Abstract
Mammalian Hedgehog (Hh) signal transduction requires the primary cilium, a microtubule-based organelle, and the Gli/Sufu complexes that mediate Hh signaling are enriched at cilia tips. KIF7, a kinesin-4 family protein, is a conserved regulator of the Hh signaling pathway and a human ciliopathy protein. Here we show that KIF7 localizes to cilia tips, the site of microtubule plus-ends, where it limits cilia length and controls cilia structure. Purified recombinant KIF7 binds the plus-ends of growing microtubules in vitro, where it reduces the rate of microtubule growth and increases the frequency of microtubule catastrophe. KIF7 is not required for normal intraflagellar transport or for trafficking of Hh pathway proteins into cilia. Instead, a central function of KIF7 in the mammalian Hh pathway is to control cilia architecture and to create a single cilia tip compartment where Gli/Sufu activation can be correctly regulated.
Introduction
Although kinesin motor proteins are best known for their roles in intracellular transport, some kinesins can shape the microtubule cytoskeleton by regulating the dynamics of tubulin polymerization. For example, KIF4A, a kinesin-4 protein, controls microtubule length during cell division 1–4 and another kinesin-4 protein, KIF21A, inhibits microtubule growth at the cell cortex 5. KIF7, a conserved regulator of Hedgehog (Hh) signaling 6–9, is also a member of the kinesin-4 family but its relationship to microtubules has not been defined.
The Hedgehog signaling pathway is an evolutionary conserved pathway responsible for many aspects of embryonic development and stem cell maintenance, and is disrupted in a spectrum of tumors10,11. Drosophila Costal2 (Cos2) and its mammalian homologue KIF7 are required to relay the signals from the membrane protein Smoothened (Smo) to the transcription factors of the Ci/Gli family. Genetic inactivation of either Cos2 or Kif7 causes a relatively mild ectopic activation of Hh signaling due to their roles in production of both activator and repressor forms of Ci/Gli proteins11,12. Recessive mutations in human KIF7 are associated with fetal hydrolethalus, and acrocallosal and Joubert syndromes; affected individuals exhibit polydactyly, brain abnormalities and cleft palate, consistent with a role for KIF7 in human Hh signaling 13,14.
A fundamental difference between the Drosophila and vertebrate Hh pathways is the dependence of vertebrate Hh signaling on a microtubule-based organelle, the primary cilium 15. Mutations that block formation of primary cilia prevent cellular responses of cells to Hedgehog ligands, and all of the proteins required for vertebrate Hh signal transduction are highly enriched in cilia and change localization in response to ligand 16. The activity of KIF7 in the mouse Hh pathway depends upon the presence of the primary cilium6.
Despite the conserved role of Cos2/KIF7 in the Hh pathway, the motor domain of Cos2 lacks residues critical for motility17 and is considered as a microtubule-associated scaffold for Hh signaling complexes 18,19. In contrast, mouse KIF7 has the sequence motifs necessary for ATP and microtubule binding and the crystal structure of its motor domain is superimposable on that of a conventional kinesin 20. How KIF7 acts within cilia and whether its motor activity is important for its function is not known.
Here we show that, unlike other core components of the mammalian Hh pathway, KIF7 is required for the normal structure of primary cilia. KIF7 localizes to the distal tips of primary cilia, the site of the plus-ends of axonemal microtubules. In the absence of KIF7, cilia are long and unstable. Using TIRF microscopy-based in vitro assays, we show that a purified KIF7 motor protein can autonomously recognize microtubule plus-ends and limit growth; these growth-limiting activities are sufficient to explain the long cilia of Kif7 mutants. Proteins that normally localize to distal cilia tips, including the Gli and Sufu proteins that mediate Hh signaling, are found in ectopic puncta along the Kif7 mutant cilium. The data suggest that KIF7 is required to organize a specialized compartment at the tip of the cilium that is necessary for Hh signal transduction.
Results
KIF7 localizes to cilia tips
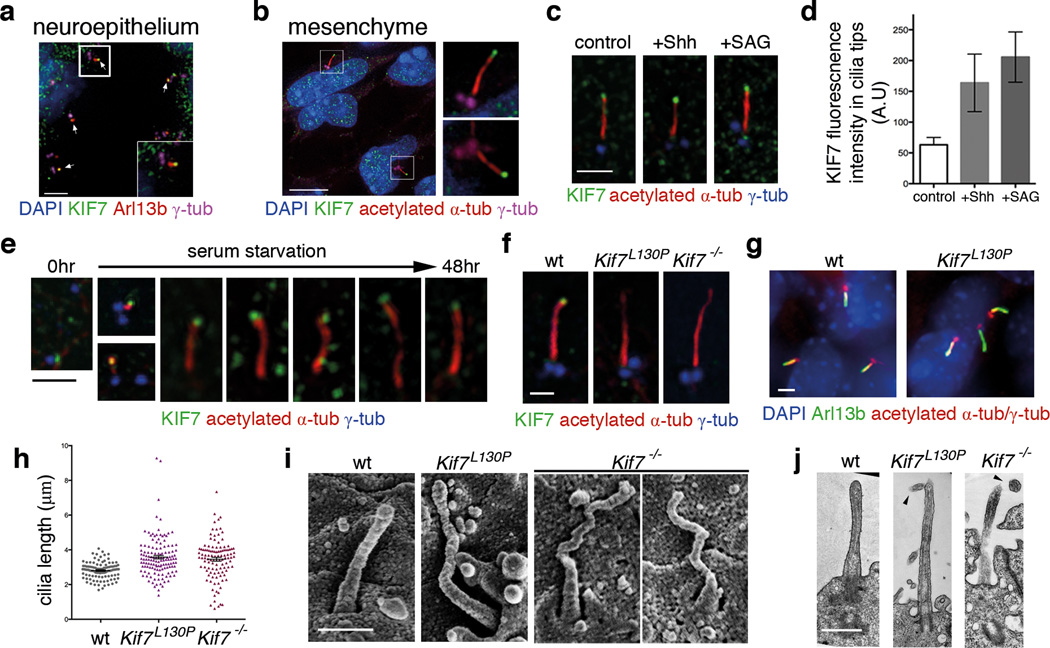
We found that endogenous KIF7 was enriched in primary cilia tips in Sonic hedgehog (Shh)-responsive cells in wild-type embryos (Fig. 1a–b) and cultured fibroblasts and was further enriched at tips in response to activation of the pathway (Fig. 1c–d; Supplementary Fig. 1b), similar to the core Hh pathway proteins Gli2, Gli3 and Sufu 21–24. KIF7 was also present at cilia tips in MEFs derived from mutant embryos that lack Smo or Gli2 and Gli3, indicating that KIF7 is targeted to cilia independently of Shh pathway proteins (Supplementary Fig. 1c–d). In wild-type MEFs, KIF7 was present at cilia tips at all stages of ciliogenesis (Fig. 1e). No KIF7 protein was detected in cilia of mouse embryonic fibroblasts (MEFs) derived from Kif7−/− embryos 8 (Fig. 1f; Supplementary Fig.1a). An allele of Kif7 with a leucine-to-proline substitution (L130P) in a conserved region of the motor domain causes a phenotype indistinguishable from that of the null allele 6. KIF7 protein level was not affected in Kif7L130P MEFs, but KIF7 was never observed at tips of Kif7L130P mutant cilia, although it was detected in the ciliary transition zone (Fig. 1f; Supplementary Fig. 1a–b).
Figure 1. KIF7 constrains the length of primary cilia.
(a–b) KIF7 (green) localizes to the tips of primary cilia in e10.5 wild-type embryos. Arl13b (red) marks cilia in (a) and acetylated-α-tubulin (red) marks microtubule axoneme in (b). γ-tubulin (magenta) marks basal bodies. DAPI (blue) labels nuclei. Scale bar = 5µm. (c) In wild-type MEFs, KIF7 (green) is enriched in tips of primary cilia upon treatment with Shh-N recombinant protein (2µg/ml) or SAG (100nM) for 24 hours. Scale bar = 2µm. (d) Quantitation of KIF7 fluorescence intensity shown in (C). n = 150 cilia pooled from 3 independent experiments were counted for each condition. The error bars represent the SD. p<0.0001 between control and SAG or Shh treatments by one-way ANOVA. (e) KIF7 (green) is associated with one of the two centrioles prior to ciliogenesis and localizes to the tips of primary cilia throughout cilia elongation induced by serum starvation in wild-type MEFs. (f) KIF7 is not detected at the tips of Kif7L130P or Kif7−/− MEF cilia. Acetylated-α-tubulin (red) marks primary cilia and γ-tubulin (blue) marks basal bodies in (c), (e) and (f). Scale bar = 1µm in (c), (e) and (f). (g) Primary cilia of wild-type and Kif7L130P MEFs stained with Arl13b (green), acetylated-α-tubulin and γ-tubulin (red). DAPI in blue. Scale bar = 2µm. (h) Measurements of MEF cilia length between wild-type and Kif7 mutants using Arl13b as cilia marker. >100 cilia pooled from 4 independent experiments were counted for each genotype (n=108 cilia for wild type, n=133 cilia for Kif7L130P mutant, and n=111 for Kif7−/− mutant). p<0.0001 between wild-type cilia and Kif7 mutant cilia by one-way ANOVA. The error bars represent the SEM. (i) Scanning electron microscopy shows neural tube cilia of e10.5 wild-type, Kif7L130P and Kif7−/− embryos. Wild-type neural cilia were 0.97±0.17 µm and Kif7L130P neural cilia were approximately 1.2±0.28µm long (n=50 for each genotype). Scale bar = 0.5µm. (j) TEM images of longitudinal sections of wild-type, Kif7L130P and Kif7−/− neural tube cilia. Arrowheads point to twisted tips of mutant cilia. Scale bar = 0.5µm.
Kif7 mutant cilia are long and twisted
Cilia were present on ~70% of Kif7L130P and Kif7−/− mutant MEFs, as in wild type, but the cilia appeared to be longer than wild type (Fig. 1f–g; Supplementary Fig. 1e–f). Using Arl13b as a cilia marker 25, wild-type MEF cilia were 2.80±0.47 µm in length, whereas mutant cilia were 3.58±1.14 µm and 3.46±1.10 µm in Kif7L130P and Kif7−/− MEFs, respectively. Thus Kif7 mutant cilia were ~30% longer, on average, and more variable in length, than wild type (Fig. 1h).
En face imaging of the embryonic neural epithelium by scanning electron microscopy (SEM) showed that neural cilia in Kif7L130P and Kif7−/− were longer and more tapered than in wild type (Fig. 1i). The diameter of neural tube cilia in Kif7L130P mutant embryos was normal near the base, but the distal region of the cilium was thin, with a diameter ~40% less than wild type, and often twisted (Supplementary Fig. 2a). Cilia in the e8.0 Kif7L130P embryonic node showed similar morphological defects (Supplementary Fig. 2b–c). Twisted distal segments of mutant cilia were also seen in longitudinal transmission electron microscopy (TEM) sections (Fig. 1j; Supplementary Fig. 2d). Transverse TEM sections of Kif7L130P cilia showed that some axonemal microtubule doublets had open B-tubules or only 8 doublets (Supplementary Fig. 2e).
Kif7 mutant axonemes are under-modified and unstable
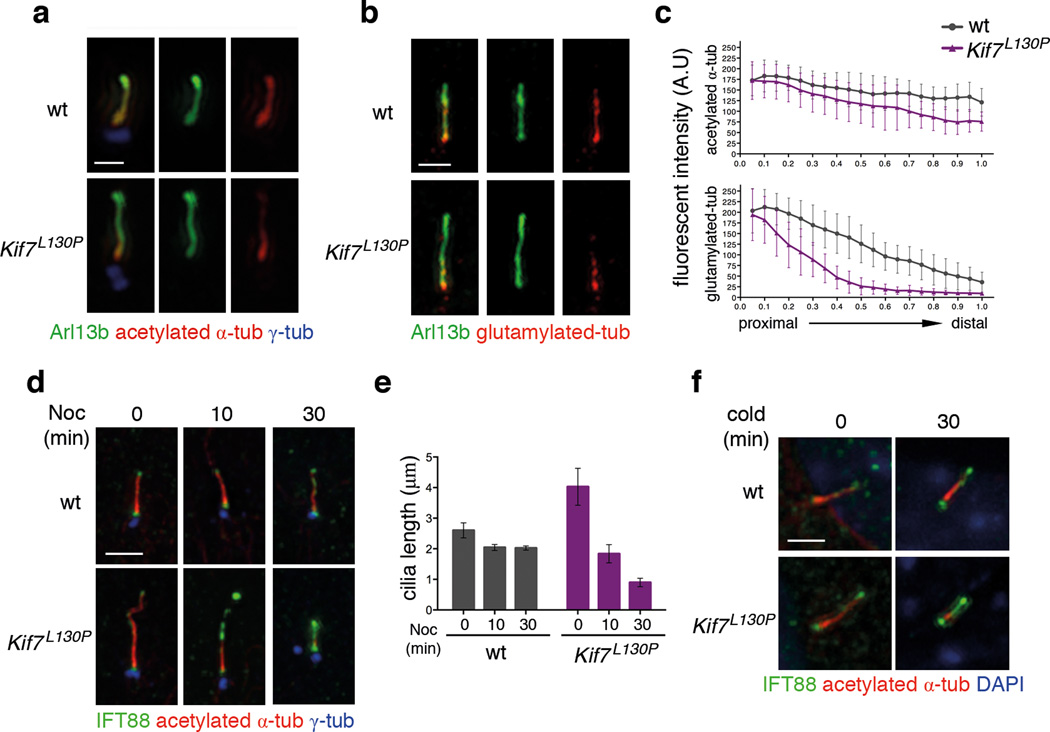
Axonemal microtubules are acetylated and glutamylated, modifications associated with stable microtubules 26. Line-scan analysis of fluorescence intensity revealed that the level of tubulin acetylation and glutamylation were comparable in wild type and Kif7L130P at the base of the ciliary axoneme (Fig. 2a–c). However both tubulin modifications faded towards the ciliary tip, and tubulin glutamylation was often undetectable distally in Kif7L130P and Kif7−/− mutant cilia (Fig. 2b–c; Supplementary Fig. 3).
Figure 2. KIF7 stabilizes primary cilia.
(a) Tubulin acetylation (red) and (b) glutamylation (red) are reduced at the distal segment of cilia in Kif7L130P MEFs compared to wild type. Arl13b (green) labels the entire cilia in (a–b) and γ-tubulin (blue) marks basal bodies in (a). Scale bar = 1µm. (b) Fluorescence intensities of acetylation and glutamylation, as shown in panel (a–b), normalized for cilia length. n= 50 cilia were measured for each genotype pooled from 3 independent experiments. The error bars represent the SD. For tubulin acetylation, p<0.001 for the distal half of the cilia. For tubulin glutamylation, p<0.0001 for the distal 80% of the cilia. (d) Time course of cilia shortening in response to nocodazole treatment in wild-type and Kif7L130P MEFs. IFT88 (green) accumulated distally Kif7L130P MEF cilia during cilia retraction. γ-tubulin (blue) marks basal bodies. Scale bar = 2µm. (e) Cilia length was measured using acetylated-α-tubulin from the time-course experiment shown in (d). n=30 for each condition and each time point pooled from 3 independent experiments. The error bars represent the SD ( p<0.001 between wild-type and Kif7L130P at 0 and 30 minutes; n.s between two genotypes at 10 minutes). (f) Time course of cilia retraction at 4°C in wild-type and Kif7L130P MEFs. Cilia are marked with acetylated-α-tubulin (red) and IFT88 (green). DAPI in blue. Scale bar = 2µm.
To test whether changes in tubulin modification were associated with decreased cilia stability, we treated cells with nocodazole to induce microtubule depolymerization. In wild-type MEFs, the length of acetylated α-tubulin-labeled axonemal microtubules was reduced by ~20% at 30 minutes after addition of nocodazole. In contrast, 10 minutes after addition of nocodazole, the Kif7L130P axoneme was half as long as at time zero and was barely detectable at 30 minutes (Fig. 2d–e). Cilia disassembly is associated with accumulation of IFT particles 27, which were first detected in wild-type cells 30 minutes after addition of nocodazole. Strong IFT88 staining was apparent in the distal half of the mutant cilia at 10 minutes, and by 30 minutes the Kif7L130P cilium appeared to be filled with IFT88 (Fig. 2e). Destabilization of axonemal microtubules by exposure to 4°C also caused more rapid loss of acetylated-α-tubulin and accumulation of IFT88 in Kif7 mutant cilia than in wild type (Fig. 2f). Thus KIF7 promotes stability of the ciliary axoneme.
KIF7 does not affect the rates of IFT
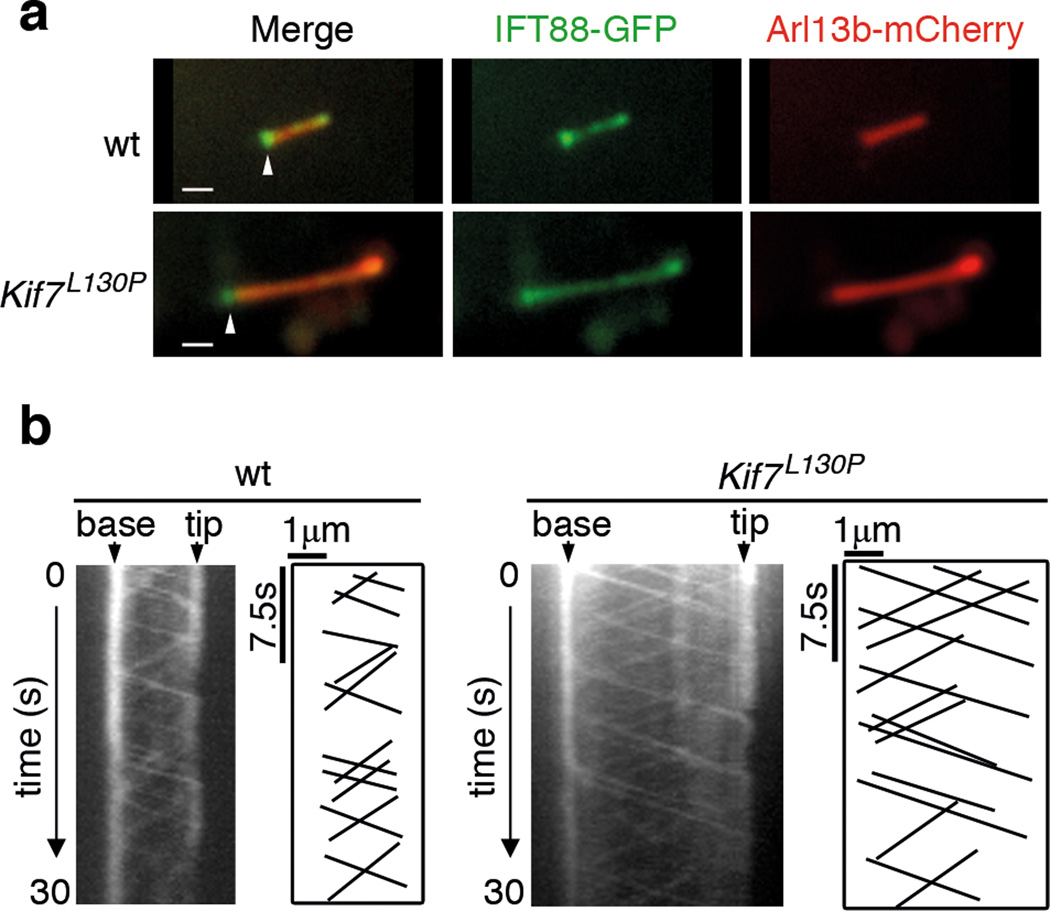
It has been proposed that cilia length can be regulated by the balance between the rates of anterograde and retrograde IFT28. Kinesin-II is the evolutionarily conserved anterograde motor required for carrying intraflagellar transport (IFT) complex from the base of cilia to the cilia tip 29; other kinesins can also promote anterograde ciliary trafficking and modify the overall rate of IFT 30–32. To test whether the long cilia in Kif7 mutants arise because KIF7 alters the rates of IFT, we expressed two fusion proteins, IFT88-GFP to label IFT particles and Arl13b-mCherry to label the ciliary membrane, in MEFs (Fig. 3a) and documented the movements of IFT using time-lapse imaging (Supplementary Movie. 1–2).
Figure 3. KIF7 does not affect the rates of intraflagellar transport (IFT).
(a) Still images of wild-type and Kif7L130P primary cilium expressing IFT88-GFP (green) and Arl13b-mCherry (red) before live imaging. Arrowheads mark the base of the ciliary axoneme. Scale bar = 1µm. (b) Kymographs generated from time lapse imaging of wild-type and Kif7L130P primary cilia. Also see Supplementary Movie. S1 and S2. The base and tip of the cilium are indicated with arrowheads. Horizontal scale bar (distance) = 1µm. Vertical scale bar (time) = 7.5 seconds.
We analyzed the speed of IFT trains of wild-type and Kif7 mutant MEF cilia by measuring the slopes of each IFT trajectory on the kymograph (Fig. 3b). The rates of anterograde IFT were 1.09±0.24 µm/s and 1.06±0.19 µm/s in wild-type and Kif7L130P cilia, respectively. The rates of retrograde IFT were 0.72±0.26 µm/s and 0.76±0.29 µm/s in wild type and Kif7L130P, respectively. Thus there was no significant difference in the rates of IFT between wild type and Kif7L130P cilia, indicating that KIF7 does not control cilia length by affecting the rates of IFT.
KIF7 can interact directly with microtubules but lacks detectable motility
Sequence analysis places KIF7 in the kinesin-4 family 33 (Supplementary Fig. 4a), which includes the homodimeric kinesins KIF4A/Xklp1 and KIF21A 1,5,34. To test whether KIF7 acts as a dimer, we expressed full-length C-terminal Flag- and GFP-tagged KIF7 constructs in HEK293T cells. KIF7-GFP co-immunoprecipitated with KIF7-Flag, and vice versa (Supplementary Fig. 4b), indicating that KIF7 can form a homodimer. When expressed in cell lines, KIF7-GFP appeared to co-localize with microtubule bundles, suggesting that KIF7 can associate with microtubules in vivo (Supplementary Fig. 4c). Domain mapping of KIF7 showed that microtubule association and KIF7 dimerization depended on both motor domain (amino acid 13–357) and the first coiled-coil of KIF7 (amino acid 487–539; Supplementary Fig. 4c–d).
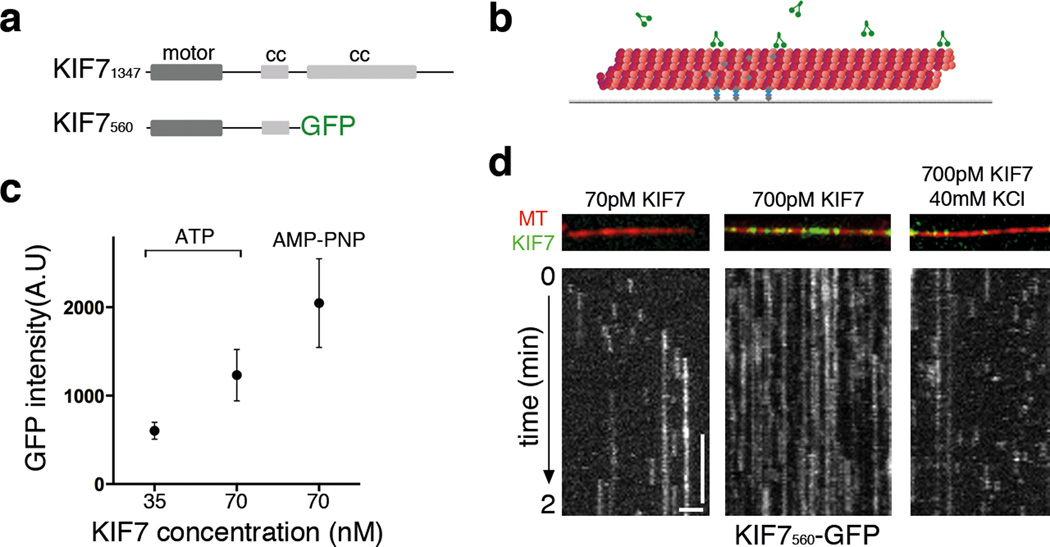
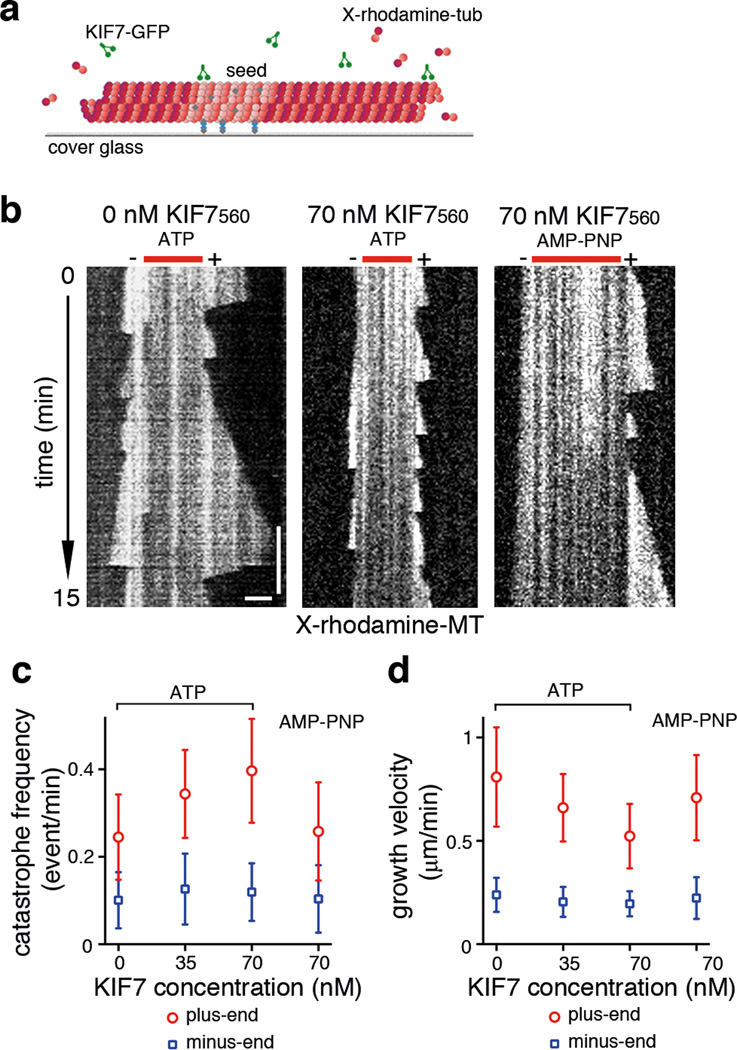
To define the biochemical functions of the KIF7 motor, we expressed and purified a recombinant construct in E. coli that included the N-terminal motor domain and the first coiled-coil of KIF7, with a C-terminal GFP tag (KIF7560-GFP) (Fig. 4a; Supplementary Fig. 5a). We used a TIRF microscopy-based assay to examine the association of KIF7560-GFP with microtubules (Fig.4b). In the presence of MgATP (1 mM), KIF7560-GFP uniformly decorated the entire microtubule (Supplementary Fig. 5b) and binding was proportional to motor protein concentration (Fig. 4c). Addition of 2 mM AMP-PNP, a non-hydrolyzable ATP analog that typically promotes tight kinesin-microtubule binding, resulted in a ~two-fold increase in filament-bound KIF7 (Fig. 4c; Supplementary Fig. 5b).
Figure 4. A purified KIF7 motor construct lacks detectable microtubule-dependent motility.
(a) Domain organizations of full-length KIF7 and the N-terminal fragment used for in vitro assays. (b) Schematic of the assay for examining KIF7-microtubule interaction. X-rhodamine labeled microtubules (red) were attached to a PEG-passivated glass surface by biotin–NeutrAvidin (gray and blue) links prior to addition of KIF7 motor protein (green). (c) Average fluorescent intensity of microtubule-bound KIF7560-GFP plotted against KIF7 protein concentration in the presence of either 1 mM MgATP or 2 mM AMP-PNP. n = 20 microtubules for each condition were analyzed. Error bars represent SD. p<0.0001 by one-way ANOVA. (d) KIF7560-GFP gliding assay (70 pM or 700 pM) under different ionic strength conditions. Representative kymographs from assays at BRB80 and BRB80 +40 mM KCl buffer conditions in the presence of 1 mM MgATP. Still images show KIF7560-GFP (green) binding to X-rhodamine labeled microtubules (red). Kymographs of KIF7560-GFP are shown in gray scale. Horizontal scale bar = 2µm. Vertical scale bar = 30s. The frame rate was 1 frame per 0.5 s.
We tested whether KIF7 can move along microtubules by analyzing the behavior of single molecules of KIF7560-GFP in our TIRF-microscopy assay (Fig. 4b and 4d). Analysis of kymographs revealed that in the presence of 1mM MgATP, KIF7560-GFP molecules were stationary when bound to microtubules, exhibiting neither unidirectional movement nor significant diffusion along the microtubule lattice (Fig. 4d), unlike most kinesins. Increasing the ionic strength of the assay buffer reduced binding life-time of KIF7560-GFP associated with microtubules (Fig. 4d), similar to other kinesins 35–37.
The KIF7 motor domain inhibits the growth of microtubule plus-ends in vitro
KIF4A/XKLP1 and KIF21A can regulate microtubule length by altering tubulin polymerization dynamics 1,5. We tested whether KIF7560-GFP could regulate microtubule dynamics using a modified TIRF microscopy-based assay to visualize microtubule polymerization in vitro, with stochastic transitions between periods of elongation (growth) and shrinkage (catastrophe) at the ends of the microtubules (Fig. 5a) 4,38. When KIF7560-GFP (1 mM MgATP) was added to the dynamic microtubule assay, there was an increase in the frequency of catastrophe at microtubule plus-ends. The catastrophe frequency (0.25±0.10 catastrophe/minute in the absence of KIF7) increased 36% in the presence of 35 nM KIF560-GFP (0.34±0.10 catastrophe/minute) and 60% in the presence of 70 nM KIF560-GFP (0.40±0.12 catastrophe/minute) (Fig. 5b–c). In the presence of AMP-PNP (2 mM), 70 nM KIF7560-GFP had no detectable effect on the catastrophe frequency (0.26±0.11 catastrophe/minute) (Fig. 5b–c). When GMP-CPP-polymerized X-rhodamine-labeled microtubules were incubated in solution with relatively high concentrations of KIF7560-GFP (500 nM) and 2mM MgATP for 2–5 minutes and then allowed to bind to a glass coverslip, the microtubules appeared fragmented, as if they had been severed in the middle (Supplementary Fig. 6), suggesting that high concentration of KIF7 might have a destabilizing effect on microtubules.
Figure 5. KIF7 alters microtubule dynamics in vitro.
(a) Schematic of the experimental setup of microtubule dynamics assay: X-rhodamine- and biotin-labeled microtubule seeds, which were polymerized in the presence of GMP-CPP, were immobilized on a glass coverslip, and then incubated with a mixture of X-rhodamine labeled tubulin (1.8 µM) and non-fluorescent tubulin (18 µM; 1 mM MgGTP). (b) Representative kymographs of X-rhodamine-labeled microtubules (show in grey scale) polymerizing in the absence or presence of KIF7560-GFP at the indicated concentrations in the presence of either 1 mM MgATP or 2 mM AMP-PNP. The frame rate was 1 frame per 5s. Microtubule seed and polarity are indicated above each kymograph. Horizontal scale bar = 2µm. Vertical scale bar = 180s. (c) The frequencies of microtubule catastrophe in the absence of KIF7 (n=272), 35 nM KIF560-GFP (n=263; p<0.0001 compared to control), 70 nM KIF560-GFP (n=492; p<0.0001 compared to control and p<0.01 compared to 35 nM KIF560-GFP) in the presence of 1 mM MgATP and 70 nM KIF560-GFP (n=179; n.s compared to control) in the presence of 2 mM AMP-PNP were plotted against the KIF7560-GFP concentration. The catastrophe frequencies of microtubule minus-ends were not affected by KIF560-GFP (n=82 for control, n=51 for 35nM KIF560-GFP with Mg ATP, n=94 for 70 nM KIF560-GFP with MgATP and n=53 for 70 nM KIF560-GFP with AMP-PNP were analyzed; n.s compared to control). (d) Microtubule growth in the absence of KIF7 (n=145), 35 nM KIF7560-GFP (n=109; p<0.0001 compared to control), 70 nM (n=263; p<0.0001 compared to control and p < 0.001 compared to 35 nM KIF7560-GFP) in the presence of 1 mM MgATP and 70nM KIF7560-GFP (n=101; p<0.01 compared to control) in the presence of 2 mM AMP-PNP were plotted against the KIF7560-GFP concentration. The growth rates of microtubule minus-ends were not affected by KIF560-GFP (n=51 for control, n=47 for 35nM KIF560-GFP with Mg ATP, n=45 for 70 nM KIF560-GFP with MgATP and n=51 for 70 nM KIF560-GFP with AMP-PNP were analyzed; n.s compared to control). Data sets pooled from 3 independent experiments were analyzed. Error bars represent the SD. p values were calculated by nonparametric test for multiple comparisons.
In addition to promoting catastrophe, KIF7 decreased the rate of microtubule growth in a dose-dependent manner (Fig. 5b). In the presence of 70nM KIF7560-GFP (1 mM MgATP), the microtubule growth rate (0.52±0.17 µm/minute) was 35% less than the rate in the absence of KIF7560-GFP (0.81±0.24 µm/min) (Fig. 5d). An intermediate growth rate (0.66±0.16 µm/minute) was observed with 35nM KIF7560-GFP. In contrast, 70nM KIF7560-GFP in the presence of AMP-PNP (2mM) only mildly suppressed microtubule growth (0.71±0.20 µm/minute), suggesting that ATP-hydrolysis contributes to the attenuation of tubulin polymerization by KIF7560-GFP. At microtubule minus-ends, neither the catastrophe frequency nor the growth rate was significantly affected by KIF7560-GFP (Fig. 5c–d), indicating that KIF7560-GFP preferentially regulates microtubule dynamics at the plus-ends.
KIF7 acts at growing microtubule plus-ends
To determine the localization of KIF7 on growing and shrinking microtubules, we analyzed snapshots from near-simultaneous imaging of KIF7560-GFP (70 nM; 1 mM MgATP) and X-rhodamine-labeled microtubules. Line scan analysis showed that KIF7560-GFP associated preferentially with the GMP-CPP-tubulin seed and the with the tip of the microtubule, while it was barely detected in the region of the newly-polymerized microtubule proximal to the tip (Fig. 6a and 6c; Supplementary Movie S3). Time lapse imaging of KIF7560-GFP localization on dynamic microtubules showed that the motor protein tracked the plus-ends of microtubules during the growth phase (Fig. 6c). During the depolymerization/catastrophe phase of the cycle, KIF7560-GFP was not detected at microtubule plus-ends (Fig. 6c), suggesting that KIF7 dissociated from the filament plus-end at the onset of catastrophe. Microtubule tip-associated KIF7560-GFP was also observed at growing minus-ends, but at <30% the frequency of plus-ends. In the presence of AMP-PNP (2 mM), KIF7560-GFP (70 nM) was observed all along the lattice of dynamic microtubules and there was no preferential association with growing microtubule tips (Fig. 6b and 6d). As microtubule plus-ends are capped by GTP-tubulin and the conformation of GMP-CPP-tubulin is similar to that of GTP-tubulin39,40, the data suggest that KIF7 associates preferentially with GTP-bound tubulin. Together, our in vitro analysis suggests that the plus-end-associated KIF7 promotes catastrophe and inhibits microtubule growth in an ATP-hydrolysis dependent manner.
Figure 6. KIF7 tracks microtubule plus-ends.
Representative images showing KIF7560-GFP (70 nM) associated with dynamic microtubules in the presence of (a) 1 mM MgATP or (b) 2 mM AMP-PNP. Arrows indicates microtubules used for line-scan based fluorescence intensity analysis. Scale bar = 2µm. Also see Supplementary Movie S3. (c) Representative kymographs of KIF7560-GFP on X-rhodamine-labeled dynamic microtubules in the presence of 1 mM MgATP (n=263; two examples shown) and (d) 2 mM AMP-PNP (n=101). Plus-ends of microtubules are to the right on the kymographs in (c) and (d). Horizontal scale bar = 2µm. Vertical scale bar = 180s. The frame rate was 1 frame per 5s. Data sets from 2–3 independent experiments were analyzed.
KIF7 is required for organization of the cilia tip compartment in vivo
The ability of KIF7 to associate directly with microtubule plus-ends and limit microtubule growth in vitro, together with the localization of KIF7 to cilia tips, suggested that the KIF7 might control the dynamics of the distal ends of axonemal microtubules. Close examination of the IFT kymographs revealed that while moving IFT88-GFP appeared as diagonal lines in the kymographs, there were also vertical lines of IFT88-GFP in the kymographs of Kif7 mutant cilia (Fig. 3b; Supplementary Fig. 7c–e), which represented stationary IFT88 particles. Retrograde tracks almost always began at distal tip in wild-type cilia, but the Kif7 kymographs showed that retrograde trafficking frequently initiated from the stationary IFT88 puncta in the middle of the axoneme (Supplementary Fig. 7a–b). Thus the stationary IFT88 puncta in Kif7 mutant cilia mark compartments that have properties of the normal distal tip.
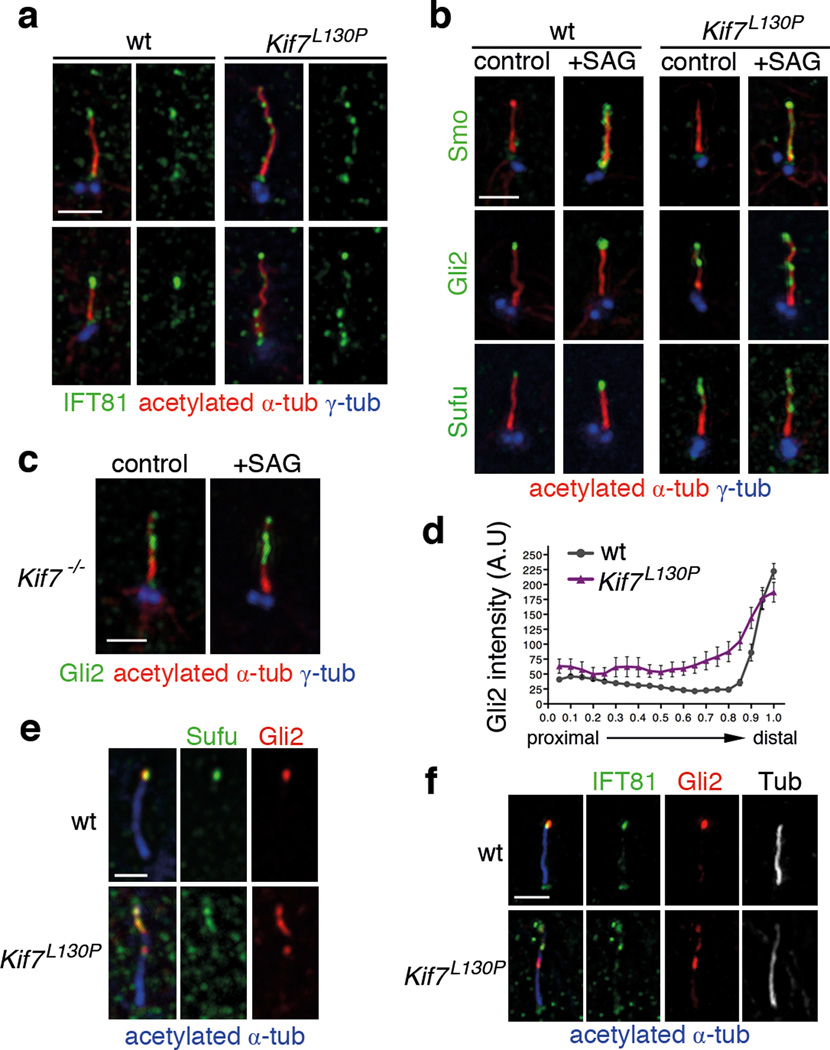
IFT81, an IFT-B complex protein, binds directly to tubulin dimers and facilitates their incorporation into the microtubule axoneme 41. In wild-type MEFs, IFT81 was enriched in the distal tip of primary cilia and was barely detected along the axoneme (Fig. 7a), consistent with the expression pattern of GFP-tagged IFT81 in ciliated cells 41. In contrast, multiple IFT81 puncta were present in Kif7L130P cilia, suggesting that the tip structure of the Kif7 mutant cilia is defective. These results suggest that some axonemal microtubules in mutant cilia do not extend to the distal tip and that ectopic microtubule plus-ends can organize cilia tip-like compartments (Fig. 7a).
Figure 7. Multiple cilia tip-like compartments form in the absence of KIF7.
(a) IFT81 (green) localization in the wild-type and Kif7L130P cilia. (b) Cilia localization of Smo, Gli2 and Sufu in wild-type and Kif7−/− MEFs in response to pathway activation by 100nM SAG treatment for 24 hr. (c) Gli2 localizes to puncta along Kif7−/− ciliary axonemes independent of pathway activation. Acetylated-α-tubulin (red) marks axonemal microtubules and γ-tubulin (red) marks basal bodies in (a) to (c). (d) Distribution of fluorescence intensity of Gli2 in SAG-treated wild-type and Kif7L130P cilia, normalized to cilia length. n=50 cilia were analyzed for each condition pooled from 3 independent experiments. p<0.0001. The error bars represent the SEM. (e) Sufu (green) and Gli2 (red) co-localization in the tip of wild-type cilia. In Kif7L130P mutant cilia, Sufu and Gli2 co-localize to the same puncta along the axoneme. (f) IFT81 (green) and Gli2 (red) co-localization in the tip of wild-type cilia. In Kif7L130P mutant cilia, IFT81 and Gli2 co-localize to the same puncta along the axoneme. Acetylated-α-tubulin shown in blue in merged panels and in gray-scale in separated channels. Scale bars = 1µm in (a)–(c) and (e)–(f).
Gli2 and Sufu colocalize with distal tip markers along the Kif7 mutant axoneme
To understand how these defects in cilia structure affected Shh signaling, we assayed the localization of Shh pathway proteins in mutant cilia. Smo moved into cilia normally in response to the small molecule agonist SAG in both wild-type and Kif7L130P mutant MEFs (Fig. 7b), indicating that KIF7 is not required for trafficking of Smo, consistent with the finding that KIF7 acts genetically downstream of Smo 6,8.
In wild-type cilia, Gli2 is present in cilia tips and is further enriched by pathway activation. In contrast, Gli2 was present in puncta along the ciliary axoneme in the absence of pathway activation in both Kif7L130P and Kif7−/− MEFs (Fig. 7b–c). When treated with SAG, additional Gli2 accumulated along the length of mutant cilia (Fig. 7b–c). Despite elevated level of Gli2 along the mutant axoneme, less Gli2 was present at the distal tip of Kif7L130P cilia than in wild type (Fig.7d). Thus KIF7 is not required for trafficking of Gli2 into cilia, but it does control proper localization of Gli2 within the cilium. Sufu, a negative regulator the Shh pathway, forms a complex with Gli proteins at cilia tips and is further enriched in response to pathway activation 24,42 (Fig. 7b and 7e). In Kif7L130P cilia, Sufu was present in multiple puncta in the distal half of the ciliary axoneme where it co-localized with Gli2 (Fig. 7b and 7e), indicating that KIF7 is required to restrict Gli-Sufu complexes to the distal end of the cilium. Double staining showed that IFT81 and Gli2 were enriched at the distal cilia tip in wild-type cilia tip and frequently colocalized in ectopic puncta along the axoneme in Kif7L130P cilia (Fig. 7f). Thus in the absence of KIF7, the Gli-Sufu complex and IFT81 localize to ectopic tip-like compartments along the axoneme.
Discussion
Mammalian KIF7 and Drosophila Cos2 are both required for the switch of the Gli/Ci from transcriptional repressors to activators in the presence of Hh ligand, but KIF7 acts through the primary cilium, which is not required for Drosophila Hh signaling. Our data show that KIF7 has an essential role in cilia structure: mouse KIF7 localizes to the tips of primary cilia, where it controls the length and structure of primary cilia and the localization of Hh signaling complexes through its ability to regulate the dynamics of microtubule plus-ends.
Mouse Kif7 mutant cilia tend to be long, and are unstable and twisted. Cilia in human KIF7 mutant fibroblasts are longer than controls 13, indicating that KIF7 has a conserved role in the control of axoneme length. Two other classes of kinesin are known limit cilia length. Chlamydomonas kinesin-13 moves into flagella via IFT and depolymerizes microtubules from the flagellar tip 43. Mouse KIF19A, a member of kinesin-8 family, is thought to use its motor activity to move along axonemes and depolymerize microtubule at the tips of motile cilia44. Thus each of these kinesins can control cilia length, but they act through distinct mechanisms.
Our in vitro reconstitution studies indicate KIF7 associates with microtubule plus-ends where it limits microtubule length. While other kinesins arrive at microtubule plus-ends either by one-dimensional diffusion (e.g. MCAK 45) or by directional motility (e.g. XKLP1 1), KIF7 neither diffuses nor moves directionally on microtubules under our assay conditions. Instead, our data suggests that KIF7 tracks the plus-ends of growing microtubules by preferential association with GTP-bound tubulin at microtubule tip (Fig. 8a). This tip-tracking behavior is ATP-hydrolysis dependent, which distinguishes it from end-binding proteins like EB1 46. At the microtubule plus-ends, KIF7 inhibits the rate of microtubule growth, an activity shared with other kinesin-4 proteins. However, unlike KIF4A/Xklp1 and KIF21A, KIF7 is a catastrophe-promoting factor.
Figure 8. Model for KIF7-dependent regulation of plus-end dynamics in primary cilia.
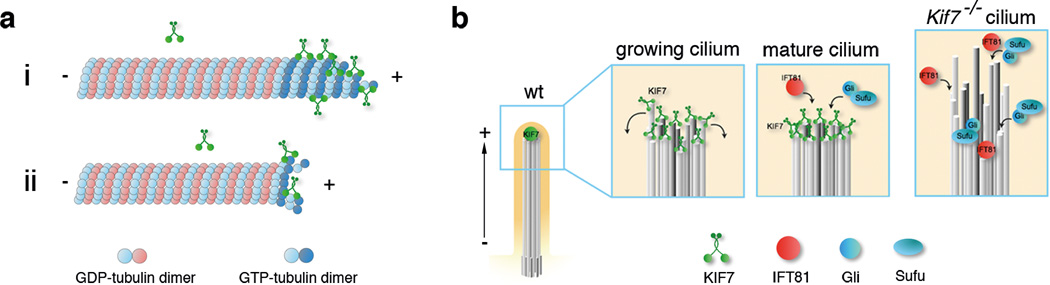
(a) Proposed model for the regulation of microtubule dynamics by KIF7. i: KIF7 preferentially binds to the GTP-bound tubulin at microtubule plus-ends, inhibiting addition of tubulin dimers and microtubule growth. ii: By destabilizing the GTP-bound tubulin cap at microtubule plus-ends, KIF7 can promote catastrophe and thereby limit microtubule growth. (b) Model for KIF7 function at the tip of primary cilia. In wild type, KIF7 acts at the distal end of the growing cilium to prevent overgrowth of individual microtubules and to coordinate the growth of 9-doublet microtubules; only organized microtubule arrays are a suitable substrate for post-translational modifications. In the mature cilium, KIF7 creates a single distal tip compartment where IFT81 and Gli-Sufu complexes are enriched. In the absence of KIF7, growth of axonemal microtubules is not limited and synchronized, leading to longer and unstable cilia and ectopic tip-like compartments that contain IFT81 along the axoneme. Gli-Sufu complexes localized to the ectopic tip compartments, where they can be inappropriately activated in the absence of Shh ligand.
The primary cilium is built on a scaffold of parallel microtubule bundles of nine doublet protofilaments, with the plus-ends at the distal cilia tip. Because KIF7 localizes to cilia tips throughout ciliogenesis and purified KIF7 motor protein can autonomously associate with the plus-ends of growing microtubules, we propose that KIF7 binds directly to the distal ends of axonemal microtubules where it limits the growth of the individual microtubules during cilia assembly. Because tip-associated proteins are mislocalized along the axoneme absence of KIF7, KIF7 also appears to help synchronize the growth of the nine doublet microtubules (Figure. 8b).
The data show that KIF7 is critical for organization of the compartment at the tip of the cilium, a topic that has long been of interest 47. The IFT complexes that control cilia elongation must disassemble and reorganize at the tip as they convert from kinesin-based to dynein-based motility in the process called tip turnaround 48. In Chlamydomonas, the mating signaling cascade is initiated from the flagellar tip 49. In mammals, key regulators of Hedgehog signaling are highly enriched at tips of primary cilia, and it has been inferred that their activity is regulated in this compartment 24,42. We propose that the ectopic activation of the pathway seen in Kif7 mutants is due to the ectopic Gli/Sufu complexes away from the cilia tip, where they become inappropriately activated in the absence of ligand (Fig. 8b). Because of its specific actions on microtubules, we propose that the ancestral function of KIF7 was to sculpt the structure of the primary cilium, and that it acted in this context in the Hedgehog pathway in the metazoan ancestor of both Drosophila and mammals.
Supplementary Material
Acknowledgements
We thank Leslie Gunther-Cummins, Peter Satir (Albert Einstein College of Medicine, Bronx, NY), Kunihiro Uryu (Rockefeller University) for assistance with TEM, and Alan Hall for use of a microscope for TIRF imaging. We thank Hisham Bazzi, Angela Parrish and Tim Bestor for helpful comments on the manuscript. We thank Bryan Tsou and Won-Jing Wang for technical support and helpful discussion. We thank C.C Hui (Sick Children’s Hospital) for providing Kif7−/− mice. We thank Jonathan Eggenschwiler (University of Georgia) and Adrian Salic (Harvard University) for antibodies. We thank Nina Lampen for help with scanning electron microscopy and the Memorial Sloan-Kettering Cancer Center core facilities and the Rockefeller University Bio-Imaging Resource Center for help with imaging. We thank Ronald Hendrickson and Hediye Erdjument-Bromage for help with mass spectrometry. This work was supported by National Institutes of Health grant NS044385 to K. V. Anderson and GM65933 to T. M. Kapoor and the MSKCC Cancer Center Support Grant (P30 CA008748).
References
- 1.Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 2.Hu C-K, Coughlin M, Field CM, Mitchison TJ. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian R, Ti S-C, Tan L, Darst SA, Kapoor TM. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 2013;154:377–390. doi: 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Vaart B, et al. CFEOM1-Associated Kinesin KIF21A Is a Cortical Microtubule Growth Inhibitor. Dev. Cell. 2013 doi: 10.1016/j.devcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Liem KF, HE M, Ocbina PJR, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. U.S. A. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tay SY, Ingham PW, Roy S. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–634. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]
- 8.Cheung HO-L, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 9.Endoh-Yamagami S, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 10.Jiang J, Hui C-C. Hedgehog signaling in development and cancer. Dev. Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 12.Ingham PW, McMahon AP. Hedgehog signalling: Kif7 is not that fishy after all. Curr. Biol. 2009;19:R729–R731. doi: 10.1016/j.cub.2009.07.060. [DOI] [PubMed] [Google Scholar]
- 13.Putoux A, et al. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat Genet. 2011;43:601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dafinger C, et al. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J. Clin. Invest. 2011;121:2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 16.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthies HJ, Baskin RJ, Hawley RS. Orphan kinesin NOD lacks motile properties but does possess a microtubule-stimulated ATPase activity. Mol. Biol. Cell. 2001;12:4000–4012. doi: 10.1091/mbc.12.12.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 19.Robbins DJ, et al. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 20.Klejnot M, Kozielski F. Structural insights into human Kif7, a kinesin involved in Hedgehog signalling. Acta Crystallogr. D Biol. Crystallogr. 2012;68:154–159. doi: 10.1107/S0907444911053042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin J, Lin Y, Norman RX, Ko HW, Eggenschwiler JT. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen X, et al. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell. Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 27.Pan J, Snell WJ. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev. Cell. 2005;9:431–438. doi: 10.1016/j.devcel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Marshall WF, Qin H, Rodrigo Brenni M, Rosenbaum JL. Flagellar length control system: testing a simple model based on intraflagellar transport and turnover. Mol. Biol. Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole DG. Kinesin-II, coming and going. J. Cell Biol. 1999;147:463–466. doi: 10.1083/jcb.147.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev. Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morsci NS, Barr MM. Kinesin-3 KLP-6 Regulates Intraflagellar Transport in Male-Specific Cilia of Caenorhabditis elegans. Curr. Biol. 2011;21:1239–1244. doi: 10.1016/j.cub.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 33.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Bringmann H, et al. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- 35.Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 36.Scholey JM, Porter ME, Grissom PM, McIntosh JR. Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature. 1985;318:483–486. doi: 10.1038/318483a0. [DOI] [PubMed] [Google Scholar]
- 37.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 39.Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422:753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- 40.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nature Publishing Group. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 41.Bhogaraju S, et al. Molecular Basis of Tubulin Transport Within the Cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piao T, et al. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niwa S, et al. KIF19A Is a Microtubule-Depolymerizing Kinesin for Ciliary Length Control. Dev. Cell. 2012 doi: 10.1016/j.devcel.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 46.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 47.Dentler WL, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. III. structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J. Cell Biol. 1977;74:747–759. doi: 10.1083/jcb.74.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedersen LB, et al. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 49.Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.