Abstract
Glioblastoma is the most common primary intrinsic brain tumor and remains incurable despite maximal therapy. Glioblastomas display cellular hierarchies with self-renewing glioma-initiating cells (GICs) at the apex. To discover new GIC targets, we used in vivo delivery of phage display technology to screen for molecules selectively binding GICs that may be amenable for targeting. Phage display leverages large, diverse peptide libraries to identify interactions with molecules in their native conformation. We delivered a bacteriophage peptide library intravenously to a glioblastoma xenograft in vivo then derived GICs. Phage peptides bound to GICs were analyzed for their corresponding proteins and ranked based on prognostic value, identifying VAV3, a Rho guanine exchange factor involved tumor invasion, and CD97 (cluster of differentiation marker 97), an adhesion G-protein-coupled-receptor upstream of Rho, as potentially enriched in GICs. We confirmed that both VAV3 and CD97 were preferentially expressed by tumor cells expressing GIC markers. VAV3 expression correlated with increased activity of its downstream mediator, Rac1 (ras-related C3 botulinum toxin substrate 1), in GICs. Furthermore, targeting VAV3 by ribonucleic acid interference decreased GIC growth, migration, invasion and in vivo tumorigenesis. As CD97 is a cell surface protein, CD97 selection enriched for sphere formation, a surrogate of self-renewal. In silico analysis demonstrated VAV3 and CD97 are highly expressed in tumors and inform poor survival and tumor grade, and more common with epidermal growth factor receptor mutations. Finally, a VAV3 peptide sequence identified on phage display specifically internalized into GICs. These results show a novel screening method for identifying oncogenic pathways preferentially activated within the tumor hierarchy, offering a new strategy for developing glioblastoma therapies.
Glioblastoma represents the most common primary brain tumor with a median survival of 15 months despite maximal surgical resection, chemoradiation and adjuvant temozolomide treatment.1 Like many solid tumors, glioblastomas display a cellular hierarchy with self-renewing cells that can initiate secondary tumors that phenocopy the parental tumors from which they were derived.2, 3, 4 Although the tumor-initiating cell (TIC) hypothesis remains controversial because of unresolved issues in TIC identification and characterization,5 these cells are functionally defined through secondary tumor initiation in vivo.2, 3, 4 The significance of glioma-initiating cells (GICs) is supported by observations that these cells are relatively resistant to conventional therapies,6 preferentially invade into normal brain7 and promote tumor angiogenesis.8 GICs express stem cell markers and undergo aberrant differentiation in vivo to form heterogeneous tumors similar to the tumors from which they were derived. The derivation of GICs has proven to be complicated because glioblastoma is a heterogeneous disease with extensive inter- and intratumoral variability in genetics and transcriptional profiles.9 The demonstration of a cellular hierarchy requires the ability to prospectively enrich or deplete tumorigenic cells, leading to substantial effort to identify cell surface markers or other biomarkers that inform the cellular hierarchy. The first GIC marker identified was CD133 (cluster of differentiation marker 133; Prominin-1) based on its expression on neural stem cells,4 but – not surprisingly in this heterogeneous cancer – CD133 is not universally informative for all tumors.10 Alternative GIC markers – CD44,11 CD15/SSEA-1,12 L1 cell adhesion molecule,13 CD49f/integrin α614 – have demonstrated utility but every marker is context dependent. Many studies have failed to consider that these molecules mediate interactions between a cell and its microenvironment, so the removal of a cell from its native environment leads to rapid loss of information. Based on this background, we adapted phage display technology as a screening method for the identification of GICs in vivo.
Originally developed as a method to study protein–ligand interactions, phage display uses randomly generated deoxyribonucleic acid (DNA) sequences to create highly diverse (>10)10 peptide libraries displayed on the surface of filamentous phages.15 Phage display has identified novel ligands and cell-specific markers on intact cells in their native conformation and environment.16 Peptides identified by phage display can form the basis of therapeutic or diagnostic reagents.15 Previous discovery studies utilizing phage display have mainly focused on targeting whole tumors or tissue types via specific peptide binding.15, 16, 17, 18 Here we describe phage display as a method for identifying peptides that selectively bind to a tumor cellular subset.
Results
In vivo phage display screen identifies a unique set of peptide sequences specific to GICs
Along with others, we have demonstrated that components of the tumor microenvironment (hypoxia, restricted nutrition, acidic stress and extracellular matrix) contribute to the maintenance of the tumor cellular hierarchy.14, 19, 20 We therefore sought to investigate potential molecular interactions in GICs within the tumor microenvironment. As noted above, CD133 has been extensively used in studies as a putative GIC enrichment,4 with some conflicting results.10 However, CD133 has proven informative to dichotomize secondary tumor initiation and sphere formation in the tumor model we selected for the current screen. To validate CD133 as a GIC marker in our model, we enriched for CD133-positive cells from primary human glioblastoma specimens and performed functional assays for self-renewal, stem cell marker expression and tumor propagation.14, 19, 20, 21 Upon tumor development, the phage library was injected intravenously to permit cellular phage binding in vivo (Figure 1a). After 24 h, the tumor was dissected and dissociated after library injection. Of note, phage display has been previously shown to remain stable for this duration.17
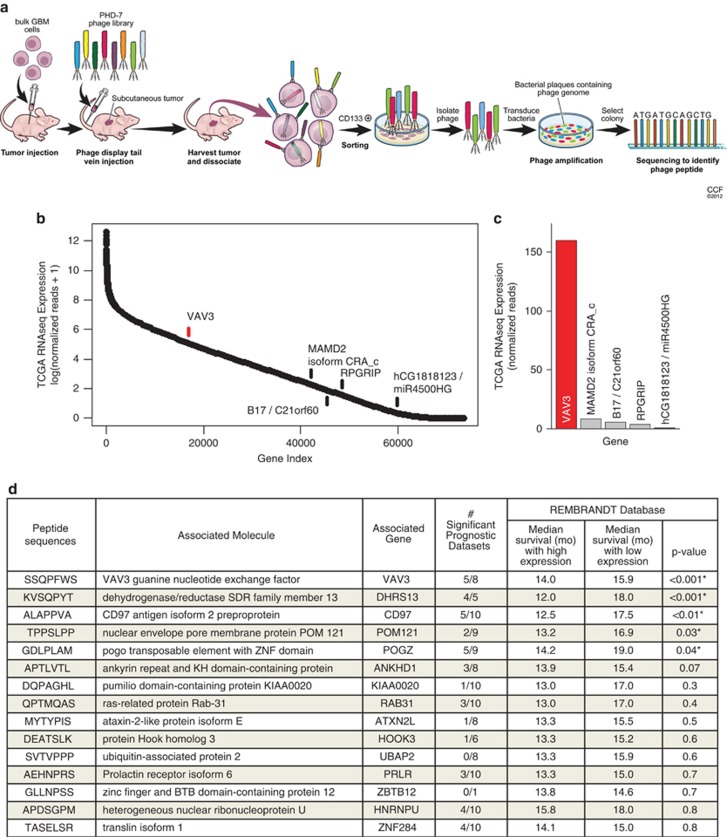
Figure 1.
In vivo phage display identifies GIC-specific peptide sequences. (a) Schematic diagram of in vivo phage display methodology. Freshly dissociated xenografted human glioblastoma cells were subcutaneously implanted into the flank of BALB/c-nu mice. Mice bearing tumors were injected with the phage library through the tail vein. GICs were purified from tumor and bound phages were isolated and sequenced. (b) Peptide sequences obtained using phage display and their corresponding genes were determined by the NCBI BLAST Search function (blast.ncbi.nlm.nih.gov). Analysis of TCGA RNAseq data reveals VAV3 as the most likely phage target for the sequence SSQPFWS. Log scale expression of 73 602 assembled isoform transcripts from TCGA RNA sequencing data were ordered by mean expression of 169 patient tumors with available data. The top scoring sequences by protein BLAST analysis are indicated. (c) Mean number of normalized RNAseq reads, representing expression, of indicated targets identified by protein BLAST is displayed for the sequence SSQPFWS. Both analyses indicate that VAV3 has markedly higher expression in patient glioblastomas than any other potential target. (d) The number of significant prognostic data sets indicates the number of data sets within Oncomine with a statistically significant prognostic survival association with the associated gene (i.e., increased gene expression is associated with significantly decreased survival). Identified genes were then queried in the Rembrandt database to determine the median survival (in months) associated with low versus high expression of the respective gene as well as the statistical significance
To identify phages bound and internalized into GICs, cellular fractions were isolated followed by sequencing to identify the corresponding peptide. Thirty-two unique (non-identical) peptides were identified (Supplementary Table 1). Querying these candidate peptides with the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) (blast.ncbi.nlm.nih.gov), we identified the genes containing peptide sequences that most closely matched those of each phage. As peptide sequences can match more than one protein, we interrogated tumors with RNA sequencing data deposited as part of the glioblastoma TCGA and compared the expression of isoform transcripts indicated by protein BLAST analysis (Figure 1b). Normalized RNAseq reads indicated that our top hit, SSQPFWS, is most likely associated with the VAV3 guanine exchange factor (Figure 1c). To prioritize the analysis of identified targets, we used bioinformatic analysis with associated survival information. We selected 10 published annotated data sets of glioblastoma expression for which survival correlation was available.22, 23, 24, 25, 26, 27, 28, 29, 30, 31 As a result of different expression profiling platforms, not all databases contained all genes for an associated peptide sequence. Owing to its breadth and high number of tumors analyzed, the REMBRANDT database was given significant weight. The top 15 targets are shown in Figure 1d. Several targets or related family members have been previously associated with cancer, including VAV3, CD97 (TM7LN1), ataxin 2-like protein (ATXN2L), ras-related protein 31 (RAB31) and ubiquitin-associated protein (UBAP2). These results suggest that in vivo phage display using patient-derived glioma xenografts can inform cancer target identification.
VAV3 is expressed more highly in GICs
Phage display peptide sequences bind to presented molecules. The targets identified in our screen represent potential binding partners (i.e., ligands) of possible primary GIC targets, not the actual receptors on the cell surface. Therefore, these molecules require direct validation to determine a potential direct relationship to GICs. We initially chose to primarily focus on VAV3 due its strong negative correlation with patient survival and frequent expression in cancers, including prostate cancer32 and glioblastoma.33, 34 In five of eight glioma expression databases, elevated VAV3 expression informed statistically significant poor prognosis. VAV3 serves as a guanine exchange factor to activate the Rho GTPase pathway to promote invasion, proliferation and survival.35 Using patient-derived xenografts (08-387 and 4121), we immunofluorescently stained for VAV3 with putative GIC markers, (sex determining region y)-box 2 (Sox2) and oligodendrocyte lineage transcription factor 2 (Olig2) (Figures 2a–d). VAV3 was consistently present in the cytoplasm of glioma cells that co-expressed Olig2 and Sox2, consistent with its expression in GICs.
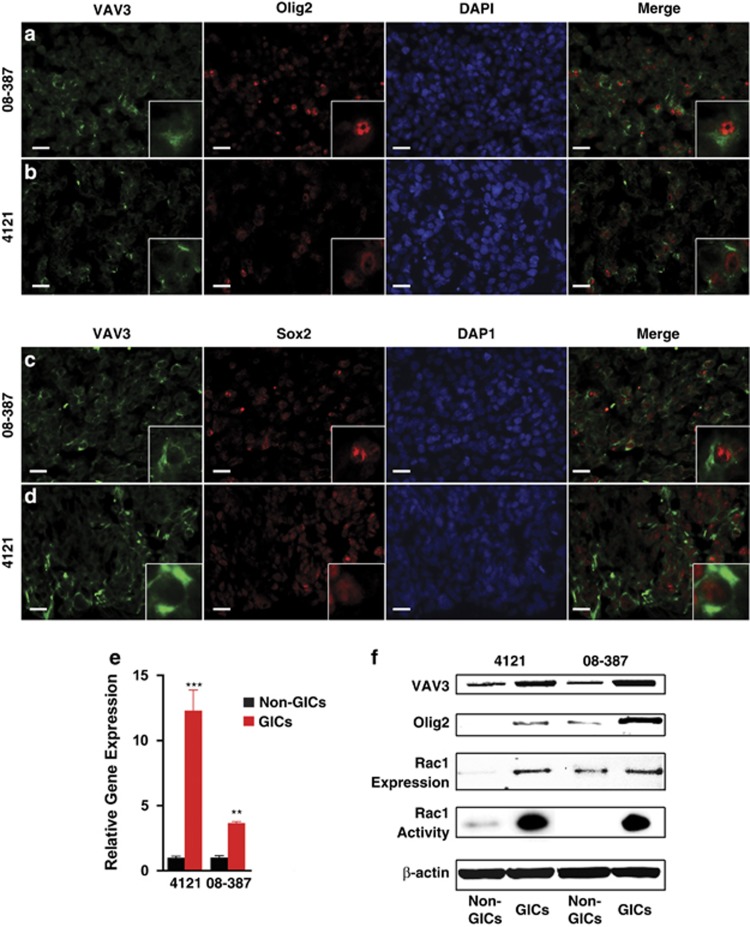
Figure 2.
GICs express higher levels of VAV3 compared with non-GICs. Immunostaining of VAV3 in intracranial xenograft glioblastoma tumors (08-387 and 4121) showing colocalization of VAV3 (green) with putative stem cell markers Olig2 (a and b) and Sox2 (c and d) in red at × 10 magnification. Inset represents a characteristic cell at × 20 magnification. Scale bars=10 μm. (e) Quantitative real-time PCR of VAV3 mRNA in GICs and non-GICs from specimens 4121 and 08-387. **P=0.002; ***P=0.0001. (f) VAV3 and Olig2 protein levels as well as Rac1 expression and activity in GICs and non-GICs from specimens 4121 and 08-387
To further ascertain relative expression levels of VAV3 within GICs as compared with matched, non-GICs, we derived populations that enriched or depleted for cells that expressed stem cell markers, formed spheres by in vitro limiting dilution and were tumorigenic in vivo by limiting dilution as previously described.14, 19, 20, 21 We then measured VAV3 gene transcript levels using quantitative real-time polymerization chain reaction (qRT-PCR). VAV3 was expressed more in GICs than non-GICs (Figure 2e), which was also demonstrated at the protein level by immunoblotting (Figure 2f).
VAV3 knockdown in GICs in vitro reduces cell growth, tumor sphere formation and migration
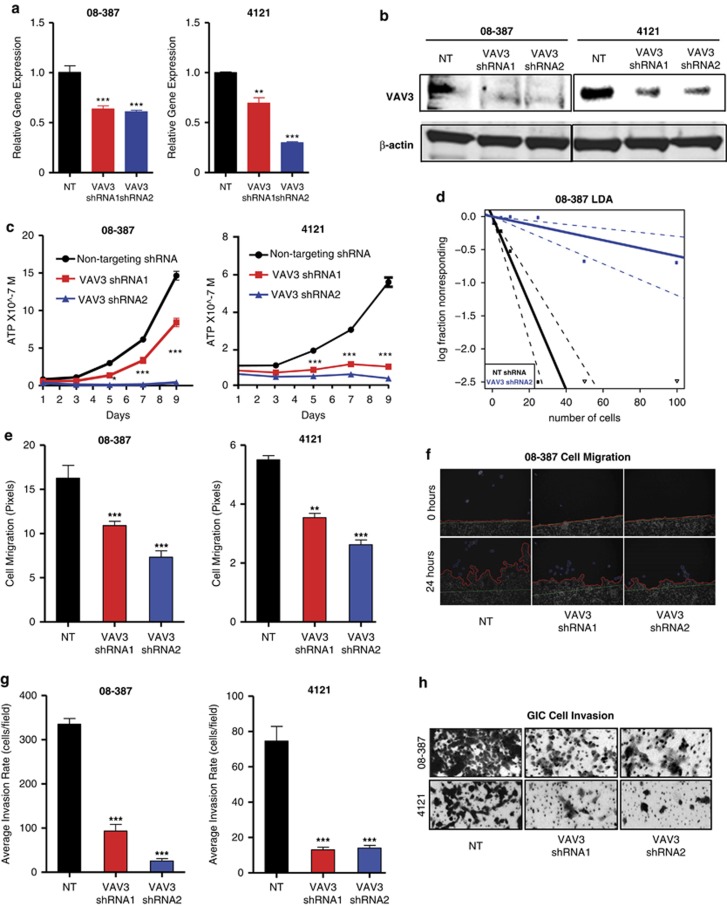
To interrogate the functional role of VAV3 in GICs, two non-overlapping short hairpin ribonucleic acid (shRNA) sequences were used to knockdown VAV3. Knockdown of VAV3 expression was confirmed at the mRNA level by qRT-PCR (Figure 3a) and protein level by immunoblotting (Figure 3b). VAV3 was critical to GIC growth as VAV3 knockdown significantly reduced cell growth beginning 5 days after knockdown (Figure 3c). Furthermore, VAV3 shRNAs also impaired tumorsphere formation by in vitro limiting dilution assay (Figure 3d).
Figure 3.
VAV3 is necessary for GIC growth, migration and invasion. VAV3 knockdown in GICs using two non-overlapping shRNA constructs results in decreased VAV3 mRNA expression quantified by qRT-PCR (a) and protein expression measured by immunoblot (b). VAV3 shRNA transduced into GICs caused decreased growth assessed by ATP cell titer assay (c) and reduced sphere formation determined by a limited dilution assay (d). VAV3 knockdown decreased migration as shown by pixel quantification (e) and representative images of 08-387 GICs at 0 and 24 h (f), as well as decreased numbers of invading GICs (g and h). *P=0.05; **P=0.002; ***P=0.0001
VAV3 stimulates RhoG-mediated formation of lamellipodia and invadopodia to increase to tumor invasion and migration.36 Therefore, we tested the hypothesis that VAV3 contributes to the invasive and migratory properties of GICs by performing a wound-healing assay. Using time-lapse microscopy, we determined that VAV3 knockdown decreased GIC migration by over twofold (Figures 3e and f). Further, VAV3 knockdown decreased the invasion rate in a Matrigel Boyden Chamber by at least fivefold compared with controls in two tumor models (Figures 3g and h).
VAV3 knockdown in GICs delays in vivo tumor development and prolongs survival
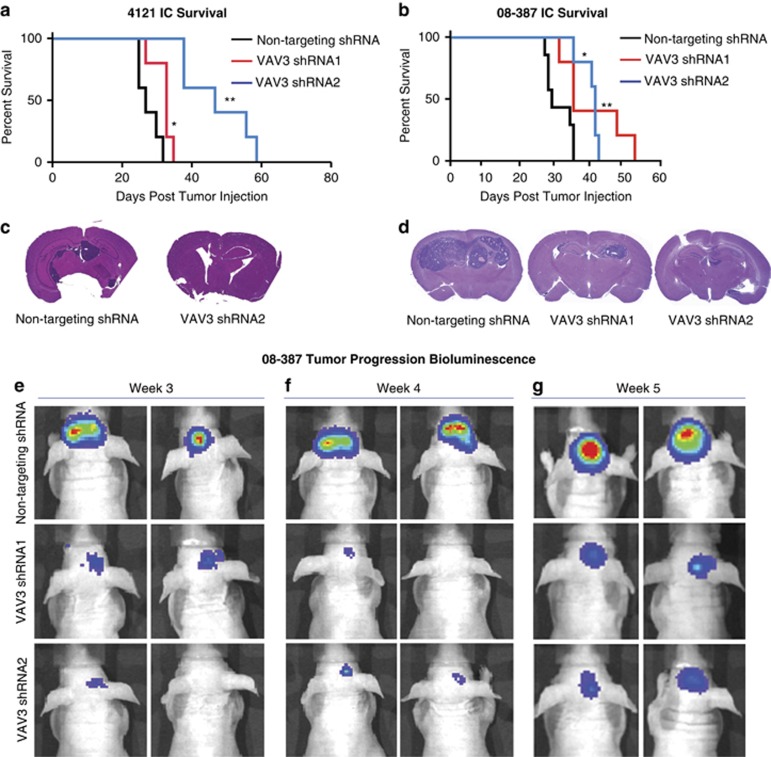
As the tumor microenvironment strongly regulates GIC maintenance and tumor growth, we investigated VAV3 function in GICs in vivo. GICs isolated from two different human specimens were transduced with non-targeting (NT) shRNA or VAV3 shRNA then implanted into the brains of immunocompromised mice. Mice that received GICs with VAV3 knockdown lived longer compared with mice injected with control GICs (Figures 4a and b). In a parallel cohort of mice, we harvested tumor-bearing brains when the first mouse displayed neurologic signs indicating tumor development. Mice that received NT shRNA developed large intracranial tumors whereas those that received GICs transduced with VAV3 shRNA had no detectable tumors (Figures 4c and d). We also monitored intracranial tumor growth during weeks 3–5 using bioluminescent tumor cells. We found more rapid growth and larger tumors among those mice that received tumor cells treated with NT shRNA relative to those with VAV3 knockdown (Figures 4e–g). Collectively, these results indicate that VAV3 contributes to in vivo GIC tumor growth.
Figure 4.
VAV3 promotes GIC tumor propagation in vivo. (a and b) Kaplan–Meier curves demonstrate an increase in median survival of mice bearing intracranial GICs transduced with VAV3 shRNA compared with mice bearing GICs transduced with NT shRNA. Median survival in mice bearing 4121 GICs with NT control shRNA was 27 days, with VAV3 shRNA1 was 33 days and VAV3 shRNA2 was 47 days; *P<0.05; **P<0.01. For mice bearing 08-387 GICs, median survival was 28 days with NT shRNA, 34 days with VAV3 shRNA1 and 40 days with VAV3 shRNA2; *P<0.05; **P<0.01. (c and d) Representative H&E sections of mice bearing intracranial GICs transduced with either NT shRNA or VAV3 shRNAs. (e–g) 08-387 Luciferase-expressing GICs transduced with either NT shRNA or VAV3 shRNAs were imaged weekly using an in vivo IVIS bioluminescence imaging system
Elevated VAV3 expression is associated with poor glioblastoma prognosis
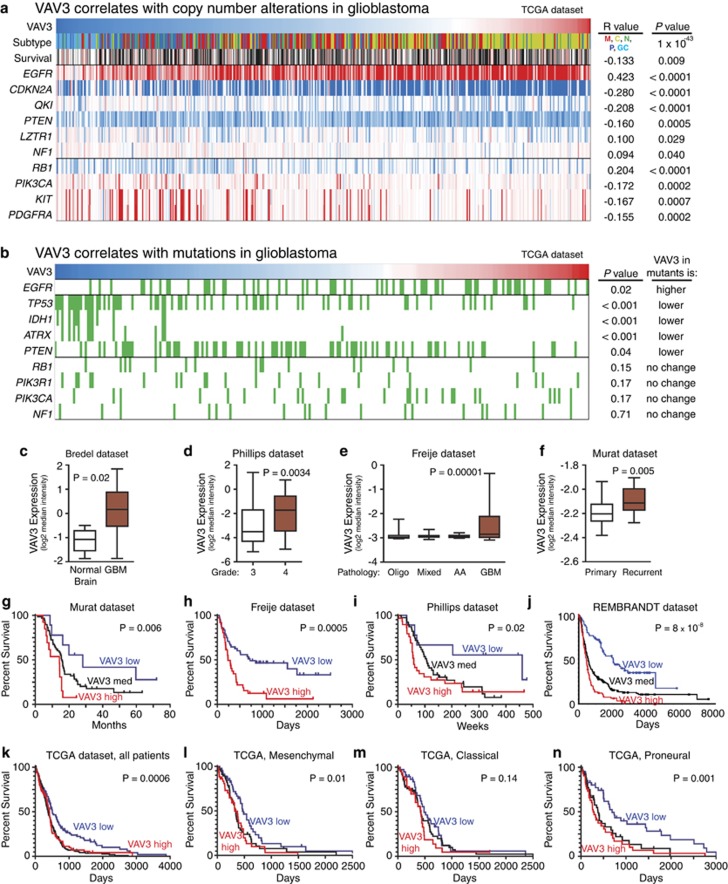
To further dissect the clinical significance of VAV3 in glioblastoma, we interrogated several databases. A strong caveat of these studies is that these data sets use a bulk tumor analysis that contains a mixture of GICs, non-GICs and stromal elements. Therefore, the expression of any gene is likely complicated in its association with GIC biology. Utilizing bioinformatics databases from TCGA, REMBRANDT and Oncomine, we correlated VAV3 expression and copy number alterations (Figure 5a), common mutations (Figure 5b), tumor type, grade or recurrence (Figures 5c–f), and patient survival (Figures 5g–n). Glioblastoma has recently been classified into four subgroups by expression profiling: proneural, neural, classical and mesenchymal.37 The proneural subgroup has been further subdivided into the glioma-CpG island methylator phenotype (G-CIMP) associated with mutations of isocitrate dehydrogenase 1 (IDH1) versus IDH1 wild type.38 VAV3 displayed an extremely strong association with the classical subtype (P=10−43) with lower association with mesenchymal tumors and the weakest with G-CIMP tumors. We consulted the gene expression signatures published used to delineated the classical subtype and found that VAV3 was a member of the signature.37 Based on this finding, we were not surprised to find that VAV3 expression strongly correlated with epidermal growth factor receptor (EGFR) copy number and mutation as these are hallmarks of the classical subtype.37 Higher VAV3 expression was significantly associated EGFR amplification (P<0.001), CDKN2A amplification (P<0.001) and EGFR mutations (P=0.048). VAV3 expression was anti-correlated with mutations common in proneural tumors (IDH1, tumor protein 53 gene (TP53) and alpha thalassemia/mental retardation syndrome x-linked (ATRX)). In addition, high VAV3 expression displayed a trend toward increased probability of neurofibromatosis type 1 mutations (P=0.065). These results suggest that VAV3 may function most strongly in classical glioblastomas, thereby informing the precise application of anti-VAV3 treatment in a specific subset of patients.
Figure 5.
Bioinformatic analysis reveals VAV3 expression correlates with clinically relevant properties of glioma. (a) Analysis of VAV3 expression and genetic copy number variation. Tumors with high VAV3 expression are more likely to be of the classical subtype, and more likely to have alterations in EGFR, CDKN2A or QKI. VAV3 high tumors are also less likely to have alterations in RB1, PIK3CA, KIT or PDGFRA. Red indicates high expression of VAV3 (top row) or copy number amplification (bottom rows), whereas blue indicates low expression of VAV3 (top row) or copy number loss (bottom rows). (b) Analysis of VAV3 expression and genetic mutation indicates that VAV3 high tumors (red) are more likely to have mutations (indicated in green) in EGFR. Conversely, VAV3 low tumors (blue) are more likely to have mutations in TP53, ATRX, IDH1 and phosphatase and tensin homolog. These results suggest that VAV3 expression is associated with the classical glioblastoma subtype, thereby potentially allowing for more effective anti-VAV3 treatment in a specific subset of patients. (c–f) VAV3 has a statistically significantly greater expression in glioblastoma versus non-malignant brain (c), in glioblastoma versus WHO grade III glioma (d), in glioblastoma versus other brain tumor histology (e) and in recurrent versus primary glioblastoma (f). Among patients with glioblastoma, high expression of VAV3 was associated with significantly shorter survival as compared with low expression as seen in Murat (g), Freije (h), Phillips (i), REMBRANDT (j) and TCGA data sets (k). Within the TCGA subgroups, VAV3 expression correlated with survival in mesenchymal (l) and proneural (n), but not classical (m) patients
Further analysis demonstrates that VAV3 has greater expression in glioblastoma versus non-malignant brain (Figure 5c), in higher tumor grade (Figure 5d) and in glioblastoma versus other glioma histologies (Figure 5e). VAV3 has higher expression in recurrent glioblastoma compared with primary glioblastoma (Figure 5f). To further examine the correlation of VAV3 expression with patient outcome, we generated Kaplan–Meier survival curves from additional data sets.23, 28, 29, 31 Consistently, high VAV3 expression was associated with significantly shorter survival (Figures 5g–k). To determine if VAV3 was more closely associated with prognosis in one of the Verhaak TCGA classified glioblastoma subtypes,37 we segregated the data and found that while VAV3 was significantly associated with survival in the proneural and mesenchymal subtypes (Figures 5l and n). As VAV3 expression is used to classify tumors in the classical subtype,37 all classical tumors have relatively high expression so no significant association occurred in the classical subtype (Figure 5m). Furthermore, among the VAV family only VAV3 expression was found to have a significant association with survival, whereas VAV1 and VAV2 did not (data not shown).
CD97 is expressed more highly in GICs and enriches for cells with GIC properties
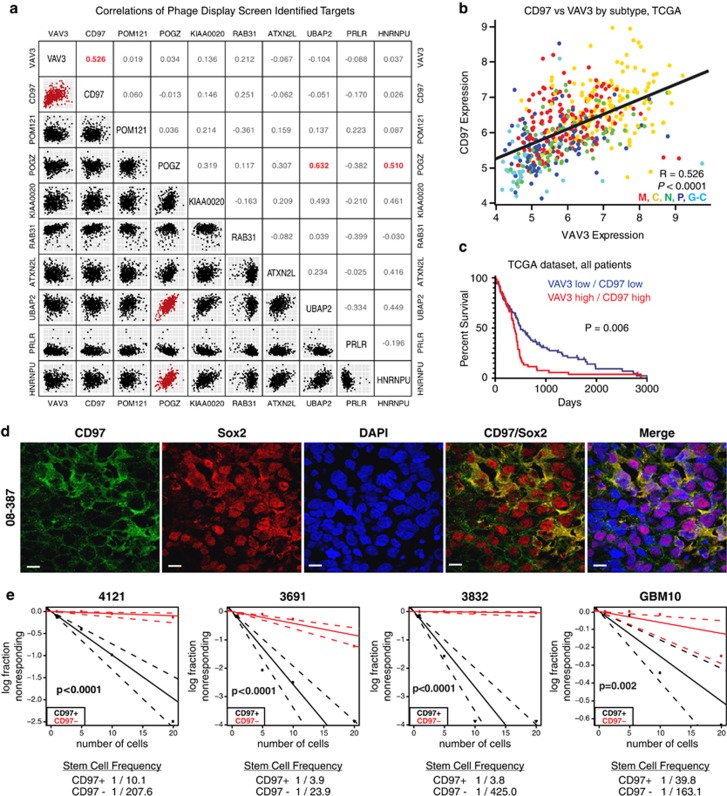
To determine if any other targets found on our screen could be functionally important, we interrogated the TCGA database and performed a correlative analysis expression between targets in individual tumors (Figure 6a). Only three correlations were found to have an R-value of >0.5 (VAV3-CD97, POGZ-UBAP2 and POGZ-HNRNPU). VAV3 and CD97 accounted for two of the top three targets found on our screen that correlated with poor patient prognosis (Figure 6b). The correlative levels of CD97 and VAV3 were strongly associated with glioblastoma subgroup (classical>mesenchymal>neural>proneural>G-CIMP). Like VAV3, CD97 is a component of the classical glioblastoma gene expression.37 The co-expression of CD97 and VAV3 also informs poor prognosis (Figure 6c).
Figure 6.
CD97 is correlated with VAV3 expression and is indicative of GIC sphere formation. (a) Pairwise correlation analysis of the top hits identified in the phage display screen. Plots indicate expression data from TCGA patients for indicated genes, and numbers represent correlation coefficient (R) values. Pairs with strong correlations (R-values >0.5) are indicated in red. (b) Detailed plot of CD97 versus VAV3 expression, colored by TCGA subtype. Data indicate that both genes are highly expressed in classical subtype tumors, and significantly (P<0.0001) and strongly (R=0.526) correlated with each other. (c) Survival analysis reveals patients with expression of both CD97 and VAV3 above each gene's average have significantly (P=0.006) worse survival than patients with below average expression of each gene. (d) Immunostaining of CD97 (green), Sox2 (putative stem cell marker; red) and nuclei (DAPI; blue) in intracranial xenograft of glioblastoma tumor 08-387. Scale bars=10 μm. (e) Bulk glioblastoma specimens (4121, 3691, 3832 and GBM10) were sorted using FACS for either CD97+ or CD97– cells into a 96-well plate for limited dilution sphere formation assay
CD97 (TM7LN1) is an adhesion G-protein-coupled receptor commonly expressed in hematopoietic cells and smooth muscle.39 Like VAV3, CD97 also activates the RhoGTPase pathway.39 CD97 has been recently shown to promote migration and invasion in glioblastoma.40, 41 To confirm that CD97 was expressed in xenograft glioblastoma tissue and preferentially expressed within the GICs, we performed immunofluorescent staining on a xenografted tumor specimen. CD97 co-stained with the GIC marker, Sox2, in glioblastoma specimens (Figure 6d). To further ascertain relative expression levels, we performed immunofluorescent staining of CD97 on GICs as compared with matched non-GICs. In two tumor specimens (08-387 and 4121), we found substantially greater CD97 expression in the GICs (data not shown). These data suggest that CD97 is found in glioblastomas localized preferentially within the GIC population.
CD97 is a cell surface protein permitting its potential use as a selection marker. We therefore interrogated CD97 as a potential method to enrich for sphere formation using four different patient-derived glioblastoma xenografts. In each of the models, cells CD97-positive displayed increased sphere formation (Figure 6e). These results further strengthened the utility of our original screen.
CD97 is associated with the classical glioblastoma subtype and poor patient prognosis
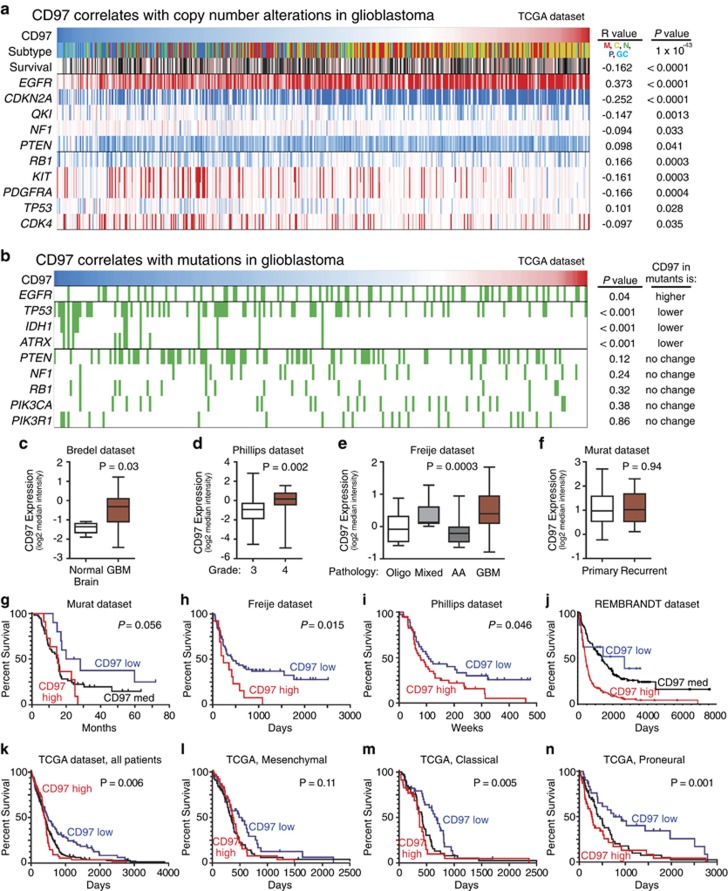
To determine the clinical significance of CD97, we performed a bioinformatics analysis (Figure 7). Like VAV3, CD97 strongly associates with the classical glioblastoma subtype and inversely with G-CIMP tumors. CD97 tumors are associated with increased EGFR and reduced CDKN2A in copy number (Figure 7a) with EGFR mutations similarly associated (Figure 7b). Owing to its anti-correlation with G-CIMP, tumors with high CD97 expression are less likely to have TP53, IDH1 or ATRX mutations (Figure 7b). CD97 is overexpressed in glioblastoma relative to normal brain (Figure 7c) and increases with tumor grade and astrocytic histology (Figures 7d and e). No difference with recurrent tumors is noted (Figure 7f). CD97 is associated with poor patient outcome, with very strong correlation in proneural glioblastoma, but not as consistently as VAV3 (Figures 7g–n). Collectively, these data support a clinical importance to CD97 in glioblastoma.
Figure 7.
Bioinformatic analysis reveals CD97 expression correlates with clinically relevant properties of glioma and alterations associated with VAV3 expression. (a) Analysis of CD97 expression and genetic copy number variation. Tumors with high CD97 expression are more likely to be classical glioblastoma subtype, and more likely to have alterations in EGFR, CDKN2A or QKI. CD97 high tumors are also less likely to have alterations in RB1, KIT or PDGFRA. Red indicates high expression of CD97 (top row) or copy number amplification (bottom rows), whereas blue indicates low expression of CD97 (top row) or copy number loss (bottom rows). (b) Analysis of CD97 expression and genetic mutation indicates that CD97 high tumors (red) are more likely to have mutations (indicated in green) in EGFR. Conversely, CD97 low tumors (blue) are more likely to have mutations in TP53, ATRX and IDH1. These results are consistent with both increased CD97 expression in classical subtype tumors and are similar to alterations found in VAV3 high-expression tumors. (c–f) CD97 was found to have statistically significantly greater expression in glioblastoma versus non-malignant brain (c), in glioblastoma versus WHO grade III glioma (d) and in glioblastoma versus other brain tumor histology (e); however, there was no association with CD97 in recurrent versus primary glioblastoma (f). Among patients with glioblastoma, high expression of CD97 was associated with significantly shorter survival as compared with low expression as seen in Freije (h), Phillips (i), REMBRANDT (j) and TCGA data sets (k); however, the analysis did not reach significance in Murat (g) data set. Within the TCGA subgroups, CD97 expression correlated with survival in classical (m) and proneural (n), but not mesenchymal (l) patients
Phage display peptide sequence identifies functional GICs
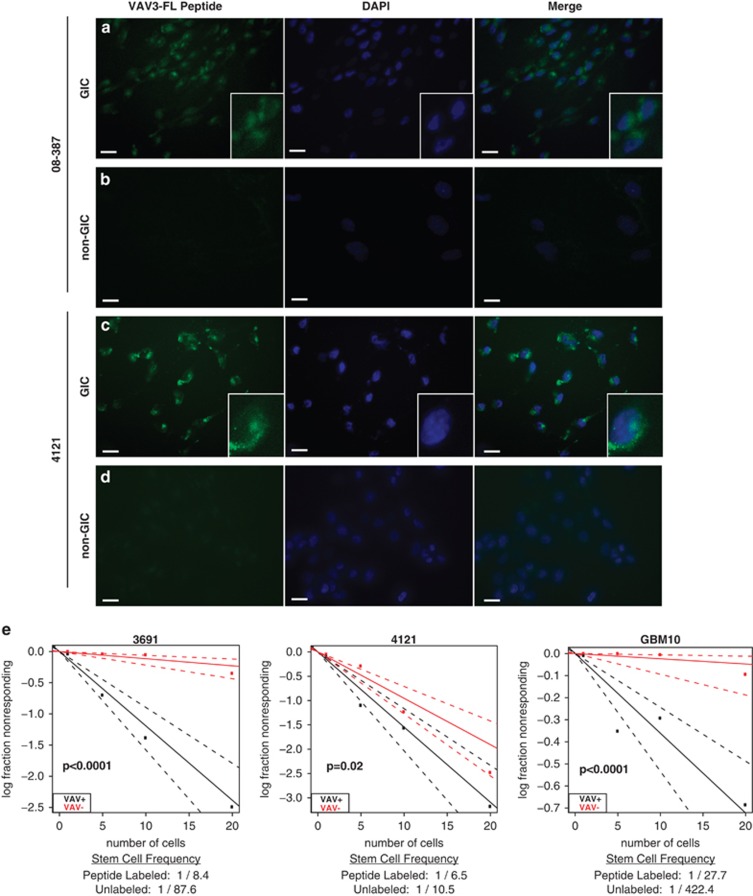
Phage display peptide sequences have been used as discovery tools and also can be used directly as tools for diagnosis and therapy.15, 17, 18, 42 Although bound phage peptide sequences can be traditional extracellular ligands, phage particles can also bind and internalize; thus, making possible identification of intracellular binding proteins to cell surface receptors. To determine the location of interaction as VAV3 is predominantly localized in the cytoplasm, we performed flow cytometric analysis of non-permeabilized GICs and detected binding of an anti-VAV3 antibody compared with an isotype control (data not shown), suggesting that VAV3 is at least transiently present on the cell surface. As VAV3 appears as a promising target, we investigated the utility of the identified VAV3 peptide as tool in GIC identification. In the first set of studies, we used functionally validated matched GIC and non-GIC cultures from two tumor models to determine peptide internalization. In both these models, the fluorescently labeled VAV3 peptide identified on phage display was internalized into GICs with minimal internalization into non-GICs (Figures 8a–d), suggesting a high specificity of the VAV3 peptide. Based on these results, we investigated the potential of the VAV3 peptide to identify GICs. We exposed bulk, unsorted glioma cultures from three tumors to the fluorescently labeled VAV3 peptide then sorted two populations using fluorescence-activated cell sorting (FACS) based on peptide internalization. Consistent with GIC functions, cells with VAV3 peptide internalization were significantly more likely to form spheres (Figure 8e). Thus, our initial phage display not only informs target discovery but also directly provides tools that may be leveraged for GIC identification.
Figure 8.
Fluorescently labeled VAV3 peptide is internalized into GICs. (a–d) GICs and non-GICs from tumor specimens 08-387 and 4121 were treated with the green fluorescent-tagged 5-Fap peptide that corresponds to the VAV3 protein. The nuclear counterstain DAPI is in blue and scale bar=10 μm. (e) Bulk, unsorted cells from three glioblastoma specimens (3691, 4121 and GBM10) were treated with green 5-Fap VAV3 peptide and then sorted based on peptide uptake by FACS into 96-well plates for a limited dilution sphere formation assay
Discussion
The limited efficacy of cytotoxic agents (ionizing radiation and chemotherapy) and failure of targeted therapeutics for the treatment of glioblastoma has prompted the development of agents that target highly resistant GIC populations. Based on the similarities between TICs and normal stem cells (embryonic and tissue-specific), TIC-targeting agents have focused on core stem cell regulatory pathways: Notch, hedgehog, wnt, phosphoinositide 3-OH kinase.43 In some cancer types, anti-TIC agents have demonstrated efficacy in early clinical trials, albeit with resistance and toxicity.44 Along with others, we have envisioned the identification of novel TIC molecular targets with limited dependence in normal cells. For example, we recently reported that GICs are dependent on inducible nitric oxide synthase (iNOS, NOS2) but iNOS is dispensable for normal cells.21 Screens for GIC regulators have included gene expression profiling, chemical screens or RNA interference.45, 46, 47 Recently, we reported the use of cell-based aptamer screening to identify several aptamers that selectively identify self-renewing, tumorigenic cells.48 Methods for identifying and subsequent targeting of target molecules are limited by cost, relatively small numbers of pre-determined antigen targets, and limited utility in cells and tissues in their native state. As the critical interactions between GICs and their microenvironment support the use of in vivo screens, here we demonstrate in vivo phage display identification of peptide sequences that bind to molecules and pathways specific to GICs. Our screening method identified known markers of GICs, including Olig2.49 As GICs have been identified as potentially invasive into normal brain, we also identified prominent regulation of mediators of cell invasion, including VAV3, CD97 and Rac1.
Phage display informs the identification of peptides that bind to specific targets, that is, potential ligands. In our screen, we identified two potentially useful targets that both regulate invasion. GIC models have demonstrated in vivo tumor cell invasion in contrast to standard cell lines, suggesting that GICs or their progeny contribute to the invasion of glioma into normal brain. To date, the direct molecular link between tumor invasion and GIC survival has not been evident so our studies have a potential to inform such interaction. In a recent report, we found that the platelet-derived growth factor receptor-β (PDGFRβ) is specifically expressed by GICs and regulates invasion through signal transducer and activator of transcription 3 (STAT3) activity.50 This study suggests that Rac1 may be another key signaling node. Further studies will directly address the utility of targeting Rac1 that may integrate several upstream pathways similar to the role of STAT3 integrating upstream kinase activity from growth factor receptors (EGFR,51 and PDGFRβ),50 cytokine receptors (interleukin-6 receptor)52 and non-receptor tyrosine kinases (bone marrow X-liked kinase)53 that can substitute for one another when a single pathway is disrupted.
No clear link between the expression of VAV3 or CD97 and the tumor hierarchy has been previously described but reports have suggested that both may mediate invasion in glioma,34, 40, 41 yet the mechanisms remain unclear. CD97 binds and amplifies the activity of the LPA receptor54 as well as the decay-accelerating factor (CD55),55 a regulator of complement activity. Unlike VAV3, CD97 is a direct cell surface protein supporting its potential utility in prospective studies of GICs. As CD97 expression has been linked to other cancer types, including thyroid, breast, gastric, and ovarian cancers, suggesting that CD97 may be useful to investigate in TICs of other tumor types.
One challenge in the use of phage display in discovery is that it generally identifies ligands for the proteins that are the direct targets of binding. Although VAV3 and CD97 appear to be strongly linked to GICs by preferential expression and function, our findings may represent only an initial discovery as further studies will be required to identify the direct target of binding on GICs that could serve as TIC selection markers. Consideration of prior molecular links may be informative in these future directions. VAV3 binds to several targets as noted above, including IQGAP, which has already been reported as a specific GIC target.56 Other potential targets include growth factor receptors, several of which promote the growth of GICs. Interacting partners of CD97 are less certain and may involve the integration of inflammatory signals (LPA or complement). As NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) has been linked to maintenance of GICs,57 our studies may provide novel upstream signals that contribute to the activation of this critical pathway.
It is useful to note that many studies in stem cell biology stress the cell autonomous aspects of stem and progenitor cells, but all cells in a multicellular organism are responsive to their environment.58 Stem cells actively maintain tissues undergoing continuous turnover, for example, the intestine, or serve as a reserve population responsive to injury. The core stem cell regulator pathways permit readout of a cellular context. Although tumors act as selfish organ systems, they generally retain features of the tissues from which they were derived, including cellular hierarchies. In particular, tumors will often display common aspects to one another in low oxygen, nutrients and pH with each of these environmental cues stimulating a stem cell-like state. These observations raise a critical need for target screens to be performed in vivo to maximize the potential for a positive and selective result in the tumor milieu. We leveraged one system to identify targets that were preferentially expressed in the stem-like tumor cells and contributed to their growth and invasion, while also being linked to poor patient outcome. Early studies suggest that the peptide sequences may also be used for the identification and perhaps selective targeting of TICs, but these studies await further development. The identification of GIC regulators of invasion may be particularly useful as the invasive front is often resistant to conventional therapies and contributes to residual disease after surgical resection. In conclusion, the development of advanced molecular discovery techniques with improved tumor models may enrich for molecular targets with greater impact.
Materials and Methods
Isolation and culture of cells
Glioblastoma tissues were obtained from excess surgical materials from patients at the Cleveland Clinic after review from a neuropathologist in accordance with an approved protocol by the Institutional Review Board. To prevent culture-induced drift, patient-derived xenografts were generated and maintained as a source of tumor cells for study.6, 14, 19, 21, 48 GICs and non-GICs were derived immediately after dissociation or after transient xenograft passage in immunocompromised mice using prospective sorting followed by assays to confirm stem cell marker expression, sphere formation and secondary tumor initiation.6, 14, 19, 21, 48 In the models used in these studies, CD133 has previously identified functional GICs. Therefore, in experiments with matched GIC or non-GIC cultures, we segregated CD133 marker-positive and marker-negative populations using CD133/2-APC-conjugated antibody (293C3, Miltenyi Biotech, Auburn, CA, USA) by magnetic bead separation, as previously described.6, 14, 19, 21, 48 The GIC phenotype of these cells was validated by stem cell marker expression (CD133, Olig2, Sox2), self-renewal (serial tumorsphere passage) and tumor propagation.14, 19, 21, 48
In vivo biopanning of phage library
The Ph.D. −7 Phage Display Peptide Library Kit (New England BioLabs, Ipswich, MA, USA) was used to complete these experiments. Methods were completed as per the manufacturer's protocol. To adapt phage display into an in vivo tumor, we selected a flank tumor model to permit the recovery of a large number relatively pure tumor cells. Bulk glioblastoma cells were implanted into the flanks of BALB/c nu/nu mice under a Cleveland Clinic Foundation Institutional Animal Care and Use Committee approved protocol as previously described.6, 14, 19, 21, 48 The flank tumor grew until it reached 1 cm in diameter, then the phage display library was injected via the tail vein of the mouse and allowed to circulate and bind to cellular targets for 24 h. The tumor was then harvested, dissociated and separated into GICs and non-GICs as described above. Tumor cells exposed to phage in vivo were then lysed to allow for isolation and purification of phage that had been internalized. Collected phages were transduced into E. coli and grown overnight to allow phage amplification. Transduced bacteria were placed on an Xgal/IPTG plate, and bacterial plaques that were successfully transduced appeared blue because of the lacZ-alpha gene in the phages. Plaques containing the phage genome were then sequenced to identify the phage peptide.
Immunofluorescent staining
Cells or 10 μm thick slices of xenografted brain tissue were fixed in 4% paraformaldehyde and immunolabeled using the following antibodies: VAV3 (Millipore, Billerica, CA, USA), CD97 (Santa Cruz Biotechnology, Dallas, TX, USA), Olig2 (R&D Systems, Minneapolis, MN, USA) and Sox2 (R&D Systems). Primary antibodies were incubated overnight at 4 °C, followed by species appropriate secondary antibodies (Alexa 488 and 568; Invitrogen Molecular Probes, Eugene, OR, USA) with incubation for 1 h. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI), and slides were then mounted using Fluoromount (Calbiochem, San Diego, CA, USA). Images were taken using a Leica DM4000 Upright microscopy (Leica, Buffalo Grove, IL, USA).
Fluorescent peptide staining/sorting
Cells were treated with the fluorescent-tagged (5-fluorogen activating protein (Fap)) peptide SSQPFWS that corresponds to the VAV3 protein. In all, 1 mg/ml of peptide was incubated with GICs or non-GICs for 15 min at 37 °C, and then washed 3–5 times with PBS. Cells were either fixed with paraformaldehyde and the nuclei counterstained with DAPI, or sorted by FACS and functionally assayed for neurosphere formation by limiting dilution.
Vectors and lentiviral transfection
Lentiviral clones to express shRNA against VAV3 (shRNA1 sequence—5′-CCGGCCAGTAGATTATTCTTGCCAACTCGAGTTGGCAAGAATAATCTACTGGTTTTTG-3′ and shRNA2 sequence—5′-CCGGCGAAGTTGTTGTCTAGCAGAACTCGAGTTCTGCTAGACAACAACTTCGTTTTTG-3′) or an NT shRNA (SHC002) were obtained from Sigma-Aldrich (St. Louis, MO, USA). The NT vector contains an shRNA insert that does not target human or mouse genes, serving as a negative control for off-target effects in experiments. Two of the five VAV3 shRNAs with non-overlapping sequences that had the best relative knockdown efficiency were used for all experiments. Lentiviral particles were generated in 293T cells with the pPACK set of helper plasmids (System Biosciences, Mountain View, CA, USA) in stem cell medium. Viral stocks were concentrated via precipitation using the PEG-8000, and then subsequently titered using the manufacturer's protocol.
Quantitative RT-PCR
Total cellular RNA was isolated using the RNeasy kit (Qiagen, Venlo, The Netherlands) and reverse transcribed into cDNA using the SuperScript III Reverse Transcription kit (Invitrogen, Carlsbad, CA, USA). Quantitative reverse transcription PCR was performed per the manufacturer's protocol on an Applied Biosystems 7900HT cycler using SYBR Green Master mix (SA Biosciences, Valencia, CA, USA) and gene-specific primers as follows: β-actin forward (5′-AGAAAATCTGGCACCACACC-3′) and reverse (5′-AGAGGCGTACAGGGATAGCA-3′), VAV3 forward (5′-ACAAGGAGCCAGAACATTCAG-3′) and reverse (5′-TTGCACAGAAGTCATACCGAG-3′).
Western blotting
Cells were collected and lysed in hypotonic buffer with nonionic detergent (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 50 mM NaF with protease inhibitors), incubated on ice for 15 min and cleared by centrifugation at 10 000 g at 4 °C for 10 min. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA). Equal amounts of protein were mixed with reducing Laemmli loading buffer, boiled and electrophoresed on NuPAGE Gels (Invitrogen), then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Blocking was performed for 30 min with 5% nonfat dry milk in tris-buffered saline and Tween 20 and blotting performed with primary antibodies for 16 h at 4 °C. Antibodies included rabbit monoclonal anti-VAV3 (Millipore, cat # 07-464) and goat polyclonal anti-Olig2 (R&D Systems, cat # AF2418).
Rac1 activation assay
Cells were cultured in adherent conditions overnight followed by lysis on ice using an Mg2+ lysis/wash buffer. Pull-down of the activated Rac1 was performed using the Rac/cdc42 Assay Reagent according to the manufacturer's protocol (Millipore, cat # 17-441). After collecting the agarose beads, and resuspended in reducing buffer, samples were then loaded into polyacrylamide gels for electrophoresis and transferred to PVDF membranes. The membranes were blocked with 3% nonfat dry milk in PBS and incubated with 1 μg/ml of anti-Rac1 antibody overnight at 4 ° C. Secondary antibody used was a goat anti-mouse horseradish peroxidase-conjugated IgG.
Proliferation and neurosphere formation assay
Cell proliferation assay using Cell-Titer Glow (Promega, Madison, WI, USA) and neurosphere formation were measured as previously described.14, 19, 21
Wound assay
To assess cellular migration, monolayer wound-healing assay was performed as previously described.59 Briefly, GIC cells were transfected with VAV3 shRNA1, VAV3 shRNA2 or NT control shRNA. Cells were plated as a confluent monolayer over a strip of polydimethylsiloxane. On removing the strip, it creates a virgin surface over which the cells are able to migrate. Cells were monitored for 24 h using time-lapse microscopy.
Invasion assay
To determine the invasiveness of GICs, we used BD BioCoat Matrigel invasion chambers (BD Biosciences, Franklin Lakes, NJ, USA). GICs were transduced with VAV3 shRNA1, VAV3 shRNA2 or NT control shRNA 48 h before analysis. Before use, the invasion chambers were rehydrated using neurobasal media (without any growth factors) for 2 h at 37 °C. Cells were then transferred to the invasion chamber (50 000 cells per well). Cells were allowed to invade through the Matrigel for 36–48 h toward neurobasal media supplemented with growth factors (epidermal growth factor and fibroblast growth factor). Chambers were then fixed in 4% paraformaldehyde for 15 min, stained with crystal violet dye and washed in PBS. Cells were counted using an AMG EVOS XL microscope (Life Technologies, Grand Island, NY, USA).
In vitro limiting dilution assay
For in vitro limiting dilution assays, cells were sorted by FACS with decreasing numbers of cells per well (20, 10, 5 and 1) plated in 96-well plates. Extreme limiting dilution analysis was performed using software available at http://bioinf.wehi.edu.au/software/elda, as previously described.60
Intracranial tumor formation in vivo
GICs were transduced with lentiviral vectors expressing VAV3 or a NT control shRNA for the knockdown experiments. Following puromycin selection, 1000 cells were engrafted intracranially into BALB/c nu/nu mice under a Cleveland Clinic Foundation Institutional Animal Care and Use Committee approved protocol. Animals were then maintained until neurological signs were apparent, then killed. The brains were harvested and fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and then cryosectioned. Sections were stained with hematoxylin and eosin. In parallel survival experiments, animals were monitored until they developed neurological signs.
Bioinformatic analysis of VAV3 expression in human gliomas
Multiple, independent brain tumor microarray databases annotated with patient survival were used to correlate survival and VAV3 gene expression in malignant glioma biopsies, including the Cancer Genome Atlas (TCGA, cancergenome.nih.gov),22 National Cancer Institute's Repository for Molecular Brain Neoplasia Data23 (REMBRANDT, https://caintegrator.nci.nih.gov/rembrandt/) as well as Oncomine22, 24, 25, 26, 27, 28, 29, 30, 31 (https://www.oncomine.org). Within REMBRANDT, the cutoff for upregulation of gene expression was defined as expression that is >2-fold relative to the mean value in normal tissue, whereas gene downregulation was defined as expression <0.5-fold relative to the mean expression. For correlations between genes, TCGA data were downloaded and analyzed using R (www.r-project.org). Pairwise correlation analysis of targets was performed using TCGA data (accessed at https://tcga-data.nci.nih.gov/tcga/ tcgaHome2.jsp) in R software (https://www.R-project.org/). The function ggpairs, available through the GGally package, was utilized to generate plots and determine pairwise correlation coefficients (http://CRAN.R-project.org/package=GGally).
Statistical analysis
All grouped data are presented as mean±S.D. Differences between groups were assessed by Student's t-test or analysis of variance using GraphPadInStat software (GraphPad Software, La Jolla, CA, USA). Kaplan–Meier curves were generated and log-rank analysis was performed using MedCalc software (Ostend, Belgium). *P<0.05; **P<0.01; ***P<0.001.
Acknowledgments
This work was supported by the National Institutes of Health (CA129958, CA154130, CA169117, CA157948 and NS083629), the Howard Hughes Medical Institute Medical Fellowship, Andrew & Lillian A Posey Foundation, the American Brain Tumor Association and the James S McDonnell Foundation. We appreciate the tissue provided by the Cleveland Clinic Foundation Tissue Procurement Service, the Flow Cytometry Core and the Imaging Core. We are grateful for the thoughtful discussions with Dr Shideng Bao.
Glossary
- ATRX
alpha thalassemia/mental retardation syndrome x-linked
- BLAST
basic local alignment search tool
- CD
cluster of differentiation
- DAPI
4',6-diamidino-2-phenylindole
- DNA
deoxyribonucleic acid
- EGFR
epidermal growth factor receptor
- FACS
fluorescence-activated cell sorting
- G-CIMP
glioma-CpG island methylator phenotype
- GIC
glioma-initiating cell
- IDH1
isocitrate dehydrogenase 1
- iNOS
inducible nitric oxide synthase
- NCBI
National Center for Biotechnology Information
- NT
non-targeting
- Olig2
oligodendrocyte lineage transcription factor 2
- PDGFRβ
platelet-derived growth factor receptor-β
- PI3K
phosphoinositide 3-OH kinase
- PVDF
polyvinylidene fluoride
- qRT-PCR
quantitative real-time polymerization chain reaction
- Rac1
ras-related C3 botulinum toxin substrate 1
- REMBRANDT
repository for molecular brain neoplasia data
- RNA
ribonucleic acid
- shRNA
short hairpin RNA
- Sox2
(sex determining region y)-box 2
- STAT3
signal transducer and activator of transcription 3
- TCGA
The Cancer Genome Atlas
- TIC
tumor-initiating cell
- TP53
tumor protein 53 gene
- UBAP2
ubiquitin-associated protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by JA Cidlowski
Supplementary Material
References
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT, Jr, Zaupa C, Aghi M, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA, et al. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam D-H, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, Li Z, Sathornsumetee S, Wang H, McLendon RE, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzazy HME, Highsmith WE., Jr Phage display technology: clinical applications and recent innovations. Clin Biochem. 2002;35:425–445. doi: 10.1016/s0009-9120(02)00343-0. [DOI] [PubMed] [Google Scholar]
- Staquicini FI, Sidman RL, Arap W, Pasqualini R. Phage display technology for stem cell delivery and systemic therapy. Adv Drug Deliv Rev. 2010;62:1213–1216. doi: 10.1016/j.addr.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler CE, Wu Q, Yan K, MacSwords JM, Chandler-Militello D, Misuraca KL, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Zenklusen J-C, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- Kawaguchi A, Yajima N, Tsuchiya N, Homma J, Sano M, Natsumeda M, et al. Gene expression signature-based prognostic risk score in patients with glioblastoma. Cancer Sci. 2013;104:1205–1210. doi: 10.1111/cas.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai R, Shi T, Kremen TJ, Horvath S, Liau LM, Cloughesy TF, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou M-F, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BKA, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65:4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Lin K-T, Gong J, Li C-F, Jang T-H, Chen W-L, Chen H-J, et al. Vav3-rac1 signaling regulates prostate cancer metastasis with elevated Vav3 expression correlating with prostate cancer progression and posttreatment recurrence. Cancer Res. 2012;72:3000–3009. doi: 10.1158/0008-5472.CAN-11-2502. [DOI] [PubMed] [Google Scholar]
- Serão NV, Delfino KR, Southey BR, Beever JE, Rodriguez-Zas SL. Cell cycle and aging, morphogenesis, and response to stimuli genes are individualized biomarkers of glioblastoma progression and survival. BMC Med Genomics. 2011;4:49. doi: 10.1186/1755-8794-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173:1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio-Real L, Kazanietz MG. Rho GEFs and cancer: linking gene expression and metastatic dissemination. Sci Signal. 2012;5:pe43. doi: 10.1126/scisignal.2003543. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska A, Didier S, Fortin S, Chuang Y, White T, Berens ME, et al. The small GTPase RhoG mediates glioblastoma cell invasion. Mol Cancer. 2012;11:65. doi: 10.1186/1476-4598-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaee M, Clark AJ, Ivan ME, Oh MC, Bloch O, Sun MZ, et al. CD97 is a multifunctional leukocyte receptor with distinct roles in human cancers (review) Int J Oncol. 2013;43:1343–1350. doi: 10.3892/ijo.2013.2075. [DOI] [PubMed] [Google Scholar]
- Safaee M, Clark AJ, Oh MC, Ivan ME, Bloch O, Kaur G, et al. Overexpression of CD97 confers an invasive phenotype in glioblastoma cells and is associated with decreased survival of glioblastoma patients. PLoS One. 2013;8:e62765. doi: 10.1371/journal.pone.0062765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidambaram A, Fillmore HL, Van Meter TE, Dumur CI, Broaddus WC. Novel report of expression and function of CD97 in malignant gliomas: correlation with Wilms tumor 1 expression and glioma cell invasiveness. J Neurosurg. 2012;116:843–853. doi: 10.3171/2011.11.JNS111455. [DOI] [PubMed] [Google Scholar]
- Lee S, Xie J, Chen X. Peptide-based probes for targeted molecular imaging. Biochemistry. 2010;49:1364–1376. doi: 10.1021/bi901135x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B-BS, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovsky Y, Kim D, Zheng S, Whyte WA, Bansal M, Bray M-A, et al. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 2014;6:313–324. doi: 10.1016/j.celrep.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Wurdak H, Zhu S, Romero A, Lorger M, Watson J, Chiang C-Y, et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 2010;6:37–47. doi: 10.1016/j.stem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Kim Y, Wu Q, Hamerlik P, Hitomi M, Sloan AE, Barnett GH, et al. Aptamer identification of brain tumor-initiating cells. Cancer Res. 2013;73:4923–4936. doi: 10.1158/0008-5472.CAN-12-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Huillard E, Kesari S, Maire CL, Golebiowski D, Harrington EP, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim E, Wu Q, Guryanova O, Hitomi M, Lathia JD, et al. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26:1247–1262. doi: 10.1101/gad.193565.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Yin J, Kim S-H, Sohn Y-W, Beck S, Lim YC, et al. EGFR-AKT-Smad signaling promotes formation of glioma stem-like cells and tumor angiogenesis by ID3-driven cytokine induction. Cancer Res. 2011;71:7125–7134. doi: 10.1158/0008-5472.CAN-11-1330. [DOI] [PubMed] [Google Scholar]
- Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells. 2009;27:2393–2404. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward Y, Lake R, Yin JJ, Heger CD, Raffeld M, Goldsmith PK, et al. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 2011;71:7301–7311. doi: 10.1158/0008-5472.CAN-11-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott RJM, Spendlove I, Roversi P, Fitzgibbon H, Knott V, Teriete P, et al. Structural and functional characterization of a novel T cell receptor co-regulatory protein complex, CD97-CD55. J Biol Chem. 2007;282:22023–22032. doi: 10.1074/jbc.M702588200. [DOI] [PubMed] [Google Scholar]
- Balenci L, Clarke ID, Dirks PB, Assard N, Ducray F, Jouvet A, et al. IQGAP1 protein specifies amplifying cancer cells in glioblastoma multiforme. Cancer Res. 2006;66:9074–9082. doi: 10.1158/0008-5472.CAN-06-0761. [DOI] [PubMed] [Google Scholar]
- Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedula SRK, Leong MC, Lai TL, Hersen P, Kabla AJ, Lim CT, et al. Emerging modes of collective cell migration induced by geometrical constraints. Proc Natl Acad Sci USA. 2012;109:12974–12979. doi: 10.1073/pnas.1119313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.