rejuvenated antigen-specific T cells by reprogramming to pluripotency and redifferentiation. Cell Stem Cell 2013; 12: 114–126. [DOI] [PubMed] [Google Scholar]
- Kobayashi E, Mizukoshi E, Kishi H, Ozawa T, Hamana H, Nagai T et al. A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nat Med 2013; 19: 1542–1546. [DOI] [PubMed] [Google Scholar]
- Gehring AJ, Haniffa M, Kennedy PT, Ho ZZ, Boni C, Shin A et al. Mobilizing monocytes to cross-present circulating viral antigen in chronic infection. J Clin Invest 2013; 123: 3766–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou HY, Wu JC, Peng WL, Chang C, Chi WK, Chu YD et al. Analysis of T cell receptor Vbeta gene usage during the course of disease in patients with chronic hepatitis B. J Biomed Sci 1998; 5: 428–434. [DOI] [PubMed] [Google Scholar]
- Maru Y, Yokosuka O, Imazeki F, Saisho H, Omata M. Analysis of T cell receptor variable regions and complementarity determining region 3 of infiltrating T lymphocytes in the liver of patients with chronic type B hepatitis. Intervirology 2003; 46: 277–288. [DOI] [PubMed] [Google Scholar]
- Piepenbrink KH, Blevins SJ, Scott DR, Baker BM. The basis for limited specificity and MHC restriction in a T cell receptor interface. Nat Commun 2013; 4: 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SN, Sommermeyer D, Piepenbrink KH, Blevins SJ, Bernhard H, Uckert W et al. Plasticity in the contribution of T cell receptor variable region residues to binding of peptide–HLA-A2 complexes. J Mol Biol 2013; 425: 4496–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows SR, Miles JJ. Immune parameters to consider when choosing T-cell receptors for therapy. Front Immunol 2013; 4: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry JA, Johansson BE, de Groot AS. A call to cellular & humoral arms: enlisting cognate T cell help to develop broad-spectrum vaccines against influenza A. Hum Vaccin 2008; 4: 148–157. [DOI] [PubMed] [Google Scholar]
- Xuan L, Wu X, Wu M, Zhang Y, Liu H, Fan Z et al. Effect of granulocyte colony-stimulating factor mobilization on the expression patterns, clonality and signal transduction of TRAV and TRBV repertoire. Immunobiology 2012; 217: 816–822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.