Abstract
Enhanced resistance to chemotherapy has been correlated with high levels of Delta-Np73 (DNp73), an anti-apoptotic protein of the p53 tumor-suppressor family which inhibits the pro-apoptotic members such as p53 and TAp73. Although genotoxic drugs have been shown to induce DNp73 degradation, lack of mechanistic understanding of this process precludes strategies to enhance the targeting of DNp73 and improve treatment outcomes. Antizyme (Az) is a mediator of ubiquitin-independent protein degradation regulated by the polyamine biosynthesis pathway. We show here that acetylpolyamine oxidase (PAOX), a catabolic enzyme of this pathway, upregulates DNp73 levels by suppressing its degradation via the Az pathway. Conversely, downregulation of PAOX activity by siRNA-mediated knockdown or chemical inhibition leads to DNp73 degradation in an Az-dependent manner. PAOX expression is suppressed by several genotoxic drugs, via selected members of the activator protein-1 (AP-1) transcription factors, namely c-Jun, JunB and FosB, which are required for stress-mediated DNp73 degradation. Finally, chemical- and siRNA-mediated inhibition of PAOX significantly reversed the resistant phenotype of DNp73-overexpressing cancer cells to genotoxic drugs. Together, these data define a critical mechanism for the regulation of DNp73 abundance, and reveal that inhibition of PAOX could widen the therapeutic index of cytotoxic drugs and overcome DNp73-mediated chemoresistance in tumors.
p73 is a member of the p53 family of transcription factors and encodes two major forms of the protein: the full-length TAp73 and the amino-terminally truncated Delta-Np73 (DNp73).1 Similar to p53, transactivation competent TAp73 exerts tumor-suppressive activities via the regulation of apoptotic signaling and cell-cycle arrest.2 However, the DNp73 form, which lacks the N-terminal transactivation domain, works as a dominant-negative inhibitor of both TAp73 and p53 and therefore inhibits their tumor-suppressor functions.3 It is noteworthy that DNp73 also possesses weak transcriptional activity and is able to transactivate several genes such as 14-3-3 and Gadd45,4 and was recently shown to induce the expression of anti-apoptotic proteins caspase-2S and hsp70,5, 6 further illustrating its anti-apoptotic potential. Consistently, DNp73−/− mouse embryonic fibroblasts were found to be sensitized to genotoxic stresses and showed enhanced p53-dependent apoptosis, indicating that DNp73 is a critical inhibitor of apoptosis.7
Not surprisingly, DNp73 expression is frequently elevated in many human cancers, including lung, neuroblastoma, colon, ovarian, liver, etc.,8, 9, 10 which suggests a role in either promoting cellular growth or inhibiting apoptosis, as well as in affecting chemosensitivity. In the latter context, TAp73 and p53 are activated by a multitude of chemotherapeutic agents,11 linking their functions to the chemosensitivity of cancer cells. Thus, DNp73 overexpression would be one mechanism of inhibiting TAp73- and p53-dependent pro-apoptotic activity in response to chemotherapeutic drugs, leading to poor clinical outcome.12, 13 Consistently, exposure to genotoxic signals leads to the upregulation of TAp73 and the downregulation of DNp73,14 hence allowing an effective apoptotic response to ensue. This indicates that the relative abundance of the two forms of p73 could be an important determinant of cellular fate during stress response, thereby regulating chemosensitivity.
Owing to their homology, TAp73 and DNp73 protein stability are regulated by many common factors, including Itch, c-Abl, PML, NQO1, Cyclin G and others, via both ubiquitin-dependent and -independent mechanisms.15, 16, 17, 18, 19 However, we have previously shown that the selective degradation of DNp73 by genotoxic stresses occur through the ubiquitin-independent, polyamine-induced antizyme (Az) pathway that is regulated by c-Jun, a member of the activator protein-1 (AP-1) family of transcription factors.20 Az1 is a small inhibitory protein that binds to its target proteins and directs them to the proteasome for degradation.21, 22 Az1 transcript exists with two overlapping open reading frames, and translation of a fully functional Az1 protein requires a unique +1 ribosomal frameshift mechanism, which is dependent on the level of polyamines, a group of cationic compounds such as putrescine, spermidine and spermine present in the cells21, 22 (Figure 1a, lower panel).
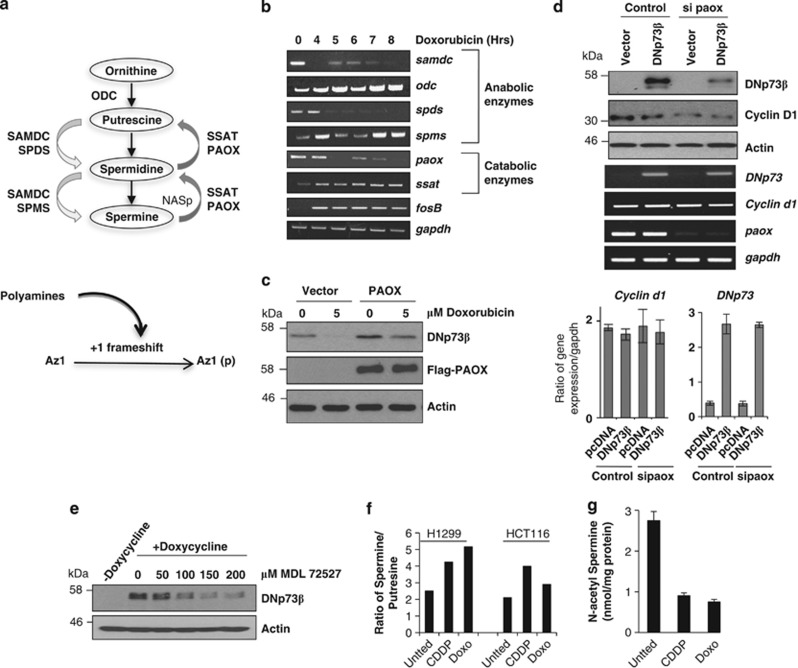
Figure 1.
Polyamine catabolic enzyme paox regulates DNp73 stability. (a) Schematic showing the polyamines (putrescine, spermidine and spermine) biosynthesis pathway and the involvement of anabolic (ODC, SAMDC, SPDS and SPMS) and catabolic enzymes (PAOX and SSAT; upper panel) and the effects of polyamines on Az1 frameshifting process (lower panel). N-acetylspermine (NASp) is the intermediate in the catabolic conversion from spermine to spermidine. (b) Semi-quantitative RT-PCR analysis of the expression of enzymes in the polyamine biosynthesis pathway in p53 null H1299 cells exposed to doxorubicin (5μM) for the indicated times. (c) Immunoblot analysis of steady-state DNp73β levels in H1299 cells transfected with or without Flag-tagged PAOX for 24 h followed by exposure to doxorubicin for 8 h. Expression of exogenous PAOX and DNp73 was detected by anti-Flag and anti-p73 (GC15) antibodies, respectively. (d) Immunoblot and semi-quantitative RT-PCR (top and middle panels), and quantitative real-time PCR (lower bar charts) analysis of overexpressed DNp73β or endogenous cyclin D1 in H1299 cells was analyzed 48 h after siRNA-mediated silencing of paox. All values are means±S.D. of duplicate experiments, reflected by error bars. (e) H1299 cells inducibly expressing DNp73β were induced by doxycycline for 24 h before treatment with the PAOX inhibitor, MDL72527, at the indicated doses for 16 h and harvested for immunodetection. All experiments were repeated at least thrice and representative data are shown throughout. (f and g) Intracellular levels of spermine and putrescine were measured in H1299 cells (left bars) or HCT116 cells (right bars) without (Untted) or with cisplatin (25 μM CDDP) or doxorubicin (5 μM Doxo) treatment for 6 h by HPLC and the ratio (see Supplementary Figure 1D for raw data) is presented (f). The levels of the intermediate product in the conversion of spermine back to spermidine, NASp was also measured and depicted (g). All values are means±S.D. of triplicate experiments for g
The balance of polyamines in cells is controlled by several key enzymes, which either synthesize or breakdown the different polyamine by-products. Ornithine is converted into diamine putrescine by the rate-limiting enzyme ornithine decarboxylase (ODC).22 A second rate-limiting enzyme S-adenosylmethionine decarboxylase (SAMDC) is required for the synthesis of both triamine spermidine and tetramine spermine by spermidine synthase (SPDS) and spermine synthase (SPMS), respectively22 (Figure 1a, upper panel). Conversely, spermine can be converted back to spermidine and the latter to putrescine by catabolic enzymes spermidine/spermine N1-acetyltransferase (SSAT) and acetylpolyamine oxidase (PAOX);22 involving the intermediates N-acetylspermine (NASp) and N-acetylspermidine. Our previous study has shown that upon genotoxic insults, c-Jun regulates the levels of functional Az to degrade DNp73, but the exact mechanistic details remain unclear.
In addition to c-Jun, the AP-1 transcription factor family also includes JunB, JunD, c-Fos, FosB, Fra1 and Fra2.23 Through the formation of heterodimers or homodimers, the latter in the case of the Jun proteins, the AP-1 members regulate diverse physiological processes in response to a plethora of stimuli, such as DNA damage, growth factors and oncogenic stimuli.24 Although AP-1 members can activate a set of common target genes, different AP-1 dimer compositions produce different biological outcomes, suggesting that they can also selectively regulate distinct sets of genes.25, 26 In addition, some biological effects of AP-1 members are the result of their ability to repress gene expression, such as inhibition of p53 by c-Jun.27
In this respect, we have investigated the mechanism of DNp73 regulation through the Az pathway with reference to the specificity of AP-1 members in regulating this process. Our results demonstrate that unlike other AP-1 factors, only c-Jun, JunB and FosB are able to specifically repress the expression of polyamine catabolic enzyme paox (the gene encoding PAOX), highlighting the functional specificity of these AP-1 factors. This effect leads to the production of active Az1 that consequently degrades DNp73. In addition, inhibition of PAOX in DNp73-overexpressing cancer cells abrogated their resistance to chemotherapeutic agents and reveals a possible avenue for effective targeted therapy in DNp73-overexpressing cancers in the presence of chemotherapeutic drugs.
Results
Polyamine catabolic enzyme PAOX positively regulates DNp73
We have utilized doxorubicin as a prototypic DNA-damaging chemotherapeutic agent to investigate the effects on the expression of the anabolic and catabolic enzymes that regulate polyamines levels in cells (Figure 1a). Treatment of p53 null H1299 cells with doxorubicin resulted in the transcriptional downregulation of anabolic enzymes samdc and spds and upregulation of odc and spms (Figure 1b). Although upregulation of odc and spms would equate to an increase in synthesis of polyamines, silencing of odc or spms did not prevent the stress-mediated degradation of DNp73 (Supplementary Figures 1A and B). Similarly, although doxorubicin induced ssat expression, siRNA-mediated silencing of this enzyme had no major effect on stress-mediated DNp73 degradation (Supplementary Figure 1B). However, downregulation of paox upon doxorubicin treatment was observed to occur concomitantly with the reduction of DNp73β protein (Supplementary Figure 1C), and exogenous expression of flag-tagged PAOX alone was able to significantly increase the steady-state levels of DNp73β after doxorubicin treatment (Figure 1c). Moreover, although doxorubicin treatment led to a reduction of DNp73β, this effect was reduced in the presence of PAOX (Figure 1c). To further explore the role of PAOX in regulating DNp73 abundance, we silenced paox expression, which resulted in a marked decrease in DNp73β steady-state levels (Figure 1d). Similar effect was observed on endogenous cyclin D1, another known target of the polyamine-Az degradation pathway (Figure 1d), with the mRNA of both DNp73 and cyclin D1 remaining unaffected by paox silencing (Figure 1d lower panel). Furthermore, treatment of cells that inducibly express DNp73β with a known chemical inhibitor of PAOX, MDL72527,28 led to a decrease in DNp73 levels in a dose-dependent manner (Figure 1e).
We further evaluated the effects of the chemotherapeutic agents on the polyamine pathway by determining the levels of polyamines upon doxorubicin or cisplatin treatment (Supplementary Figure 1D). Although spermine levels were generally unaltered, levels of putrescine were reduced upon treatment (Supplementary Figure 1D), resulting in an increase in the ratio of spermine to putrescine (Figure 1f). Importantly, the levels of NASp were significantly reduced after drug treatment (Figure 1g), consistent with the reduction of paox by treatment with these agents. These data together, demonstrate that the polyamine catabolic enzyme paox that is downregulated upon genotoxic stress is critical in functioning as a regulator of DNp73 levels.
PAOX is regulated by selected AP-1 members
Treatment of cells with doxorubicin or cisplatin led to the downregulation of paox, which paralleled an increase in the expression of AP-1 factors c-Jun and FosB, concomitant to an increase of processed Az [Az1(p)] (Figure 2a). We therefore examined whether c-Jun and other AP-1 members are able to regulate paox expression. Overexpression of only c-Jun, JunB and FosB led to a dose-dependent downregulation of paox expression, whereas no significant effects were observed with c-Fos, Fra1 (Figure 2b) and JunD (data not shown). Conversely, silencing their expression resulted in increased basal paox expression as well as reduced its downregulation by stress (Figure 2c), thus confirming the requirement of c-Jun, JunB and FosB for the downregulation of paox upon doxorubicin stress.
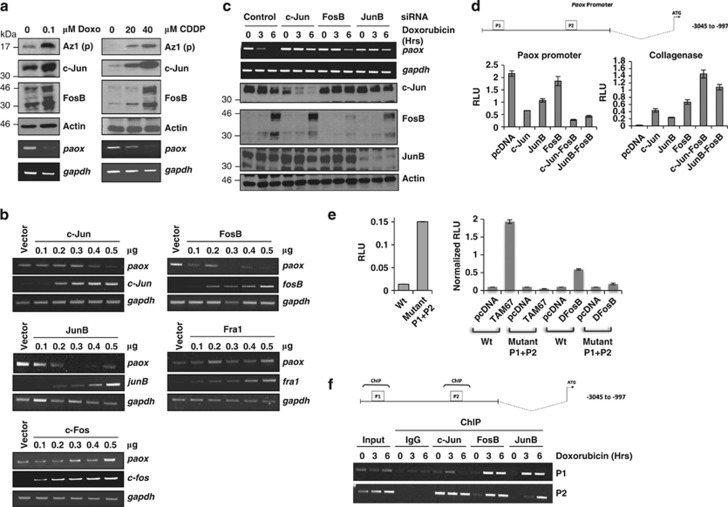
Figure 2.
Polyamine oxidase (paox) expression is selectively regulated by AP-1 family members. (a) Expression of processed antizyme, Az1(p), and the indicated AP-1 members were analyzed by immunoblotting, and mRNA expression of paox was monitored by semi-quantitative RT-PCR, after exposure of H1299 cells to different concentration of doxorubicin (doxo) or cisplatin (CDPP), for 16 h. (b) Expression of paox was determined by RT-PCR 24 h after transfection of increasing amounts of indicated AP-1 members in H1299 cells. (c) Effect of siRNA-mediated silencing of c-jun, fosB or junB on the expression of paox mRNA upon doxorubicin treatment at the indicated times was analyzed by semi-quantitative PCR in H1299 cells (upper panels). Knockdown efficiency after 48 h was assessed by immunoblotting (lower panels). (d) Schematic showing human paox promoter with two potential AP-1-binding sites (P1 and P2) predicted in silico (upper panel). The effect of overexpresssing c-Jun, JunB, FosB or the c-Jun/FosB and JunB/FosB dimers on the promoter activity of paox (left) or collagenase (right) was assessed by luciferase reporter assay in H1299 cells. RLU: relative luciferase units, which are ratios of luciferase and β-galactosidase activities. All values are means±S.D. of duplicate experiments. (e) The effect of ablating the predicted AP-1-binding sites on paox promoter activity was assessed using the wild-type and mutated (Mutant P1+P2) promoters (left panel) in the absence or presence of dominant-negative c-Jun (TAM67) or FosB (DNFosB) (right panel). All values are means±S.D. of duplicate experiments. (f) H1299 cells exposed to doxorubicin for the indicated time periods were used for chromatin immunoprecipitation (ChIP) using antibodies against the indicated AP-1 members (lower panel). Total DNA collected before immunoprecipitation was used as input control and binding specificity was demonstrated by the negative signal in anti-rabbit IgG control. Schematic shows sites of enrichment for AP-1 on the human paox promoter (upper panel)
We next investigated whether all three AP-1 transcription factors were able to repress paox expression by directly acting on its promoter. For this purpose, we cloned a portion of the paox promoter (−3045 to −997) predicted by TESS (Transcription Element Search System online software) to contain two potential AP-1-binding sites (indicated as P1 and P2 in Figure 2d upper panel) into a luciferase reporter construct. Overexpression of c-Jun and JunB led to a significant decrease, and FosB led to a marginal decrease in paox promoter activity (Figure 2d, lower left panel) and in parallel, upregulated collagenase promoter activity, a classical target of AP-1 transcription factors27 (Figure 2d, lower right panel). Members of the AP-1 transcription factors form heterodimers with each other to regulate many of their target genes25, 27 and expectedly, AP-1 monomers that were tethered to force specific pairing of c-Jun/FosB and JunB/FosB26 suppressed paox or activated collagenase to a higher level relative to c-Jun, JunB and FosB alone (Figure 2d, lower panels). In order to locate the binding sites for AP-1 on the paox promoter, the two predicted repressor sites (P1 and P2) were mutated. As expected, mutations of both P1 and P2 sites increased the basal promoter activity by >10-fold relative to the wild-type paox promoter, indicating that P1 and P2 were indeed the repression sites of these selective AP-1 factors (Figure 2e, left panel). Furthermore, expression of transactivation-deficient mutants of c-Jun (TAM67) or FosB (DFosB), which exert a dominant negative effect on AP-1 transcriptional activity,29, 30 led to the de-repression of the paox promoter, which was not observed upon mutation of the predicted binding sites of AP-1 factors, P1 and P2 (Figure 2e, right panel). Moreover, the mutant promoter was not suppressed by the AP-1 members (Supplementary Figure 2A), altogether confirming the requirement of these sites for AP-1-mediated repression of paox.
To determine whether these three AP-1 transcription factors could bind to paox promoter in vivo, we performed chromatin immunoprecipitation (ChIP) of c-Jun, JunB and FosB on regions corresponding to P1 and P2. All three AP-1 members were highly enriched at the predicted sites (Figure 2f), but significant loading of JunB and FosB only occurred after doxorubicin treatment, whereas c-Jun was generally constitutively bound on P2. Altogether, the data show that selected members of the AP-1 family, namely c-Jun, JunB and FosB, bind to the paox promoter after genotoxic stress to repress its expression.
Selectivity in PAOX regulation by AP-1 members corresponds to DNp73 degradation
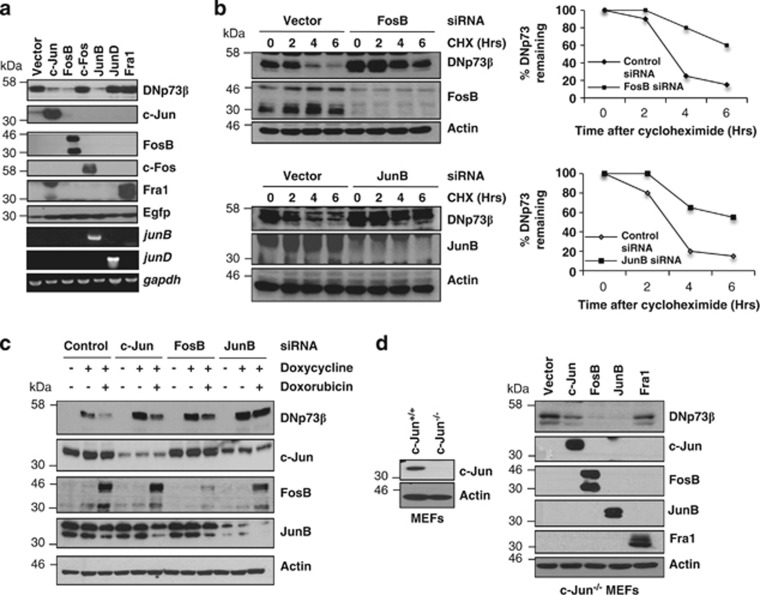
We next assessed the ability of the various members of the AP-1 family to degrade DNp73. Co-expression of DNp73β with several AP-1 members showed that besides c-Jun, only JunB and FosB were equally competent at decreasing the steady-state levels of DNp73 (Figure 3a). This effect was further confirmed using the protein synthesis inhibitor cycloheximide (CHX), which revealed that the half-life of DNp73β was significantly extended from 3.5 h to ∼6 h upon silencing the expression of FosB, JunB (Figure 3b) or c-Jun (Supplementary Figure 2B). Conversely, overexpression of these AP-1 members led to a significant reduction in the half-life of DNp73 (Supplementary Figures 2C and D). Furthermore, individual silencing of c-Jun, JunB or FosB in DNp73β-inducible H1299 cells before doxorubicin treatment led to the stabilization of DNp73β levels at the basal level, and significantly inhibited the doxorubicin-mediated degradation of DNp73β (Figure 3c). We further also assessed whether JunB and FosB could degrade DNp73 independently of c-Jun. Overexpression of FosB or JunB, but not Fra1, in mouse embryonic fibroblasts deficient in c-Jun was able to degrade DNp73β to a similar extent as exogenously expressed c-Jun (Figure 3d), illustrating a clear functional redundancy of these three AP-1 members in regulating DNp73. These data together with the observation that only c-Jun, JunB and FosB can regulate paox expression (Figure 2), highlight the specificity of these AP-1 members in degrading DNp73 independently.
Figure 3.
Selectivity in AP-1-mediated regulation of paox correlates with selectivity in AP-1-dependent DNp73 degradation. (a) Effect of co-expressing DNp73β with the indicated AP-1 members on the steady-state levels of DNp73 were assessed by immunoblotting 24 h after transfection in H1299 cells. Expression of AP-1 members was analyzed by immunoblotting and semi-quantitative PCR. Egfp was co-transfected as a control for transfection efficiency. (b) Half-life of transfected DNp73β after siRNA-mediated knockdown of FosB and JunB were assessed by the addition of 25 μg/ml cycloheximide for the indicated times before immunoblotting. The adjacent graphs show the quantification of the remaining DNp73 (arbitrary values). (c) H1299 cells inducibly expressing DNp73β were transfected with control scramble, c-jun-, fosB- or junB-specific siRNA for 24 h before induction of DNp73β expression for a further 24 h by doxycycline addition. Cells were then treated with doxorubicin for 4 h and harvested for immunodetection. (d) c-jun−/− mouse embryonic fibroblasts (MEFs) were transfected with DNp73β and the AP-1 members for 36 h before analysis (right panel). Status of endogenous c-Jun in wild-type and knockout MEFs is shown by immunoblotting (left panel)
AP-1-mediated degradation of DNp73 occurs via the PAOX-Az pathway
As besides c-Jun, JunB and FosB also have the ability to downregulate paox and subsequently degrade DNp73 levels, it is highly likely that Az1(p) processing is a common mechanism regulated by all three AP-1 members. To test this hypothesis, we examined the effects of AP-1 members on Az1(p) production, with addition of putrescine being used as a positive control to induce Az1(p) (Figure 4a, last lane from left). Overexpression of c-Jun, JunB or FosB, but not Fra1, was able to upregulate Az1(p) expression, with no significant changes in the levels of Az1 mRNA. To validate the role of Az1 in mediating the degradation of DNp73 by JunB and FosB, we used the natural inhibitor of Az1, AZI, to block the function of this pathway. Degradation of DNp73 by JunB or FosB (Figure 4b), or c-Jun20 (data not shown) was significantly reduced in the presence of AZI. Similarly, knockdown of Az1 mRNA reduced c-Jun and FosB-mediated DNp73 degradation (Supplementary Figure 3a). Together, these data confirm the role of Az1 as a downstream effector of all three AP-1 transcription factors to degrade DNp73.
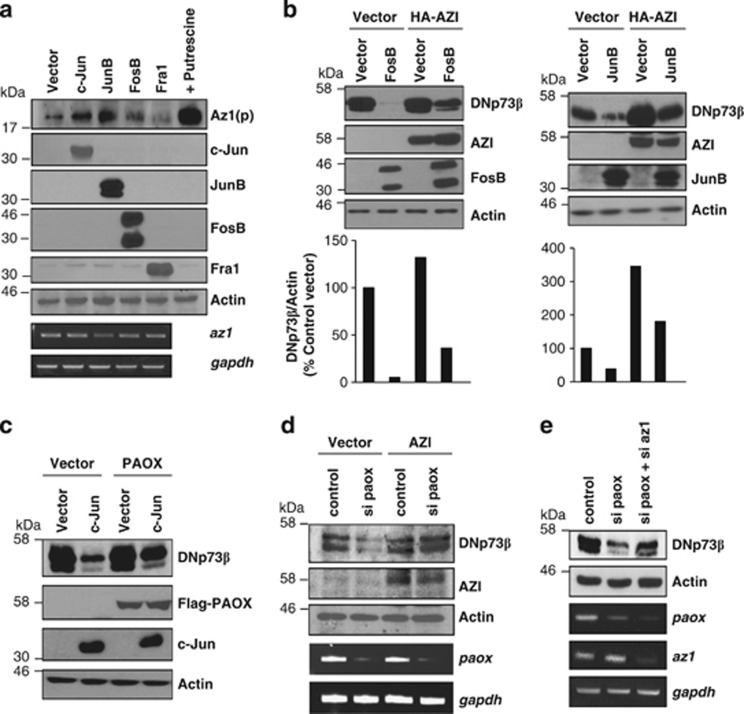
Figure 4.
AP-1-mediated DNp73 depletion occurs via the PAOX-antizyme pathway. (a) Effect of overexpressing AP-1 members on the expression of Az1 mRNA and processed Az, [Az1(p)], was assessed by semi-quantitative RT-PCR and immunodetection, respectively, 24 h after transfection in H1299 cells. (b and c) Effect of the AP-1 members on transfected DNp73β in the presence of the naturally occurring antizyme inhibitor, AZI (b) or Flag-tagged PAOX (c) was determined by immunoblot analysis, 24 h after transfection in H1299 cells. Lower panels show quantification of DNp73 from the respective lanes from the blots above (b). (d and e) Effect of siRNA-mediated silencing of paox in the presence of AZI (d) or Az1 siRNA (e) on co-transfected DNp73β levels was determined by immunoblotting 24 h and 48 h after transfection in H1299 cells, respectively. Knockdown efficiency was determined by semi-quantitative RT-PCR
Given that the production of Az1(p) is governed by polyamine biosynthesis, downregulation of catabolic enzyme PAOX by stress signals could lead to elevated polyamine content, thus enhancing Az1(p) production. Inhibition of PAOX activity by treatment with MDL72527 indeed led to enhanced production of Az1(p) (Supplementary Figure 3B). We thus assessed whether suppression of paox is the mechanism employed by these three AP-1 factors to degrade DNp73. To verify this, PAOX was overexpressed to counteract the downregulation of endogenous paox mediated by c-Jun. Co-expression of Flag-tagged PAOX with c-Jun inhibited the reduction of DNp73 levels (Figure 4c), suggesting that transcriptional repression of paox is indeed mediating the effect of AP-1-dependent degradation of DNp73.
To further confirm the role of PAOX-Az axis in regulating DNp73, we examined whether Az1 was acting downstream of paox. Overexpression of AZI, which inhibits Az1, rescued DNp73 degradation caused by paox knockdown (Figure 4d). Similarly, silencing of Az1 partially inhibited DNp73 degradation induced by paox silencing (Figure 4e). Taken together, these data demonstrate that suppression of the polyamine catabolic enzyme paox by c-Jun, JunB and FosB leads to the degradation of DNp73 via the Az pathway.
PAOX inhibition reverses DNp73-dependent chemoresistance
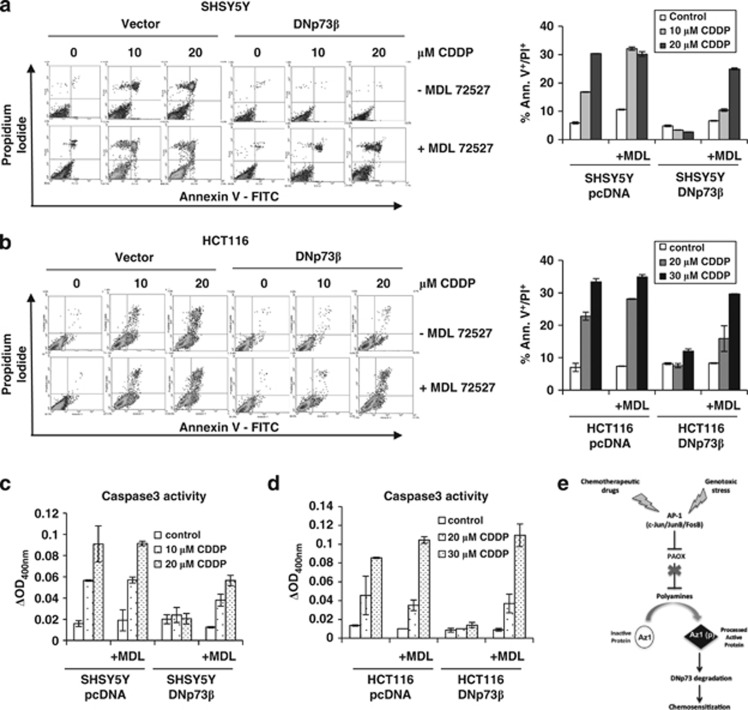
Analysis of human cancer data sets using Oncomine showed that paox was indeed highly expressed in several types of tumors relative to their normal counterparts (Supplementary Figure 4A), emphasizing a potential clinical relevance of PAOX. Given that DNp73 overexpression leads to resistance to chemotherapeutic drugs such as cisplatin,5 and that PAOX expression regulates the abundance of DNp73, we assessed whether inhibition of PAOX with MDL72527 can reverse the chemoresistance mediated by DNp73. Treatment with MDL72527 was sufficient for the reduction of DNp73β levels in the SH-SY5Y neuroblastoma cell line overexpressing DNp73β in a dose-dependent manner (Supplementary Figure 4B). Concomitant cell death analysis revealed that cisplatin treatment resulted in increased cell death only in vector SHSY-5Y cells but not in DNp73β overexpressing cells (% dead cells 0/10/20 μM cisplatin – vector versus DNp73β cells: 5.85/16.8/30.35 versus 4.8/3.35/2.65; Figure 5a). Treatment with MDL72527 alone caused a slight but consistent increase in cell death in SHSY5Y cells (% dead vector cells ±MDL72527: 5.85 versus 9.5). However, treatment of MDL72527 in combination with cisplatin potentiated cell death in SH-SY5Y-DNp73β (% dead cells 0/10/20 μM cisplatin – vector versus DNp73β cells: 10.6/32.1/30.2 versus 6.6/10.4/24.95; Figure 5a). Similar results were observed when the DNp73β overexpressing cells were treated with doxorubicin (% dead cells 0/0.25/0.5/1 μM doxorubicin – vector versus DNp73β cells: 2/22.6/29.45 versus 2/1.45/1.1), and the same potentiation effect of MDL72527 on cell death was observed specifically in DNp73β cells (% dead cells 0/0.25/0.5/1 μM doxorubicin – with versus without MDL72527: 4.85/17/24 versus 2.75/4.75/17.25; Supplementary Figure 5A). Furthermore, resistance induced by stable DNp73β overexpression in another cell line, HCT116 (% dead cells 0/20/30 μM cisplatin – vector versus DNp73β cells: 7/22.8/33.4 versus 8.2/7.55/12.05) was equally reversed in the presence of MDL72527 (% dead cells 0/20/30 μM cisplatin – vector versus DNp73β cells: 7.35/28.1/34.9 versus 8.3/15.9/29.65; Supplementary Figure 5B and Figure 5b). The chemosensitization effect of MDL72527 upon cisplatin or doxorubicin was verified to be through apoptotic activation as demonstrated by enhancement in cleaved-caspase-3 activity (Figures 5c and d, and Supplementary Figure 5C). In addition, siRNA-mediated knockdown of paox before cisplatin treatment also led to sensitization to cell death in DNp73β overexpressing HCT116 (Supplementary Figure 5D). These data therefore indicate that the protective effect of DNp73 against exposure to chemotherapeutic drugs could be reversed by inhibition of the polyamine catabolic enzyme PAOX.
Figure 5.
PAOX inhibition reverses chemoresistance by inducing apoptosis. (a and b) SHSY5Y-DNp73β or SHSY5Y-pcDNA cells were treated with the indicated doses of cisplatin (CDDP) (a) in the absence or presence of 200 μM of MDL72527 for 24 h, and the proportion of cell death was determined by Annexin V/propidium iodide (PI) staining and flow cytometry. Raw data are shown as density plots (left) and percentage of Annexin V+/PI+ cells are represented in a graphical format (right). The same experiment as a was repeated with HCT116-pcDNA and HCT-116-DNp73β (b). All values are means±S.D. of duplicate experiments. (c and d) SHSY5Y-DNp73β or SHSY5Y-pcDNA cells were treated with the indicated doses of cisplatin (CDDP) in the absence or presence of 200 μM of MDL72527 for 24 h and caspase-3 activity in cell lysates were measured using a colorimetric protease assay. The values were calculated by subtracting background signal (with no substrate) from duplicate experiment (c). The same experiment as c was repeated in HCT116-pcDNA and HCT116-DNp73β cells (d). All values are means±S.D. of duplicate experiments. (e) Schematic illustrates the proposed mechanism by which genotoxic stresses can induce the degradation of anti-apoptotic DNp73 by activating c-Jun, FosB and JunB. Upon stress, these three AP-1 family members are induced and inhibit the expression of the polyamine catabolic enzyme paox, resulting in the eventual increase in the expression of Az1(p), which leads to the degradation of DNp73, and consequently, chemosensitizes to genotoxic drugs
Discussion
High levels of DNp73 have been correlated with increased chemoresistance and poor clinical outcome in many cancers.8, 9, 10 Therefore, understanding mechanisms of how DNp73 expression is regulated in cancers and subsequently, devising strategies to specifically target DNp73 are critical for improving treatment.31 In this respect, we have previously shown that DNp73 is degraded by c-Jun upon exposure to genotoxic stress via the Az-mediated degradation pathway.20 However, the exact mechanisms for this process as well as its relevance to DNp73 in mediating chemoresistance was unclear. Az has been shown to degrade several cell-cycle regulatory proteins, including cyclin D1, Smad1 and Aurora A kinase, as well as Mps1, a protein that regulates centrosome duplication,32, 33, 34, 35 supporting its role as an inhibitor of tumor cell proliferation and transformation. The expression of Az is tightly controlled at the post-transcriptional level by a +1 frameshift translation mechanism, which is regulated by the polyamine biosynthesis pathway.21 Polyamine homeostasis in cells is modulated by the interplay of several anabolic and catabolic enzymes22 and we have now shown that paox is specifically targeted by selected AP-1 factors that were activated upon exposure to chemotherapeutic drugs. PAOX is a FAD-dependent oxidase that preferentially retro-converts NASp to spermidine and N-acetylspermidine to putrescine.22 Putrescine has been shown to be less effective at inducing Az expression compared with spermidine and spermine36 and therefore inhibition of PAOX would likely lead to higher levels spermine and spermidine relative to putrescine, thus enabling a more pronounced induction of Az in response to genotoxic stress. Many studies have shown that MDL72527, a chemical inhibitor of PAOX, can potentiate apoptosis in several cancer cells both in vitro and in vivo.37 Conversely, expression of another polyamine catabolic enzyme SSAT under the keratinocyte-specific K6 promoter was shown to promote skin tumors.38 Interestingly, treatment of these mice with MDL72527 partially counteracted tumor growth,38 indicating that inhibition of polyamine catabolism can be a potential therapeutic tool to suppress tumor formation. It is noteworthy that enhanced polyamine synthesis has also been shown to promote tumor growth and progression.38 For example, overexpression of the polyamine biosynthesis enzymes ODC or SAMDC induced malignant transformation.38, 39 All these observations together suggest that disruption of the homeostatic balance of polyamine ratios and not their overall cellular increase could support tumorigenesis. Our current findings that inhibition of PAOX in DNp73-overexpressing cancer cells increases their sensitivity to apoptosis-inducing chemotherapeutic drugs are consistent with these observations. Interestingly, several studies targeting the polyamine pathways in conjunction with other drugs are currently in clinical trials.40 However, the direct role of PAOX in tumor formation in vivo and its effects on the levels of the different polyamines warrants further investigation in the future.
PAOX promoter is repressed upon exposure to genotoxic stresses through selective members of the AP-1 transcription family, namely c-Jun, JunB and FosB. c-Jun is known to have a dual role in regulating cell fate where it can either enable apoptotic responses or support growth and proliferation, depending on cellular context.41 These distinct functions of c-Jun might be determined by the interplay with additional factors such as the interaction with other AP-1 members. Similar to c-Jun, other AP-1 members can also be induced by DNA-damaging signals,42, 43 indicating their potential involvement in promoting apoptosis with c-Jun. The data presented here show that c-Jun, JunB and FosB are activated by chemotherapeutic drugs to regulate DNp73 degradation. Although JunB is classically known to antagonize some biological functions of c-Jun such as cell proliferation,27 both also share redundancy on certain target genes, which include Egfr and Proliferin.44, 45 On the other hand, FosB has been shown to bind in a complex with c-Jun, which was associated with FasL induction and cell death, indicating that c-Jun/FosB dimer has a pro-apoptotic function.46 Consistently, our data indicate that similar to c-Jun, expression of JunB and FosB could also induce the suppression of paox, increase the level of Az and lead to the subsequent degradation of DNp73. Moreover, JunB and FosB had similar effects in c-Jun-deficient cells, suggesting a level of in-built redundancy to perhaps ensure the efficient regulation of DNp73 levels upon genotoxic insults.
In conclusion, we have unraveled a mechanism through which selective AP-1 transcription factors c-Jun, JunB and FosB induce the degradation of anti-apoptotic DNp73 by Az, via the suppression of polyamine catabolic enzyme PAOX (Figure 5e). We have therefore identified PAOX as a positive regulator of DNp73 that defines a potential strategy to improve the therapeutic index of cytotoxic drugs in DNp73-overexpressing chemoresistant tumors.
Materials and Methods
Cells and reagents
Cell lines used are as follows: human cell lines – p53 null H1299 lung cancer cells – parental and with the inducible DNp73β-construct-tet-on system; p53 proficient neuroblastoma cell line SHSY5Y and colorectal cell line HCT116 stably expressing DNp73β; mouse fibroblasts – wild-type and cJun−/−. 2 μg/ml of doxycycline was added to H1299 and HCT116 DNp73β inducible cells to induce DNp73β. Chemicals and inhibitors used: doxorubicin (Sigma, St. Louis, MO, USA), Cisplatin (Sigma), putrescine (Sigma), MDL72527 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and CHX (Sigma).
Plasmids, siRNAs and transfection
Plasmids used in this study are as follows: pCDNA3.1, DNp73β, c-Jun, Flag-Tam67, JunB, FosB, Flag-Delta Fosb, c-Fos, Fra1, JunD, HA-AZI, Flag-PAOX (kind gift from Dr. Robert Casero Jr., John Hopkins University School of Medicine), β-gal, Egfp, pGL3-luc basic, various constructs of paox-luc (as indicated in the results section) and collagenase-luc. 2 × 105 cells were transfected with the various plasmids using Lipofectamine Plus-Reagent. Total amount of transfected DNA were equalized with appropriate amounts of pCDNA3 vector in all cases. Cells were collected 24–48 h after transfection for immunoblot analyses. siRNA transfections were performed using Transmessenger transfection kit (Qiagen, Hilden, Germany) or HiPerfect (Qiagen) and cells were harvested 48 h post transfection. Target sequence of siRNAs used are as follows: human c-jun: 5′-AAGAACGTGACAGATGAGCAG-3′ human junb: 5′-GAACGTGACAGATGAGCAG-3′ human fosB: 5′-GAGGAAGAGGAGAAGCGAA-3′ human paox: 5′-CGGAGATGATGGACACATA-3′ human Az1: 5′-GCTGAATGTAACAGAGGAA-3′ and scramble control: 5′ AATTCTCCGAACGTGTCACGT 3′.
Immunoblot analysis and half-life determination
Immunoblot analysis was performed essentially as described.47 A measure of 50 μg cell lysate was used for analyses and detected using enhanced Chemiluminescent reagent (Amersham Biosciences, Buckinghamshire, UK) or Super signal West Dura (Pierce Biotechnology, Rockford, IL, USA). Antibodies used in this study are: anti-p73 (ER15; Calbiochem, San Diego, CA, USA), anti-p73 (GC15; Calbiochem), anti-HA (Y-11; Santa Cruz Biotechnology), anti-FLAG (M2; Stratagene, La Jolla, CA, USA), anti-c-Jun (Cell Signaling Technology, Danvers, MA, USA), anti-FosB (5G4; Cell Signaling Technology), anti-JunB (P169; Cell Signaling Technology), anti-Fra1 (R20; Santa Cruz Biotechnology), anti-Actin (Sigma), anti-Egfp (Santa Cruz Biotechnology), anti-cyclin D1 (H295; Santa Cruz Biotechnology), anti-Az1(p) (a gift from Dr. John Mitchell, Northern Illinois University), anti-AZI (a gift from Dr. Senya Matsufuji, The Jikei University School of Medicine). For measuring half-life of proteins, cells were treated with 50 μg/ml of CHX and collected at indicated time points for immunoblotting.
Semi-quantitative RT-PCR analyses
Total RNA was prepared from cells using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) and converted into single-strand cDNA using Superscript II Reverse Transcriptase (Invitrogen). Semi-quantitative PCR was performed using Taq polymerase (Qiagen) in a 25-μl solution according to manufacturers' protocols. The reactions were carried under the following conditions: initial 94 °C for 3 min, 94 °C for 35 s, 55 °C for 35 s and 72 °C for 35 s for indicated cycles. Primers specific to both human (h) and mouse (m) genes are:
samdc (h) forward 5′GGTGATGGAAGCTGCACATTT3′, reverse 5′-GGTGGTACCACATGTCTTCAA-3′ odc (h) forward 5′-GACTGAAATACAGTTGGTG-3′, reverse 5′-TCAGATCCAGGAAAGCCAC-3′ spds (h) forward 5′-GAGTTCTCCTACCAGGAGATG-3′, reverse 5′-GTAATAGGACTCCTTGAAGAG-3′ spms (h) forward 5′-GTGTTCTGGACCTTCAGAG-3′, reverse 5′-AAACTGCTTCGAGTGTAG-3′ paox (h) forward 5′-AAGCAATCAGGAAGATAGGC-3′, reverse 5′-CAGAAGTACTTCTTCATCCG-3′, ssat (h) forward 5′-GAAGAGGTGCTTCTGATCTGTC-3′, reverse 5′-ACTCTCCAAACTGCATCTGAG-3′ fosb (h) forward 5′-CAAGACTTGGAAACTTGATTG-3′, reverse 5′-TACAGAATCAGCTTCAGCTAC-3′ gapdh (m/h) forward 5′-CATTGACCTCAACTACATG-3′, reverse 5′-CTTCTCCATGGTGGTGAAG-3′ cyclin D1 (h) forward 5′-CTGGCCATGAACTACCTGGA-3′, reverse 5′-GTCACACTTGATCACTCTGG-3′ cjun (m/h) forward 5′-CTGCTCGTCGGTCACGTTCTTGGG-3′, reverse 5′-ATGCCCTCAACGCCTCGTTCCTCC-3′ junb (m) forward 5′-ACCTGGCGGATCCCTATCGG-3′, reverse 5′-GCTCCGGACCAGCATAGACG-3′ fra1 (m/h) forward 5′- GGCTGCAGCGAGAGATTGA-3′, reverse 5′-TGGTCATGAGTGTGGGGGT-3′ cfos (m/h) forward 5′-CCAACCTGCTGAAGGAGAAG-3′, reverse 5′-GCTGCTGATGCTCTTGACAG-3′ junD (m) forward 5'-GATTGAAACCAGGGTCGG-3', reverse 5′-CTGCTCAGGTTGGCGTAGAC-3′.
Site-directed mutagenesis (SDM) and luciferase reporter assays
The –3045 to –997 paox promoter fragment was cloned into in pGL3 basic or enhancer vector (Promega, Fitchburg, WI, USA). H1299 cells were transiently transfected with 0.2 μg of the relevant plasmids along with the reporter (paox-luc or collagenase-luc) and β-galactosidase constructs to normalize for transfection efficiency. Luciferase assays were performed as described.47
Potential AP-1-binding sites were found using TESS (Transcription Element Search System online software). SDM of the AP-1 sites P1 and P2 were carried out as described.5 Primers used are: SDM-P1 5′-ATGAACAAGCCAAGTTACAGTCAAAGCCAC-3′ SDM-P2 5′-ACCGCCCAGAGAATAAGCCCGACTC-3′.
ChIP
H1299 cells treated with 5 μM doxorubicin for 0, 3 and 6 h were crosslinked with 1% formaldehyde for 10 min at room temperature. The reaction was stopped by the addition of ice-cold PBS and washed twice with the same buffer. Cells were lysed for 10 min at 4 °C with Triton X-100 lysis buffer (0.25% triton X-100, 10 mM EDTA, 10 mM Tris pH8, 100 mM NaCl and protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). Samples were centrifuged at 1500 r.p.m. at 4 °C for 5 min and the pellet containing the chromatin fraction was lysed in SDS lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris pH8 and protease inhibitor cocktail). Chromatin extracts containing genomic DNA were sonicated on ice to an average size of 500 bp and subsequently immunoprecipitated using control anti-IgG, anti c-Jun, anti-FosB or anti-JunB (all from Cell Signalling as previously described) at 4 °C. The immunocomplex was incubated with 50 μl of 50% slurry Protein A-Sepharose beads (Invitrogen) for a further 2 h at 4 °C. Samples were washed with, 1 × HS buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8, 500mM NaCl), 1 × LS buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8, 150 mM NaCl), 1 × 250 mM LiCl buffer at 4 °C for 10 min, followed by 2 × in TE buffer. DNA/protein complex was eluted three times with 100 μl of extraction buffer (1%SDS and 100 mM NaHCO3) for 15 min. To reverse DNA/protein crosslink, eluted samples were incubated with 200 mM NaCl for 4 h at 65 °C, followed by proteinase K treatment at final concentration of 500 μg/ml at 45 °C for 2 h. DNA was purified for PCR using DNA Qiaquick columns (Qiagen).
Semi-quantitative PCR analyses were performed to assess the ChIP-enriched DNA, and non-immunoprecipitated input DNA was included as loading control. The following primers were used: P1 paox promoter forward 5′-GGAGAAGTCTTACAGCATGAT-3′, P1 paox promoter reverse 5′- ACAAGGTATGTGCTTCTCCTA-3′ P2 paox promoter forward 5′- TCCTTAGTCCTGGCATGTCGG-3′, P2 paox promoter reverse 5′- GTGACCTGGTGTCGCTGGATT-3′.
Cell death assay
Cell death was measured by propidium iodide (PI) exclusion assay. SHSY5Y stably transfected with pcDNA or DNp73β cells were treated with indicated doses of doxorubicin or cisplatin with or without MDL72527. Both floating and attached cells were harvested and washed 2 × with PBS before re-suspension in 2 μg/ml PI. Percentage of Annexin V+/PI+ cells was measured by flow cytometry (BD Biosciences, FACScalibur, San Jose, CA, USA).
Polyamine measurements
Polyamine levels were measured by HPLC as described previously.48 Cells treated with doxorubicin or cisplatin, or untreated controls (6 × 105 cells) were harvested and polyamines were extracted from cell lysate with 5% trichloroacetic acid and the supernatant after centrifugation at 27 000 × g for 15 min at 4 °C was used for polyamine measurement by HPLC. The precipitate was used for measurement of protein content.
Caspase activity assay
Caspase-3 activity was measured by colorimetric assay according to manufacturer's manual (ApoTarget Caspase-3 Kit, Life Sciences, Invitrogen). In brief, cells were lysed in Tris-buffered saline containing reagent and protein concentrations were measured using Comassie Bradford's reagent. 200 μg of the extracted protein were mixed with 2X extraction buffer and 200 μM of DEVD-pNA caspase-3 substrate and incubated for 2 h away from light at 37 °C. Developed reaction was measured at the optical density of 400 nm using spectrophotometry, and the absorbance reading for caspase activity was calculated by subtracting the absorbance of blank samples (without substrate).
Acknowledgments
We thank the National Medical Research Council of Singapore for financial support to KS and Drs John Mitchell, Senya Matsufuji, Robert Casero Jr. and Leah Vardy for reagents and advise on their use.
Glossary
- AP-1
activator protein-1
- Az
Antizyme
- DNp73
Delta-N p73
- PAOX
polyamine oxidase
- ODC
ornithine decarboxylase
- SAMDC
S-adenosylmethionine decarboxylase
- SPDS
spermidine synthase
- SPMS
spermine synthase
- SSAT
spermidine/spermine N1-acetyltransferase
- CHX
cycloheximide
- NAsp
acetylspermine
- PI
propidium iodide
- ChIP
chromatin immunoprecipitation
- SDM
site-directed mutagenesis
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino.
Supplementary Material
References
- Melino G, De Laurenzi V, Vousden KH.p73: friend or foe in tumorigenesis Nat Rev Cancer 20022605–615.Review. [DOI] [PubMed] [Google Scholar]
- Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- Stiewe T, Theseling CC, Pützer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- Liu G, Nozell S, Xiao H, Chen X. DeltaNp73beta is active in transactivation and growth suppression. Mol Cell Biol. 2004;24:487–501. doi: 10.1128/MCB.24.2.487-501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh WH, Logette E, Corcos L, Sabapathy K. TAp73beta and DNp73beta activate the expression of the pro-survival caspase-2S. Nucleic Acids Res. 2008;36:4498–4509. doi: 10.1093/nar/gkn414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kameoka M, Itaya A, Ota K, Yoshihara K. Regulation of HSF1-responsive gene expression by N-terminal truncated form of p73alpha. Biochem Biophys Res Commun. 2004;317:865–872. doi: 10.1016/j.bbrc.2004.03.124. [DOI] [PubMed] [Google Scholar]
- Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- Buhlmann S, Pützer BM. DNp73 a matter of cancer: mechanisms and clinical implications. Biochim Biophys Acta. 2008;1785:207–216. doi: 10.1016/j.bbcan.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Lin KW, Nam SY, Toh WH, Dulloo I, Sabapathy K. Multiple stress signals induce p73beta accumulation. Neoplasia. 2004;6:546–557. doi: 10.1593/neo.04205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di C, Yang L, Zhang H, Ma X, Zhang X, Sun C, et al. Mechanisms, function and clinical applications of DNp73. Cell Cycle. 2013;12:1861–1867. doi: 10.4161/cc.24967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiewe T, Pützer BM. Role of p73 in malignancy: tumor suppressor or oncogene. Cell Death Differ. 2002;9:237–245. doi: 10.1038/sj.cdd.4400995. [DOI] [PubMed] [Google Scholar]
- Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell Death Differ. 2004;11:685–687. doi: 10.1038/sj.cdd.4401376. [DOI] [PubMed] [Google Scholar]
- Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KK, Yuan ZM. c-Abl stabilizes p73 by a phosphorylation-augmented interaction. Cancer Res. 2003;63:3418–3424. [PubMed] [Google Scholar]
- Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, et al. Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med. 2004;199:1545–1557. doi: 10.1084/jem.20031943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ryu H, Minamishima YA, Ryo A, Lee SW. Modulation of p53 and p73 levels by cyclin G: implication of a negative feedback regulation. Oncogene. 2003;22:1678–1687. doi: 10.1038/sj.onc.1206306. [DOI] [PubMed] [Google Scholar]
- Dulloo I, Gopalan G, Melino G, Sabapathy K. The antiapoptotic DeltaNp73 is degraded in a c-Jun-dependent manner upon genotoxic stress through the antizyme-mediated pathway. Proc Natl Acad Sci USA. 2010;107:4902–4907. doi: 10.1073/pnas.0906782107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, et al. Autoregulatory frameshifting in decoding mammalian ornithinedecarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol. 2002;22:4952–4964. doi: 10.1128/MCB.22.13.4952-4964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Coffino P. Regulation of cellular polyamines by antizyme. Nat Rev Mol Cell Biol. 2001;2:188–194. doi: 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- Young MR, Li JJ, Rincón M, Flavell RA, Sathyanarayana BK, Hunziker R, et al. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M, Qu S, Moore C, Wisdom R. Transcriptional activation and transformation by FosB protein require phosphorylation of the carboxyl-terminal activation domain. Mol Cell Biol. 1997;17:2372–2380. doi: 10.1128/mcb.17.5.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- Newman RM, Mobascher A, Mangold U, Koike C, Diah S, Schmidt M, et al. Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J Biol Chem. 2004;279:41504–41511. doi: 10.1074/jbc.M407349200. [DOI] [PubMed] [Google Scholar]
- Lin Y, Martin J, Gruendler C, Farley J, Meng X, Li BY, et al. A novel link between the proteasome pathway and the signal transduction pathway of the bone morphogenetic proteins (BMPs) BMC Cell Biol. 2002;3:15. doi: 10.1186/1471-2121-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26:6593–6603. doi: 10.1038/sj.onc.1210482. [DOI] [PubMed] [Google Scholar]
- Kasbek C, Yang CH, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell. 2010;21:3878–3889. doi: 10.1091/mbc.E10-04-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ray RM, Viar MJ, Johnson LR. Polyamine regulation of ornithine decarboxylase and its antizyme in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2001;280:G130–G138. doi: 10.1152/ajpgi.2001.280.1.G130. [DOI] [PubMed] [Google Scholar]
- Seiler N, Renault J, Gossé F, Roussi S, Raul F. Cytotoxicity of the polyamine oxidase inactivator MDL 72527 to cancer cells: comparison with a saturated structural analogue. Int J Oncol. 2005;27:1669–1676. [PubMed] [Google Scholar]
- Wang X, Feith DJ, Welsh P, Coleman CS, Lopez C, Woster PM, et al. Studies of the mechanism by which increased spermidine/spermine N1-acetyltransferase activity increases susceptibility to skin carcinogenesis. Carcinogenesis. 2007;28:2404–2411. doi: 10.1093/carcin/bgm162. [DOI] [PubMed] [Google Scholar]
- Chen Y, Megosh LC, Gilmour SK, Sawicki JA, O'Brien TG. K6/ODC transgenic mice as a sensitive model for carcinogen identification. Toxicol Lett. 2000;116:27–35. doi: 10.1016/s0378-4274(00)00196-x. [DOI] [PubMed] [Google Scholar]
- Nowotarski SL, Woster PM, Casero RA., Jr Polyamines and cancer: implications for chemotherapy and chemoprevention. Expert Rev Mol Med. 2013;15:e3. doi: 10.1017/erm.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A, Sibilia M, Wagner EF. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M.AP-1 as a regulator of cell life and death Nat Cell Biol 20024E131–E136.Review. [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Le Panse R, Cano E, Mahadevan LC. Anisomycin selectively desensitizes signalling components involved in stress kinase activation and fos and jun induction. Mol Cell Biol. 1998;18:1844–1854. doi: 10.1128/mcb.18.4.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4:879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- Toft DJ, Rosenberg SB, Bergers G, Volpert O, Linzer DI. Reactivation of proliferin gene expression is associated with increased angiogenesis in a cell culture model of fibrosarcoma tumor progression. Proc Natl Acad Sci USA. 2001;98:13055–13059. doi: 10.1073/pnas.231364798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S, Hess J, Eichhorst ST, Krueger A, Angel P, Krammer PH, et al. An unexpected role for FosB in activation-induced cell death of T cells. Oncogene. 2003;22:1333–1339. doi: 10.1038/sj.onc.1206126. [DOI] [PubMed] [Google Scholar]
- Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. J Biol Chem. 2004;279:44713–44722. doi: 10.1074/jbc.M407672200. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K, Hamasaki H, Miura A, Kakegawa T, Hirose S, et al. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol. 1986;166:128–134. doi: 10.1128/jb.166.1.128-134.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.