DNA double-strand breaks and chromosomal aberrations after treatment with N-alkylating agents likely arise as a result of replication fork collision with single-strand breaks generated during base excision repair.
Abstract
Exposures that methylate DNA potently induce DNA double-strand breaks (DSBs) and chromosomal aberrations, which are thought to arise when damaged bases block DNA replication. Here, we demonstrate that DNA methylation damage causes DSB formation when replication interferes with base excision repair (BER), the predominant pathway for repairing methylated bases. We show that cells defective in the N-methylpurine DNA glycosylase, which fail to remove N-methylpurines from DNA and do not initiate BER, display strongly reduced levels of methylation-induced DSBs and chromosomal aberrations compared with wild-type cells. Also, cells unable to generate single-strand breaks (SSBs) at apurinic/apyrimidinic sites do not form DSBs immediately after methylation damage. In contrast, cells deficient in x-ray cross-complementing protein 1, DNA polymerase β, or poly (ADP-ribose) polymerase 1 activity, all of which fail to seal SSBs induced at apurinic/apyrimidinic sites, exhibit strongly elevated levels of methylation-induced DSBs and chromosomal aberrations. We propose that DSBs and chromosomal aberrations after treatment with N-alkylators arise when replication forks collide with SSBs generated during BER.
Introduction
Most chemicals exert their genotoxic effects when the primary DNA damage interferes with replication (Evans and Scott, 1969). This is true for chemicals inducing bulky lesions or DNA cross-links, for simple methylating agents, and also for UV light (Bender et al., 1974; Kaina, 1998). Methylating agents are powerful genotoxicants that induce chromosomal breaks and translocations (Pimpinelli et al., 1977; Natarajan et al., 1983). As chromosomal aberrations are caused by DNA double-strand breaks (DSBs; Natarajan and Obe, 1984) and methylating agents do not induce DSBs, per se, it is thought that DSBs arise during S phase when replication forks encounter methylated bases. Thus, the current paradigm states that critical minor lesions such as N3-methyladenine (N3-MeA) block replication, which can lead to DSB formation by replication fork collapse and the generation of chromosomal aberrations (Kaina, 2004). Methyl methanesulfonate (MMS) is a model compound for methylating agents, which produces a wide spectrum of DNA methyl adducts. Methylations of nitrogens in the DNA are the key primary lesions, with N7-methylguanine (N7-MeG) comprising 82% of all DNA adducts, followed by N3-MeA with 11% and N3-methylguanine (N3-MeG) with 1.2% (Beranek, 1990; Wyatt and Pittman, 2006).
The most important pathway for the repair of N-methylated bases is base excision repair (BER; Dianov and Hübscher, 2013; Parsons and Dianov, 2013). BER is initiated by the N-methylpurine DNA glycosylase (MPG; also known as AAG), which recognizes and removes N7-MeG, N3-MeA, and N3-MeG (O’Brien and Ellenberger, 2004). The resulting apurinic (AP) site is cleaved by apurinic endonuclease 1 (APE1), leading to the formation of a single-strand break (SSB; Bennett et al., 1997; Erzberger et al., 1998). The next step of BER is performed by DNA polymerase β (Polβ), which inserts a nucleotide and creates 3′ and 5′ ends that can be ligated by DNA ligase III (Srivastava et al., 1998). X-ray cross-complementing protein 1 (XRCC1) interacts with nearly all important BER factors and is believed to exert a regulatory or mediator function (Vidal et al., 2001; Whitehouse et al., 2001; Caldecott, 2003, 2008; Campalans et al., 2005; Nazarkina et al., 2007; Chou et al., 2008). XRCC1- and Polβ-deficient cells have been established that fail to complete BER and accumulate unrepaired SSBs at BER intermediates (Thompson et al., 1982; Zdzienicka et al., 1992; Pascucci et al., 2005; Horton et al., 2008). Poly (ADP-ribose) polymerase 1 (PARP1) interacts with XRCC1 and Polβ, and PARP1-deficient cells also show unrepaired SSBs after MMS treatment (Caldecott et al., 1996, Masson et al., 1998, Trucco et al., 1998).
Here, we show that the formation of DSBs after MMS requires, in addition to S-phase progression, the induction of SSBs during BER. First, cells deficient in XRCC1, Polβ, or PARP1 activity, which all accumulate SSBs during BER, show strongly elevated levels of γH2AX foci compared with wild-type (WT) cells. Second, and most importantly, the formation of γH2AX foci immediately after MMS treatment is essentially abolished in MPG-deficient cells and in cells unable to cleave AP sites, which both do not induce SSBs during BER. We further demonstrate that XRCC1-deficient cells exhibit very high levels of chromosomal aberrations (chromatid breaks and translocations) and sister chromatid exchanges (SCEs) and show an impressive delay in S-phase progression. In contrast, MPG-deficient cells exhibit markedly reduced levels of chromosomal aberrations, although SCE levels and the degree of S-phase delay were similar to WT cells. This suggests that the formation of chromosomal aberrations after DNA methylation damage involves DSB induction at BER intermediates, most likely at SSBs, whereas SCE formation and delayed S-phase progression do not necessarily require BER and DSB induction. We propose that BER represents a double-edged sword; it is essential for removing potentially genotoxic base lesions, but also harbors the risk of forming DNA breakage and chromosomal aberrations when it collides with replication.
Results
Disrupted repair of MMS-induced base damage leads to DSBs in S phase
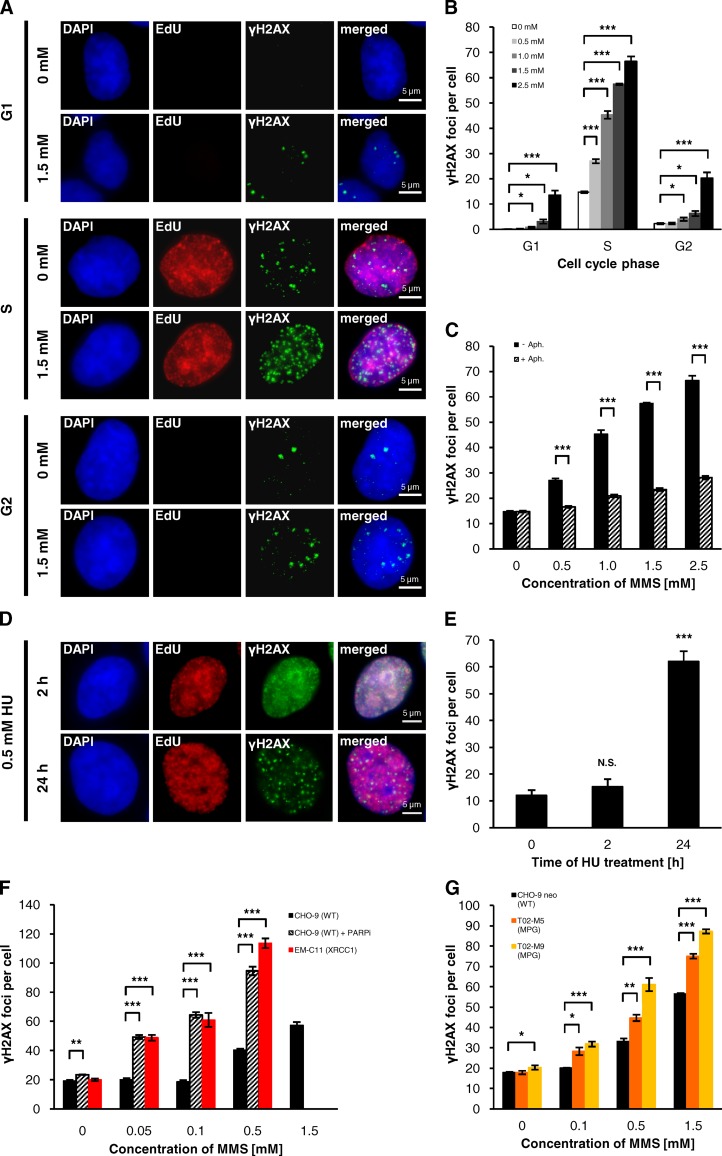
We analyzed the cell cycle dependence of DSB formation after the model DNA alkylator MMS by combining the technique of γH2AX immunofluorescence analysis with labeling of replicating A549 cells using the thymidine analogue 5-ethynyl-2′-desoxyuridine (EdU). EdU-positive S-phase cells exhibited high numbers of γH2AX foci that increased with increasing MMS concentrations. In contrast, γH2AX foci in EdU-negative G1- and G2-phase cells were only observed after high MMS concentrations (Fig. 1, A and B). We then investigated if foci formation depends on replication and treated A549 cells with a low concentration of the DNA polymerase inhibitor aphidicolin before adding MMS to the medium (Groth et al., 2012). Under these replication-inhibiting conditions, γH2AX foci formation was strongly diminished (Fig. 1 C). Thus, γH2AX foci after MMS arise primarily in S-phase cells in a manner dependent on ongoing replication.
Figure 1.
Disrupted repair of MMS-induced base damage leads to DSBs in S phase. (A and B) γH2AX foci in MMS-treated A549 cells. Cells were treated with 10 µM EdU and different concentrations of MMS for 1 h. 15 min after removing EdU/MMS, cells were fixed and stained against γH2AX and EdU. γH2AX foci were assessed in EdU-positive S-phase cells and EdU-negative G1- and G2-phase cells (B, ±SEM from three experiments); G1- and G2-phase cells were distinguished by quantifying the DAPI signal using the Metafer Scanning System (Metasystems). Representative images for cells treated with 0 or 1.5 mM MMS are shown in A. (C) γH2AX foci under replication-inhibiting conditions. After EdU labeling for 1 h, A549 cells were treated with 1 µM aphidicolin (Aph.) for 1 h before MMS was added for an additional hour. 15 min after removing MMS, cells were fixed and stained against γH2AX and EdU. γH2AX foci were assessed in EdU-positive S-phase cells (±SEM from three experiments). (D and E) γH2AX foci in HU-treated A549 cells. After labeling with 10 µM EdU for 1 h, cells were treated with 0.5 mM HU for the indicated times, fixed, and stained against γH2AX and EdU. γH2AX foci were assessed in EdU-positive S-phase cells (E, ±SEM from at least three experiments). For statistical analysis, γH2AX foci after HU treatment were compared with the untreated control. Representative images of the γH2AX signals after 2 and 24 h of HU treatment are shown in D. (F) γH2AX foci in CHO-9 WT cells treated with a specific PARP inhibitor (PARPi) and in XRCC1-deficient EM-C11 cells. Cells were treated with 10 µM EdU and different concentrations of MMS for 1 h. 15 min after removing EdU/MMS, cells were fixed and stained against γH2AX and EdU. Where indicated, cells were treated with 15 µM PARPi 1 h before EdU and MMS were added and PARPi was present until fixation. γH2AX foci were assessed in EdU-positive S-phase cells (±SEM from 3–4 experiments). In CHO-9 cells with PARPi and in EM-C11 cells, γH2AX foci numbers after 1.5 mM MMS were too high for exact enumeration. (G) γH2AX foci in CHO-9 neo (WT) and two MPG-overexpressing cell lines (T02-M5 and T02-M9) that show a twofold or 12-fold elevated MPG activity compared with WT cells. Cells were treated and analyzed as in F (±SEM from three experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We then wished to investigate whether MMS-induced γH2AX foci represent DSBs or other structures such as stalled replication forks and first analyzed ATR-deficient cells. Single-stranded regions at stalled replication forks activate ATR, whereas ATM and DNA-PK are activated by DSBs (Andegeko et al., 2001; Zou and Elledge, 2003; Löbrich et al., 2010). Thus, γH2AX foci in ATR-deficient cells do not arise from stalled replication forks. Significantly, γH2AX foci induction was identical in WT and ATR-deficient cells and was abolished when ATR-deficient cells were treated with specific inhibitors of ATM and DNA-PK (Fig. S1, A and B). We also analyzed ATM activation at MMS-induced lesions by Western blotting and observed phosphorylation of the ATM substrates KAP1 and Chk2 (Fig. S1 C). We next performed experiments with hydroxyurea (HU), an agent that depletes the cellular nucleotide pool and blocks cells within S phase (Bianchi et al., 1986). After HU treatment times of a few hours, replication forks are transiently stalled, and replication is resumed upon HU removal. If HU treatment and replication stalling is extended over longer time periods (24 h), one-ended DSBs are generated by the action of the endonuclease complex Mus81–Eme1 (Saintigny et al., 2001; Hanada et al., 2007; Petermann et al., 2010). After 2 h of HU, we observed a pan-nuclear γH2AX signal but no distinct γH2AX foci in S-phase cells (Fig. 1, D and E). In contrast, after 24 h, high numbers of γH2AX foci were observed (Fig. 1, D and E), which were significantly reduced after depletion of Mus81 by siRNA (Fig. S1 D). Mus81 depletion had no effect on the level of γH2AX foci after MMS treatment, indicating that MMS-induced foci are not generated by a Mus81-mediated process (Fig. S1 D). Collectively, these data demonstrate that γH2AX foci observed in S-phase cells at early times after MMS treatment mark DSBs.
We next analyzed the formation of γH2AX foci in XRCC1-deficient EM-C11 cells and CHO-9 WT cells treated with a specific PARP inhibitor (PARPi), both of which display a defect in repairing SSBs during BER. Low concentrations of 0.05 and 0.1 mM MMS induced similar levels of γH2AX foci in EM-C11 and PARPi-treated CHO-9 cells as 0.5 and 1.5 mM MMS in WT cells (Fig. 1 F), suggesting that SSBs at disrupted BER sites cause DSBs. We then studied T02-M5 and T02-M9 cells, which overexpress MPG after transfection with an MPG expression vector. In these cells, the initial step of BER is up-regulated, leading to elevated SSB frequencies (Ibeanu et al., 1992; Coquerelle et al., 1995). T02-M5 and T02-M9 cells showed higher levels of MMS-induced γH2AX foci than control cells (Fig. 1 G). The increase was higher in T02-M9 than in T02-M5 cells, consistent with their higher MPG level (12-fold vs. twofold elevated compared with WT cells; Ibeanu et al., 1992). XRCC1-deficient EM-C11 and MPG-overexpressing T02-M5 and T02-M9 G2-phase cells also showed higher γH2AX foci numbers than their corresponding WT cells in G2 (Fig. S2, A and B), consistent with a model that two disrupted or imbalanced BER processes close to each other on opposite DNA strands can cause DSBs (Coquerelle et al., 1995; Ma et al., 2011). Together, these findings demonstrate that imbalanced BER causes DSB formation in S phase and, to a minor extent, in G2 phase.
The formation of DSBs after MMS requires MPG activity
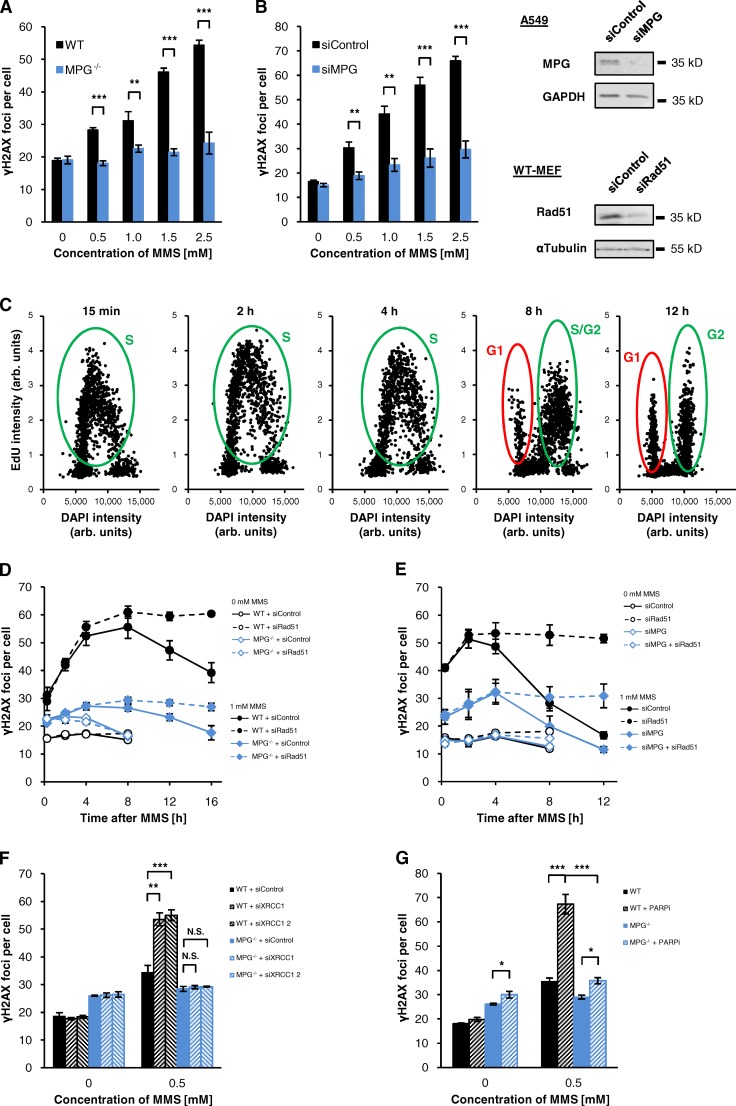
Although the findings above establish that DSBs are formed when disrupted or imbalanced BER processes interfere with replication, they do not reveal the process by which DSBs are generated in WT cells with normal, unperturbed BER activity. To address this question, we analyzed MPG-defective cells, which do not remove MMS-induced N-methylpurines from DNA and, therefore, do not execute BER. Strikingly, MPG−/− MEFs showed almost no γH2AX foci induction after MMS in S phase (Fig. 2 A) or G2 phase (Fig. S2 C). The same result was obtained after MPG depletion by siRNA in S-phase (Fig. 2 B) and G2-phase (Fig. S2 D) A549 cells and was confirmed with several independent MPG siRNAs (Fig. S3 A). We then asked if γH2AX foci arise slowly in MPG-deficient cells and analyzed the formation and repair of γH2AX foci for up to 16 h after MMS treatment (cell cycle progression of EdU-positive cells after MMS treatment is monitored in Fig. 2 C). We had previously established that γH2AX foci kinetics after MMS show a biphasic course with an increase in foci number during the first hours after MMS removal followed by a period with continuously decreasing foci levels (Nikolova et al., 2010). Whereas this time course was clearly observed in WT cells, only a small increase in foci number was detected in MPG−/− MEFs and MPG-depleted A549 cells, and foci levels clearly stayed below those of WT cells during the entire post-incubation period (Fig. 2, D and E). The lower level of MMS-induced DSBs in MPG−/− MEFs was confirmed by the neutral comet assay (Fig. S3 B). Because repair of MMS-induced DSBs requires homologous recombination (HR; Nikolova et al., 2010), we analyzed γH2AX foci after depletion of Rad51 to determine the total number of DSBs formed in WT and MPG-deficient cells. After Rad51 depletion, both WT and MPG-deficient cells failed to repair γH2AX foci, and MPG-deficient cells exhibited substantially fewer γH2AX foci than WT cells (Fig. 2, D and E). The same was observed after treatment with a specific Rad51 inhibitor (Fig. S3, D and E).
Figure 2.
The formation of DSBs after MMS requires MPG activity. (A) γH2AX foci in WT and MPG−/− MEFs. Cells were treated with 10 µM EdU and different concentrations of MMS for 1 h. 15 min after removing EdU/MMS, cells were fixed and stained against γH2AX and EdU. γH2AX foci were assessed in EdU-positive S-phase cells (±SEM from three experiments). (B) γH2AX foci in A549 cells after siRNA for 72 h. Cells were treated and analyzed as in A (±SEM from three experiments). Efficient siRNA depletion was confirmed by Western blotting; GAPDH was used as a loading control. (C) Cell cycle distribution of A549 cells treated with siControl after 1 mM MMS monitored by measuring the EdU and the DAPI signal with the Metafer Scanning System (Metasystems). The data presented are from a single representative experiment out of three repeats; in each blot the distribution of 2,000 cells is shown. In exponentially growing A549 cells, a fraction of ∼40% was EdU positive; this fraction was neither affected by transfection with siRNA nor by the MMS treatment. In WT and MPG−/− MEFs the fraction of EdU-positive cells was 20–25%. (D) γH2AX foci kinetics in WT and MPG−/− MEFs. Cells were treated with siRNA and, 48 h later, treated with EdU and 1 mM MMS for 1 h. After removing EdU/MMS, cells were fixed at the indicated times and γH2AX foci were analyzed in EdU-positive S/G2 cells (C, green ovals). EdU-positive G1 cells (red ovals) were excluded from analysis (±SEM from three experiments). Efficient depletion of Rad51 was confirmed by Western blotting (B). α-Tubulin was used as a loading control. (E) γH2AX foci kinetics in siRNA-treated A549 cells. Cells were treated with siRNA and, 48 h later, analyzed as in D (±SEM from three experiments). Western blots for Rad51 depletion and for co-depletion of Rad51 and MPG are shown in Fig. S3 C. (F) γH2AX foci in XRCC1-depleted WT and MPG−/− MEFs. 48 h after transfection with siRNA, cells were treated with EdU and 0.5 mM MMS for 1 h. Cells were fixed 4 h after removing EdU/MMS and γH2AX foci were analyzed in EdU-positive S-phase cells (±SEM from four experiments). Efficient depletion of XRCC1 was confirmed by Western blotting (Fig. S3 C). (G) γH2AX foci in WT and MPG−/− MEFs treated with PARPi. Cells were treated with 15 µM PARPi and 1 h later EdU and 0.5 mM MMS were added for an additional hour. 4 h after removing the drugs, the cells were fixed and γH2AX foci were analyzed in EdU-positive S-phase cells (±SEM from three experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We then analyzed the formation of γH2AX foci in XRCC1-depleted MPG−/− MEFs. Although depletion of XRCC1 increased foci numbers in WT MEFs treated with MMS, this was not observed in MPG−/− MEFs (Fig. 2 F). Similar results were obtained for WT and MPG−/− MEFs treated with PARPi (Fig. 2 G), indicating that a XRCC1 or PARP1 deficiency has no impact in the absence of MPG. Together, these results establish that DSBs after MMS treatment arise when normal, unperturbed BER processes interfere with replication.
MMS-induced S-phase delay involves distinct processes in WT and XRCC1-deficient cells
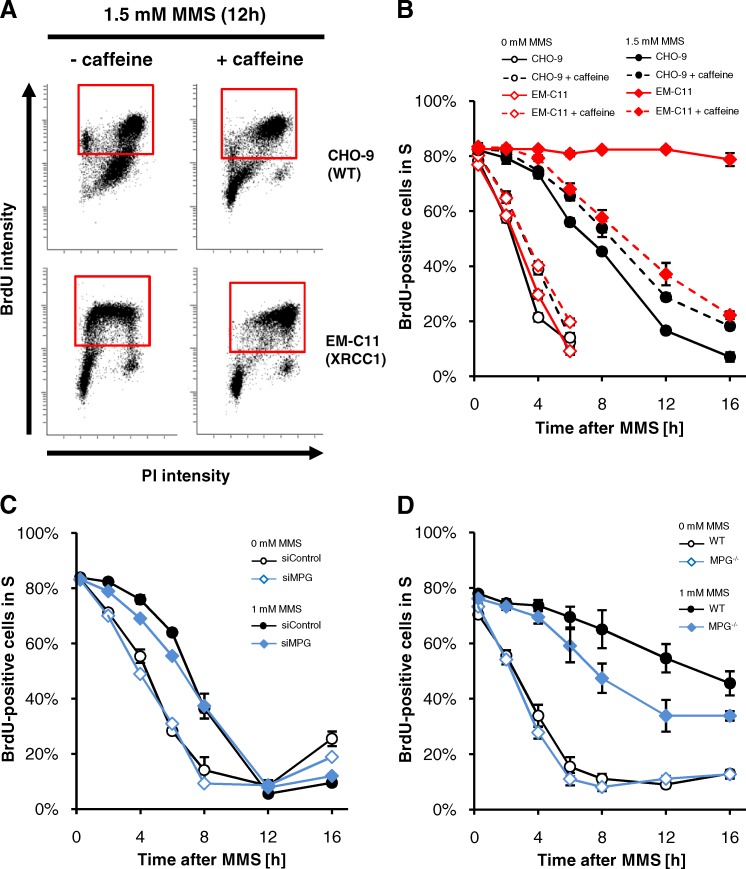
A characteristic effect of MMS treatment is delayed S-phase progression. However, it is unclear if the delay represents an active checkpoint-mediated process (Brem et al., 2008) or rather a passive process caused by replication fork blockage at MMS-induced base lesions (Groth et al., 2010). We used flow cytometry to compare the cell cycle progression of XRCC1-deficient EM-C11 and CHO-9 WT cells. We treated cells for 1 h with 1.5 mM MMS and BrdU to label S-phase cells and followed the progression of these cells through the cell cycle over 16 h (Fig. 3 A). Consistent with previous findings (Groth et al., 2010), we observed a significant S-phase delay in WT cells after 1.5 mM MMS. The delay was much stronger in EM-C11 cells (Fig. 3 B), demonstrating that persisting BER intermediates can induce a strong cell cycle arrest. To investigate if the S-phase delay represents an active checkpoint response, we treated cells with 5 mM of caffeine before MMS, which inhibits the checkpoint signaling kinases ATM and ATR. Consistent with previous studies (Groth et al., 2010), we detected a small delay in S-phase progression by caffeine alone. MMS-treated CHO-9 cells were unaffected by caffeine treatment, indicating that the MMS-induced S-phase delay in WT cells is largely independent of checkpoint signaling. In contrast, caffeine exerted a strong effect on MMS-treated EM-C11 cells, which progressed through S-phase similar to WT cells treated with MMS and caffeine (Fig. 3 B). Similar results were obtained after treatment with 0.1 mM MMS (Fig. S4 A). The observation that the pronounced S-phase delay in XRCC1-deficient cells is abolished after ATM/ATR inhibition indicates that it represents an active checkpoint response, likely arising from the high amount of induced DSBs. We also analyzed the cell cycle progression of MPG-deficient cells, which did not show significant DSB induction upon MMS treatment. MPG-depleted A549 cells showed an S-phase delay similar to WT cells (Fig. 3 C), and MPG−/− MEFs were only slightly less delayed compared with WT MEFs (Fig. 3 D). In conclusion, the strong checkpoint response in XRCC1-deficient cells is likely caused by the high amount of DSBs, whereas the comparatively lower level of DSBs formed in WT cells does not seem to contribute significantly to their S-phase delay.
Figure 3.
MMS-induced S-phase delay involves distinct processes in WT and XRCC1-deficient cells. (A and B) Cell cycle progression in CHO-9 (WT) and EM-C11 (XRCC1-deficient) cells analyzed by FACS. Cells were treated with 10 µM BrdU and 1.5 mM MMS for 1 h and fixed at the indicated times after removing BrdU/MMS. 5 mM caffeine was added 1 h before MMS addition and was present until fixation. Representative FACS blots are shown in A. Red rectangles mark BrdU-positive cells. The frequency of BrdU-positive cells in S phase was calculated (B, ±SEM from three experiments). (C) S-phase progression in A549 cells treated with siRNA for 72 h. Cells were treated with 10 µM BrdU and 1 mM MMS for 1 h and fixed at the indicated times after removing BrdU/MMS (±SEM from three experiments). (D) S-phase progression in WT and MPG−/− MEFs. Cells were treated and analyzed as in C (±SEM from three experiments).
SCEs after MMS arise from BER-dependent and -independent processes
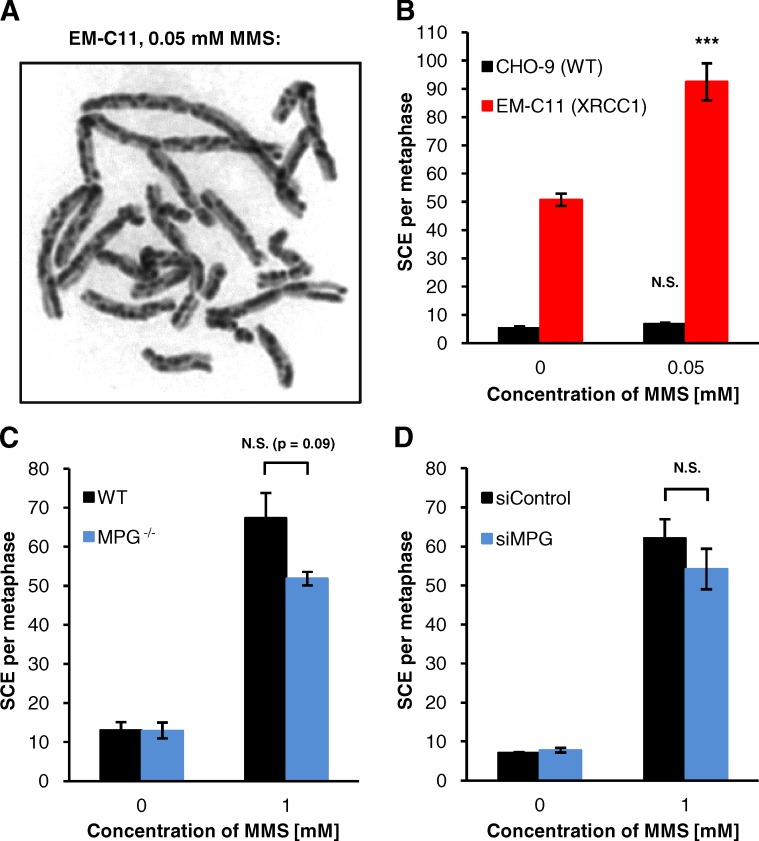
DNA base methylation by MMS induces SCEs, but the underlying process is unclear (Kaina, 2004). We have previously shown that DSBs induced by MMS are repaired via HR (Nikolova et al., 2010), a process that might result in SCEs (Natarajan et al., 1985). Another model is that MMS-induced methylated bases block replication forks (Larson et al., 1985; Engelward et al., 1998; Groth et al., 2010), which are then reactivated by HR (Petermann et al., 2010) and form SCEs. To characterize SCE formation after MMS, we first analyzed SCEs in CHO-9 WT and XRCC1-deficient EM-C11 cells after 0.05 mM MMS. This concentration of MMS, which induced a significant number of DSBs in EM-C11 but not in CHO-9 cells (see Fig. 1 F), induced only a few SCEs in CHO-9 but many SCEs in EM-C11 cells (Fig. 4, A and B). Notably, even untreated EM-C11 cells showed high SCE levels, which likely result from the incubation with BrdU, an essential technical step to visualize SCEs (Carrano et al., 1986; Caldecott, 2003). These data suggest that SCEs induced by MMS in EM-C11 cells arise from the high level of SSBs. We next analyzed SCEs in WT and MPG-deficient cells after 0.5 and 1 mM MMS. Interestingly, MPG−/− MEFs and MPG-depleted A549 cells showed SCE levels only slightly lower than WT cells (Fig. 4, C and D; and Fig. S4 B), although their γH2AX foci levels were dramatically reduced (see Fig. 2). We conclude from the similar (or only slightly different) SCE levels in WT and MPG−/− MEFs that SCE formation does not necessarily require BER, and from the high SCE level in XRCC1-deficient cells that SSBs induced during BER can also trigger SCE formation.
Figure 4.
SCEs after MMS arise from BER-dependent and -independent processes. (A and B) SCEs in CHO-9 (WT) and EM-C11 (XRCC1-deficient) cells. After BrdU labeling, cells were treated with 0.05 mM MMS for 1 h. Caffeine and colcemid were added at 12 h after MMS and samples were harvested 2 h later (B; ±SEM from 3–4 experiments). A representative image of a mitotic EM-C11 cell after 0.05 mM MMS is shown in A. For statistical analysis, SCEs after MMS were compared with the untreated controls. (C) SCEs in WT and MPG−/− MEFs. After BrdU labeling, cells were treated with 1 mM MMS for 1 h. Caffeine and colcemid were added at 24 h after MMS and samples were harvested 2 h later (±SEM from 3–5 experiments). (D) SCEs in A549 cells after siRNA for 72 h. After BrdU labeling, cells were treated with 1 mM MMS for 1 h. Caffeine and colcemid were added at 20 h after MMS and samples were harvested 2 h later (±SEM from three experiments). The analysis times after MMS treatment (24–26 h for MEFs and 20–22 h for A549 cells) were chosen such that cells in early S phase or late G1 phase at the time of MMS treatment had progressed into mitosis. Thus, this approach analyzed cells that had traversed a full S phase. ***, P < 0.001.
DSBs formed after MMS at BER intermediates cause chromosomal aberrations
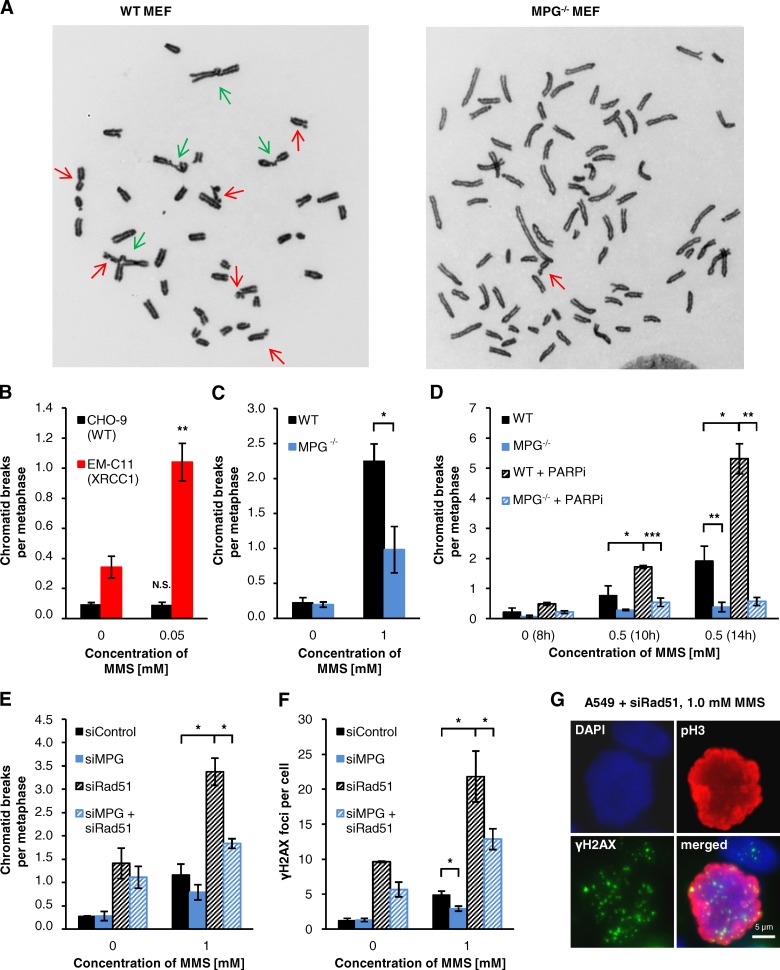
We have shown that DSBs after MMS arise in a replication-dependent manner when replication forks encounter BER intermediates. We next asked whether these DSBs cause chromosome breakage, examples of which are shown in Fig. 5 A. After 0.05 mM MMS, XRCC1-deficient EM-C11 cells showed a substantial level of chromatid breaks, whereas no significant break induction was detected in WT cells at this low concentration (Fig. 5 B). This indicates that chromatid breaks arise from unrepaired DSBs, which are much more frequent in EM-C11 than in CHO-9 cells (see Fig. 1 F). Consistent with the lower level of γH2AX foci, we detected a decreased frequency of chromatid breaks in MPG−/− MEFs and MPG-depleted A549 cells treated with 0.5 or 1 mM MMS (Fig. 5, C–E), demonstrating that the initiation of BER is required for chromosome breakage after MMS. To further substantiate this finding, we assessed chromatid breaks in MPG−/− MEFs treated with PARPi. Although WT MEFs showed highly elevated break levels after treatment with MMS and PARPi reminiscent of XRCC1-deficient cells, PARPi had only a modest impact in MPG−/− MEFs (Fig. 5 D). We further analyzed Rad51-depleted cells showing elevated levels of MMS-induced chromatid breaks that were substantially rescued by co-depletion of MPG (Fig. 5 E). We finally analyzed γH2AX foci after MMS in mitotic cells and observed results similar to chromatid breaks (Fig. 5, F and G).
Figure 5.
DSBs formed after MMS at BER intermediates cause chromatid breaks. (A) Representative images of MMS-induced chromosomal aberrations in PARPi-treated WT and MPG−/− MEFs (14 h after 0.5 mM MMS). Chromatid breaks and chromatid-type translocations are marked by red and green arrows, respectively. Chromatid breaks were analyzed in (B) CHO-9 (WT) and EM-C11 (XRCC1-deficient) cells, (C and D) WT and MPG−/− MEFs with or without PARPi, and (E) siRNA-treated A549 cells. Data in B, C, and E were obtained from the experiments performed for Fig. 4, B, C, and D. For D, 15 µM PARPi was added 1 h before MMS and was only present until the end of the MMS treatment. Therefore, PARPi had only an impact on cells in S phase. The analysis times for D (6–8 h for untreated cells and 8–10 h or 12–14 h for MMS-treated cells) were chosen such that cells in S phase at the time of MMS treatment had progressed into mitosis. (F and G) γH2AX foci in mitotic A549 cells after siRNA for 72 h. Cells were treated with 1 mM MMS for 1 h and caffeine was added at 20 h after MMS. 2 h later, samples were fixed and stained against γH2AX and pH3. γH2AX foci were assessed in mitotic cells (F); a representative image for foci in mitotic cells is shown in G (±SEM from 3–5 experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
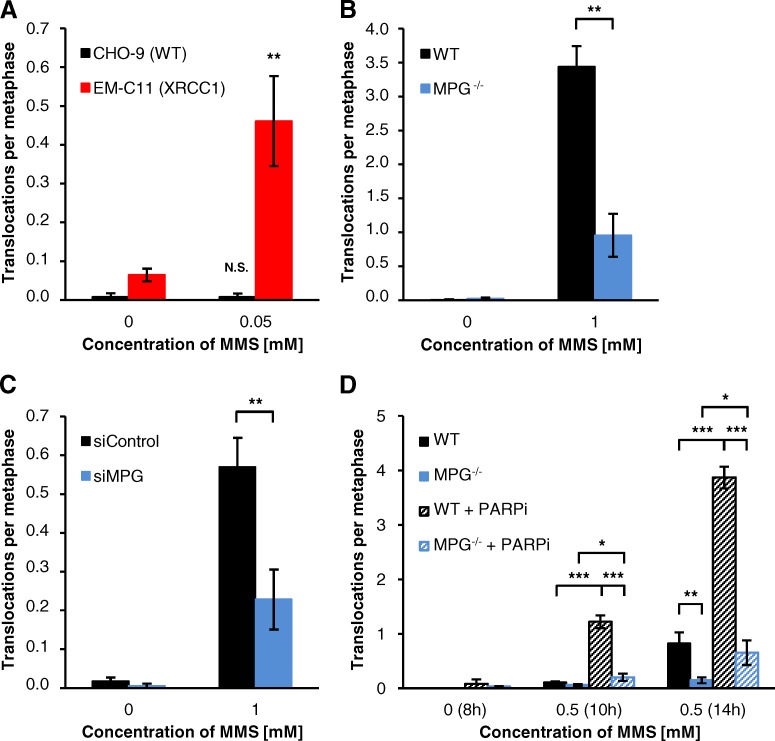
Because unrepaired DSBs can illegitimately rejoin to form chromosomal rearrangements, a hallmark of induced clastogenicity (Boei et al., 2002), we finally analyzed the occurrence of chromatid-type translocations in the first mitosis after MMS treatment (examples shown in Fig. 5 A). Similar to chromatid breaks and γH2AX foci in mitotic cells, we observed substantially elevated levels of MMS-induced translocations in XRCC1-deficient EM-C11 cells (Fig. 6 A) and decreased levels of translocations in MPG−/− MEFs and MPG-depleted A549 compared with WT cells (Fig. 6, B–D). WT MEFs treated with PARPi showed highly elevated translocation levels reminiscent of XRCC1-deficient cells, but PARPi had only a modest impact in MPG−/− MEFs (Fig. 6 D). Overall, our data demonstrate that the formation of chromatid breaks and translocations after MMS depends on DSBs that arise in a BER- and replication-dependent manner.
Figure 6.
DSBs formed after MMS at BER intermediates cause translocations. Chromatid-type translocations in (A) CHO-9 (WT) and EM-C11 (XRCC1-deficient) cells, (B) WT and MPG−/− MEFs, (C) siRNA-treated A549 cells, and (D) PARPi-treated WT and MPG−/− MEFs. Data in panels A–C were obtained from the experiments performed for Fig. 4, B–D. Data in D were obtained from the experiments performed for Fig. 5 D (±SEM from 3–5 experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DSB formation after MMS requires SSB induction
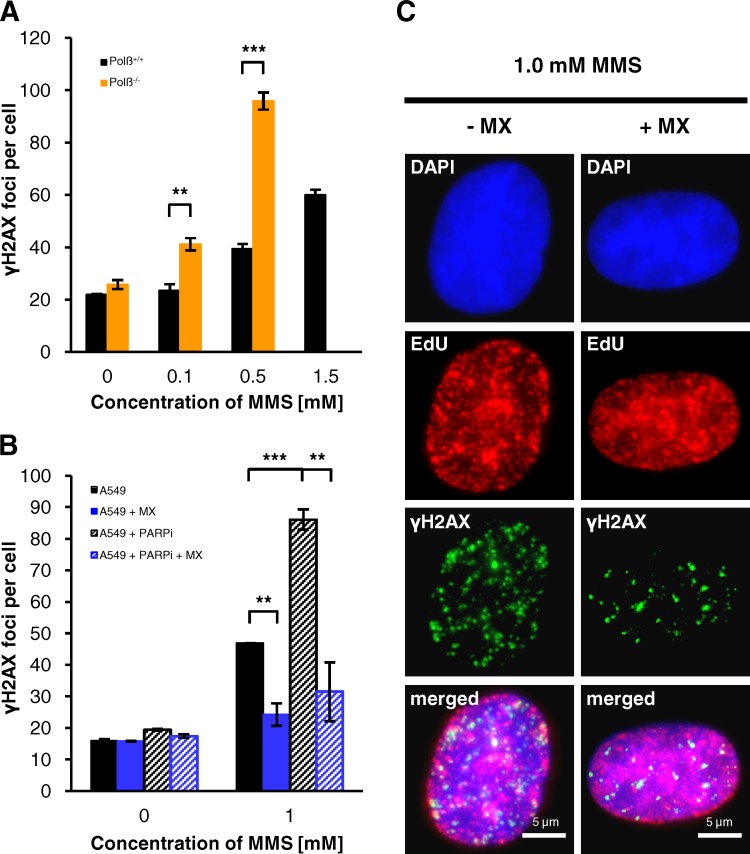
Having established that DSBs after MMS arise when normal, unperturbed BER processes interfere with replication, we finally wished to characterize the BER intermediate(s) causing DSB formation. Because XRCC1-deficient and PARP1-inhibited cells exhibited both elevated unrepaired SSBs and high levels of DSBs, we addressed the possibility that SSB formation during normal BER processes might underlie the induction of DSBs in WT cells. SSB formation during BER is mediated by APE1 cleavage at AP sites, and Polβ is the first enzyme operating on the SSB lesion. Thus, we investigated cells unable to cleave AP sites and cells deficient in Polβ. Strikingly, DSB induction immediately after MMS treatment was nearly abolished in WT cells treated with methoxyamine (MX), an agent that binds AP sites to prevent them from cleavage by APE1 (Liu et al., 1999), but highly elevated in Polβ−/− MEFs (Fig. 7, A and B). Moreover, even the highly elevated level of γH2AX foci in PARP1-inhibited cells was almost completely rescued by MX (Fig. 7, B and C). These data provide strong evidence that SSB formation during BER underlies the induction of DSBs. Further, it shows that AP sites, which accumulate in cells treated with APE1 inhibitors (Rai et al., 2012), do not cause immediate DNA breakage. Of note, we and others have observed that persistent inhibition of APE1 cleavage results in delayed expression of DSBs (unpublished data; Taverna et al., 2001). However, this likely occurs by replication fork collapse after prolonged stalling at AP sites, whereas WT cells show DSB formation immediately after MMS treatment.
Figure 7.
DSB formation after MMS requires SSB induction. (A) γH2AX foci in Polβ+/+ and Polβ−/− MEFs. Cells were treated with 10 µM EdU and different concentrations of MMS for 1 h. 15 min after removing EdU/MMS, cells were fixed and stained against γH2AX and EdU. γH2AX foci were assessed in EdU-positive S-phase cells (±SEM from three experiments). In Polβ−/− MEFs treated with 1.5 mM MMS, γH2AX foci numbers were too high for exact enumeration. (B and C) γH2AX foci after treatment with MMS and methoxyamine (MX). Where indicated, A549 cells were treated with 6 mM MX, 15 µM PARPi, or both for 1 h before 10 µM EdU and 1 mM MMS were added for an additional hour. 15 min after removing the drugs, cells were fixed and stained against γH2AX and EdU. γH2AX foci were assessed in EdU-positive S-phase cells (B, ±SEM from two experiments). Representative images of MMS-treated A549 cells with and without MX are shown in C. **, P < 0.01; ***, P < 0.001.
Discussion
Despite intensive research since the first reports on clastogenic effects of MMS (Frei and Venitt, 1975; Natarajan et al., 1983), it is still unclear how DNA and chromosome breakage arise after exposure to methylating agents. As DNA methylation does not directly cause DNA breakage, it was proposed that methylated base adducts lead to the formation of DSBs during replication, but the mechanism by which this occurs remained elusive. Evidence for the formation of DSBs after methylation damage was provided with the neutral comet assay (Angelis et al., 1999), an approach with limited suitability for a cell cycle–specific quantification of DSBs. This can be achieved with the γH2AX assay, and first studies provided evidence for H2AX phosphorylation (Pascucci et al., 2005; Zhou et al., 2006) and γH2AX foci formation after MMS treatment (Nikolova et al., 2010). However, it remained unclear if γH2AX foci represent DSBs or other structures such as stalled replication forks. Here, we demonstrate that MMS-induced γH2AX foci arise during S phase in a manner dependent on replication, and provide evidence that these foci exclusively mark DSBs. Most importantly, we show that the occurrence of DSBs, chromatid breaks, and translocations after methylation damage depends on the level of BER activity: if SSBs are formed but not repaired, DSBs and chromosomal aberrations increase dramatically; if BER is not initiated, they are strongly reduced. We propose that DSBs after methylation damage arise when replication forks encounter SSBs induced during BER. The data leading to this model are discussed below.
γH2AX foci after methylation damage represent DSBs and not stalled replication forks
Experiments using the DNA fiber assay revealed that all active replication forks are stalled after MMS concentrations similar to those used in our study (Groth et al., 2010). Interestingly, the number of replication forks active at any given time during S phase largely exceeds the number of 40–50 γH2AX foci induced under these treatment conditions (Méchali, 2010), suggesting that γH2AX foci form at structures other than stalled replication forks. Moreover, the generation of γH2AX foci is largely abolished in cells defective in the initial step of BER, although such cells are expected to show the same level of (or even more) replication fork stalling at methylated base adducts as control cells. Furthermore, we observed that the majority of γH2AX foci colocalize with pATM foci (Nikolova et al., 2010) and that ATM is activated after MMS treatment (Fig. S1, A–C), a finding consistent with a previous study (Chou et al., 2008). Because ATM responds to DSBs and stalled replication forks do not directly activate ATM (Löbrich et al., 2010), these results support the notion that MMS-induced γH2AX foci represent DSBs. This was finally confirmed by studying γH2AX foci formation after treatment with HU. In these experiments, we observed a strong pan-nuclear γH2AX signal but no formation of γH2AX foci in S-phase cells after 2 h of HU, a treatment period known to transiently arrest replication forks without DSB induction (Saintigny et al., 2001; Petermann et al., 2010). In contrast, we detected robust γH2AX foci formation in a Mus81-dependent manner after 24 h of HU, which is known to cause replication fork collapse and DSB formation (Saintigny et al., 2001; Hanada et al., 2007; Groth et al., 2010). Together, these results demonstrate that stalled replication forks produce a pan-nuclear γH2AX signal but no γH2AX foci, the latter being exclusively formed at DSBs.
DSBs and chromosomal aberrations arise after methylation damage when replication forks encounter BER intermediates
The prevailing model of how replication-dependent DSBs and chromosomal aberrations arise after methylation damage involves replication fork stalling and collapse at methylated base adducts, similar to the mechanism of DSB formation after prolonged HU treatment (Saintigny et al., 2001; Petermann et al., 2010). However, our observation that γH2AX foci form within minutes after MMS treatment argues against this model. Moreover, XRCC1-deficient and PARPi-treated cells, both of which accumulate SSBs during BER, showed highly elevated levels of γH2AX foci and chromosomal aberrations compared with WT cells (Zdzienicka et al., 1992; Trucco et al., 1998). But most importantly, γH2AX foci formation is nearly absent if BER initiation is suppressed by MPG depletion or if SSB formation is abolished by preventing cleavage of AP sites. Under such conditions, even a deficiency in XRCC1 or PARP1 activity does not significantly increase foci levels. The reduced γH2AX foci level in MPG-depleted cells is observed from S phase through G2 and into mitosis, and parallels the reduced level of DSBs measured by the comet assay as well as the reduction in chromosomal aberrations. The elevated levels of γH2AX foci and chromosomal aberrations in Rad51-depleted cells are reduced after MPG depletion, demonstrating that HR is involved in repairing DSBs generated at BER sites. The few MMS-induced γH2AX foci and chromosomal aberrations detected in MPG-deficient cells likely result from spontaneous hydrolysis of the methylated bases and subsequent AP endonuclease cleavage at the abasic sites (Wyatt and Pittman, 2006). Finally, increased γH2AX foci levels are observed in MPG-overexpressing cells. In summary, our data with MPG-defective and -overexpressing cells show that DSBs and chromosomal aberrations arise at BER intermediates. Based on (1) the elevated SSB and DSB levels in cells deficient in XRCC1, Polβ, or PARP1 activity and (2) the lack of SSB and DSB formation at uncleaved AP sites, we suggest that DSBs after methylation damage might form by replication fork run-off at SSB sites generated during BER, similar to the mechanism suggested for the generation of DSBs at topoisomerase I cleavage complexes (Strumberg et al., 2000). We would like to note in this context that SSBs induced during BER might arise within a complex of BER proteins and could therefore be different from SSBs induced by ionizing radiation or reactive oxygen species. It is possible that such an SSB–protein complex represents a structure that is particularly vulnerable upon collision with the replication fork.
We would like to note that retinal cell degeneration as well as toxicity in other organs was reported to be reduced in MPG knockout compared with WT mice, suggesting that BER drives the process of toxicity in specific organs after treatment of animals with MMS (Meira et al., 2009; Calvo et al., 2013). However, the observed tissue-specific toxicity is not directly related to cell proliferation because it was observed in proliferating as well as in nonproliferating tissue. Further, PARP1 depletion was shown to reduce toxicity, consistent with a model that PARP1 activation during BER leads to ATP depletion and cell death by necrosis (Calvo et al., 2013). In contrast, we observed that DSBs arise almost exclusively in replicating cells and show that PARP1-inhibited cells exhibit elevated levels of DSBs and chromosomal aberrations. Thus, the mechanism by which chromosomal instability arises appears to be different from the mechanism leading to organ-specific toxicity in the mice models referred to above.
SCE formation and S-phase delay after methylation damage does not require BER
Although the formation of chromosomal aberrations after MMS requires BER, this is not necessarily the case for SCE formation. MPG-deficient cells unable to initiate BER showed SCE levels only slightly reduced compared with WT cells, demonstrating that SCEs can form by BER-independent processes. We propose that SCEs arise from replication forks stalled at replication-blocking adducts such as N3-MeA (Larson et al., 1985; Engelward et al., 1998). It has been suggested that the two main pathways to overcome a replication block are translesion synthesis (TLS) and fork regression, the latter process leading to the formation of a chicken foot structure (Heyer et al., 2010). When replication is resumed, a subset of these chicken foot structures is converted into active replication forks by an HR-mediated process (Petermann et al., 2010). HR-mediated repair of single-stranded gaps generated by a blocking lesion on the lagging strand might additionally contribute to SCE formation independently of BER (Heller and Marians, 2006; Heyer et al., 2010). However, SCEs can also arise at SSBs induced during BER, as suggested by the high SCE frequency in XRCC1-deficient cells (Thompson et al., 1982; Saleh-Gohari et al., 2005).
Similar to SCEs but in contrast to chromosomal aberrations, the characteristic S-phase delay of MMS-treated cells does not require the generation of DSBs. First, the ATM/ATR inhibitor caffeine, which completely abolishes checkpoint signals from DSBs, did not significantly impact on S-phase progression after MMS. Second, we observed a similar S-phase delay in MPG-deficient and WT cells. The lack of an impact of DSBs on S-phase progression after DNA methylation damage is consistent with a recent study using the DNA fiber assay, which demonstrated a checkpoint-independent S-phase delay after MMS treatment (Groth et al., 2010). In contrast to WT cells, we observed a pronounced S-phase arrest in XRCC1-deficient cells, which was abolished in the presence of caffeine. Together, this indicates that DSBs can cause S-phase delay by an active checkpoint response, but direct replication stalling by methylated bases covers the impact of DSB checkpoint signaling in WT cells.
Conclusion
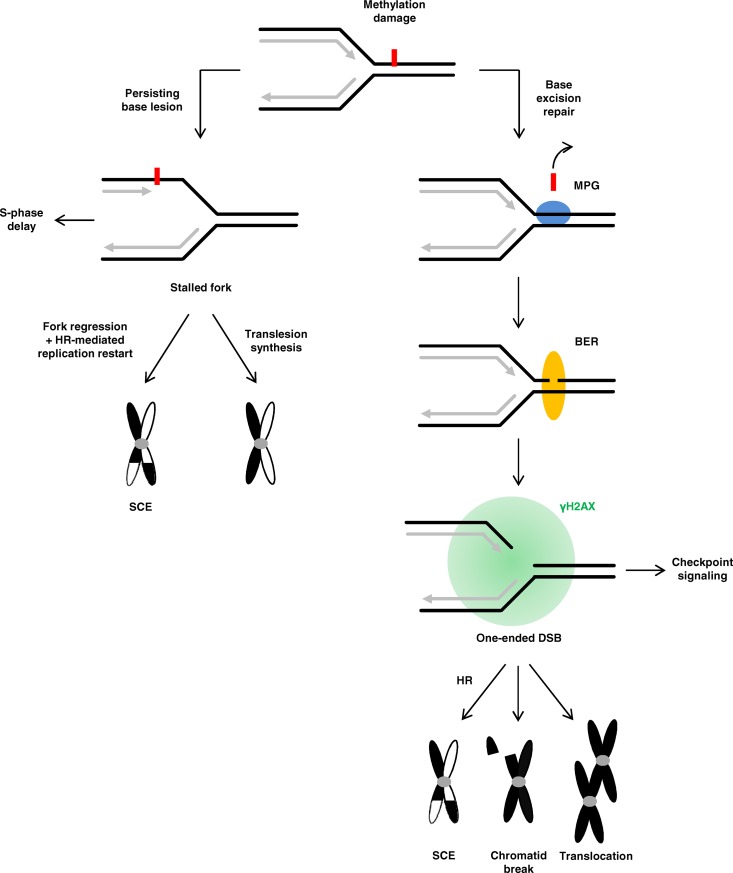
Here, we rigorously modulated different BER steps and found that γH2AX foci arise after DNA methylation damage when replication forks interfere with SSBs induced during BER. We present evidence that these γH2AX foci mark DSBs and demonstrate that they are responsible for the formation of chromosomal aberrations. In contrast, SCEs and delayed S-phase progression do not require DSB formation and can arise independently of BER, likely as a result of the presence of small replication-blocking lesions such as 3-MeA (Fig. 8). In extension of our findings to other genotoxic agents, we propose that the process of replication fork collision at SSBs induced during BER is not restricted to DNA methylation damage but might occur at many if not all DNA lesions repaired by BER, including oxidative base damage. According to this model, BER is a double-edged sword: it is required for removing deleterious lesions from DNA and, after collision with the replication process, it can result in DSBs and chromosomal aberrations. A tight regulation between BER and replication is therefore important to minimize the collision of replication forks with BER intermediates. If BER is de-regulated, chromosomal instability can arise, which is frequently observed in transgenic mouse models and cancer cells.
Figure 8.
Methylated base adducts and BER intermediates interfering with replication cause distinct effects. MMS-induced N3-MeA can block replication forks (here shown for a lesion on the leading strand). Such stalled forks are mainly responsible for the strong S-phase delay after MMS. Different mechanisms exist to overcome such a blockage, with translesion synthesis being one of the predominant pathways that is not associated with SCE formation. Another process is fork regression, which leads to the formation of a chicken foot structure that can be resolved by an HR-mediated process associated with SCE formation. HR-mediated repair of single-stranded gaps, which are formed at methylated base adducts blocking lagging strand synthesis, might also contribute to SCE formation (not depicted in the model). The majority of methylated base lesions induced by MMS is repaired by BER, which is initiated by MPG. Upon interference of replication forks with SSBs induced during BER, one-ended DSBs are generated, which give rise to the formation of γH2AX foci. If unrepaired or mis-repaired, such one-ended DSBs cause chromatid breaks or translocations, respectively. Thus, one-ended DSBs arising in a BER-dependent manner are responsible for the formation of chromosomal aberrations after methylation damage. BER-induced SSBs can also lead to SCE formation.
Materials and methods
Cell lines and culture
XRCC1-deficient EM-C11 cells were derived from CHO-9 cells and do not express functional XRCC1 proteins due to a missense mutation in the XRCC1 gene (Shen et al., 1998). MPG-overexpressing T02-M5 and T02-M9 cells were obtained using the expression vector pEZ-1 that was constructed from a pGB4 vector and the human MPG cDNA sequence in the plasmid pPG23 (Chakravarti et al., 1991; Ibeanu et al., 1992). pGB4 is a high-level expression vector for mammalian cells under the control of LTR promoter of WNB5 ectopic mouse leukemia virus (Ch’ang et al., 1989). MPG−/− MEFs were generated from MPG knockout mouse embryos. The MPG knockout mice strains were generated using gene targeting in mouse embryonic stem cells and harboring an MPG gene lacking exon 1 and 2 (Elder et al., 1998). Polβ−/− MEFs were provided by Sobol et al. (1996), who generated this cell line using transgenic mice with a heterozygous germline deletion mutation of the promoter and the first exon of the Polβ gene (Gu et al., 1994). The transformed cell lines were established by transfection with an expression vector for the SV40 large-T antigen (Sobol et al., 1996). ATR-deficient F02-98 hTert cells are hTert-immortalized fibroblasts that were derived from a patient with Seckel syndrome. These cells harbor a point mutation in exon 9, leading to the expression of a truncated ATR protein (lacking exon 9). To a small amount, functional ATR is expressed as well (O’Driscoll et al., 2003).
XRCC1-deficient EM-C11 cells and their corresponding parental WT cell line CHO-9, as well as MPG-overexpressing T02-M5 and T02-M9 cells with their corresponding WT cell line CHO-9 neo, were cultured in RPMI-1640 supplemented with 10% FBS and penicillin/streptomycin. G418 was added at a concentration of 1.5 mg/ml for CHO-9 neo, T02-M5, and T02-M9 cells. Human A549 cells and WT, MPG−/−, and Polβ−/− MEFs were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin. The ATR-deficient cell line F02-98 hTert was cultured in MEM Earle’s medium supplemented with 20% FBS and penicillin/streptomycin. All cell lines were incubated at 37°C in a humidified atmosphere with 5% CO2.
siRNA transfection was performed using HiPerFect Transfection Reagent (QIAGEN) following the manufacturer’s instructions. The following siRNA oligonucleotides were obtained from QIAGEN: 5′-AATTCTCCGAACGTGTCACGT-3′ for siControl, 5′-CACCGCCAGCCGTGTCCTCAA-3′ for siMPG, 5′-CAACCGAGGCATGTTCATGAA-3′ for siMPG 2, 5′-ACCGCAGCATCTATTTCTCAA-3′ for siMPG 3, 5′-CCTGTACGTGTACATCATTTA-3′ for siMPG 4, 5′-AAGGGAATTAGTGAAGCCAAA-3′ for siRad51, 5′-CCCGGTGGATCTACAGTTGTA-3′ for siXRCC1, 5′-TACACACAAGAGTTTAATAAA-3′ for siXRCC1 2, and 5′-AACAGCCCTGGTGGATCGATA-3′ for siMus81.
Genotoxic treatment
MMS (Sigma-Aldrich) was diluted in sterile distilled water to 100 mM and added to the cell culture medium to a final concentration between 0.05 and 2.5 mM. Cells were treated with MMS for 1 h, the medium was removed, and cells were washed with PBS before fresh medium was added. HU (Sigma-Aldrich) was diluted in sterile distilled water to 100 mM and used at a final concentration of 0.5 mM. Aphidicolin (EMD Millipore) was diluted in DMSO and used at a final concentration of 1 µM. The ATM and ATR inhibitor caffeine (Roth) was diluted in PBS and used at a final concentration of 5 mM. The specific ATM inhibitor KU55933 and DNA-PK inhibitor NU7026 (Kudos Pharmaceuticals) were diluted in DMSO and used at a final concentration of 10 µM or 20 µM, respectively. The Rad51 inhibitor B02 (EMD Millipore) was diluted in DMSO and used at a final concentration of 60 µM. The PARP inhibitor (PJ34; EMD Millipore) was diluted in DMSO and used at a final concentration of 15 µM. PARPi was added 1 h before MMS and was removed together with MMS. Methoxyamine (Sigma-Aldrich) was diluted in sterile distilled water and used at 6 mM.
Immunofluorescence microscopy
For γH2AX/EdU immunofluorescence analysis, cells were seeded on coverslips and cultured for 2 d. Then, cells were treated for 1 h with 10 µM EdU (Invitrogen) and with different concentrations of MMS when indicated. After recovery times of 15 min to 16 h, cells were fixed in 2% formaldehyde as described previously (Quennet et al., 2011). For γH2AX staining, samples were incubated with a primary mouse anti–phospho-H2AX antibody (05-636; EMD Millipore) overnight at 4°C and with an Alexa Fluor 488–conjugated secondary antibody (Molecular Probes) for 1 h at room temperature. To stain EdU, the Invitrogen EdU staining kit was used following the manufacturer’s instructions. If γH2AX foci were analyzed in mitosis, cells were additionally stained against pH3 using a primary rabbit anti–phospho-H3 antibody (EMD Millipore) and an Alexa Fluor 594–conjugated secondary antibody (Molecular Probes). In all assays, nuclei were stained with DAPI (0.2 µg/ml in PBS). In at least three independent experiments, γH2AX foci were counted in 40 EdU-positive cells. If γH2AX foci were analyzed in EdU-negative G1- and G2-phase cells, these cells were identified by quantifying the DAPI signal using the Metafer Scanning System (Metasystems). Analysis was performed on an inverted fluorescence microscope (Axiovert 200M; Carl Zeiss). As an objective, an EC Plan-Neofluar (100×; Carl Zeiss) with an NA of 1.3 was used with immersion oil. Imaging was performed at room temperature using a camera (AxioCam MRm; Carl Zeiss) and ISIS acquisition software (Metasystems). Images were processed using ImageJ (National Institutes of Health).
Flow cytometric analysis
For the analysis of cell cycle progression after MMS treatment, exponentially growing cells were treated with 10 µM BrdU (Roche) and different concentrations of MMS for 1 h. Where indicated, 5 mM caffeine was added 1 h before the BrdU treatment. Cells were analyzed according to standard protocols. Cells were fixed with ice-cold 70% ethanol and permeabilized with 2 M HCl in PBS for 20 min. For BrdU staining, cells were incubated with 20 µl FITC-conjugated anti-BrdU monoclonal antibody (mouse; BD) for 30 min, followed by an incubation with 20 µg/ml propidium iodide containing 0.5 mg/ml RNase in PBS for 30 min at room temperature. Analysis was performed on a flow cytometer (Cytomics FC500; Beckman Coulter) using CXP software (Beckman Coulter). Regarding the BrdU-positive cells, the amount of cells in S phase was calculated for times up to 16 h after MMS treatment.
Chromosomal studies
To analyze SCEs and chromosomal aberrations, cells were grown for two cell cycles in BrdU-containing medium before they were treated with MMS. Here, 1 µM BrdU was used for CHO cells and 10 µM for MEFs and A549 cells. 2 h before cells were fixed, 2 mM caffeine and 100 ng/ml colcemid (Sigma-Aldrich) were added. SCE staining was performed according to standard protocols for the FPG technique. In at least three independent experiments the number of SCEs, chromatid breaks (including chromatid gaps), and chromatid-type translocations (triradials, quatriradials, and ring chromosomes) was analyzed in at least 40 metaphase spreads. Because of the variable numbers of chromosomes in the different cell lines used, we normalized the data to 22 chromosomes in CHO cells, 60 chromosomes in A549 cells, and 70 chromosomes in MEFs.
Western blotting
Cells were washed with ice-cold PBS, collected with a cell scraper, and 150 µl lysis buffer (20 mM Tris-HCl, pH 8.5, 1 mM EDTA, 5% glycine, 20 mM PMSF, and 0.2 mM sodium orthovanadate) was added. Lysates were sonificated and protein concentration was determined by the Bradford assay (Bio-Rad Laboratories). Equal amounts of total protein lysate (5–15 µg) were resolved by SDS-PAGE using 11% gels. Proteins were then transferred to a nitrocellulose membrane and blocked for 1 h in 5% BSA/TBS-T or 5% low-fat milk in TBS-T. The incubation with primary antibodies was performed overnight at 4°C in 5% BSA/TBS-T or in 1% or 2.5% low-fat milk in TBS-T against the following proteins: MPG (rabbit, 155092, 1:10,000; Abcam), Rad51 (rabbit, 63801, 1:2,000; Abcam), XRCC1 (mouse, 1838, 1:1,000; Abcam), pKAP1 (rabbit, 3640-1, 1:10,000; Epitomics), pChk2 (rabbit, 2661, 1:1,000; Cell Signaling Technology), Mus81 (mouse, 53382, 1:1,000; Santa Cruz Biotechnology, Inc.), GAPDH (rabbit, 25778, 1:1,000; Santa Cruz Biotechnology, Inc.) and α-tubulin (mouse, 8035, 1:2,000; Santa Cruz Biotechnology, Inc.). Anti–mouse (goat, 1:1,000, in 5% low-fat milk in TBS-T; Santa Cruz Biotechnology, Inc.) or anti–rabbit (donkey, 1:1,000, in 5% low-fat milk in TBS-T; Dianova) IgG conjugated with horseradish peroxidase was used as a secondary antibody for 1 h at room temperature. The detection was performed using an ECL Western blot detection system (Roche) and a Chemi-Smart system (Vilber Lourmat).
Neutral comet assay
Neutral version of the comet assay was performed as described previously with slight modifications (Nikolova et al., 2010). In brief, exponentially growing WT- and MPG-deficient mouse fibroblasts were exposed to 1.0 mM MMS for 60 min, postincubated in fresh complete medium for the indicated time periods, trypsinized, resuspended in fresh medium, and centrifuged. The pellets were resuspended in ice-cold PBS. Cell suspension (15 µl) was embedded in 120 µl of low-melting point agarose (0.5% in dH2O at 37°C) onto agarose-coated (1.5% in PBS) and dried slides, then covered with coverslips to form microgels. After hardening, the coverslips were removed and the microgels were submersed for 1 h in precooled lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris-HCl, and 1% sodium laurylsarcosine, pH 7.5), to which 1 h before use 1% Triton X-100 was added. Slides were electrophoresed in precooled neutral buffer (90 mM Tris, 90 mM boric acid, and 2 mM EDTA, pH 7.5) at 0.7 V/cm for 20 min at 4°C. Slides were rinsed in water, fixed in 100% ethanol, dried, and stained with 50 µg/ml propidium iodide. Stained slides were evaluated using a fluorescence microscope and Comet IV software (Perceptive Instruments). Data were expressed as tail factor, which represents the intensity of DNA in the tail. In each experiment 50 cells were analyzed. The data were normalized to the amount of replicating cells in the two tested cells lines.
Statistical analysis
All data shown represent the mean values of at least three independent experiments; the error bars show the SEM between the experiments. Data were compared by t test with: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and N.S., not significant.
Online supplemental material
Fig. S1 shows that ATM is activated after MMS treatment and that MMS-induced DSBs are formed in a Mus81-independent manner. Fig. S2 shows the formation of MMS-induced γH2AX foci in G2-phase cells of different cell lines. Fig. S3 shows the formation of MMS-induced DSBs measured by γH2AX foci and by the comet assay in S-phase WT and MPG-deficient cells, as well as MMS-induced γH2AX foci in WT and MPG-deficient cells after Rad51 inhibition. Fig. S4 shows the cell cycle progression in CHO-9 (WT) and EM-C11 (XRCC1-deficient) cells after 0.1 mM MMS and SCE frequencies in WT and MPG-deficient cells after 0.5 mM MMS. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201312078/DC1.
Supplementary Material
Acknowledgments
We thank Georg Nagel for technical assistance.
Work in the M. Löbrich laboratory is supported by the Deutsche Forschungsgemeinschaft (Lo 677/4-3 and GRK1657) and the Bundesministerium für Bildung und Forschung (02S8355, 02NUK016D). Work in the B. Kaina laboratory is funded by Ka 724 and Ni 1319/1-1.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APE1
- apurinic endonuclease 1
- AP site
- apurinic
- BER
- base excision repair
- DSB
- double-strand break
- EdU
- 5-ethynyl-2’-desoxyuridine
- HU
- hydroxyurea
- MPG
- N-methylpurine DNA glycosylase
- MMS
- methyl methanesulfonate
- N3-MeA
- N3-methyladenine
- PARPi
- PARP inhibitor
- Polβ
- polymerase β
- SCE
- sister chromatid exchange
- SSB
- single-strand break
- WT
- wild type
- XRCC1
- x-ray cross-complementing protein 1
References
- Andegeko, Y., Moyal L., Mittelman L., Tsarfaty I., Shiloh Y., and Rotman G.. 2001. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276:38224–38230 [DOI] [PubMed] [Google Scholar]
- Angelis, K.J., Dusinská M., and Collins A.R.. 1999. Single cell gel electrophoresis: detection of DNA damage at different levels of sensitivity. Electrophoresis. 20:2133–2138 [DOI] [PubMed] [Google Scholar]
- Bender, M.A., Griggs H.G., and Bedford J.S.. 1974. Mechanisms of chromosomal aberration production. 3. Chemicals and ionizing radiation. Mutat. Res. 23:197–212 10.1016/0027-5107(74)90140-7 [DOI] [PubMed] [Google Scholar]
- Bennett, R.A.O., Wilson D.M. III, Wong D., and Demple B.. 1997. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl. Acad. Sci. USA. 94:7166–7169 10.1073/pnas.94.14.7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek, D.T.1990. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 231:11–30 10.1016/0027-5107(90)90173-2 [DOI] [PubMed] [Google Scholar]
- Bianchi, V., Pontis E., and Reichard P.. 1986. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 261:16037–16042 [PubMed] [Google Scholar]
- Boei, J.J.W.A., Vermeulen S., Moser J., Mullenders L.H.F., and Natarajan A.T.. 2002. Intrachanges as part of complex chromosome-type exchange aberrations. Mutat. Res. 504:47–55 10.1016/S0027-5107(02)00078-7 [DOI] [PubMed] [Google Scholar]
- Brem, R., Fernet M., Chapot B., and Hall J.. 2008. The methyl methanesulfonate induced S-phase delay in XRCC1-deficient cells requires ATM and ATR. DNA Repair (Amst.). 7:849–857 10.1016/j.dnarep.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Caldecott, K.W.2003. XRCC1 and DNA strand break repair. DNA Repair (Amst.). 2:955–969 10.1016/S1568-7864(03)00118-6 [DOI] [PubMed] [Google Scholar]
- Caldecott, K.W.2008. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9:619–631 [DOI] [PubMed] [Google Scholar]
- Caldecott, K.W., Aoufouchi S., Johnson P., and Shall S.. 1996. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 24:4387–4394 10.1093/nar/24.22.4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, J.A., Moroski-Erkul C.A., Lake A., Eichinger L.W., Shah D., Jhun I., Limsirichai P., Bronson R.T., Christiani D.C., Meira L.B., and Samson L.D.. 2013. Aag DNA glycosylase promotes alkylation-induced tissue damage mediated by Parp1. PLoS Genet. 9:e1003413 10.1371/journal.pgen.1003413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans, A., Marsin S., Nakabeppu Y., O’connor T.R., Boiteux S., and Radicella J.P.. 2005. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst.). 4:826–835 10.1016/j.dnarep.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Carrano, A.V., Minkler J.L., Dillehay L.E., and Thompson L.H.. 1986. Incorporated bromodeoxyuridine enhances the sister-chromatid exchange and chromosomal aberration frequencies in an EMS-sensitive Chinese hamster cell line. Mutat. Res. 162:233–239 10.1016/0027-5107(86)90090-4 [DOI] [PubMed] [Google Scholar]
- Ch’ang, L.Y., Yang W.K., Myer F.E., and Yang D.M.. 1989. Negative regulatory element associated with potentially functional promoter and enhancer elements in the long terminal repeats of endogenous murine leukemia virus-related proviral sequences. J. Virol. 63:2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti, D., Ibeanu G.C., Tano K., and Mitra S.. 1991. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J. Biol. Chem. 266:15710–15715 [PubMed] [Google Scholar]
- Chou, W.C., Wang H.C., Wong F.H., Ding S.L., Wu P.E., Shieh S.Y., and Shen C.Y.. 2008. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 27:3140–3150 10.1038/emboj.2008.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquerelle, T., Dosch J., and Kaina B.. 1995. Overexpression of N-methylpurine-DNA glycosylase in Chinese hamster ovary cells renders them more sensitive to the production of chromosomal aberrations by methylating agents—a case of imbalanced DNA repair. Mutat. Res. 336:9–17 10.1016/0921-8777(94)00035-5 [DOI] [PubMed] [Google Scholar]
- Dianov, G.L., and Hübscher U.. 2013. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 41:3483–3490 10.1093/nar/gkt076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder, R.H., Jansen J.G., Weeks R.J., Willington M.A., Deans B., Watson A.J., Mynett K.J., Bailey J.A., Cooper D.P., Rafferty J.A., et al. . 1998. Alkylpurine-DNA-N-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol. Cell. Biol. 18:5828–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelward, B.P., Allan J.M., Dreslin A.J., Kelly J.D., Wu M.M., Gold B., and Samson L.D.. 1998. A chemical and genetic approach together define the biological consequences of 3-methyladenine lesions in the mammalian genome. J. Biol. Chem. 273:5412–5418 10.1074/jbc.273.9.5412 [DOI] [PubMed] [Google Scholar]
- Erzberger, J.P., Barsky D., Schärer O.D., Colvin M.E., and Wilson D.M. III. 1998. Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 26:2771–2778 10.1093/nar/26.11.2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, H.J. Wilson, and Scott D.. 1969. The induction of chromosome aberrations by nitrogen mustard and its dependence on DNA synthesis. Proc. R. Soc. Lond. B Biol. Sci. 173:491–512 10.1098/rspb.1969.0073 [DOI] [PubMed] [Google Scholar]
- Frei, J.V., and Venitt S.. 1975. Chromosome damage in the bone marrow of mice treated with the methylating agents methyl methanesulphonate and N-methyl-N-nitrosourea in the presence or absence of caffeine, and its relationship with thymoma induction. Mutat. Res. 30:89–96 10.1016/0027-5107(75)90257-2 [DOI] [PubMed] [Google Scholar]
- Groth, P., Ausländer S., Majumder M.M., Schultz N., Johansson F., Petermann E., and Helleday T.. 2010. Methylated DNA causes a physical block to replication forks independently of damage signalling, O(6)-methylguanine or DNA single-strand breaks and results in DNA damage. J. Mol. Biol. 402:70–82 10.1016/j.jmb.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Groth, P., Orta M.L., Elvers I., Majumder M.M., Lagerqvist A., and Helleday T.. 2012. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res. 40:6585–6594 10.1093/nar/gks315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H., Marth J.D., Orban P.C., Mossmann H., and Rajewsky K.. 1994. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 265:103–106 10.1126/science.8016642 [DOI] [PubMed] [Google Scholar]
- Hanada, K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., and Kanaar R.. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096–1104 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Heller, R.C., and Marians K.J.. 2006. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 7:932–943 10.1038/nrm2058 [DOI] [PubMed] [Google Scholar]
- Heyer, W.D., Ehmsen K.T., and Liu J.. 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44:113–139 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, J.K., Watson M., Stefanick D.F., Shaughnessy D.T., Taylor J.A., and Wilson S.H.. 2008. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 18:48–63 10.1038/cr.2008.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeanu, G., Hartenstein B., Dunn W.C., Chang L.Y., Hofmann E., Coquerelle T., Mitra S., and Kaina B.. 1992. Overexpression of human DNA repair protein N-methylpurine-DNA glycosylase results in the increased removal of N-methylpurines in DNA without a concomitant increase in resistance to alkylating agents in Chinese hamster ovary cells. Carcinogenesis. 13:1989–1995 10.1093/carcin/13.11.1989 [DOI] [PubMed] [Google Scholar]
- Kaina, B.1998. Critical steps in alkylation-induced aberration formation. Mutat. Res. 404:119–124 10.1016/S0027-5107(98)00103-1 [DOI] [PubMed] [Google Scholar]
- Kaina, B.2004. Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go. Cytogenet. Genome Res. 104:77–86 10.1159/000077469 [DOI] [PubMed] [Google Scholar]
- Larson, K., Sahm J., Shenkar R., and Strauss B.. 1985. Methylation-induced blocks to in vitro DNA replication. Mutat. Res. 150:77–84 10.1016/0027-5107(85)90103-4 [DOI] [PubMed] [Google Scholar]
- Liu, L., Taverna P., Whitacre C.M., Chatterjee S., and Gerson S.L.. 1999. Pharmacologic disruption of base excision repair sensitizes mismatch repair-deficient and -proficient colon cancer cells to methylating agents. Clin. Cancer Res. 5:2908–2917 [PubMed] [Google Scholar]
- Löbrich, M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A.A., Barton O., and Jeggo P.A.. 2010. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 9:662–669 10.4161/cc.9.4.10764 [DOI] [PubMed] [Google Scholar]
- Ma, W.J., Westmoreland J.W., Gordenin D.A., and Resnick M.A.. 2011. Alkylation base damage is converted into repairable double-strand breaks and complex intermediates in G2 cells lacking AP endonuclease. PLoS Genet. 7:e1002059 10.1371/journal.pgen.1002059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., and de Murcia G.. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchali, M.2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 11:728–738 10.1038/nrm2976 [DOI] [PubMed] [Google Scholar]
- Meira, L.B., Moroski-Erkul C.A., Green S.L., Calvo J.A., Bronson R.T., Shah D., and Samson L.D.. 2009. Aag-initiated base excision repair drives alkylation-induced retinal degeneration in mice. Proc. Natl. Acad. Sci. USA. 106:888–893 10.1073/pnas.0807030106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan, A.T., and Obe G.. 1984. Molecular mechanisms involved in the production of chromosomal aberrations. III. Restriction endonucleases. Chromosoma. 90:120–127 10.1007/BF00292448 [DOI] [PubMed] [Google Scholar]
- Natarajan, A.T., Tates A.D., Meijers M., Neuteboom I., and de Vogel N.. 1983. Induction of sister-chromatid exchanges (SCEs) and chromosomal aberrations by mitomycin C and methyl methanesulfonate in Chinese hamster ovary cells. An evaluation of methodology for detection of SCEs and of persistent DNA lesions towards the frequencies of observed SCEs. Mutat. Res. 121:211–223 10.1016/0165-7992(83)90206-3 [DOI] [PubMed] [Google Scholar]
- Natarajan, A.T., Mullenders L.H.F., Meijers M., and Mukherjee U.. 1985. Induction of sister-chromatid exchanges by restriction endonucleases. Mutat. Res. 144:33–39 10.1016/0165-7992(85)90121-6 [DOI] [PubMed] [Google Scholar]
- Nazarkina, ZhK., Khodyreva S.N., Marsin S., Radicella J.P., and Lavrik O.I.. 2007. Study of interaction of XRCC1 with DNA and proteins of base excision repair by photoaffinity labeling technique. Biochemistry Mosc. 72:878–886 10.1134/S000629790708010X [DOI] [PubMed] [Google Scholar]
- Nikolova, T., Ensminger M., Löbrich M., and Kaina B.. 2010. Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate. DNA Repair (Amst.). 9:1050–1063 10.1016/j.dnarep.2010.07.005 [DOI] [PubMed] [Google Scholar]
- O’Brien, P.J., and Ellenberger T.. 2004. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 279:9750–9757 10.1074/jbc.M312232200 [DOI] [PubMed] [Google Scholar]
- O’Driscoll, M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., and Goodship J.A.. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33:497–501 10.1038/ng1129 [DOI] [PubMed] [Google Scholar]
- Parsons, J.L., and Dianov G.L.. 2013. Co-ordination of base excision repair and genome stability. DNA Repair (Amst.). 12:326–333 10.1016/j.dnarep.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Pascucci, B., Russo M.T., Crescenzi M., Bignami M., and Dogliotti E.. 2005. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res. 33:280–288 10.1093/nar/gki168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann, E., Orta M.L., Issaeva N., Schultz N., and Helleday T.. 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 37:492–502 10.1016/j.molcel.2010.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli, S., Pignone D., Santini G., Gatti M., and Olivieri G.. 1977. Mutagen specificity in the induction of chromosomal aberrations in somatic cells of Drosophila melanogaster. Genetics. 85:249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quennet, V., Beucher A., Barton O., Takeda S., and Löbrich M.. 2011. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 39:2144–2152 10.1093/nar/gkq1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, G., Vyjayanti V.N., Dorjsuren D., Simeonov A., Jadhav A., Wilson D.M. III, and Maloney D.J.. 2012. Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J. Med. Chem. 55:3101–3112 10.1021/jm201537d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny, Y., Delacôte F., Varès G., Petitot F., Lambert S., Averbeck D., and Lopez B.S.. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20:3861–3870 10.1093/emboj/20.14.3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari, N., Bryant H.E., Schultz N., Parker K.M., Cassel T.N., and Helleday T.. 2005. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 25:7158–7169 10.1128/MCB.25.16.7158-7169.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, M.R., Zdzienicka M.Z., Mohrenweiser H., Thompson L.H., and Thelen M.P.. 1998. Mutations in hamster single-strand break repair gene XRCC1 causing defective DNA repair. Nucleic Acids Res. 26:1032–1037 10.1093/nar/26.4.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobol, R.W., Horton J.K., Kühn R., Gu H., Singhal R.K., Prasad R., Rajewsky K., and Wilson S.H.. 1996. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 379:183–186 10.1038/379183a0 [DOI] [PubMed] [Google Scholar]
- Srivastava, D.K., Berg B.J., Prasad R., Molina J.T., Beard W.A., Tomkinson A.E., and Wilson S.H.. 1998. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 273:21203–21209 10.1074/jbc.273.33.21203 [DOI] [PubMed] [Google Scholar]
- Strumberg, D., Pilon A.A., Smith M., Hickey R., Malkas L., and Pommier Y.. 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20:3977–3987 10.1128/MCB.20.11.3977-3987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna, P., Liu L., Hwang H.S., Hanson A.J., Kinsella T.J., and Gerson S.L.. 2001. Methoxyamine potentiates DNA single strand breaks and double strand breaks induced by temozolomide in colon cancer cells. Mutat. Res. 485:269–281 10.1016/S0921-8777(01)00076-3 [DOI] [PubMed] [Google Scholar]
- Thompson, L.H., Brookman K.W., Dillehay L.E., Carrano A.V., Mazrimas J.A., Mooney C.L., and Minkler J.L.. 1982. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res. 95:427–440 10.1016/0027-5107(82)90276-7 [DOI] [PubMed] [Google Scholar]
- Trucco, C., Oliver F.J., de Murcia G., and Ménissier-de Murcia J.. 1998. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26:2644–2649 10.1093/nar/26.11.2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, A.E., Boiteux S., Hickson I.D., and Radicella J.P.. 2001. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 20:6530–6539 10.1093/emboj/20.22.6530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse, C.J., Taylor R.M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D.D., Weinfeld M., and Caldecott K.W.. 2001. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 104:107–117 10.1016/S0092-8674(01)00195-7 [DOI] [PubMed] [Google Scholar]
- Wyatt, M.D., and Pittman D.L.. 2006. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem. Res. Toxicol. 19:1580–1594 10.1021/tx060164e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzienicka, M.Z., van der Schans G.P., Natarajan A.T., Thompson L.H., Neuteboom I., and Simons J.W.I.M.. 1992. A Chinese hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis. 7:265–269 10.1093/mutage/7.4.265 [DOI] [PubMed] [Google Scholar]
- Zhou, C.X., Li Z.X., Diao H.L., Yu Y.K., Zhu W., Dai Y.Y., Chen F.F., and Yang J.. 2006. DNA damage evaluated by gamma H2AX foci formation by a selective group of chemical/physical stressors. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 604:8–18 10.1016/j.mrgentox.2005.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., and Elledge S.J.. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.