Abstract
Various proteins are involved in the generation and maintenance of the membrane complex known as the Golgi apparatus. We have used mutant Chinese hamster ovary (CHO) cell lines Lec4 and Lec4A lacking N-acetylglucosaminyltransferase V (GlcNAcT-V, MGAT5) activity and protein in the Golgi apparatus to study the effects of the absence of a single glycosyltransferase on the Golgi apparatus dimension. Quantification of immunofluorescence in serial confocal sections for Golgi α-mannosidase II and electron microscopic morphometry revealed a reduction in Golgi volume density up to 49 % in CHO Lec4 and CHO Lec4A cells compared to parental CHO cells. This reduction in Golgi volume density could be reversed by stable transfection of Lec4 cells with a cDNA encoding Mgat5. Inhibition of the synthesis of β1,6-branched N-glycans by swainsonine had no effect on Golgi volume density. In addition, no effect on Golgi volume density was observed in CHO Lec1 cells that contain enzymatically active Glc-NAcT-V, but cannot synthesize β1,6-branched glycans due to an inactive GlcNAcT-I in their Golgi apparatus. These results indicate that it may be the absence of the GlcNAcTV protein that is the determining factor in reducing Golgi volume density. No dimensional differences existed in cross-sectioned cisternal stacks between Lec4 and control CHO cells, but significantly reduced Golgi stack hits were observed in cross-sectioned Lec4 cells. Therefore, the Golgi apparatus dimensional change in Lec4 and Lec4A cells may be due to a compaction of the organelle.
Keywords: Golgi apparatus; N-acetylglucosaminyltransferase V; N-acetylglucosaminyltransferase I; CHO Lec4 cells; CHO Lec1 cells; β1,6-branched N-glycans; Golgi volume density; Golgi α-mannosidase II
Introduction
In higher eukaryotic cells, the Golgi apparatus forms a continuous branching and anastomosing ribbon consisting of stacks of cisternae bridged by non-compact, fenestrated regions, and tubular networks at its cis and trans face (Day et al. 2013; Klumperman 2011; Martínez-Alonso et al. 2013). Depending on cell-type, the number of cisternae making up the stack and the area comprising the cis and trans Golgi networks may vary (Han et al. 2013; Rambourg and Clermont 1997; Sengupta and Linstedt 2011; Uemura and Nakano 2013). While structurally complex, the Golgi apparatus is a highly dynamic organelle (Boncompain and Perez 2013; Colanzi et al. 1997; Cole et al. 1996; Polishchuk and Lutsenko 2013; Presley et al. 1998, 2002; Sciaky et al. 1997; Tillmann et al. 2013; Willett et al. 2013) and most important for cellular traffic (Boncompain and Perez 2013; Chia et al. 2013; Farquhar and Hauri 1997; Machamer 2013; Polishchuk and Lutsenko 2013; Sandvig et al. 2013; Tillmann et al. 2013; Warren 2013; Willett et al. 2013). Not surprisingly therefore, variations in the size of the Golgi apparatus or parts thereof have been reported to depend on the functional state of the cell and to become altered under disease conditions (Clermont et al. 1995; Fujita et al. 2002; Griffiths et al. 1989; Maeda et al. 2008; Noske et al. 2008; Rambourg et al. 1993; Sengupta and Linstedt 2011; Stieber et al. 1998).
There is much interest in understanding the molecular mechanisms responsible for generating and maintaining the integrity of the Golgi apparatus, and various types of proteins involved in this process have been identified. While microtubules and associated proteins are important for positioning the Golgi apparatus (Kreis et al. 1997; Presley et al. 1997; Zhu and Kaverina 2013), microtubule disassembly results in Golgi apparatus vesiculation (Thyberg and Moskalewski 1999). Cytoplasmic dynein and probably other motor proteins as well as actin filaments seem to be additionally involved in the formation and maintenance of Golgi apparatus structure (Allan 1996; Burkhardt 1998; Dippold et al. 2009; Egea et al. 2006, 2013; Harada et al. 1998; Yadav et al. 2012). Much information about the Golgi stack reassembly has been obtained through studies on the Golgi apparatus during mitosis (Acharya and Malhotra 1996; Barr and Warren 1996; Rabouille and Kondylis 2007; Shorter and Warren 2002). Golgi reassembly stacking proteins (Barr et al. 1997; Feinstein and Linstedt 2008; Puthenveedu et al. 2006; Sengupta et al. 2009; Shorter et al. 1999; Xiang and Wang 2010), a cis Golgi matrix protein, GM130 (Lowe et al. 1998; Marra et al. 2007; Nakamura et al. 1995, 1997), an NSF-like ATPase, p97, and NSF together with SNAPs and p115, a vesicle docking protein (Nelson et al. 1998; Rabouille et al. 1995b, 1998), seem to be important for the formation of the cisternal stack. In interphase cells, proteins cycling between the endoplasmic reticulum and the Golgi apparatus, such as Rab1b (Haas et al. 2007; Monetta et al. 2007; Romero et al. 2013; Tomas et al. 2012; Wilson et al. 1994), Arf1 (Boal et al. 2010; Lin et al. 2011; Manolea et al. 2008; Zhang et al. 1994) and TAP/p115 (Nelson et al. 1998; Puthenveedu and Linstedt 2001; Radulescu et al. 2011), are involved in maintaining Golgi apparatus morphology. Furthermore, the spectrin membrane skeleton (Nelson et al. 1998) is required for Golgi apparatus architecture.
Glycosyltransferases are Golgi residential proteins (Dunphy and Rothman 1983; Goldberg and Kornfeld 1983; Roth and Berger 1982; Roth et al. 1985), and the activity of a particular subset results in the synthesis of complex N-glycans (Kornfeld and Kornfeld 1985; Zuber and Roth 2009). Together with other proteins, glycosyltransferases seem to be involved in the formation and maintenance of the Golgi apparatus architecture. An illustrative example for this function was provided by studies on the parasitic protozoan Giardia lamblia (Lujan et al. 1995). During differentiation from throphozoites to cysts, the developmental induction of Golgi enzyme activities correlated with the appearance of a morphologically identifiable Golgi apparatus, which was absent in non-encysting cells. There are also data that N-acetylglucosaminyltransferase I (GlcNAcT-I) is involved in maintaining the structure of the cisternal stack (Nilsson et al. 1996). Replacement of part or the entire membrane-spanning domain of this type II membrane protein (Kumar et al. 1990) with leucine residues transformed the part of the Golgi stack housing the mutated GlcNAcT-I from flat cisternae into tubulo-vesicular membranes.
A number of Chinese hamster ovary (CHO) cells defective in glycosylation, due to mutated glycosyltransferases, have been isolated and characterized (Stanley and Ioffe 1995). In the CHO Lec4 and Lec4A cell mutants, N-acetylglucosaminyltransferase V (GlcNAcT-V) activity is affected (Chaney et al. 1989; Weinstein et al. 1996). Glc NAcT-V adds a β1,6 N-acetylglucosamine (GlcNAc) branch to the core α1,6-mannose (Man) in complex N-linked glycans attached to proteins. Lec4 cells lack GlcNAcT-V activity, due to a base insertion at nucleotide 822 of the Magt5 gene that shifts the open reading frame. A 155 amino acid truncated GlcNAcT-V (instead of a full length 740 amino acid enzyme) may be synthesized, which consists of the cytosolic and transmembrane domains and a short piece of the stem region. The fate of this truncated GlcNAcT-V is not known, but recognition by the protein quality control and subsequent proteolysis by ER-associated protein degradation may occur, as shown for other proteins (Roth et al. 2010). By contrast, CHO Lec4A cells have GlcNAcT-V activity equivalent to that of parental CHO cells in cellular homogenates. However, a single-point mutation, changing leucine to arginine at position 188, causes GlcNAcT-V to mislocalize to the endoplasmic reticulum and consequently to be functionally inactive in vivo. In both CHO Lec4 and Lec4A cells, complex N-glycans lack a β1,6 GlcNAc branch. In addition, the Golgi apparatus in Lec4A, and probably also in Lec4 mutant cells, lacks the residential protein GlcNAcT-V.
The structure of the Golgi apparatus in cells stably transfected and therefore overexpressing different glycosyltransferases appears normal (Lee et al. 1989; Nilsson et al. 1993; Rabouille et al. 1995a). However, more recent quantitative studies (Guo and Linstedt 2006) provided evidence that overexpression of the Golgi glycosyltransferase GlcNAcT-II results in a 1.3-fold increase in the size of the Golgi apparatus and that this change is sustained depending on interaction of GlcNAcT-II with COPII-coated transport carriers.
Currently, there is no information on the effects on Golgi apparatus structure and size of eliminating a Golgi residential protein such as a glycosyltransferase. Here, we demonstrate that the absence of the Golgi apparatus residential protein GlcNAcT-V causes a reduction in the dimensional size of the Golgi apparatus without affecting its general architecture.
Materials and methods
Cell lines
Chinese hamster ovary mutant cell lines Pro−Lec4.7b, Pro−Lec4A.12.2, Pro−5, and Pro−5 Lec1.3c were previously characterized (Chaney et al. 1989; Puthalakath et al. 1996; Stanley et al. 1982; Weinstein et al. 1996). Cells were maintained in α-MEM supplemented with 10 % fetal calf serum, glutamine and DNA, and RNA precursors.
Transfection of Lec4 cells with Mgat5 cDNA
Rat Mgat5 cDNA (Shoreibah et al. 1993) was subcloned into the EcoRI site of the pcDNA3 expression vector (Invitrogen, San Diego, CA). Lec4 cells were plated on 35 mm cell culture dishes and grown in α-MEM medium supplemented with 10 % fetal calf serum. Plasmid DNA (1 μg per 35 mm culture dish) was transfected into Lec4 cells, using Fugene™ 6 transfection reagent (Boehringer Mannheim, Germany) according to the manufacturer's protocol. Following transfection, clonal cell lines were established by selection for G418 resistance and tested for cell surface expression of β1,6 GlcNAc-branched N-glycans using digoxigenin-conjugated leukoagglutinating Phaseolus vulgaris lectin (dig L-PHA; Boehringer Mannheim, Germany) as described below. The positive clonal cell lines were designated Lec4 GnTV-N5, Lec4 GnTV-N10, and Lec4GnTVN30. Lec4 cells were mock-transfected with the pcDNA3 expression vector as described above, and clonal cell lines were established and designated Lec4 pcDNA3.
GlcNAcT-V assay
Cells were grown to confluence, harvested in 50 mM PBS, and concentrated to a pellet by centrifugation in microfuge tubes. Cell pellets were frozen on dry ice and shipped to Michael Pierce (Athens, GA, USA). For the assay, an approximately equal volume of ice-cold buffer (0.1 M MES, pH 6.5) was added to each pellet, followed by rapid thawing and sonication as described (Palcic et al. 1990). Each assay tube contained 106 cpm of UDP-[3H]-GlcNAc (25 cpm/pmol) and 10 nmol of synthetic trisaccharide acceptor (octyl 6–O–[2–O–(2-acetamido-2-deoxy-β-d-glucosyl-pyranosyl)-α-d-mannopyranosyl[-β-d-glucopyranoside) that were dried under vacuum in a 1.5-ml tube. The dried contents of each tube were resuspended in 0.05 ml of assay buffer (0.05 mM MES, pH 6.5, 2.0 % Triton X-100). Next, 5 μl of cell lysate was added to the tube, and the contents gently mixed by pipetting. Tubes were incubated for 6 h at 37 °C. Reactions were quenched by adding 0.5 ml water. Radiolabeled product was isolated using a C18 Sep-Pak (Waters) cartridge, eluted with 2 ml of methanol, and subjected to scintillation counting. Assay values for each extract were conducted in duplicate or triplicate and averaged. Aliquots of cell lysates were removed prior to enzymatic assay for protein determination in triplicate using the Bio-Rad protein assay for calculation of GlcNAcT-V specific activities.
Detection of β1,6-branched N-glycans with L-PHA
For lectin blots (Zuber et al. 1998), cells grown to 50–80 % confluence were removed by EDTA treatment (0.1 % EDTA-PBS at 37 °C for 10 min), pelleted by centrifu gation (1,500×g, 5 min at 4 °C) and lysed in three vol umes of 1 % Triton X-100 containing 1 mM AEBSF and 1 % aprotinin on ice for 30–40 min. After centrifugation (8,000×g, 10 min at 4 °C), the supernatant was denatured by boiling in Laemmli buffer. Samples were electrophoretically resolved in 3–10 % gradient polyacrylamide gels (120 μg protein per lane) and transferred to nitrocellu-lose. After blocking in 0.05 % Tween 20 and 1 % BSA in PBS, nitrocellulose strips were incubated with dig L-PHA (2 μg/ml for 2 h), rinsed, and incubated in alkaline phosphatase-conjugated polyclonal sheep anti-dig (Fab)2 fragments (5,000-fold diluted, Boehringer Mannheim, Germany) for 1 h. Alkaline phosphatase activity was revealed by the NBT-BCIP color reaction according to the manufacturer's instructions.
For light microscopic cell surface staining, cells grown on coverslips were fixed in 2 % paraformaldehyde in Earle's balanced salt solution (EBSS) (pH 7.2–7.4) for 5 min at 37 °C, and then at ambient temperature for 25 min, and the fixation was stopped by immersing the cover slips in 50 mM NH4Cl in PBS for 30 min (Roth et al. 1989). The coverslips were blocked in 2 % BSA in PBS for 30 min, incubated with digoxigenated (dig) L-PHA (2 μg/ml) for 2 h, followed by three rinses in PBS containing 0.1 % BSA (5 min each) and rhodamine-conjugated sheep anti-digoxigenin (Fab)2 fragments (200-fold diluted in 2 % BSA in PBS) for 1 h. After rinses, coverslips were embedded in Mowiol and mounted on glass slides. Cells were observed with a Leica TCS4D confocal laser scanning microscope.
For scanning electron microscopy, cells grown on glass coverslips were fixed in 2 % paraformaldehyde–0.1 % glutaraldehyde in EBSS as described above. Cells were incubated with directly 8 nm gold particle-labeled L-PHA (absorbance at 525 nm = 0.12) for 1 h. Following rinses and critical point drying, samples were coated and observed in a scanning electron microscope under standard and backscattering mode (Hermann et al. 1996).
Golgi α-mannosidase II immunostaining and morphometric analysis
Cells grown on coverslips were fixed as described above and permeabilized with PBS containing 0.15 % saponin and 0.1 % BSA for 15 min. After rinses with buffer, the cover-slips were incubated with rabbit anti-Golgi α-mannosidase II (1,000-fold diluted in PBS containing 0.5 % saponin and 0.1 % BSA; antibody kindly provided by Dr. Kelley More-men, University of Georgia, Athens, GA) for 2 h, rinsed in PBS containing 0.1 % BSA, and incubated in rhodamineconjugated affinity-purified goat anti-rabbit IgG (1,000-fold diluted in 0.1 % BSA-PBS; Jackson ImmunoResearch Laboratories Inc., West Grove, PA) for 1 h, rinsed with 0.1 % BSA in PBS and distilled water and embedding in Mowiol. After storage overnight at 4 °C, the cells were observed under a Leica TCS4D confocal laser scanning microscope. Cells grown in the presence of swainsonine (see below) were processed in the same way.
We estimated Golgi apparatus volume density by confocal laser scanning microscopy based on differential voxel counting of consecutive serial optical sections. As per each experimental set, three different views containing 30–40 cells were recorded randomly with a 40× objective and consecutive serial optical sections (0.4-μm optical resolution and optical section distance) were taken from the top to the bottom of each cell. The digitized images were analyzed using Photoshop®. Since Golgi α-mannosidase II has a broad Golgi distribution in CHO cells (Velasco et al. 1993), it was used to delineate the Golgi apparatus by immunofluorescence. The entire cells could be discerned by the presence of a faint cytoplasmic fluorescence. Using a histogram program, the sum of pixels for Golgi α-mannosidase II immunofluorescence and for the cytoplasmic fluorescence was obtained separately. From the ratio of the sum of pixels for Golgi α-mannosidase II immunofluorescence and the sum of pixels for the remaining cytoplasm, the volume density of the Golgi apparatus was calculated using the following equation:
In this equation, v/V (G/C) represents the volume density of the Golgi apparatus; m and n represent the initial gray level for the Golgi apparatus (G) and cytoplasm (C), respectively; and gi represents the number of pixels at each gray level i.
Transmission electron microscopy and morphometric analysis
Cells grown on Thermanox coverslips (Nalge Nunc. International, Roskilde, Denmark) were fixed in 2 % paraformaldehyde–1 % glutaraldehyde in EBSS (pH 7.2–7.4) for 5 min at 37 °C and then at ambient temperature for 25 min. After rinses with buffer and immersion in 50 mM NH4Cl in PBS for 30 min (Roth et al. 1989), cells were postfixed in 1 % reduced osmium tetroxide (Karnovsky 1971) for 30 min, rinsed in distilled water and stained with 2 % aqueous uranyl acetate for 15 min. The cells were embedded in Epon according to standard protocol. Ultrathin sections were contrasted with uranyl acetate and lead citrate. From each cell line, photographs were taken randomly from 30 to 40 cross-sectioned cells. In addition, photographs at the original magnification of 25,000 were taken from each cell containing the Golgi apparatus in the cross section. The number of cisternae per Golgi stack, the length and thickness of cisternal stacks, as well as the area occupied by the Golgi stacks and the size of the cell profile in the cross sections were measured on digitized photographic negatives. The volume density of the Golgi apparatus was calculated by the ratio of the surface area of the Golgi stacks, and the surface size of the cell profile in the cross sections (Weibel 1979).
Swainsonine treatment of cell lines and lectin spot plots
Swainsonine (1 μg/ml; Sigma Chemical Co, Ltd., St. Louis, MO) was added to the culture medium of cells growing at 30–40 % confluence, and the cells were analyzed 12 and 24 h after addition of the drug. For L-PHA spot blot analysis, cells were harvested and lysed as described above. About 1 μl aliquots of serially diluted cell lysates were pipetted onto nitrocellulose strips and air-dried, and β1,6-branched N-glycans detected with dig L-PHA as described above. The quantitative analysis was performed with Wincam® software (Camtek Oy, Vantaa, Finland). Using the gray levels of the serial dilutions of the zero time-point homogenates as reference, the gray levels (relative percentage) of the spots from the homogenates of the cells treated with swainsonine for 12 and 24 h were estimated.
Results
Expression of β1,6-branched N-glycans in various CHO cell lines
Parent and mutant CHO cell lines were studied by confocal laser scanning microscopy and lectin-gold scanning electron microscopy as well as by lectin blotting using the leukoagglutinating Phaseolus vulgaris lectin (L-PHA) to detect β1,6-branched complex N-glycans (Cummings and Kornfeld 1982), the product of GlcNAcT-V. While wild-type CHO Pro−5 cells were reactive with L-PHA, CHO Lec4 and Lec4A mutant cells lacking GlcNAcT-V and mock-transfected Lec4pcDNA3 cells were unreactive (Fig. 1a, b, e, f). The three clonal cell lines stably expressing GlcNAcT-V exhibited cell surface L-PHA staining undistinguishable from that observed for CHO Pro−5 cells (Fig. 1c, g). In L-PHA blots, two major bands at 140 and 85 kDa and several minor bands were evident in parent CHO and Lec4 GlcNAcT-V cells lines (Fig. 1d), but CHO Lec4, CHO Lec4A, and mock-transfected CHO Lec4 cell glycoproteins did not bind L-PHA (data not shown). Different GlcNAcT-V activity levels were found in the transfected cell lines (data not shown). The Lec4 GnTV-N5 transfectants had enzyme activity similar to CHO Pro−5 cells and, based on densitometric evaluation of spot blots, synthesized similar amounts of β1,6-branched N-glycans. Therefore, CHO Lec4 GnTV-N5 cells were used in most comparative analyses.
Fig. 1.
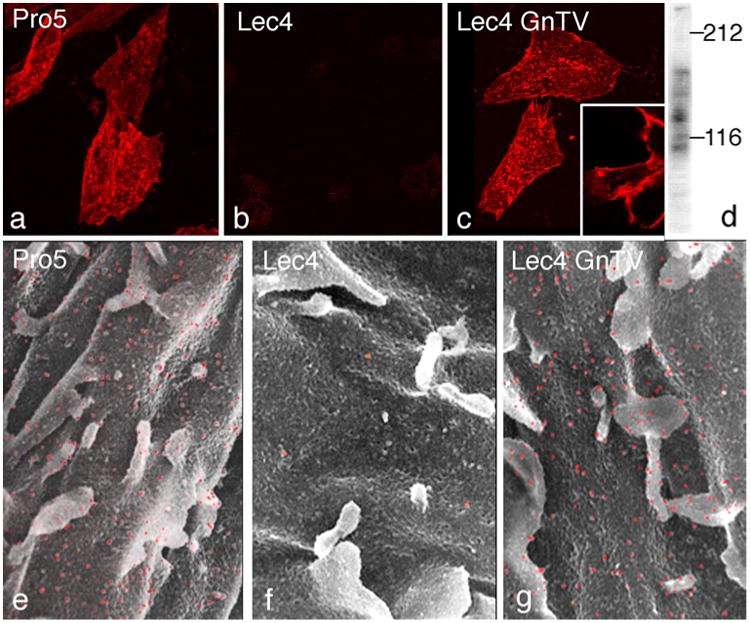
Demonstration of β1,6-branched N-glycans on the CHO cell surface using dig L-PHA and confocal laser scanning microscopy (a–c) and lectin-gold scanning electron microscopy (e–g). The CHO Pro−5 cells (a, e) and CHO Lec4 GnTV-N5 cells (c, g) showed positive cell surface staining with microvilli being strongly positive. In contrast, CHO Lec4 cells (b, f) showed no specific cell surface labeling. ×1,100 (a–c), ×9,000 (e–g). Dig L-PHA—blot of homogenates from CHO Lec4 GnTV cells revealed two major bands at 140 and 85 kDa and several minor bands (d)
Golgi apparatus dimension is reduced in the absence of GlcNAcT-V
To measure the dimension of the Golgi apparatus at the light microscopic level, immunofluorescence for Golgi α-mannosidase II, a cis/medial Golgi resident protein in CHO cells (Velasco et al. 1993), was quantified by confocal laser scanning microscopy (Fig. 2). As detailed in Table 1, the volume density of the Golgi apparatus of CHO Lec4 and CHO Lec4A cells, as well as of CHO Lec4 pcDNA3 cells, was significantly smaller to that of CHO parental cells and of the three clonal CHO Lec4 GnTV cell lines. By electron microscopy, no differences in the structure of the Golgi apparatus were apparent between CHO Pro−5 (Fig. 3a), CHO Lec4 (Fig. 3b), and CHO Lec4 GnTV (Fig. 3c). The results of the electron microscopic morpho-metric analyses are presented in Table 2 and confirm the subjective impression. The mean number of Golgi stack hits per cross-sectioned CHO Lec4 cells was reduced by 31–42 % as compared to CHO Pro−5 and CHO Lec4 GnTV cells. Therefore, the volume density of the Golgi apparatus was significantly smaller in CHO Lec4 cells (reduced by 37–49 %) as compared to CHO Pro−5 and CHO Lec4 GnTV cells. Interestingly, the mean number of cisternae per stack, as well as the horizontal and vertical dimension of the Golgi cisternal stacks, was similar in all cell lines studied (Table 2). Therefore, the most important contribution for the reduced volume density of the Golgi apparatus in CHO Lec4 cells came from the reduced Golgi stack hits in cross-sectioned cells. In order to obtain more insight into this phenomenon, we analyzed the distribution pattern of several other parameters. We found that the number of cisternae per stack, as well as the horizontal and vertical dimension of the Golgi stacks, had a Poisson distribution for each cell line (not shown). However, the distribution of the Golgi apparatus volume density was considerably different between CHO Lec4 cells and CHO Pro−5 as well as CHO Lec4 GnTV cells (Fig. 4). The number of cross-sectioned cells not exhibiting Golgi stacks was much higher in Lec4 cells than in Pro−5 and Lec4 GnTV. N5 cells. This parameter was the main contributor to differences in the mean value of Golgi apparatus volume density (Table 3).
Fig. 2.
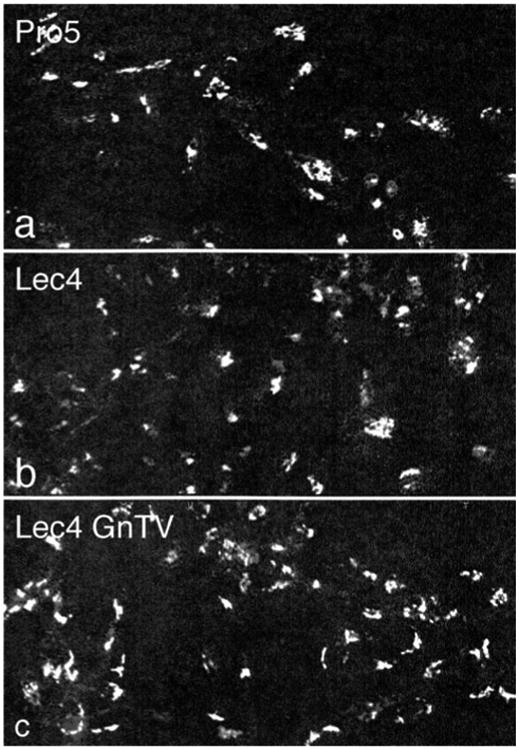
Confocal laser scanning immunofluorescence for Golgi α-mannosidase II. CHO Pro−5 cells (a), CHO Lec4 cells (b), and CHO Lec4 GnTV-N5 cells (c) exhibit a typical perinuclear Golgi staining pattern in a single confocal optical section (×560)
Table 1. Morphometric analysis of the Golgi mannosidase II immunofuorescence.
| Cells | GlcNAcT-V protein | Exp. 1 | Exp. 2 | Exp. 3 | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| v/Va | R (%)b | v/Va | R (%)b | v/Va | R (%)b | ||
| Pro−5 | + | 30 ± 1c | 43 | 27 ± 4c | 44 | 28 ± 1c | 36 |
| Lec4 | − | 17 ± 1 | – | 15 ± 1 | – | 18 ± 1 | – |
| Lec4 GnT-N5 | + | 25 ± 1c | 32 | 26 ± 1c | 42 | 27 ± 3c | 33 |
| Lec4 GnT V-N10 | + | 25 ± 5d | 32 | – | – | ||
| Lec4 GnT V-N30 | + | 30 ± 5c | 43 | – | – | ||
| Lec4 PcDNA3 | − | 17 ± 4 | – | – | – | ||
| Lec4A | +e | 18 ± 1 | 6 | – | – | ||
Volume density of Golgi apparatus, data are expressed as mean ± SD. Three different views were evaluated in each experiment
Reduction rate of the volume density of the Golgi apparatus in Lec4 cells as compared to other cell lines
Indicates significant difference as compared with Lec4, Lec4 PcDNA3, or Lec4A cells (p < 0.01)
Indicates significant difference as compared with Lec4, Lec4 PcDNA3, or Lec4A cells (p < 0.05)
GlcNAcT-V protein mislocated in the endoplasmic reticulum
Fig. 3.
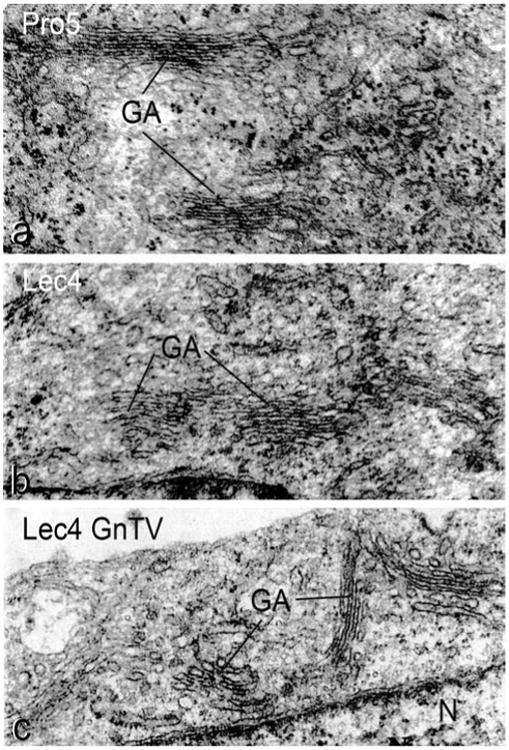
Transmission electron micrographs showing the morphology of the Golgi apparatus (GA) in CHO Pro−5 cells (a), CHO Lec4 cells (b), and CHO Lec4 GnTV-N5 cells (c). Note the structural similarity in appearance of the Golgi apparatus in the different cells lines. N part of the nucleus (×41,300)
Table 2. Morphometric analysis of the Golgi apparatus by electron microscopy.
| Cells | GlcNAcT-V protein | Golgi cisternal stacka | Number of stacks/cell cross section | Size of cell profle in cross sectionsa (μm2) | Volume density of GAa (×10−3) | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Number of cisternae | Length (μm) | Thickness (μm) | |||||
| Pro−5 | |||||||
| Exp. 1 (n = 35) | + | 5.1 ± 0.1 | 0.71 ± 0.03 | 0.19 ± 0.01 | 1.7 | 54.7 ± 4.0b | 5.1 ± 0.6c |
| Exp. 2 (n = 67) | + | 4.7 ± 0.1 | 0.68 ± 0.03 | 0.21 ± 0.01 | 1.6 | 61.9 ± 4.5b | 4.1 ± 0.5c |
| Lec4 | |||||||
| Exp. 1 (n = 30) | - | 5.0 ± 0.1 | 0.64 ± 0.04 | 0.18 ± 0.01 | 1.1 | 54.9 ± 3.4 | 3.2 ± 0.7 |
| Exp. 2 (n = 50) | - | 4.5 ± 0.1 | 0.61 ± 0.03 | 0.20 ± 0.01 | 1.1 | 65.4 ± 2.9 | 2.4 ± 0.3 |
| Lec4 GnTV-N5 | |||||||
| Exp. 1 (n = 36) | + | 5.0 ± 0.1 | 0.61 ± 0.03 | 0.17 ± 0.01 | 1.9 | 43.1 ± 2.4b | 5.5 ± 0.7c |
| Exp. 2 (n = 71) | + | 4.7 ± 0.1 | 0.57 ± 0.02 | 0.22 ± 0.01 | 1.7 | 49.3 ± 3.4b | 4.7 ± 0.5c |
Data are expressed as mean ± SEM
Indicates significant difference as compared with Lec4 cells (p < 0.01, Wilcoxon test) within the same experiment
Indicates significant difference as compared with Lec4 cells (p < 0.05, Wilcoxon test) within the same experiment
Fig. 4.
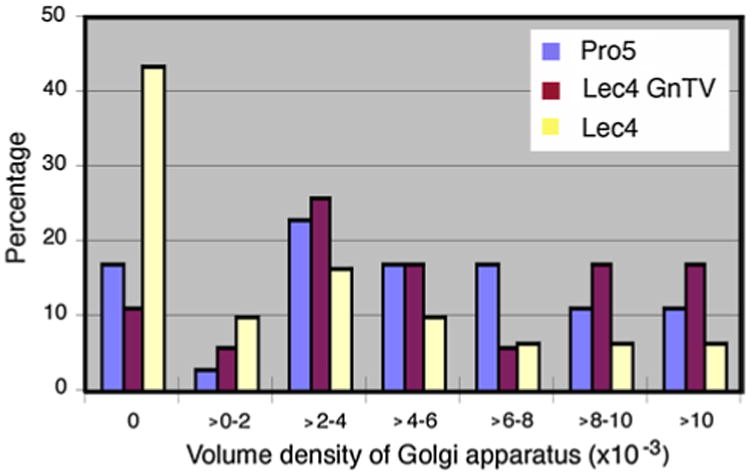
The distribution pattern of volume density of the Golgi apparatus in the various CHO cell lines. The number of cross-sectioned cells containing no cisternal stacks was much higher in CHO Lec4 cells than in CHO Pro−5 as well as CHO Lec4 GnTV-N5 cells
Table 3. Mean values of Golgi apparatus volume density with or without Golgi stack hits in cross-sectioned cells.
| Cells | Mean volume density of Golgi apparatusa × 10−3 | Mean volume density of Golgi apparatusb × 10−3 |
|---|---|---|
| Pro−5 | 5.1 | 6.1 |
| Lec4 | 3.2 | 5.6 |
| Lec4 GnTV | 5.5 | 6.4 |
Calculations were performed by considering only cross sections containing Golgi apparatus
Calculations were performed by including cross sections with and without Golgi apparatus hits
Golgi apparatus dimension is not influenced by the absence of GlcNAcT-V glycosylation product
At this point of the study, the question arose: Is the physical absence of the GlcNAcT-V protein in the Golgi apparatus, and/or the lack of its enzymatic activity and glycosylation products, the basis for the changes in Golgi apparatus dimension in Lec4 cells? Unfortunately, no specific GlcNAcT-V inhibitor was available for in vivo studies with intact cells. Thus, to address this question indirectly, we investigated CHO Lec1 cells, which contain an enzymatically inactive GlcNAcT-I protein in the Golgi apparatus (Kumar et al. 1990; Puthalakath et al. 1996; Stanley et al. 1975). As a consequence, no acceptor substrate for GlcNAcT-V is synthesized in Lec1 cells. Thus, although they contain enzymatically active GlcNAcT-V in their Golgi apparatus, β1,6-branched N-glycans cannot be synthesized. We found that the volume density of the Golgi apparatus in CHO Lec1 cells, based on the evaluation of Golgi α-mannosidase II immunofluorescence, was not different from that of Pro−5 or Lec4 GnTV cells (Table 4). Second, we used swainsonine, an inhibitor of Golgi α-mannosidase II activity, to block the synthesis of acceptor substrate for GlcNAcT-V. Both CHO Pro−5 cells and CHO Lec4 GnTV cells treated for 24 h with 1 μg/ml of swainsonine contained less than 10 % L-PHA reactivity, as compared to control or swainsonine-treated CHO Lec4 cells (Fig. 5). However, the swainsonine treatment had no influence on the dimension of the Golgi apparatus in CHO Pro−5 and CHO Lec4 GnTV cells, or in CHO Lec4 cells (Table 4).
Table 4. The volume density of the Golgi apparatus in various CHO cell lines following swainsonine treatment.
| Cells | GlcNAcT-V protein | Volume density (×10−3)a | |
|---|---|---|---|
|
| |||
| Control | Swainsonine treatmentb | ||
| Pro−5 | + | 27 ± 4c | 26 ± 4c |
| Lec4 | – | 15 ± 3 | 16 ± 3 |
| Lec4 GnTV-N5 | + | 27 ± 3c | 26 ± 3c |
| Lec1 | + | 26 ± 3c | – |
Data are expressed as mean ± SD and based on three different evaluations each comprising three different randomly taken views containing 30–40 cells
Cells were treated for 24 h with 1 μg/ml swainsonine
Indicates significant difference as compared with Lec4 or Lec4A cells (p < 0.01, x2 test, t test)
Fig. 5.
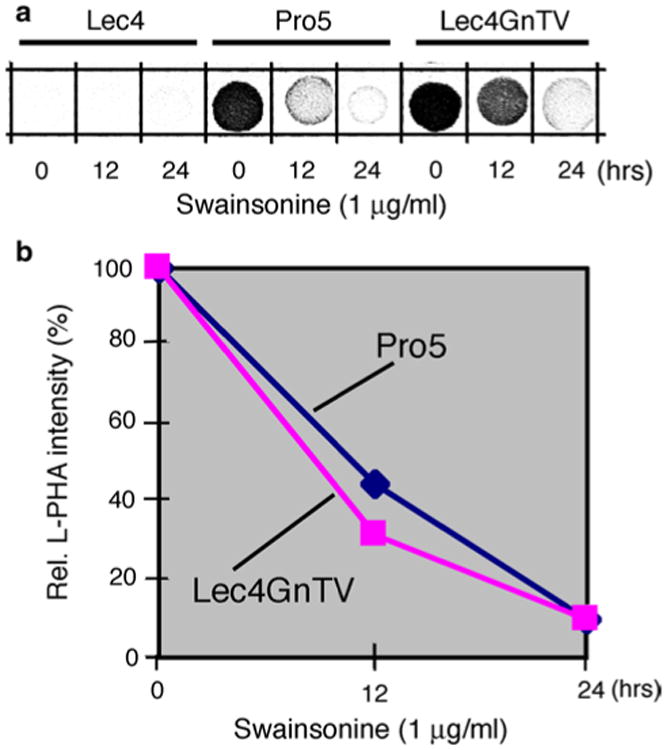
Reduction in the β1,6-branched N-glycans in CHO cells after swainsonine treatment. Swainsonine (1 μg/ml) was added to the cell culture medium, and cells were collected after 0, 12, and 24 h of treatment. Homogenates were processed for L-PHA spot blots, and the results densitometrically analyzed. The CHO Lec4 cells served as control
Discussion
The present studies were undertaken to investigate whether the absence of a single glycosyltransferase protein, and/or the product of its transferase activity, has an influence on the morphology of the Golgi apparatus. For this, we have made use of CHO cells lines with mutations in glycosyltransferase genes (Patnaik and Stanley 2006) and have focused on GlcNAcT-V. GlcNAcT-V with a molecular mass of 95 kDa is a comparatively large glycosyltransferase of the Golgi apparatus (Shoreibah et al. 1993). It initiates the synthesis of tri- and tetra-antennary complex N-glycans, whose expression is associated with malignancy and the metastatic potential of tumor cells (Dennis 1999; Dennis et al. 1987; Granovsky et al. 2000; Partridge et al. 2004; Seelentag et al. 1998). The absence of tri- and tetraantennary, β1,6-branched N-glycans has no influence on the viability of CHO Lec4 and CHO Lec4A cells although they differ in gross morphology from the parental cells (Stanley and Sudo 1981). The present study demonstrates that the defect in CHO Lec4 and CHO Lec4A cells is associated with a significant decrease in the volume density of their respective Golgi apparatus. This defect could be fully compensated by stable transfection of a cDNA encoding GlcNAcT-V into CHO Lec4 cells. The analysis of Golgi apparatus dimension in cell lines treated with swainso-nine to block the synthesis of β1,6-branched N-glycans, or in CHO Lec1 cells, which cannot synthesize the acceptor substrate for GlcNAcT-V, provides strong evidence that the observed change of volume density in CHO Lec4 and CHO Lec4A cells does not simply result from the synthesis of truncated N-glycans. Rather it seems that the absence of the GlcNAcT-V protein in these cell lines leads to the change in Golgi apparatus volume density. This assumption is also supported by our analysis of CHO Lec1 cells, which contain enzymatically inactive Golgi apparatus located Glc-NAcT-I (Puthalakath et al. 1996) and which have the same Golgi apparatus dimension as the parental CHO Pro−5 cell line. In this context, it is interesting to note that the Golgi apparatus in wild-type yeast Saccharomyces cerevisiae lacks clearly defined stacks of cisternae and appears in the form of single cisternae or individual networks of tubules (MorinGanet et al. 1998; Preuss et al. 1992; Rambourg et al. 1995). It is tempting to speculate that the structurally simple Golgi apparatus of yeast cells may be related to the low number of Golgi glycosyltransferases, essentially five mannosyltransferases (Herscovics and Orlean 1993; Lehle and Tanner 1995), as compared to the large families of glycosyltransferases existing in the Golgi apparatus of higher eukaryotic cells.
The reduction in volume density of the Golgi apparatus by up to 49 % in the absence of GlcNAcT-V is surprising even if one considers its molecular mass of 95 kDa (Shoreibah et al. 1993). We noticed from electron microscopic analyses that the number of cisternae per Golgi stacks, as well as the length and the thickness of the Golgi cisternal stacks, was the same in parental and the CHO Lec1 cell mutant. Thus, in contrast to the replacement of part or the entire membrane-spanning domain of GlcNAcT-I by leucine, which disrupted part of cisternal structure (Nilsson et al. 1996), the absence of GlcNAcTV protein was compatible with normal cisternal structure. However, our quantitative analysis revealed that the mean number of Golgi stack hits per cell cross section was clearly reduced in CHO Lec4 cells as compared to CHO Pro−5 cells and CHO Lec4 cells stably expressing GlcNAcT-V.
How could these data be related to the measured reduction in the volume density of the Golgi apparatus in CHO Lec4 cells? All current evidence indicates that the inter-phase Golgi apparatus in higher eukaryotic cells can be regarded as a single organelle composed of numerous dynamic and interrelated units (Hermo and Smith 1998; Rambourg and Clermont 1997). Non-compact regions composed of tubules link these units, which correspond to compact cisternal regions, to each other laterally, and therefore forming a twisting ribbon that bifurcates and rejoins. Although there can be variability in the compact cisternal regions, the mean number of cisternae in one stack, and the size of the units, can be considered to be cell-type specific and relatively constant under steady-state conditions. One possible explanation for the present results could be that the unit size of the Golgi apparatus is reduced in the absence of GlcNAcT-V. Although a reduction in the number of Golgi apparatus units cannot be ruled out, this seems rather unlikely. A more likely explanation, which would account for the reduction in Golgi apparatus volume density, could be a change in the overall three-dimensional shape of the organelle, resulting in a more compact structure in CHO Lec4 and CHO Lec4A cells. The absence of GlcNAcT-V may result in secondary effects on, for example, interactions with intercisternal matrix proteins and other protein– protein interactions, which have been proposed to be of importance for Golgi apparatus structure (Sengupta and Linstedt 2011).
In summary, our results indicate that the absence of the Golgi residential protein GlcNAcT-V results in the adaptive reduction in volume density of the Golgi apparatus without altering its overall architecture. Although the mechanism leading to this phenomenon remains to be clarified, this finding adds a novel aspect to the structural flexibility of this dynamic steady-state organelle.
Acknowledgments
We are grateful to Kelley Moremen for the Golgi α-mannosidase II antibody. This work was supported by the Swiss National Science Foundation (to J. R.) and the National Cancer Institute RO1 36434 (to P. S.) and partially supported by the Albert Einstein Cancer Center NCI PO1 13330 (to P. S.). P. S. thanks Subha Sundaram for technical assistance.
Abbreviations
- GlcNAcT-I
N-acetylglucosaminyltransferase I
- GlcNAcT-V, MGAT5
N-acetylglucosaminyltransferase V
- CHO
Chinese hamster ovary cells
- GlcNAc
N-acetylglucosamine
- Man
Mannose
- L-PHA
Leukoagglutinating Phaseolus vulgaris lectin
- Dig
Digoxigenin
Contributor Information
Zhizhong Dong, Division of Cell and Molecular Pathology, Department of Pathology, University of Zürich, 8091 Zürich, Switzerland.
Christian Zuber, Division of Cell and Molecular Pathology, Department of Pathology, University of Zürich, 8091 Zürich, Switzerland.
Michael Pierce, Complex Carbohydrate Research Center, Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA 30602, USA.
Pamela Stanley, Department of Cell Biology, Albert Einstein College of Medicine, New York, NY 10461, USA.
Jürgen Roth, Email: jurgen.roth@bluewin.ch, Division of Cell and Molecular Pathology, Department of Pathology, University of Zürich, 8091 Zürich, Switzerland.
References
- Acharya U, Malhotra V. Reconstitution of Golgi stacks from vesiculated Golgi membranes in permeabilized cells. Semin Cell Dev Biol. 1996;7:511–516. doi: 10.1101/sqb.1995.060.01.059. [DOI] [PubMed] [Google Scholar]
- Allan V. Role of motor proteins in organizing the endoplasmic reticulum and Golgi apparatus. Semin Cell Dev Biol. 1996;7:335–342. [Google Scholar]
- Barr FA, Warren G. Disassembly and reassembly of the Golgi apparatus. Semin Cell Dev Biol. 1996;7:505–510. [Google Scholar]
- Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Boal F, Guetzoyan L, Sessions RB, Zeghouf M, Spooner RA, Lord JM, Cherfils J, Clarkson GJ, Roberts LM, Stephens DJ. LG186: an inhibitor of GBF1 function that causes Golgi disassembly in human and canine cells. Traffic. 2010;11:1537–1551. doi: 10.1111/j.1600-0854.2010.01122.x. [DOI] [PubMed] [Google Scholar]
- Boncompain G, Perez F. The many routes of Golgi-dependent trafficking. Histochem Cell Biol. 2013;140:251–260. doi: 10.1007/s00418-013-1124-7. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK. The role of microtubule-based motor proteins in maintaining the structure and function of the Golgi complex. Biochim Biophys Acta Mol Cell Res. 1998;1404:113–126. doi: 10.1016/s0167-4889(98)00052-4. [DOI] [PubMed] [Google Scholar]
- Chaney W, Sundaram S, Friedman N, Stanley P. The Lec4A CHO glycosylation mutant arises from miscompartmentalization of a Golgi glycosyltransferase. J Cell Biol. 1989;109:2089–2096. doi: 10.1083/jcb.109.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P, Gunn P, Gleeson P. Cargo trafficking between endosomes and the trans-Golgi network. Histochem Cell Biol. 2013;140:307–315. doi: 10.1007/s00418-013-1125-6. [DOI] [PubMed] [Google Scholar]
- Clermont Y, Rambourg A, Hermo L. Trans-Golgi network (TGN) of different cell types: three-dimensional structural characteristics and variability. Anat Rec. 1995;242:289–301. doi: 10.1002/ar.1092420302. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Mironov A, Weigert R, Limina C, Flati S, Cericola C, Di Tullio G, Di Girolamo M, Corda D, De Matteis MA, Luini A. Brefeldin A-induced ADP-ribosylation in the structure and function of the Golgi complex. Adv Exp Med Biol. 1997;419:331–335. doi: 10.1007/978-1-4419-8632-0_43. [DOI] [PubMed] [Google Scholar]
- Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- Cummings R, Kornfeld S. Characterization of the structural determinants required for the high affinity interaction of asparagine-linked oligosaccharides with immobilized Phaseolus vulgaris leukoagglutinating and erythroagglutinating lectins. J Biol Chem. 1982;257:11230–11234. [PubMed] [Google Scholar]
- Day KJ, Staehelin LA, Glick BS. A three-stage model of Golgi structure and function. Histochem Cell Biol. 2013;140:239–249. doi: 10.1007/s00418-013-1128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J. Protein glycosylation in development and disease. BioEssays. 1999;21:412–421. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Laferte S, Waghorne C, Breitman ML, Kerbel RS. Beta 1-6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science. 1987;236:582–585. doi: 10.1126/science.2953071. [DOI] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peter-man MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, Fuchs GJ, Meerloo T, Farquhar MG, Zhou H, Field SJ. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy WG, Rothman JE. Compartmentation of asparagine-linked oligosaccharide processing in the Golgi apparatus. J Cell Biol. 1983;97:270–275. doi: 10.1083/jcb.97.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea G, Lazaro-Dieguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Egea G, Serra-Peinado C, Salcedo-Sicilia L, Gutiérrez-Martínez E. Actin acting at the Golgi. Histochem Cell Biol. 2013;140:347–360. doi: 10.1007/s00418-013-1115-8. [DOI] [PubMed] [Google Scholar]
- Farquhar M, Hauri HP. Protein sorting and vesicular traffic in the Golgi apparatus. In: Berger E, Roth J, editors. The Golgi apparatus. Birkhäuser Verlag; Basel: 1997. pp. 63–129. [Google Scholar]
- Feinstein TN, Linstedt AD. GRASP55 regulates Golgi ribbon formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Okamoto K, Sakurai A, Kusaka H, Aizawa H, Mihara B, Gonatas NK. The Golgi apparatus is fragmented in spinal cord motor neurons of amyotrophic lateral sclerosis with basophilic inclusions. Acta Neuropathol. 2002;103:243–247. doi: 10.1007/s004010100461. [DOI] [PubMed] [Google Scholar]
- Goldberg DE, Kornfeld S. Evidence for extensive subcellular organization of asparagine-linked oligosaccharide processing and lysosomal enzyme phosphorylation. J Biol Chem. 1983;258:3159–3165. [PubMed] [Google Scholar]
- Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6:306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- Griffiths G, Fuller S, Back R, Hollinshead M, Pfeiffer S, Simons K. The dynamic nature of the Golgi complex. J Cell Biol. 1989;108:277–29. doi: 10.1083/jcb.108.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Linstedt AD. COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. J Cell Biol. 2006;174:53–63. doi: 10.1083/jcb.200604058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AK, Yoshimura S, Stephens DJ, Preisinger C, Fuchs E, Barr FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- Han HM, Bouchet-Marquis C, Huebinger J, Grabenbauer M. Golgi apparatus analyzed by cryo-electron microscopy. Histochem Cell Biol. 2013 doi: 10.1007/s00418-013-1136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann R, Walther P, Muller M. Immunogold labeling in scanning electron microscopy. Histochem Cell Biol. 1996;106:31–39. doi: 10.1007/BF02473200. [DOI] [PubMed] [Google Scholar]
- Hermo L, Smith C. The structure of the Golgi apparatus: a sperm's eye view in principal epithelial cells of rat epididymis. Histochem Cell Biol. 1998;109:431–447. doi: 10.1007/s004180050246. [DOI] [PubMed] [Google Scholar]
- Herscovics A, Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Karnovsky M. Use of ferrocyanide-reduced osmium tetroxide in electron microscopy. Abstracts of Fourteenth Annual Meeting American Society of Cell Biology. 1971:114. [Google Scholar]
- Klumperman J. Architecture of the Mammalian Golgi. Cold Spring Harb Perspect Biol. 2011;3:a005181. doi: 10.1101/cshperspect.a005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Ann Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kreis T, Goodson H, Perez F, Rönnholm R. Golgi apparatus-cytoskeleton interactions. In: Berger E, Roth J, editors. The Golgi apparatus. Birkhäuser Verlag; Basel: 1997. pp. 179–193. [Google Scholar]
- Kumar R, Yang J, Larsen R, Stanley P. Cloning and expression of N-acetylglucosaminyltransferase I, the medial Golgi transferase that initiates complex N-linked carbohydrate formation. Proc Natl Acad Sci USA. 1990;87:9948–9952. doi: 10.1073/pnas.87.24.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EU, Roth J, Paulson JC. Alteration of terminal glycosylation sequences on N-linked oligosaccharides of Chinese hamster ovary cells by expression of β-galactoside α2,6 sialyltransferase. J Biol Chem. 1989;264:13848–13855. [PubMed] [Google Scholar]
- Lehle L, Tanner W. Protein glycosylation in yeast. In: Montreuil J, Vliegenthart JFG, Schachter H, editors. Glycoproteins. 29a. Elsevier; Amsterdam: 1995. pp. 475–509. [Google Scholar]
- Lin YC, Chiang TC, Liu YT, Tsai YT, Jang LT, Lee FJ. ARL4A acts with GCC185 to modulate Golgi complex organization. J Cell Sci. 2011;124:4014–4026. doi: 10.1242/jcs.086892. [DOI] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJC, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Lujan HD, Marotta A, Mowatt MR, Sciaky N, Lippincott-Schwartz J, Nash TE. Developmental induction of Golgi structure and function in the primitive eukaryote Giardia lamblia. J Biol Chem. 1995;270:4612–4618. doi: 10.1074/jbc.270.9.4612. [DOI] [PubMed] [Google Scholar]
- Machamer C. Accommodation of large cargo within Golgi cisternae. Histochem Cell Biol. 2013;140:261–269. doi: 10.1007/s00418-013-1120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Ide T, Koike M, Uchiyama Y, Kinoshita T. GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat Cell Biol. 2008;10:1135–1145. doi: 10.1038/ncb1773. [DOI] [PubMed] [Google Scholar]
- Manolea F, Claude A, Chun J, Rosas J, Melancon P. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol Biol Cell. 2008;19:523–535. doi: 10.1091/mbc.E07-04-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P, Salvatore L, Mironov A, Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Alonso E, Tomás M, Martínez-Menárguez J. Golgi tubules: their structure, formation and role in intra-Golgi transport. Histochem Cell Biol. 2013;140:327–339. doi: 10.1007/s00418-013-1114-9. [DOI] [PubMed] [Google Scholar]
- Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell. 2007;18:2400–2410. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MorinGanet MN, Rambourg A, Clermont Y, Kepes F. Role of endoplasmic reticulum-derived vesicles in the formation of Golgi elements in sec23 and sec18 Saccharomyces cerevisiae mutants. Anat Rec. 1998;251:256–264. doi: 10.1002/(SICI)1097-0185(199806)251:2<256::AID-AR15>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nelson D, Alvarez C, Gao Y, Garcia-Mata R, Fialkowski E, Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J Cell Biol. 1998;143:319–331. doi: 10.1083/jcb.143.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Pypaert M, Hoe MH, Slusarewicz P, Berger EG, Warren G. Overlapping distribution of two glycosyltransferases in the Golgi apparatus of HeLa cells. J Cell Biol. 1993;120:5–13. doi: 10.1083/jcb.120.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Rabouille C, Hui N, Watson R, Warren G. The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J Cell Sci. 1996;109:1975–1989. doi: 10.1242/jcs.109.7.1975. [DOI] [PubMed] [Google Scholar]
- Noske AB, Costin AJ, Morgan GP, Marsh BJ. Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J Struct Biol. 2008;161:298–313. doi: 10.1016/j.jsb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palcic MM, Ripka J, Kaur KJ, Shoreibah M, Hindsgaul O, Pierce M. Regulation of N-acetylglucosaminyltransferase V activity. Kinetic comparisons of parental, Rous sarcoma virus-trans-formed BHK, and l-phytohemagglutinin-resistant BHK cells using synthetic substrates and an inhibitory substrate analog. J Biol Chem. 1990;265:6759–6769. [PubMed] [Google Scholar]
- Partridge EA, LeRoy C, DiGuglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- Patnaik SK, Stanley P. Lectin-resistant CHO glycosylation mutants. Meth Enzymol. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- Polishchuk R, Lutsenko S. Golgi in copper homeostasis: a view from the membrane trafficking field. Histochem Cell Biol. 2013;140:285–295. doi: 10.1007/s00418-013-1123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJM, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Presley JF, Smith C, Hirschberg K, Miller C, Cole NB, Zaal KJM, Lippincott-Schwartz J. Golgi membrane dynamics. Mol Biol Cell. 1998;9:1617–1626. doi: 10.1091/mbc.9.7.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Ward TH, Pfeifer AC, Siggia ED, Phair RD, Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Sacharomyces cerevisiae Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Burke J, Gleeson PA. Glycosylation defect in Lec1 Chinese hamster ovary mutant is due to a point mutation in N-acetylglucosaminyltransferase I gene. J Biol Chem. 1996;271:27818–27822. doi: 10.1074/jbc.271.44.27818. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Linstedt AD. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J Cell Biol. 2001;155:227–237. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondylis V. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle. 2007;6:2723–2729. doi: 10.4161/cc.6.22.4896. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T. Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci. 1995a;108:1617–1627. doi: 10.1242/jcs.108.4.1617. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995b;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Radulescu AE, Mukherjee S, Shields D. The Golgi protein p115 associates with gamma-tubulin and plays a role in Golgi structure and mitosis progression. J Biol Chem. 2011;286:21915–21926. doi: 10.1074/jbc.M110.209460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y. Three-dimensional structure of the Golgi apparatus in mammalian cells. In: Berger E, Roth J, editors. The Golgi apparatus. Birkhäuser Verlag; Basel: 1997. pp. 37–61. [Google Scholar]
- Rambourg A, Clermont Y, Chretien M, Olivier L. Modulation of the Golgi apparatus in stimulated and nonstimulated prolactin cells of female rats. Anat Rec. 1993;235:353–362. doi: 10.1002/ar.1092350304. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Ovtracht L, Kepes F. Three-dimensional structure of tubular networks, presumably Golgi in nature, in various yeast strains: a comparative study. Anat Rec. 1995;243:283–293. doi: 10.1002/ar.1092430302. [DOI] [PubMed] [Google Scholar]
- Romero N, Dumur CI, Martinez H, García IA, Monetta P, Slavin I, Sampieri L, Koritschoner N, Mironov AA, De Matteis MA, Alvarez C. Rab1b overexpression modifies Golgi size and gene expression in HeLa cells and modulates the thyrotrophin response in thyroid cells in culture. Mol Biol Cell. 2013;24:617–632. doi: 10.1091/mbc.E12-07-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Berger EG. Immunocytochemical localization of galactosyltransferase in HeLa cells: codistribution with thia-mine pyrophosphatase in trans—Golgi cisternae. J Cell Biol. 1982;93:223–229. doi: 10.1083/jcb.93.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Taatjes DJ, Lucocq JM, Weinstein J, Paulson JC. Demonstration of an extensive trans-tubular network continuous with the Golgi apparatus stack that may function in glycosylation. Cell. 1985;43:287–295. doi: 10.1016/0092-8674(85)90034-0. [DOI] [PubMed] [Google Scholar]
- Roth J, Taatjes DJ, Warhol MJ. Prevention of non-specific interactions of gold-labeled reagents on tissue sections. Histochemistry. 1989;92:47–56. doi: 10.1007/BF00495015. [DOI] [PubMed] [Google Scholar]
- Roth J, Zuber C, Park S, Jang I, Lee Y, Kysela KG, Le Fourn V, Santimaria R, Guhl B, Cho JW. Protein N-glycosylation, protein folding, and protein quality control. Mol Cells. 2010;30:497–506. doi: 10.1007/s10059-010-0159-z. [DOI] [PubMed] [Google Scholar]
- Sandvig K, Skotland T, van Deurs B, Klokk TI. Retrograde transport of protein toxins through the Golgi apparatus. Histochem Cell Biol. 2013;140:317–326. doi: 10.1007/s00418-013-1111-z. [DOI] [PubMed] [Google Scholar]
- Sciaky N, Presley J, Smith C, Zaal KJM, Cole N, Moreira JE, Terasaki M, Siggia E, Lippincott-Schwartz J. Golgi tubule traffic and the effects of Brefeldin A visualized in living cells. J Cell Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelentag WK, Li WP, Schmitz SF, Metzger U, Aeberhard P, Heitz PU, Roth J. Prognostic value of β1,6-branched oligosaccharides in human colorectal carcinoma. Cancer Res. 1998;58:5559–5564. [PubMed] [Google Scholar]
- Sengupta D, Linstedt AD. Control of organelle size: the Golgi complex. Ann Rev Cell Dev Biol. 2011;27:57–77. doi: 10.1146/annurev-cellbio-100109-104003. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Truschel S, Bachert C, Linstedt AD. Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J Cell Biol. 2009;186:41–55. doi: 10.1083/jcb.200902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoreibah M, Perng GS, Adler B, Weinstein J, Basu R, Cupples R, Wen DZ, Browne JK, Buckhaults P, Fregien N, Pierce M. Isolation, characterization, and expression of a cDNA encoding N-acetylglucosaminyltransferase V. J Biol Chem. 1993;268:15381–15385. [PubMed] [Google Scholar]
- Shorter J, Warren G. Golgi architecture and inheritance. Ann Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Ioffe E. Glycosyltransferase mutants: key to new insights in glycobiology. FASEB J. 1995;9:1436–1444. doi: 10.1096/fasebj.9.14.7589985. [DOI] [PubMed] [Google Scholar]
- Stanley P, Sudo T. Microheterogeneity among carbohydrate structure at the cell surface may be important in recognition phenomena. Cell. 1981;23:763–769. doi: 10.1016/0092-8674(81)90440-2. [DOI] [PubMed] [Google Scholar]
- Stanley P, Caillibot V, Siminovitch L. Selection and characterization of eight phenotypically distinct lines of lectin-resistant Chinese hamster ovary cell. Cell. 1975;6:121–128. doi: 10.1016/0092-8674(75)90002-1. [DOI] [PubMed] [Google Scholar]
- Stanley P, Vivona G, Atkinson P. Carbohydrate heterogeneity of vesicular stomatitis virus G glycoprotein allows localization of the defect in a glycosylation mutant of CHO cells. Arch Biochem Biophys. 1982;219:128–139. doi: 10.1016/0003-9861(82)90141-2. [DOI] [PubMed] [Google Scholar]
- Stieber A, Chen YJ, Wei S, Mourelatos Z, Gonatas J, Okamoto K, Gonatas NK. The fragmented neuronal Golgi apparatus in amyotrophic lateral sclerosis includes the trans-Golgi-net-work: functional implications. Acta Neuropathol. 1998;95:245–253. doi: 10.1007/s004010050794. [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Tillmann K, Millarte V, Farhan H. Regulation of traffic and organelle architecture of the ER-Golgi interface by signal transduction. Histochem Cell Biol. 2013;140:297–306. doi: 10.1007/s00418-013-1118-5. [DOI] [PubMed] [Google Scholar]
- Tomas M, Marin MP, Martinez-Alonso E, Esteban-Pretel G, DiazRuiz A, Vazquez-Martinez R, Malagon MM, Renau-Piqueras J, Martinez-Menarguez JA. Alcohol induces Golgi fragmentation in differentiated PC12 cells by deregulating Rab1-dependent ER-to-Golgi transport. Histochem Cell Biol. 2012;138:489–501. doi: 10.1007/s00418-012-0970-z. [DOI] [PubMed] [Google Scholar]
- Uemura T, Nakano A. Plant TGNs: dynamics and physiological functions. Histochem Cell Biol. 2013;140:341–345. doi: 10.1007/s00418-013-1116-7. [DOI] [PubMed] [Google Scholar]
- Velasco A, Hendricks L, Moremen KW, Tulsiani DRP, Touster O, Farquhar MG. Cell type-dependent variations in the subcellular distribution of alpha-mannosidase-I and alpha-mannosidase-II. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. Transport through the Golgi in Trypanosoma brucei. Histochem Cell Biol. 2013;140:235–238. doi: 10.1007/s00418-013-1112-y. [DOI] [PubMed] [Google Scholar]
- Weibel E. Stereological methods 1 Practical methods for biological morphometry. Academic Press; New York: 1979. [Google Scholar]
- Weinstein J, Sundaram S, Wang XH, Delgado D, Basu R, Stanley P. A point mutation causes mistargeting of Golgi GlcNAcTV in the Lec4A Chinese hamster ovary glycosylation mutant. J Biol Chem. 1996;271:27462–27469. doi: 10.1074/jbc.271.44.27462. [DOI] [PubMed] [Google Scholar]
- Willett R, Ungar D, Lupashin V. The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem Cell Biol. 2013;140:271–283. doi: 10.1007/s00418-013-1117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BS, Nuoffer C, Meinkoth JL, Mccaffery M, Feramisco JR, Balch WE, Farquhar MG. A rab1 mutant affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J Cell Biol. 1994;125:557–571. doi: 10.1083/jcb.125.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Puthenveedu MA, Linstedt AD. Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev Cell. 2012;23:153–165. doi: 10.1016/j.devcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rosenwald A, Willingham M, Skuntz S, Clark J, Kahn R. Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J Cell Biol. 1994;124:289–300. doi: 10.1083/jcb.124.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Kaverina I. Golgi as an MTOC: making microtubules for its own good. Histochem Cell Biol. 2013;140:361–367. doi: 10.1007/s00418-013-1119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C, Roth J. N-Glycosylation. In: Gabius H, editor. The sugar code. Wiley-VCH; Weinheim: 2009. pp. 87–110. [Google Scholar]
- Zuber C, Li WP, Roth J. Blot analysis with lectins for the evaluation of glycoproteins in cultured cells and tissues. In: Rhodes JM, Milton JD, editors. Methods in molecular biology series. Humana Press; Totowa: 1998. pp. 159–166. [DOI] [PubMed] [Google Scholar]


