Abstract
The process of autophagy has been described in detail at the molecular level in normal cells, but less is known of its regulation in cancer cells. Aplasia Ras homolog member I (ARHI; DIRAS3) is an imprinted tumor suppressor gene that is downregulated in multiple malignancies including ovarian cancer. Re-expression of ARHI slows proliferation, inhibits motility, induces autophagy and produces tumor dormancy. Our previous studies have implicated autophagy in the survival of dormant ovarian cancer cells and have shown that ARHI is required for autophagy induced by starvation or rapamycin treatment. Re-expression of ARHI in ovarian cancer cells blocks signaling through the PI3K and Ras/MAP pathways, which, in turn, downregulates mTOR and initiates autophagy. Here we show that ARHI is required for autophagy-meditated cancer cell arrest and ARHI inhibits signaling through PI3K/AKT and Ras/MAP by enhancing internalization and degradation of the epidermal growth factor receptor. ARHI-mediated downregulation of PI3K/AKT and Ras/ERK signaling also decreases phosphorylation of FOXo3a, which sequesters this transcription factor in the nucleus. Nuclear retention of FOXo3a induces ATG4 and MAP-LC3-I, required for maturation of autophagosomes, and also increases the expression of Rab7, required for fusion of autophagosomes with lysosomes. Following the knockdown of FOXo3a or Rab7, autophagolysosome formation was observed but was markedly inhibited, resulting in numerous enlarged autophagosomes. ARHI expression correlates with LC3 expression and FOXo3a nuclear localization in surgical specimens of ovarian cancer. Thus, ARHI contributes to the induction of autophagy through multiple mechanisms in ovarian cancer cells.
Autophagy is a dynamic intracellular process that degrades organelles and long-lived cytosolic proteins by sequestration within double-membrane enclosed vesicles termed autophagosomes. Lysosomes fuse with autophagosomes to produce autolysosome. Within acidified autolysosome, hydrolases cleave proteins and lipids, releasing amino acids and fatty acids that can provide energy for cells in a nutrient-poor environment.1, 2 While many of the steps involved in autophagy have been well described at a molecular level, regulation of these events in malignant mammalian cells is less well understood.
Our group has identified a maternally imprinted tumor suppressor gene, DIRAS3, which regulates several steps in autophagy including induction and membrane elongation. Aplasia Ras homolog member I (ARHI) is an imprinted gene that is expressed from a single paternal allele in most normal tissues and that is downregulated in a fraction of carcinomas of the ovary, breast, lung, prostate, thyroid and pancreas. ARHI's downregulation in >60% of ovarian cancers is associated with decreased progression-free survival.3 Downregulation is achieved through multiple mechanisms, including loss of heterozygosity, DNA methylation, transcriptional regulation and shortened RNA half-life.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 ARHI encodes a 26 kDa GTPase with 50–60% homology to Ras and Rap.5 In contrast to the oncogenic activity of Ras, re-expression of ARHI at physiologic levels slows proliferation, inhibits motility, produces tumor dormancy and induces autophagy. ARHI not only induces autophagy but is also required for autophagy induced by treatment with rapamycin.6 ARHI-induced autophagy can also sustain survival of dormant ovarian cancer cells in xenografts. When dormancy was induced by re-expression of ARHI, treatment of dormant xenografts with chloroquine, a functional inhibitor of autophagy, significantly delayed outgrowth of tumors when dormancy was interrupted by subsequent downregulation of ARHI.6
In this report, we have explored mechanism(s) by which ARHI inhibits PI3K and Ras/MAP signaling, as well as the induction of the cysteine protease ATG4, which cleaves the MAP-LC3-I precursor during formation of MAP-LC3-II. FOXo3a has been found to regulate autophagy in skeletal and cardiac muscles.7 FOXo3a activity is downregulated by AKT- and ERK-mediated phosphorylation when phosphorylated FOXo3a is exported from the nucleus and degraded in the cytoplasm.7 Inhibition of AKT or ERK activity restricts FOXo3a to the nucleus and encourages transcription of autophagy-related genes, including ATG4, MAP-LC3 and Rab7. As ARHI inhibits both the PI3K/AKT and Ras/ERK signaling pathways and induces autophagy, we hypothesized that FOXo3a might mediate some of the proautophagic activities of ARHI.
Results
ARHI produces cell growth arrest and autophagy-mediated cell death
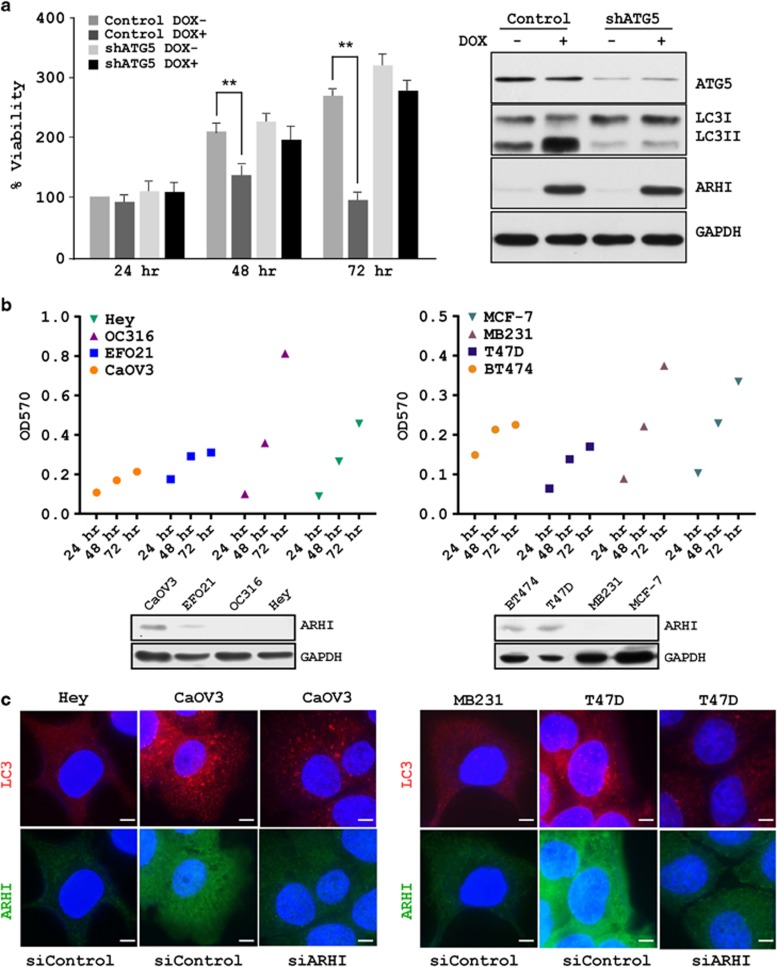
Our previous study demonstrated that re-expression of ARHI in cell culture produced autophagic cell death within 3–4 days and re-expression of ARHI in xenografts induced dormancy with persistent suppression of viable tumor growth.6 To test whether ARHI-induced ovarian cancer cell growth arrest depends upon autophagy, we established a stable knockdown of ATG5 in SKOv3-ARHI cells by the lentiviral infection. As expected, re-expression of ARHI in SKOv3-ARHI-shATG5 cells produced significantly less conversion of LC3I to LC3II, when compared with SKOv3-ARHI-shControl cells (Figure 1a). ARHI expression significantly inhibited tumor cell growth in a time-dependent manner in SKOv3-ARHI-shControl cells (Figure 1a), but exhibited little growth inhibitory effect on SKOv3-ARHI-shATG5 cells (Figure 1a), suggesting that ARHI induces autophagy-mediated cancer cell death. To evaluate the effect of endogenous expression of ARHI on growth of ovarian and breast cancer cells, we measured the growth rates of ovarian and breast cancer cell lines that expressed different levels of ARHI protein. CaOv3, EFO21, BT474 and T47D expressed higher level of ARHI and exhibited slower growth, whereas Hey, OC316, MB231 and MCF-7 expressed lower levels of ARHI had relatively higher rates of growth (Figure 1b). Interestingly, ARHI expression positively correlated with the formation of basal autophagy in the ovarian and breast cancer cells (Figure 1c) and knock down of ARHI expression with siRNA transfection reduced basal autophagy in the cancer cells, as determined by immunofluorescence staining (Figure 1c). Taken together, we demonstrated that ARHI expression induced cell growth arrest and autophagy-mediated cell death.
Figure 1.
ARHI produces cell growth arrest and autophagy-mediated cell death. (a) Knockdown of ATG5 rescues ARHI-induced cancer cell growth inhibition. SKOv3-ARHI-shControl and SKOv3-ARHI-shATG5 ovarian cancer cells were cultured (2000 cells per well) in 96-well plates. Cells were incubated overnight, and then treated with DOX (to induce ARHI) or diluent for the indicated intervals. Cell viability was assessed using an SRB assay. Data were obtained from three independent experiments. ATG5 and LC3I/II protein levels were measured by western blot analysis. The columns indicate the mean, and the bars indicate the S.E. (**P<0.01). (b) Endogenous expression of ARHI in ovarian and breast cancer cells negatively correlates with tumor cell growth. Growth over time of four ovarian cancer cell lines (Hey, OC316, EFO21 and CaOV3) and four breast cancer cell lines (MCF-7, MB231, T47D and BT474) was plotted with the GraphPad software (La Jolla, CA, USA), and endogenous ARHI level of the cell lines were examined by western blot analysis. (c) ARHI expression positively correlates with formation of basal autophagy in the ovarian and breast cancer cells. Cells with low ARHI expression were treated with siControl, whereas cells with high ARHI expression were treated with siControl and siARHI for 48 h, fixed and then stained with anti-LC3 (red) and anti-ARHI (green). Scale bars: 10 μm
ARHI inhibits AKT and ERK activation by EGF
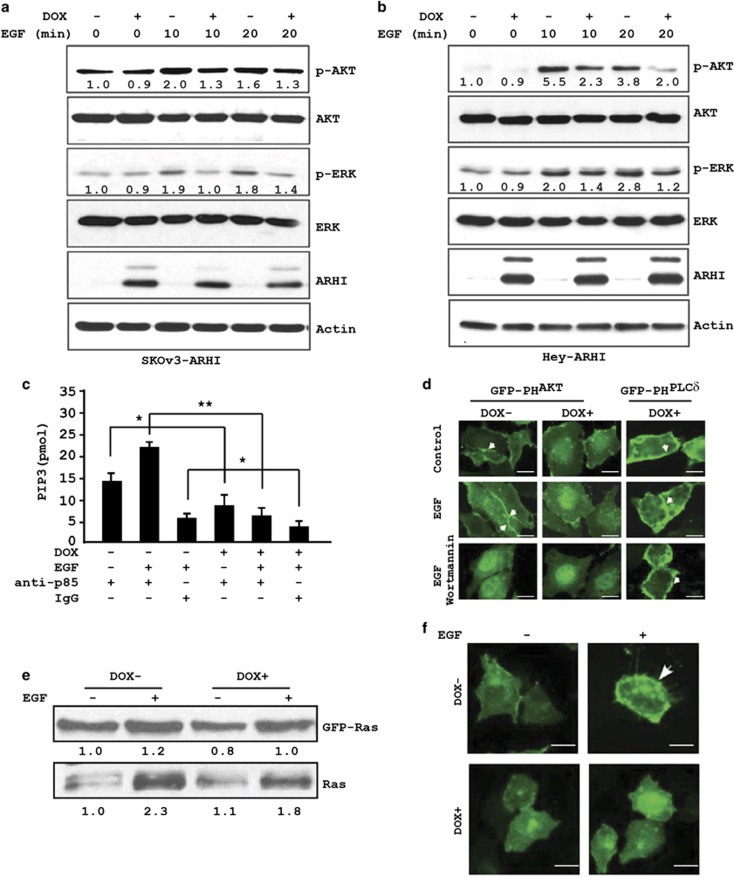
In an earlier study, we reported that re-expression of ARHI in SKOv3-ARHI cells inhibited both basal and lysophosphatidic acid-induced activation of AKT6 To determine if ARHI inhibits growth factor-induced activation of the PI3K/AKT and Ras/ERK signaling pathways, SKOv3-ARHI cells were treated with doxycycline (DOX) for 24 h to induce ARHI expression and were then stimulated with EGF before measurement of pAKT and pERK levels by western blotting. Treatment with EGF stimulated rapid AKT and ERK phosphorylation in uninduced SKOV3-ARHI cells, whereas ARHI re-expression inhibited both the activation of AKT and ERK by EGF, as indicated by reduced levels of pAKT and pERK (Figure 2a). In addition, we examined a second ARHI-inducible ovarian cancer cell line, Hey-ARHI. Similar to the SKOv3-ARHI cells, ARHI re-expression in Hey-ARHI cells also reduced EGF-mediated AKT and ERK phosphorylation (Figure 2b).
Figure 2.
ARHI re-expression inhibits the PI3K/AKT and Ras/ERK signaling pathways. ARHI re-expression in (a) SKOv3-ARHI and (b) Hey-ARHI decreases basal and EGF-induced increases in levels of pAKT and pERK. Ovarian cancer cells were treated with DOX for 24 h to induce ARHI expression. EGF (10 ng/ml) was added for 10 or 20 min. Cell lysates were prepared and probed with the indicated antibodies by western blot analysis. Band intensities were quantified and compared with that of control untreated cells to which a value of 1.0 was assigned. (c) ARHI inhibits EGF-mediated induction of Class I PI3K activity. Cell lysates were harvested from SKOv3-ARHI cells treated with DOX (24 h) and EGF (15 min) and were immunoprecipitated with anti-p85 PI3K or IgG as control. Immune complexes were assayed for the product of PI3K activity PIP3. *P<0.05; **P<0.01. Data were obtained from two independent experiments. (d) Expression of ARHI reduces the levels of membrane-associated PIP3. SKOv3-ARHI cells were transfected with a GFP-PHAKT or a GFP-PHPLCδ reporter and cultured in the presence or absence of DOX for 24 h before they were treated with EGF (10 ng/ml for 8 min) alone or with EGF and wortmannin (100 nM for 8 min). Scale bars: 5 μm. Arrows indicate membrane accumulation of PIP3-associated GFP-PHAKT or PIP2-associated GFP-PHPLCδ. (e) Expression of ARHI reduces the levels of GTP-bound Ras. SKOv3-ARHI cells were treated with or without DOX and EGF. Active, GTP-bound Ras was pulled down with RBD-agarose beads and quantified by western blot analysis. (f) Expression of ARHI reduces the levels of membrane-associated Ras. SKOv3-ARHI cells were transfected with GFP-Ras and treated with DOX for 24 h. Cells were then fixed and GFP-Ras was detected by confocal microscopy. The arrowheads indicate the membrane localization of GFP-Ras. Scale bars: 5 μm
ARHI inhibits EGF-stimulated PI3K activity and membrane localization of AKT
Activation and phosphorylation of AKT by peptide growth factors such as EGF occur through activation of tyrosine kinase growth factor receptors. These receptors, in turn, stimulate PI3K lipid kinase activity, which increases production of PIP3. Finally, increased PIP3 results in the recruitment of cytosolic AKT to the plasma membrane, where it is phosphorylated by PDK1.8 Furthermore, we sought to determine if reduced AKT activation was due to inhibition of upstream PI3K activity. SKOv3-ARHI cells were treated with DOX for 24 h to express ARHI and then stimulated with EGF for 15 min. Whole-cell lysates were prepared, immunoprecipitated with anti-PI3K p85 antibody and assayed for PI3K activity. While EGF increased PI3K activity in control cells, expression of ARHI significantly reduced both basal and EGF-stimulated PI3K activity (Figure 2c). To confirm inhibition of PI3K activity by ARHI, we examined the cell membrane localization of AKT in response to EGF in the presence or absence of ARHI. Using the PH domain of AKT (PHAKT) as a probe to detect PIP3, we found that the presence of ARHI in SKOv3-ARHI cells markedly reduced EGF-induced membrane accumulation of GFP-PHAKT, a result that is similar to cells treated with the PI3K inhibitor wortmannin (Figure 2d). To demonstrate that ARHI-induced inhibition was specific to PIP3, we also imaged the PH domain from PLCδ that binds PIP2. As shown in Figure 2d, neither ARHI nor wortmannin inhibited PHPLCδ membrane localization. Taken together, our results demonstrate that ARHI inhibits the PI3K/AKT signaling pathway by inhibiting PI3K activity, resulting in reduced PIP3 production and AKT phosphorylation.
ARHI suppresses EGF-stimulated Ras activation and membrane localization
A key upstream regulator of ERK activation is Ras, which exists in a GTP-bound active state or GDP-bound inactive state.9, 10 To determine if reduced pERK levels may be due to the inhibition of Ras activation by ARHI, we performed Ras-GTP pull-down experiments in SKOv3-ARHI cells treated with DOX or diluent followed by EGF exposure. In cells stimulated with EGF and expressing ARHI, binding of GTP both by endogenous Ras and transfected GFP-Ras was decreased (Figure 2e). Consistent with these observations, confocal image analysis of transiently transfected GFP-Ras also showed a marked reduction in EGF-stimulated membrane localization of GFP-Ras in DOX-treated SKOv3-ARHI cells (Figure 2f).
ARHI downregulates EGFR
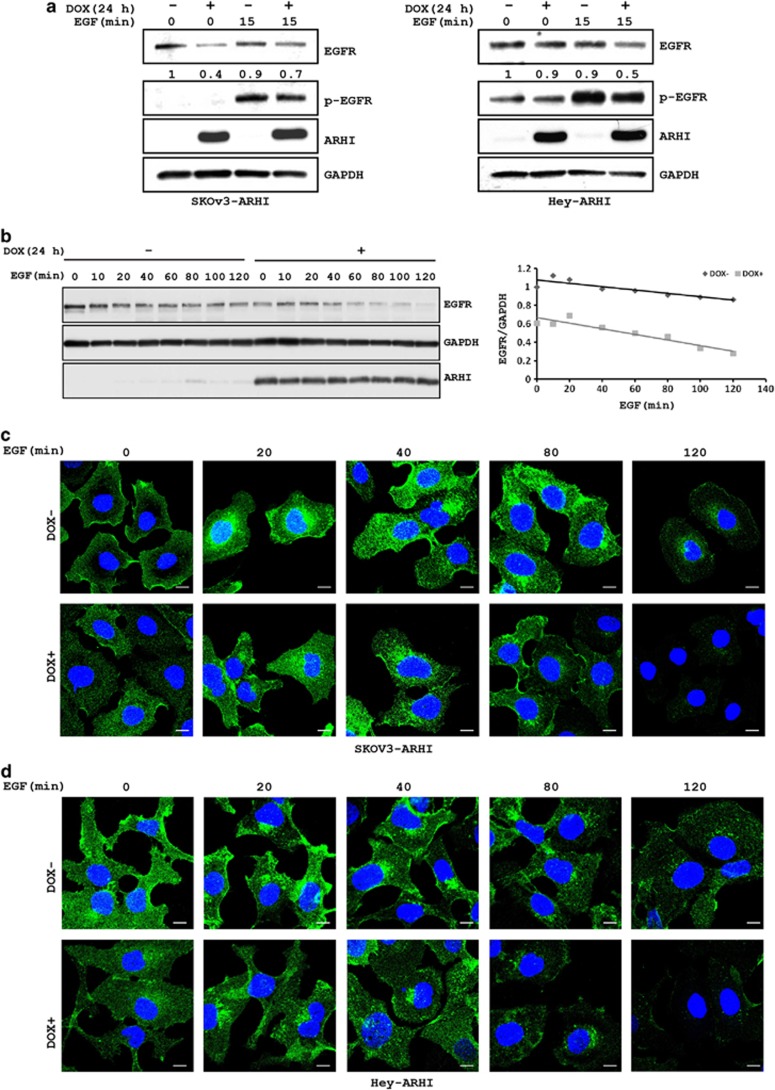
As expression of ARHI in SKOv3-ARHI and Hey-ARHI affected EGF-mediated stimulation of both the PI3K/AKT and Ras/ERK signaling pathways, we hypothesized that ARHI might alter the activation or function of tyrosine kinase growth factor receptors. To test this hypothesis, we examined EGF-stimulated epidermal growth factor receptor (EGFR) phosphorylation and the steady-state levels of total EGFR. In the presence of ARHI in both SKOv3-ARHI and Hey-ARHI cells, inhibition of both EGF-stimulated EGFR phosphorylation and steady-state levels of total EGFR was observed (Figure 3a). Upon binding to their ligands, growth factor receptors are rapidly internalized into the multivesicular endosomes, which can then be recycled back to the plasma membrane or be delivered to lysosomes for degradation.11, 12, 13, 14 To determine whether ARHI affects the recycling process that contributes to the reduction in EGFR, cell lysates were prepared at different intervals after EGF stimulation in control and ARHI-expressing SKOv3-ARHI cells and the levels of EGFR were measured. As shown in Figure 3b, EGFR levels decreased in a time-dependent manner in control cells following EGF treatment. Expression of ARHI significantly accelerated this process, suggesting that ARHI may affect the recycling or the stability of EGFR upon stimulation with EGF.
Figure 3.
ARHI expression facilitates the internalization and degradation of EGFR. (a) SKOv3-ARHI and Hey-ARHI cells were treated with or without DOX and EGF. Lysates were used to determine the levels of EGFR and pEGFR. GAPDH was used as a loading control. (b) SKOv3-ARHI cells were treated with DOX for 24 h before they were treated with EGF for the indicated intervals. Left panel shows western blot blots of EGFR, GAPDH and ARHI. Right panel shows the time-dependent reduction in EGFR relative to GAPDH. (c) SKOv3-ARHI and (d) Hey-ARHI cells were treated with or without DOX and EGF for the indicated time periods. Cells were then fixed and stained with anti-EGFR antibody and visualized with confocal microscopy. Scale bars: 5 μm
ARHI shortens the half-life of EGFR
To determine if ARHI accelerates the endocytic recycling of EGFR, ARHI-expressing and control SKOv3-ARHI cells (Figure 3c) and Hey-ARHI cells (Figure 3d) were treated with EGF for different intervals. The amount and cellular localization of EGFR were then assessed by immunofluorescent staining with anti-EGFR. In unstimulated control cells, EGFR is enriched at the cell surface (Figures 3c and d). Within 20 min of EGF stimulation, EGFR accumulated in the cytoplasm (Figures 3c and d). Maximum accumulation was observed at 80 min and then decreased rapidly. ARHI-expressing cells resulted in an even more rapid and greater cytoplasmic accumulation of EGFR (Figures 3c and d) as seen by the substantial reduction of membrane staining at only 20 min. In addition, the decrease of intracellular EGFR occurred earlier and at a significantly faster rate than in control cells (Figures 3c and d). This is in agreement with western blot analysis that demonstrated a shorter half-life of EGFR in ARHI-expressing cells (T1/2=65 min) than in control cells (T1/2=100 min) (Figure 3b).
ARHI reduces FOXo3a phosphorylation and induces its nuclear localization
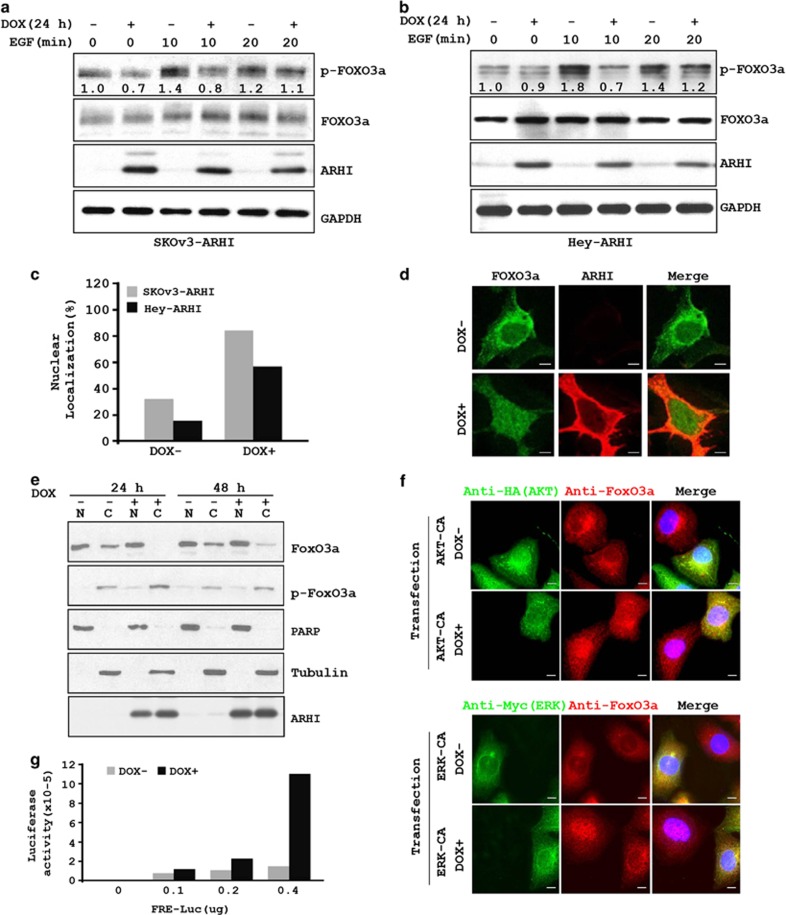
The transcription factor FOXo3a has been reported to contribute to the induction of autophagy in muscle cells.15 FOXo3a is negatively regulated by several growth-promoting signaling molecules, including AKT and ERK.16, 17, 18 Phosphorylation of FOXo3a at Ser318 by AKT and phosphorylation of FOXo3a at Ser294, Ser344 and Ser425 by ERK target pFOXo3a for rapid degradation in the cytoplasm via the proteasome pathway.18, 19, 20 Consequently, we asked if ARHI-induced inhibition of AKT and ERK can inhibit FOXo3a phosphorylation and enhance its transcriptional activity. SKOv3-ARHI cells were treated with or without DOX and EGF, as well as with an AKT inhibitor and an ERK inhibitor, before measuring the phosphorylation status, nuclear localization and transcriptional activity of FOXo3a. Treatment of SKOv3-ARHI cells with EGF stimulated FOXo3a phosphorylation within 10 min (Figure 4a). ARHI reduced the levels of EGF-stimulated pFOXo3a, consistent with an inhibition of AKT activity (Figure 4a). Similar results were also observed in Hey-ARHI cells (Figure 4b). Concomitant with the reduced FOXo3a phosphorylation, ARHI expression induced a marked increase in the nuclear localization of FOXo3a, as determined by immunohistochemical staining (Figure 4c), immunofluorescence staining (Figure 4d) and cell fractionation followed by western blotting (Figure 4e). In addition, ARHI-induced nuclear localization of FOXo3a was not further enhanced by incubation with AKT and ERK inhibitors (Supplementary Figure S4). Transient transfection of constitutively active AKT plasmid and constitutively active AKT prevented ARHI-induced nuclear localization of Foxo3a, suggesting that FOXo3a nuclear localization is mediated by ARHI-induced inhibition of both AKT and ERK signaling (Figure 4f). Finally, we determined that the nuclear localized FOXo3a is transcriptionally active as upregulation of ARHI increased the luciferase activity of a transfected FOXo3a-luciferase reporter gene (Figure 4g).
Figure 4.
ARHI expression reduces FOXo3a phosphorylation resulting in increased nuclear localization of FOXo3a. (a) SKOv3-ARHI and (b) Hey-ARHI cells were treated with DOX for 24 h before the addition of EGF for 10 or 20 min. Cell lysates were probed with anti-pFOXo3a and anti-FOXo3a antibodies. Band intensities were quantified and shown relative to that of control untreated cells to which a value of 1.0 was assigned. (c) Cells were treated with DOX for 24 h, fixed and then stained with anti-FOXo3a. Cells with nuclear staining of FOXo3a were counted and expressed as fraction of total cells counted with and without DOX. (d) Cells were treated with DOX and stained with anti-FOXo3a and anti-ARHI and visualized using confocal microscopy. Scale bars: 10 μm. (e) SKOv3-ARHI cells were treated with DOX for 24 or 48 h. Nuclear and cytoplasmic fractions were isolated and probed with antibodies against FOXo3a, pFOXo3a, tubulin and PARP. (f) ARHI-mediated FOXo3a nuclear localization is regulated by both AKT and ERK signaling. SKOv3-ARHI cells were treated with DOX and transfected with plasmid AKT-CA (Myr-HA-AKT1-DD) or ERK-CA (pCMV-myc-ERK2-L4A-MEK1-fusion) simultaneously for 24 h. Cells were then stained with anti-HA (AKT) or anti-Myc (ERK) and anti-FOXo3a antibodies and examined with confocal microscopy. Scale bars: 10 μm. (g) SKOv3-ARHI cells were transfected with an FRE-luciferase reporter and then treated with DOX to induce ARHI expression. Cell lysates were harvested 24 h later and assayed for luciferase activity
Knockdown of FOXo3a reduces ARHI-mediated induction of ATG4 and MAP-LC3-I expression
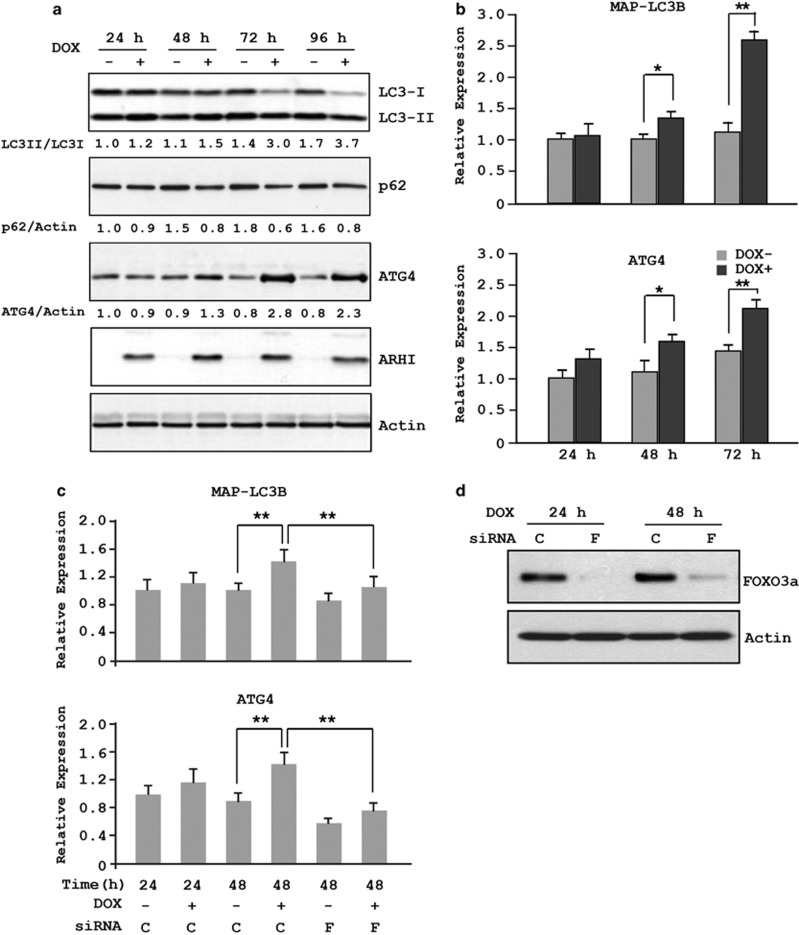
We previously observed that induction of ARHI expression in SKOv3-ARHI cells leads to increased levels of ATG4.6 To examine in greater detail the kinetics of ATG4 expression in the presence of ARHI, cell lysates were collected at different intervals after DOX treatment to measure levels of ATG4, p62 and LC3. As shown in Figure 5a, ATG4 expression was elevated 48 h after DOX treatment and peaked at 72 h, whereas the increase in LC3 could easily be detected by 24 h. More importantly, the ratio of LC3 II to LC3 I, an indication of autophagy, was significantly increased at 48 h and was maximal at 72–96 h, consistent with cells undergoing ARHI-induced autophagy. To determine whether increased ATG4 and LC3 expression is due, at least in part, to increased gene transcription, mRNA levels of ATG4 and LC3 were quantified by RT-PCR. Indeed, the presence of ARHI resulted in increased ATG4 and LC3 mRNAs (Figure 5b). Moreover, this increase is mediated by the transcriptional activity of FOXo3a, as a knockdown of FOXo3a abolished the ARHI-mediated increase of ATG4 and LC3 mRNAs (Figures 5c and d).
Figure 5.
ARHI induces ATG4 and MAPLC3 expression and is mediated through FOXo3a. (a) SKOv3-ARHI cells were treated with or without DOX for the indicated intervals before cells were harvested. Cell lysates were probed with antibodies against LC3, ATG4, ARHI and actin. Band intensities were quantified and expressed relative to the untreated controls in which a value of 1.0 was assigned. (b) Cells were treated as in a and the levels of LC3B and ATG4 mRNAs were quantified by real-time RT-PCR. The columns indicate the mean, and the bars indicate the S.E. (*P<0.05; **P<0.01). Data were obtained from three independent experiments. (c) SKOv3-ARHI cells were transfected with siControl (C) or siFOXo3a (F) and treated with or without DOX. The columns indicate the mean, and the bars indicate the S.E. (**P<0.01). Data were obtained from three independent experiments. (d) Efficient knockdown of FOXo3a
ARHI induces Rab7 expression and promotes the formation of autolysosome
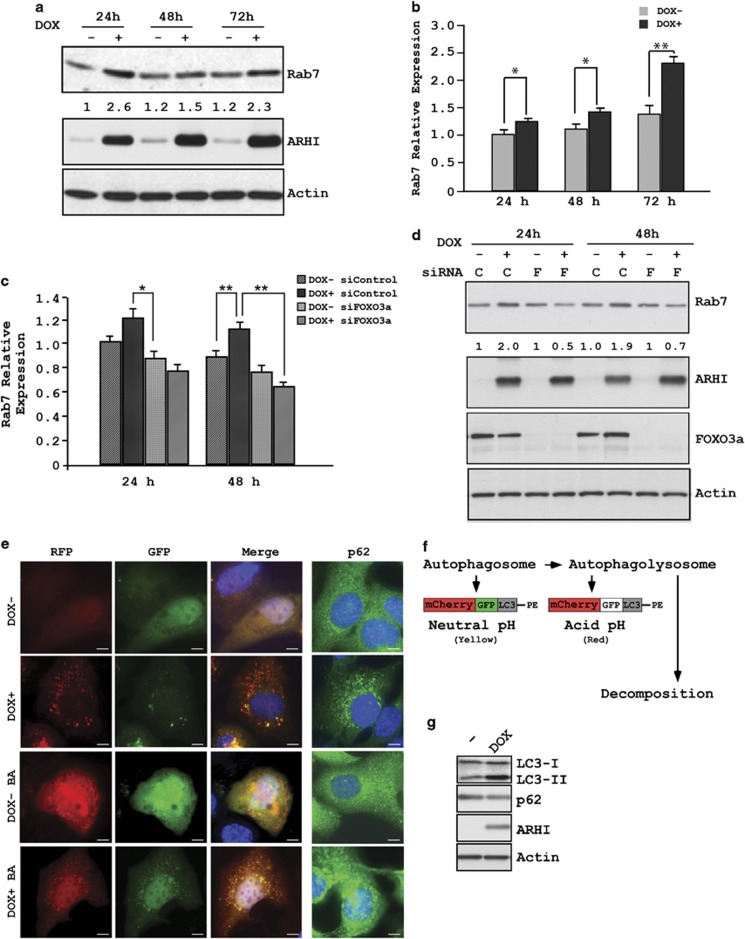
Rab7 has a role in the late endocytic pathway and lysosome biogenesis and participates in the late stages of autophagy by facilitating the fusion between autophagosomes and lysosomes to form autolysosome.21, 22, 23 Therefore, we asked whether ARHI might also regulate Rab7 expression and influence autolysosome formation. SKOv3-ARHI cells were treated with DOX for 24, 48 and 72 h to induce ARHI expression. Analysis of the cell lysates by western blot analysis showed a significant increase in Rab7 after 24 h incubation with DOX (Figure 6a). Elevated Rab7 expression was maintained at 72 h, albeit at a reduced level. Concomitant with increased Rab7 protein levels, the Rab7 mRNA level was also increased after 24 h of incubation with DOX (Figure 6b). ARHI expression had no effect on mRNA levels of EEA1, an early endosome marker.24, 25 (Supplementary Figure S1). To examine whether increased Rab7 mRNA related to increased FOXo3a transcriptional activity, SKOv3-ARHI cells were transfected with siFOXo3a for 24 h before treatment with DOX to induce ARHI expression. As shown in Figures 6c and d, knockdown of FOXo3a completely abolished ARHI-mediated induction of Rab7 at the mRNA and protein levels. To confirm that ARHI induces autophagic flux, SKOv3-ARHI cells were transfected with mCherry-GFP-LC3 plasmid and treated with or without DOX for 24 h to induce ARHI expression, followed by treatment with bafilomycin-a1 (100 nM) for the last 16 h of transfection. Analysis of fluorescence images showed a significant increase in the formation of mCherry LC3 puncta (red) and reduced diffuse green p62 staining after 24 h incubation with DOX (Figures 6e and f). A significant increase in mCherry-GFP puncta (yellow dots) was observed following treatment with bafilomycin-a1 (Figures 6e and f). Concomitant with the induced autophagic flux, ARHI expression increased LC3II and decreased p62 on western blot analysis (Figure 6g).
Figure 6.
ARHI expression induces Rab7 and is mediated through FOXo3a. (a) Cell lysates from SKOv3-ARHI cells were examined for Rab7 expression at indicated intervals after treatment with DOX. (b) Cells were treated as in a and the level of Rab7 mRNA was quantified by real-time RT-PCR. The columns indicate the mean, and the bars indicate the S.E. (*P<0.05; **P<0.01). Data were obtained from three independent experiments. (c) SKOv3-ARHI cells were transfected with siControl (C) or siFOXo3a (F) and treated with or without DOX. Rab7 expression was determined by RT-PCR and (d) western blot analysis. (e) ARHI expression induces autophagosome formation. SKOv3-ARHI cells were transfected with mCherry-GFP-LC3 for 24. Bafilomycin-a1 (BA, 100 nM) was added to the plate for the last 16 h of transfection. Scale bars: 10 μm. (f) A schematic diagram of mCherry-GFP-LC3 degradation. The double-tagged LC3 protein (mCherry-GFP-LC3) will emit yellow (green merged with red) fluorescence in non-acidic structures and appear as red only in the autolysosome owing to quenching of GFP in these acidic structures, (g) ARHI increase LC3-II and decreases p62. Western bot analysis of LC3 and p62 were examined
Knockdown of FOXo3a reduces the formation of autolysosome leading to an accumulation of autophagosomes in SKOv3 ovarian cancer cells
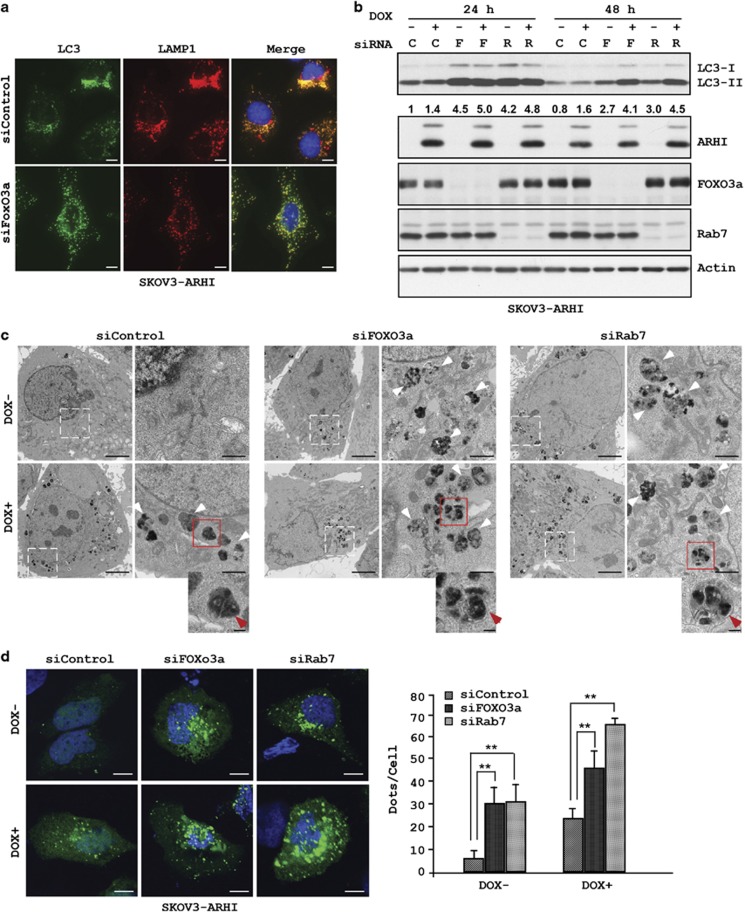
As Rab7 has a central role in the fusion of autophagosomes with lysosomes, we sought to examine the effect of FOXo3a knockdown on the formation of autolysosome. SKOv3-ARHI cells were transfected with siFOXo3a or siControl for 24 h and then induce the expression of ARHI before the cells were fixed and stained for LC3 and for the lysosomal biomarkers LAMP1 and LAMP2. Significant colocalization of LC3 with LAMP1 and LAMP2 was observed in control cells, but was markedly reduced in cells after FOXo3a knockdown (Figure 7a and Supplementary Figure S5). Consistent with the reduced fusion of autophagosomes and lysosomes, we observed an increase in the levels of LC3 in both DOX-treated and -untreated lysates (Figure 7b and Supplementary Figure S2), presumably related to a reduced degradation of LC3 by proteases in the lysosomes.
Figure 7.
FOXo3a knockdown reduces the fusion between autophagosomes and lysosomes and leads to an accumulation of LC3. (a) SKOv3-ARHI cells were transfected with siControl or siFOXo3a for 48 h before they were fixed and stained with anti-LC3 or anti-LAMP1 antibodies. The yellow color indicates colocalization of LC3 and LAMP1. Scale bars: 10 μm. (b) Cell lysates from FOXo3a (F) and Rab7 (R) knockdown SKOV3-ARHI cells were probed for LC3 levels. Band intensities were quantified and expressed relative to the untreated controls in which a value of 1.0 was assigned. (c) Hey-ARHI cells were transfected with siControl or siFOXo3a for 24 h and then treated with or without DOX for 24 h. TEM images of autophagosomes are indicated by the white arrows. The red arrows indicate the double-layered membranes in the siFOXo3a and siRab7 cells but absent in the siControl cells. Scale bars: 0.2 μm. Higher magnification of an indicated region is shown. Scale bar: 1 μm. (d) SKOv3-ARHI cells were transfected with siControl, siFOXo3a or siRab7 for 24 h before they were transfected with GFP-LC3 and treated with or without DOX for 24 h. Cells were fixed and GFP-LC3 dots were analyzed by confocal microscopy and quantified. The columns indicate the mean, and the bars indicate the S.E. (**P<0.01). Scale bars: 10 μm. Data were obtained from two independent experiments
To confirm that knockdown of FOXo3a leads to increased accumulation of autophagosomes owing to reduced fusion of autolysosome, we performed transmission electron microscopy (TEM) to observe the autophagosomes directly in Hey-ARHI cells. As expected, very few autophagosomes were present in untreated cells; however, the presence of ARHI markedly increased the number of autophagosomes (Figure 7c). In contrast, many autophagosomes were present in cells with FOXo3a knockdown even in the absence of ARHI (Figure 7c), suggesting that basal FOXo3a activity is required in the autophagic flux that regulates proper fusion and degradation of autophagosomes. Consistent with its role in autophagy fusion, knockdown of Rab7 in Hey-ARHI cells resulted in a similar accumulation of autophagosomes in the absence of ARHI (Figure 7c), although the expression of ARHI further increased autophagosome accumulation. At high magnification, vesicles in the FOXo3a and Rab7 knockdown cells are frequently enclosed by double-layered membranes, a characteristic feature of autophagosomes (Figure 7c), whereas ARHI-expressing cells include more vesicles enclosed by single-layered membranes, consistent with the presence of successfully fused autolysosomes.
In a third approach to determine whether knockdown of FOXo3a or Rab7 leads to accumulation of autophagosomes that are unable to fuse with lysosomes, we compared the effect of FOXo3a knockdown to Rab7 knockdown on the formation of GFP-LC3 puncta in SKOv3-ARHI cells with and without ARHI. In control cells, the number of GFP-LC3 puncta was relatively low but increased significantly following ARHI expression (Figure 7d). In sharp contrast, both FOXo3a and Rab7 knockdown markedly increased GFP-LC3 puncta in the absence of ARHI and the number of puncta was further increased by ARHI expression (Figure 7d). Taken together, these results further support that FOXo3a has an important role in maintaining the steady-state flux of autophagy in ovarian cancer cells.
Expression of ARHI correlates positively with FOXo3a nuclear staining and LC3 staining in surgical specimens of ovarian cancer
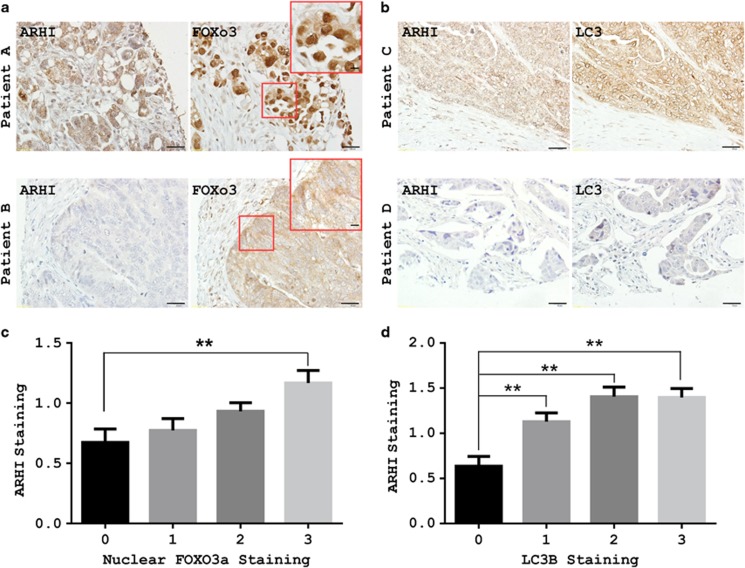
Our studies in cell culture indicate that ARHI expression induced an increase in the nuclear localization of FOXo3a and in autophagic flux. To assess the clinical relevance of these observations, immunohistochemical methods have been developed to measure association of ARHI, FOXo3a and LC3 in formalin-fixed, paraffin-embedded ovarian cancer tissues obtained at surgery. Expression of ARHI, FOXo3a and LC3 was measured by immunohistochemical staining of tissue microarray (TMA) sections that contained cores from more than 200 primary ovarian cancers. Immunohistochemical staining was measured on a scale of 0–3 for staining intensity in comparison with staining observed in normal ovarian surface epithelial cells (Supplementary Figure S3). ARHI expression was correlated positively with FOXo3a nuclear staining (Spearman's ρ, 0.2011, P=0.001) (Figures 8a and c) and LC3 staining (Spearman's ρ, 0.322, P=0.0001) (Figures 8b and d). The remarkable correlation of staining for ARHI with LC3 and FOXo3a suggests that these molecules are colocalized in human ovarian cancers undergoing autophagy and that ARHI and FOXo3a are important for inducing autophagy in ovarian cancer.
Figure 8.
Correlation of ARHI, FOXo3a and LC3 in an ovarian cancer TMA. (a and b) Representative TMA images show different combinations of ARHI, FOXo3a and LC3 expression in different ovarian cancers. Patient A: Positive for ARHI, FOXo3a with dormant nuclear staining. Patient B: Negative for ARHI, FOXo3a with dormant cytoplasmic staining. Patient C: Positive ARHI with positive LC3 staining. Patient D: Negative ARHI with negative LC3 staining. Lower magnification, scale bars: 10 μm. Higher magnification (inset), scale bars: 10 μm. (c) ARHI staining correlates with FOXo3a nuclear staining. Figure was established with GraphPad Prism column analysis. (d) ARHI staining correlates with LC3 staining. Analysis is similar to c (for intensity of staining, 0: negative; 1: + 2: ++ 3: +++). The columns indicate the mean, and the bars indicate the s.e. (**P<0.01)
Discussion
Our observations indicate that the expression of ARHI (DIRAS3) at physiologic levels regulates autophagy in ovarian cancer cells at several steps, including initiation, membrane elongation and fusion of autophagosomes with lysosomes. Decreased mTOR activity has long been known to initiate autophagy.26, 27, 28 Decreased signaling through the PI3K pathway can inhibit mTOR activity.29, 30 In this report, we have shown that the expression of ARHI decreases both PI3K and Ras/MAP signaling by downregulating EGFR through enhanced internalization and degradation leading to a shortened half-life.
Both transcription-dependent and -independent mechanisms have been described that regulate autophagy.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 In this report, we have identified a novel transcription-dependent mechanism regulating autophagy through FOXo3a. Different posttranslational modifications regulate the function of FOXo3a protein. Treatment with EGF in the absence of ARHI activates AKT and ERK. FOXo3a is phosphorylated at multiple sites and translocated from the nucleus to cytosol, and FOXo3a-mediated transcription is attenuated.16 In response to the expression of ARHI, FOXo3a protein is dephosphorylated and retained in the nucleus, inducing transcription of genes that mediate autophagy (ATG4, LC3 and Rab7). Previous studies have shown that FOXo3a regulates autophagy in skeletal muscle cells by transcriptional activation of genes that contribute to autophagosome formation, including MAP-LC3-I, ATG12, ATG4, Beclin-1, ULK1 and Bnip3.15, 32, 33 These previous studies did not detect the induction of Rab7 and have not resolved how FOXo3a activates autophagy or the consequence of enhanced expression of autophagy-related genes.
In this report, we found that ARHI increased FOXo3a-mediated expression of Rab7. Rab7 is required for fusion of autophagosomes with lysosomes to form autolysosomes, where proteins and lipids are hydrolyzed, releasing amino acids and fatty acids to provide energy for cells under nutrient poor conditions. When FOXo3a was knocked down, autophagosomes still formed, but fusion with lysosomes was inhibited. This suggests that ATG4 and MAP-LC3-I were long-lived, not in limiting supply or regulated by additional transcription factors. FOXo3a induction of Rab7 was required for successful fusion of autophagosomes and lysosomes to form functional autolysosome. Rab7 is required for maturation of late autophagosomes and fusion of mature autophagosomes with lysosomes.21, 34
Inactivation of FOXO proteins has been associated with a fraction of breast cancers, prostate cancers, glioblastomas, rhabdomyosarcomas and leukemias.35 In ovarian cancer, low expression of FOXo3a protein correlates with advanced disease stage and poor prognosis, particularly when combined with high expression of Skp2, which is required for its proteasomal degradation.36 In addition to ovarian cancer, downregulation of ARHI has been associated with cancers at other sites, including breast, prostate, pancreatic, thyroid and lung carcinomas.4, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46 Consequently, the observations made in ovarian cancer are likely to be relevant to many other types of cancer. In the future, it will be of interest to explore the interaction of ARHI and FOXo3a across cancers that originate from non-ovarian sites.
Taken together with our earlier studies,6 ARHI regulates autophagy at several different levels including (1) induction through a decrease in AKT/mTOR signaling, (2) enhanced expression of LC3 and ATG4 during membrane elongation and (3) upregulation of Rab7 to permit fusion of autophagosomes and lysosomes. Cancer cells with low levels of ARHI may be less able to cope with nutrient deprivation. Conversely, high levels of ARHI have been found in dormant autophagic ovarian cancer cells in the peritoneal cavity detected during second look operations after primary chemotherapy. These persistent cells may prove susceptible to inhibitors of autophagy potentially improving clinical outcomes.
Materials and Methods
Antibodies and reagents
Bafilomycin-a1 and AKT1/2 inhibitor (A6730) were purchased from Sigma (St. Louis, MO, USA) and an ERK inhibitor (CAS 1049738-54-6) was obtained from Millipore (Billerica, MA, USA). Antibodies against AKT, pAKTT308, EGFR, pEGFRY1148, IGFR, pIGFRY1131, FOXo3a, pFOXo3aS318/321 and MAP-LC3 were obtained from Cell Signaling Technology (Beverley, MA, USA). Anti-ATG4 was purchased from MBL (Woburn, MA, USA) and anti-actin antibody was from Sigma. Antibodies against pERK were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-LAMP1 was purchased from Abcam (Cambridge, MA, USA). LAMP2 (H4B4) antibody was from Developmental Study Hybridoma Bank (The University of Iowa, Iowa City, IA, USA) and antibodies against Rab7 were from Sigma and Rab11a was from Millipore. Murine monoclonal antibodies against ARHI were generated in our laboratory.
Cell culture
The Tet-on-inducible SKOv3-ARHI cells (p53 null) were grown in McCoy's medium supplemented with 10% FBS, 200 μg/ml G418 and 0.12 μg/ml puromycin. The Tet-on-inducible Hey-ARHI cells (p53 wild type) were cultured in RPMI-1640 medium supplemented with 10% FBS, 25 μ g/ml blastocidin and 1 μg/ml puromycin. ARHI expression was induced by adding 1 μg/ml DOX to the culture medium. We have shown in previous studies that induced ARHI expression is maximal at 24 h and is in the same range as that found in normal ovarian surface epithelial cells.6, 47 SKOv3-ARHI-shControl and SKOv3-ARHI-shATG5 cells were established by stable knockdown using a scrambled sequence shRNA and ATG5 shRNA, respectively, with lentiviral infection of SKOv3-ARHI cells followed by flow cytometric cell sorting. GIPZ non-silencing lentiviral shRNA Control and lentiviral shATG5 were obtained from Fisher Scientific (Pittsburgh, PA, USA). The expression of ATG5 and the formation of autophagy of the cell lines were confirmed by western blot analysis.
Plasmids
GFP-PHAKT and GFP-PHPLCδ were a gift from Dr. G Mills, MD Anderson Cancer Center. GFP-Ras was obtained from Dr. H Wang. FOXo3a response element (FRE)-luciferase reporter plasmid was a gift from Dr. MC Hung, MD Anderson Cancer Center. AKT-CA (Myr-HA-AKT1-DD) was provided by Dr. Gordon B Mills, MD Anderson Cancer Center and plasmid pCMV-myc-ERK2-L4A-MEK1-fusion was purchased from Addgene (Cambridge, MA, USA).
Cell proliferation assay
Cells (2000 cells per well) were plated in 96-well plates in 100 μl media per well. The cells were cultured overnight and then with or without DOX (for ARHI induction) for the time intervals indicated in the figures. Cell viability was assessed by sulforhodamine B (SRB) assay.48 Briefly, 50 μl of 30% TCA was added to each well and plates were incubated at 4 °C for 1 h. Plates were rinsed with distilled H2O, before the addition of 100 μl of 0.4% SRB in 1% acetic acid to each well. Plates were incubated for 30 min at room temperature, and then rinsed with 1% acetic acid. SRB was solubilized with 100 μl of 10 mM Tris for 5 min with shaking. Absorbance values were read on a microplate reader at 570 nm.
Immunofluorescent staining
SKOv3-ARHI cells were grown on coverslips and fixed in 4% formaldehyde in PBS for 10 min, rinsed two times in PBS and then permeabilized in 100% ethanol for 1 h at −20 °C. Coverslips were rinsed two times in PBS and blocked with 5% BSA in PBS for 1 h at room temperature, followed by incubation with primary antibodies diluted in 1.5% BSA in PBS for overnight at 4 °C. Cells were then washed four times for 5 min in PBS and incubated with secondary antibodies diluted in 1.5% BSA for 1 h at room temperature. Coverslips were then washed four times for 5 min in PBS, mounted on glass slides with Vectashield fluorescent mounting medium (Vector Labs, Burlingame, CA, USA) and examined using confocal microscopy (Olympus FluoView 500 or 1000; Olympus Inc., Melville, NY, USA). Goat anti-mouse or goat anti-rabbit secondary antibodies were conjugated to Alexa Fluor 350, 488 or 594 and were purchased from Invitrogen (Grand Island, NY, USA).
Western blot analysis
SKOv3-ARHI cells were incubated in lysis buffer (50 mM HEPES, pH 7.0, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10 mM NaF, 10 mM sodium pyrophosphate, 10% glycerol, 1% Triton X-100) plus protease and phosphatase inhibitors (1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml aprotinin and 1 mM Na3VO4). Cells were lysed for 30 min on ice, and then centrifuged at 17 000 × g for 30 min at 4 °C. Proteins were separated on 12 or 15% SDS-PAGE and transferred to PVDF membranes. Immunoblot analysis was performed with the indicated antibodies and visualized with an ECL enhanced chemiluminescence detection kit (GM Healthcare Bio-Sciences, Pittsburgh, PA, USA). Quantification of protein band intensities were completed using the Java-based image processing program ImageJ developed at the National Institutes of Health (Rockville Pike, MD, USA).
Ras-GTP assay
Ras-GTP assay was performed with cell lysates prepared from SKOv3-ARHI with or without DOX treatment using the small GTPase Activation Assay kit from Cell Biolabs Inc. (San Diego, CA, USA), according to manufacturer's protocols. Briefly, Raf1 agarose beads were used to isolate and pull-down the active form of Ras from the lysate. Subsequently, the precipitated GTP-Ras is detected by western blot analysis using an anti-Ras antibody.
siRNA transfection
SKOv3-ARHI cells were transfected with control or targeted siRNAs using the Transfectin no. 4 reagent (Dharmacon Research, Lafayette, CO, USA). Briefly, a mixture of siRNA (100 nM final concentration) and transfection reagents were incubated for 20 min at room temperature. This mixture was then added to cells and allowed to incubate for 48 h before cells were harvested and analyzed for protein and RNA expression. For experiments with serial transfections, cells were transfected with siRNA using the Transfectin no. 4 reagent on day 1 and with GFP-LC3 plasmid using Lipofectamine 2000 (Invitrogen) on day 2. Cells were harvested 24 h later for protein expression measurements or fixed for fluorescence microscopy analysis.
Confocal microscopic analysis
SKOv3-ARHI cells were transiently transfected with pGFP-LC3 (24 h) or mCherry-GFP-LC3 and fixed with 4% paraformaldehyde. Cells were then washed with PBS, mounted and examined using a confocal microscope (Olympus FluoView 1000; Olympus Inc.). Digital images were obtained using FluoView 1000 software (Olympus Inc.). LC3 puncta of cells were counted and the mean of puncta per cell was calculated.
Real-time quantitative reverse transcription-PCR
Expression of FOXo3a target genes was measured using real-time quantitative reverse transcription-PCR (RQ RT-PCR). Total cDNA was synthesized using 2 μg of total RNA. The reverse transcriptase reaction was carried out according to the manufacturer's instructions using oligo(dT)16 and SuperScript II reverse transcriptase (Invitrogen), followed by SYBR Green RT-PCR (ABI Prism 7000 Sequence Detection System; Applied Biosystems, Foster City, CA, USA). Oligonucleotide sequences of the primer sets used were: human ATG4 (forward, 5′-CAGACCCCGTTGGATACTG-3′ reverse, 5′-TCTTCCTTTGTGTCCACCTC-3′); human MAP-LC3 (forward, 5′-GAAGCAGCTTCCTGTTCTGG-3′ reverse, 5′-TCATCCCGAACGTCTCCTGG-3′); human Rab7 (forward, 5′- CTCTAGGAAGAAAGTGTTGC-3′ reverse, 5′-TTGTGACTAGCCTGTCATCC-3′); human Rab11a (forward, 5′-GAAAGCAAGAGCACCATTGG-3′ reverse, 5′-ATCTCTCAGTTCTTTCAGCC-3′) and human GAPDH (forward, 5′-ATGGAATCCATCACCATCTT-3′ reverse, 5′-CGCCCCACTTGATTTTGG-3′). The melting curves were used to ensure there was not any nonspecific amplification. mRNA levels were normalized with a concurrent determination for GAPDH mRNA.
Transient transfection and luciferase reporter assay
For transfection, 1 × 105 cells were seeded in 6-well plates and transfected using Lipofectamine 2000 (Invitrogen) with 0.5 μg of p(FOXo3a)3/luciferase reporter construct containing three copies of the FOXo3a binding site in front of a minimal promoter. Twenty hours later, the cells were treated with or without 1 μg/ml DOX for 24 h. The cells were harvested, and the cell extracts were assayed, in triplicate, for luciferase activity using a luciferase assay kit (Pharmingen, San Diego, CA, USA).
Transmission electron microscopy
For TEM examination of autophagosomes and autolysosome, SKOv3 cells were transfected with siControl, siFOXo3a or siRab7 siRNAs for 24 h before they were treated with or without DOX for 48 h. Cells were washed in PBS and fixed with 2.5% glutaraldehyde in 0.1 M PBS buffer and further fixed with 1% osmium tetroxide in 0.1 M cacodylate buffer. Specimens were stained with aqueous uranyl acetate and lead citrate before being observed with a Jeol-100 CX II (JEOL, Peabody, MA, USA) TEM at 80 kV.
Immunohistochemistry
A formalin-fixed, paraffin-embedded TMA samples of primary ovarian cancers were obtained from the Yale University Pathology Department and additional specimens were obtained from the MD Anderson Pathology Department. Specimens and associated clinical information were collected under informed consent under the ethics guidelines and approval of the Yale and MDACC Human Investigation Committee. Six micron sections were cut from each TMA. Incubation at 60 °C for 20 min was used to restore antigenic reactivity, followed by two 20 min incubations in xylene. After slides were rehydrated, antigen retrieval was performed in 6.5 mM sodium citrate buffer (pH 6.0) for 10 min. Three percent bovine serum albumin in 0.1 M Tris-buffered saline was used for blocking. Sections were incubated at 4 °C overnight with each of the primary antibodies, including anti-ARHI murine monoclonal (1 : 500; RC Bast Laboratory, Houston, TX, USA), anti-LC3B rabbit monoclonal (1 : 250; Cell Signaling Technologies, Danvers, MA, USA) and an anti-FOXo3a rabbit monoclonal (1 : 400; Cell Signaling Technologies). Anti-mouse or -rabbit immunoglobulin secondary antibodies were then applied for 1 h at room temperature, followed by washingthree times in PBS for 10 min. DAB chromagen was added for 1 min per slide, followed by three additional washes in PBS for 10 min and then hematoxylin staining was performed for 1 min per slide, followed by three additional washes in PBS for 10 min. Serial sections of cell line test arrays and IgG staining served as positive and negative controls and were stained alongside TMAs to confirm assay reproducibility. A TMA serial section stained for mouse IgG1 served as an additional negative control. Total staining intensity was determined as 0 (no staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining).
Statistics
All experiments were repeated independently at least two times and the data (bar graphs) were expressed as mean±s.e. Statistical analysis was performed using a Student's t-test (two-sample assuming unequal variances). The criterion for statistical significance was taken as P<0.05 (two-sided). Staining expression values for ARHI, LC3B and FOXo3a were obtained from each TMA. Spearman's ρ was used to assess the direction and strength of association between expression scores for ARHI, LC3B and FOXo3a (JMP Statistical Discovery Software, Version 7.0.1; SAS Institute Inc., Cary, NC, USA).
Acknowledgments
These studies were supported in part by grants from the National Cancer Institute P01 CA064602 and R01 CA135354, by the MD Anderson SPORE in Ovarian Cancer NCI P50 CA83639, the Shared Resources of the MD Anderson CCSG NCI P30 CA16672, the Ovarian Cancer Research Fund, the National Foundation for Cancer Research and philanthropic support from the Zarrow Foundation and Stuart and Gaye Lynn Zarrow.
Glossary
- ARHI
aplasia Ras homolog member I
- LC3
myosin-associated protein light chain 3
- DOX
doxycycline
- TEM
transmission electron microscopy
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Piacentini
Supplementary Material
References
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Levine B. Autophagy in C. elegans. WormBook. 2009;24:1–26. doi: 10.1895/wormbook.1.147.1. [DOI] [PubMed] [Google Scholar]
- Rosen DG, Wang L, Jain AN, Lu KH, Luo RZ, Yu Y, et al. Expression of the tumor suppressor gene ARHI in epithelial ovarian cancer is associated with increased expression of p21WAF1/CIP1 and prolonged progression-free survival. Clin Cancer Res. 2004;10:6559–6566. doi: 10.1158/1078-0432.CCR-04-0698. [DOI] [PubMed] [Google Scholar]
- Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci USA. 1999;96:214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RZ, Fang X, Marquez R, Liu SY, Mills GB, Liao WS, et al. ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene. 2003;22:2897–2909. doi: 10.1038/sj.onc.1206380. [DOI] [PubMed] [Google Scholar]
- Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–3929. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofler A, Nichols T, Grant S, Lingardo L, Esposito EA, Gridley S, et al. Study of the PDK1/AKT signaling pathway using selective PDK1 inhibitors, HCS, and enhanced biochemical assays. Anal Biochem. 2011;414:179–186. doi: 10.1016/j.ab.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Rosenberger G, Meien S, Kutsche K. Oncogenic HRAS mutations cause prolonged PI3K signaling in response to epidermal growth factor in fibroblasts of patients with Costello syndrome. Hum Mutat. 2009;30:352–362. doi: 10.1002/humu.20855. [DOI] [PubMed] [Google Scholar]
- Gremer L, De Luca A, Merbitz-Zahradnik T, Dallapiccola B, Morlot S, Tartaglia M, et al. Duplication of Glu37 in the switch I region of HRAS impairs effector/GAP binding and underlies Costello syndrome by promoting enhanced growth factor-dependent MAPK and AKT activation. Hum Mol Genet. 2010;19:790–802. doi: 10.1093/hmg/ddp548. [DOI] [PubMed] [Google Scholar]
- Clague MJ. Met receptor: a moving target. Sci Signal. 2011;4:pe40. doi: 10.1126/scisignal.2002422. [DOI] [PubMed] [Google Scholar]
- Jacob C, Cottrell GS, Gehringer D, Schmidlin F, Grady EF, Bunnett NW. c-Cbl mediates ubiquitination, degradation, and down-regulation of human protease-activated receptor 2. J Biol Chem. 2005;280:16076–16087. doi: 10.1074/jbc.M500109200. [DOI] [PubMed] [Google Scholar]
- Tanowitz M, von Zastrow M. A novel endocytic recycling signal that distinguishes the membrane trafficking of naturally occurring opioid receptors. J Biol Chem. 2003;278:45978–45986. doi: 10.1074/jbc.M304504200. [DOI] [PubMed] [Google Scholar]
- Futter CE, Collinson LM, Backer JM, Hopkins CR. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J Cell Biol. 2001;155:1251–1264. doi: 10.1083/jcb.200108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A, Mickle A, Medda B, Zhang Z, Phillips RJ, Tipnis N, et al. Altered mechanosensitive properties of vagal afferent fibers innervating the stomach following gastric surgery in rats. Neuroscience. 2009;162:1299–1306. doi: 10.1016/j.neuroscience.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Wang H, Zhou L, Yu D. Akt-FOXO3a signaling axis dysregulation in human oral squamous cell carcinoma and potent efficacy of FOXO3a-targeted gene therapy. Oral Oncol. 2011;47:16–21. doi: 10.1016/j.oraloncology.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Chen J, Gomes AR, Monteiro LJ, Wong SY, Wu LH, Ng TT, et al. Constitutively nuclear FOXO3a localization predicts poor survival and promotes Akt phosphorylation in breast cancer. PLoS One. 2010;5:e12293. doi: 10.1371/journal.pone.0012293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278:12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117 (Part 20:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117 (Part 13:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Thi EP, Lambertz U, Reiner NE. Class IA phosphatidylinositol 3-kinase p110alpha regulates phagosome maturation. PLoS One. 2012;7:e43668. doi: 10.1371/journal.pone.0043668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawe DC, Patki V, Heller-Harrison R, Lambright D, Corvera S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J Biol Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- Huang S, Yang ZJ, Yu C, Sinicrope FA. Inhibition of mTOR kinase by AZD8055 can antagonize chemotherapy-induced cell death through autophagy induction and down-regulation of p62/sequestosome 1. J Biol Chem. 2011;286:40002–40012. doi: 10.1074/jbc.M111.297432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-ATG13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Gu Y, Li F, Wang Q, Huang B, Jin X, et al. The PI3K/AKT/mTOR signaling pathway is overactivated in primary aldosteronism. PLoS One. 2013;8:e62399. doi: 10.1371/journal.pone.0062399. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen J, Crawford R, Xiao Y. Vertical inhibition of the PI3K/Akt/mTOR pathway for the treatment of osteoarthritis. J Cell Biochem. 2013;114:245–249. doi: 10.1002/jcb.24362. [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WJ, Deng R, Feng GK, Zhu XF. [A G-quadruplex ligand SYUIQ-5 induces autophagy by inhibiting the Akt-FOXO3a pathway in nasopharyngeal cancer cells] Ai Zheng. 2009;28:1049–1053. doi: 10.5732/cjc.009.10203. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Ray D, Lekli I, Bak I, Tosaki A, Das DK. Effects of Longevinex (modified resveratrol) on cardioprotection and its mechanisms of action. Can J Physiol Pharmacol. 2010;88:1017–1025. doi: 10.1139/y10-082. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal–lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Hung MC. Deciphering the role of forkhead transcription factors in cancer therapy. Curr Drug Targets. 2011;12:1284–1290. doi: 10.2174/138945011796150299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhao Y, Xu F, Wang Y, Xiang J, Chen D. The expression and prognosis of FOXO3a and Skp2 in human ovarian cancer. Med Oncol. 2012;29:3409–3415. doi: 10.1007/s12032-012-0275-z. [DOI] [PubMed] [Google Scholar]
- Wu X, Liang L, Dong L, Yu Z, Fu X. Effect of ARHI on lung cancer cell proliferation, apoptosis and invasion in vitro. Mol Biol Rep. 2012;40:2671–2678. doi: 10.1007/s11033-012-2353-x. [DOI] [PubMed] [Google Scholar]
- Field JK, Liloglou T, Warrak S, Burger M, Becker E, Berlin K, et al. Methylation discriminators in NSCLC identified by a microarray based approach. Int J Oncol. 2005;27:105–111. doi: 10.3892/ijo.27.1.105. [DOI] [PubMed] [Google Scholar]
- Li Y, Shi L, Han C, Wang Y, Yang J, Cao C, et al. Effects of ARHI on cell cycle progression and apoptosis levels of breast cancer cells. Tumour Biol. 2012;33:1403–1410. doi: 10.1007/s13277-012-0388-1. [DOI] [PubMed] [Google Scholar]
- Chen MY, Liao WS, Lu Z, Bornmann WG, Hennessey V, Washington MN, et al. Decitabine and suberoylanilide hydroxamic acid (SAHA) inhibit growth of ovarian cancer cell lines and xenografts while inducing expression of imprinted tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. Cancer. 2011;117:4424–4438. doi: 10.1002/cncr.26073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen EA, Ovestad IT, Skaland I, Soiland H, Gudlaugsson E, Kjellevold KH, et al. LOH at 1p31 (ARHI) and proliferation in lymph node-negative breast cancer. Cell Oncol. 2009;31:335–343. doi: 10.3233/CLO-2009-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zaman MS, Deng G, Majid S, Saini S, Liu J, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev Res (Phila) 2011;4:76–86. doi: 10.1158/1940-6207.CAPR-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu Q, Cho AH, Rondeau G, Welsh J, Adamson E, et al. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia. 2005;7:748–760. doi: 10.1593/neo.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YQ, Si LJ, Ye ZS, Lin ZH, Zhou JP. Inhibitory effect of ARHI on pancreatic cancer cells and NF-kappaB activity. Mol Med Rep. 2013;7:1180–1184. doi: 10.3892/mmr.2013.1342. [DOI] [PubMed] [Google Scholar]
- Yang H, Lu X, Qian J, Xu F, Hu Y, Yu Y, et al. Imprinted tumor suppressor gene ARHI induces apoptosis correlated with changes in DNA methylation in pancreatic cancer cells. Mol Med Rep. 2010;3:581–587. doi: 10.3892/mmr_00000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Aldred MA, Morrison CD, Plass C, Frilling A, Broelsch CE, et al. Silencing of the maternally imprinted tumor suppressor ARHI contributes to follicular thyroid carcinogenesis. J Clin Endocrinol Metab. 2005;90:1149–1155. doi: 10.1210/jc.2004-1447. [DOI] [PubMed] [Google Scholar]
- Badgwell DB, Lu Z, Le K, Gao F, Yang M, Suh GK, et al. The tumor-suppressor gene ARHI (DIRAS3) suppresses ovarian cancer cell migration through inhibition of the Stat3 and FAK/Rho signaling pathways. Oncogene. 2011;31:68–79. doi: 10.1038/onc.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt W. Sulforhodamine B assay and chemosensitivity. Methods Mol Med. 2005;110:39–48. doi: 10.1385/1-59259-869-2:039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.