Accurate regulation of kinetochore–microtubule affinity is driven by incremental phosphorylation of an NDC80 molecular “lawn,” in which NDC80–microtubule bonds reorganize dynamically in response to the number and stability of microtubule attachments.
Abstract
Accurate chromosome segregation relies on dynamic interactions between microtubules (MTs) and the NDC80 complex, a major kinetochore MT-binding component. Phosphorylation at multiple residues of its Hec1 subunit may tune kinetochore–MT binding affinity for diverse mitotic functions, but molecular details of such phosphoregulation remain elusive. Using quantitative analyses of mitotic progression in mammalian cells, we show that Hec1 phosphorylation provides graded control of kinetochore–MT affinity. In contrast, modeling the kinetochore interface with repetitive MT binding sites predicts a switchlike response. To reconcile these findings, we hypothesize that interactions between NDC80 complexes and MTs are not constrained, i.e., the NDC80 complexes can alternate their binding between adjacent kinetochore MTs. Experiments using cells with phosphomimetic Hec1 mutants corroborate predictions of such a model but not of the repetitive sites model. We propose that accurate regulation of kinetochore–MT affinity is driven by incremental phosphorylation of an NDC80 molecular “lawn,” in which the NDC80–MT bonds reorganize dynamically in response to the number and stability of MT attachments.
Introduction
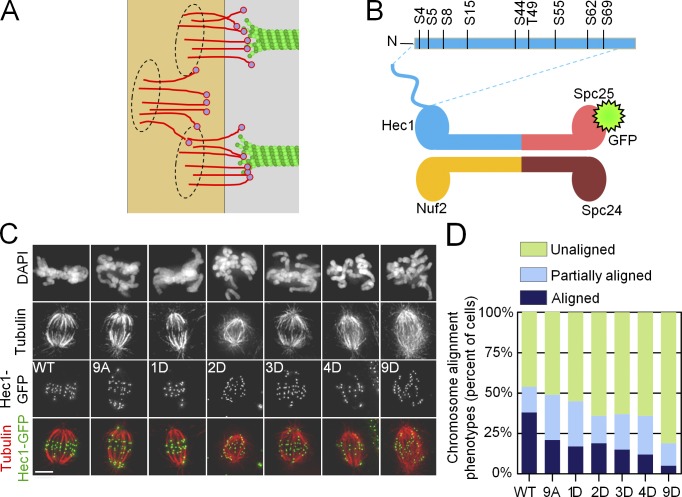
Proper chromosome segregation requires that all chromosomes become tethered at their kinetochores to the plus ends of dynamic microtubules (MTs; Santaguida and Musacchio, 2009). In the budding yeast Saccharomyces cerevisiae, a single MT is captured by the kinetochore; the organization and composition of this site has been determined through high-resolution light and electron microscopy (Joglekar et al., 2009; Gonen et al., 2012). In vertebrate cells, the number of kinetochore-bound MTs (KMTs) increases during prometaphase, and by metaphase, the kinetochore MT fiber (K fiber) contains 20–30 MTs (McDonald et al., 1992; McEwen et al., 1997). The individual MT binding site of a vertebrate kinetochore appears to have a similar molecular makeup and organization to the budding yeast site (Joglekar et al., 2008). It is therefore plausible that the vertebrate KMT interface is composed of repetitive MT-binding units (Fig. 1 A; Zinkowski et al., 1991).
Figure 1.
Regulation of kinetochore–MT interactions by Hec1 phosphorylation. (A) Schematic of the KMT interface with repetitive MT binding sites (circles), each containing multiple NDC80 complexes. (B) Schematic of the NDC80 complex and location of Aurora B phosphorylation sites in the Hec1 tail. N, N terminus. (C) Fluorescence images of PtK1 cells depleted of endogenous Hec1 and rescued with phosphomimetic versions of Hec1 in an A background fused to GFP. Cells were fixed after a brief incubation in ice-cold media to reduce the number of nonkinetochore spindle MTs. Bar, 10 µm. (D) Quantification of chromosome alignment phenotypes for the Hec1 silence/rescue experiment. At least 108 cells were analyzed for each Hec1 mutant from three separate experiments.
Consistent with this view, prevailing theoretical models of the kinetochore assume that one KMT end binds to a defined set of MT-binding proteins, which are clustered in one site, structured as a sleeve or a ring (Hill, 1985; Efremov et al., 2007). The essential feature of such a theoretical interface is that one MT end can interact only with the MT-binding proteins that belong to that site, so these molecular interactions are constrained, and there is no cross-binding by kinetochore proteins from different sites (Joglekar and Hunt, 2002). Electron microscopy of vertebrate kinetochores, however, has not yet revealed structural repeated units that could correspond to such protein clusters (Dong et al., 2007; McIntosh et al., 2013). This opens a possibility that the KMT interface represents an unstructured, random assortment of MT-associated proteins (MAPs), which we will refer to as a molecular “lawn.” The rules of engagement between the structurally unconstrained MAPs of the molecular lawn and the MTs, and whether such interactions can explain observed mitotic phenotypes, have not been examined. For example, it is possible that one MT end may interact only with a defined set of “dedicated” MAPs within the molecular lawn (Powers et al., 2009; Tooley and Stukenberg, 2011), such that the resulting interface remains similar to the traditionally described interface with repetitive sites, despite different structural organization. Such restricted interactions, for instance, can occur if the kinetochore MAPs interact with each other on the MT surface, so their binding may become biased to the same MT (Alushin et al., 2010; Tooley and Stukenberg, 2011). Alternatively, the kinetochore MAPs may retain their independence, and their interactions with MTs may be unconstrained throughout mitosis. In this case, these MAPs may dissociate from one KMT and bind to a nearby KMT, thereby engaging dynamically with different KMTs.
The importance of the rules of engagement between kinetochore MAPs and KMTs, as reported here, became apparent during our quantitative study of the regulation of kinetochore–MT binding affinity by phosphorylation of the NDC80 protein complex. This complex is the major MT-binding component of eukaryotic kinetochores, and it is comprised of two heterodimers: Spc24-Spc25 and Hec1-Nuf2 (Fig. 1 B). The Hec1-Nuf2 dimer directly binds MTs, and Hec1 also has an unstructured “tail,” which is required for KMT attachment in vivo and contributes to MT binding in vitro (Wei et al., 2007; Guimaraes et al., 2008; Miller et al., 2008). This tail is an established target for Aurora kinases (Cheeseman et al., 2002; Santaguida and Musacchio, 2009); nine phosphorylation sites have been mapped in vitro, and six have been confirmed to be phosphorylated in cells (DeLuca et al., 2006, 2011; Nousiainen et al., 2006; Ciferri et al., 2008; Malik et al., 2009). Previous work suggested that phosphorylation of Hec1 is a powerful regulator of MT binding affinity of the kinetochore: whereas there were virtually no stable KMT attachments in cells expressing a Hec1 mutant in which all nine phosphorylation sites were substituted with phosphomimetic aspartic acid, cells expressing a nonphosphorylatable Hec1 mutant had hyperstable KMT attachments (Guimaraes et al., 2008; DeLuca et al., 2011). In vitro, phosphorylation of the Hec1 tail modulates NDC80–MT affinity (Cheeseman et al., 2006; Umbreit et al., 2012), and MT binding affinity decreases incrementally with an increasing number of Hec1 phosphomimetic substitutions (Table 1). However, whether overall kinetochore–MT binding affinity in cells responds to small incremental changes in Hec1 phosphorylation has remained unknown.
Table 1.
Kinetic constants used for modeling NDC80–MT interactions in both models
| Symbol | Model parameter | Construct/model | Value | Reference |
| s−1 | ||||
| kon | MT association rate for NDC80 complex | Site model | 1.52 | Based on model calibration (Fig. S3 A) |
| Lawn model | 0.80 | |||
| koff | MT dissociation rate for NDC80 complex | noD | 2.40 | Based on in vitro measurements with purified NDC80 proteins |
| 1D | 4.17 | |||
| 2D | 7.00 | |||
| 3D | 8.56 | |||
| 4D | 10.66 | |||
| 9D | 46.30 | |||
| ω | Cooperativity parameter for NDC80 complexes | 2.5a | Based on in vitro measurements with purified NDC80 proteins (unpublished data) |
No dimension is associated with this value.
To create an integrated view of the phosphoregulation of MT binding affinity via NDC80 complexes, we have combined quantitative approaches in vivo and in silico. Our experiments in live cells demonstrate that changing the number of phosphomimetic substitutions on Hec1 in a dynamic range of one to four residues exerts a smooth, graded tuning of KMT binding affinity. The increased levels of phosphorylation in prometaphase drive high KMT turnover, which is required to correct KMT attachment errors, whereas low but persistent levels of phosphorylation in metaphase facilitate both stabilization of KMT attachment and coordinated oscillations of metaphase chromosomes. Importantly, increasing the number of phosphomimetic substitutions leads to a gradual loss of the number of KMTs. Surprisingly, this behavior cannot be explained by a mathematical model in which the kinetochore is represented as a traditional ensemble of repetitive sites. However, a model in which the KMT interface contains a molecular lawn of NDC80 complexes and MAPs, which are not constrained and can interact simultaneously with different KMTs, provides a good match to our findings in cells. This study demonstrates for the first time that the rules of molecular interactions between MTs and kinetochore MAPs, not just their structural and molecular characteristics, play a critical role in ensuring proper regulation of the overall MT binding affinity at the kinetochore.
Results
Proper chromosome segregation requires dynamic Hec1 tail phosphorylation
To allow for precise control of the phosphomimetic state of the Hec1 tail, we used a silence and rescue system in PtK1 cells in which endogenous Hec1 was depleted and rescued with either wild-type (WT) or mutant Hec1 fused to GFP. We generated a series of Hec1 mutants in which one, two, three, four, or all nine of the in vitro–mapped Aurora B kinase target sites were mutated to Asp (D) to mimic phosphorylation, whereas all the remaining target sites were mutated to Ala (A), so no endogenous phosphorylation could occur (Fig. 1 B; DeLuca et al., 2006; Ciferri et al., 2008). Aspartic acid is often used for phosphomimetic substitution because it is chemically similar to phospho-Ser, although at physiological pH, native phospho-Ser carries a negative charge of ∼1.9 compared with ∼1.0 for Asp in a peptide chain (Zachariou et al., 1996). All fusion proteins localized correctly to kinetochores and supported formation of normal bipolar spindles in mitosis (Fig. 1 C). For all Hec1 mutants, their expression levels were similar to WT, and the residual amount of endogenous Hec1 was <4% (Fig. S1, A–D). We tested whether preventing phosphorylation of Hec1 perturbed the phosphorylation status of other kinetochore components, but there was no significant change in phosphorylation of KNL1 or Dsn1 in human cells expressing 9A Hec1 (Fig. S1, E–H). We gradually increased the phosphomimetic status of the Hec1 tail by introducing constructs with an increasing number of D substitutions and found that none permitted cells to align chromosomes as efficiently as cells expressing WT Hec1 (Fig. 1 D). Thus, dynamic phosphorylation of the Hec1 tail is required for cells to proceed normally through mitosis. This is not surprising because Hec1 phosphorylation levels change during mitosis: multiple sites are highly phosphorylated in early mitosis, and phosphorylation decreases as mitosis progresses (DeLuca et al., 2011).
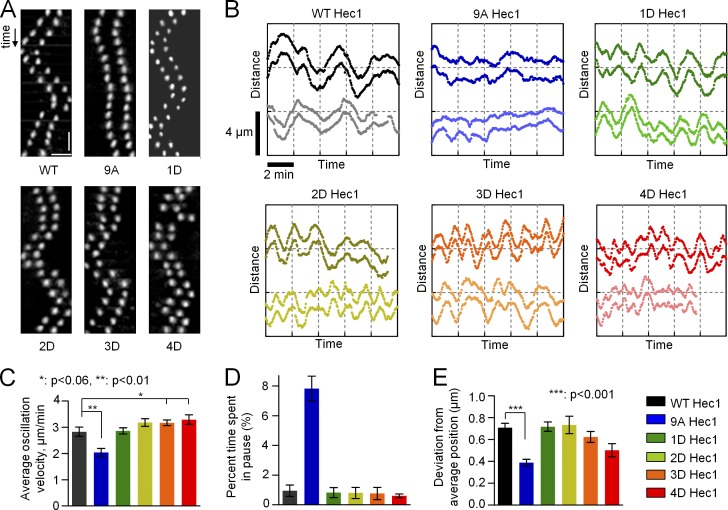
A single Hec1 phosphomimetic substitution supports WT oscillations of metaphase chromosomes
We then hypothesized that constant levels of Hec1 phosphorylation may nonetheless facilitate KMT interactions during specific mitotic stages. In metaphase, kinetochores must stabilize attachments to MTs but still fluidly track dynamic MT ends. We asked what level of Hec1 tail domain phosphomimetic substitution is required to support WT metaphase kinetochore movement. Kinetochore oscillations were analyzed in cells depleted of Hec1 and rescued with different Hec1 mutants. In cells rescued with WT Hec1, kinetochores oscillated normally, whereas oscillations in cells expressing 9A Hec1 were significantly dampened (Fig. 2; DeLuca et al., 2011). Interestingly, cells expressing Hec1 with a single phosphomimetic substitution regained WT oscillatory motion (Fig. 2, A and B). On bioriented sister kinetochores in 1D Hec1-expressing cells, the mean velocity, pausing, and oscillation amplitude did not differ from WT Hec1-expressing cells (Fig. 2, C–E). As the number of phosphomimetic substitutions increased, the oscillations became more erratic (Fig. 2, A and B), reminiscent of cells with defective KMT attachment. These results correlated with the number of phosphomimetic substitutions but not their exact positions. Indeed, we found no statistical difference in oscillation characteristics for three different 1D mutants: S8D, S15D, and S55D (Fig. S2, A–C). Together, these data suggest that normal oscillations of congressed chromosomes do not require dynamic Hec1 phosphorylation and that a single phosphomimetic residue at any of several sites is sufficient for fluid tracking of dynamic MT ends.
Figure 2.
Phosphoregulation of metaphase chromosome oscillations. (A) Kymographs (time and distance scale bars are 24 s and 2 µm, respectively) of representative sister kinetochore pairs from bioriented chromosomes in cells rescued with WT Hec1-GFP or the indicated phosphomimetic mutants generated in an A background. (B) Representative tracings for two sister kinetochore pairs from the indicated backgrounds. The y axis shows the relative position along the spindle axis; time and distance scale bars are the same for all graphs. The vertical offset between each set of pairs is for easier visualization. (C) Mean velocity of kinetochore movement along the spindle axis. See legend on the right for color coding. Data for each bar in C–E are based on 22 kinetochore tracks from at least seven cells. Here and in D and E, the results for 1D Hec1 are the means for three different mutants (Fig. S2, A–C). (D) Time spent with no motion for two sequential frames, or 6 s, normalized to the total time of the time lapse. (E) Deviation from average position, a measure of oscillation amplitude (Stumpff et al., 2008). Error bars are SEMs.
A single Hec1 phosphomimetic substitution facilitates formation of normal-sized K fibers in metaphase
To study how Hec1 phosphorylation regulates KMT binding affinity, we measured interkinetochore distances on aligned chromosomes in metaphase cells with various backgrounds (Fig. 3 A). This distance was enlarged in cells expressing 9A Hec1, indicative of hyperstable KMT attachments (Fig. 3 B; Guimaraes et al., 2008). As the number of phosphomimetic substitutions increased, interkinetochore distances decreased, consistent with destabilization of KMT attachments. Three different 1D phosphomutants showed highly similar results (Fig. S2 D). None of the mutants precisely matched the interkinetochore distance in WT cells, but 2D and 1D constructs provided the closest fit. Next, we determined the size of K fibers on bioriented chromosomes in these cells by quantifying the brightness of KMTs with an antitubulin antibody (Fig. 3 A). The mean K-fiber intensity in 1D Hec1 cells was similar to that measured in WT Hec1 cells (Fig. 3 C), suggesting that in metaphase the MT-binding properties of 1D Hec1 correspond well to that of WT Hec1. We also found that K-fiber intensities in cells expressing several different 1D constructs were not statistically different from each other or from those measured in cells expressing WT Hec1 (Fig. S2 E), further supporting our conclusion that the number of substitutions, rather than their exact location, is important for regulating K-fiber size.
Figure 3.
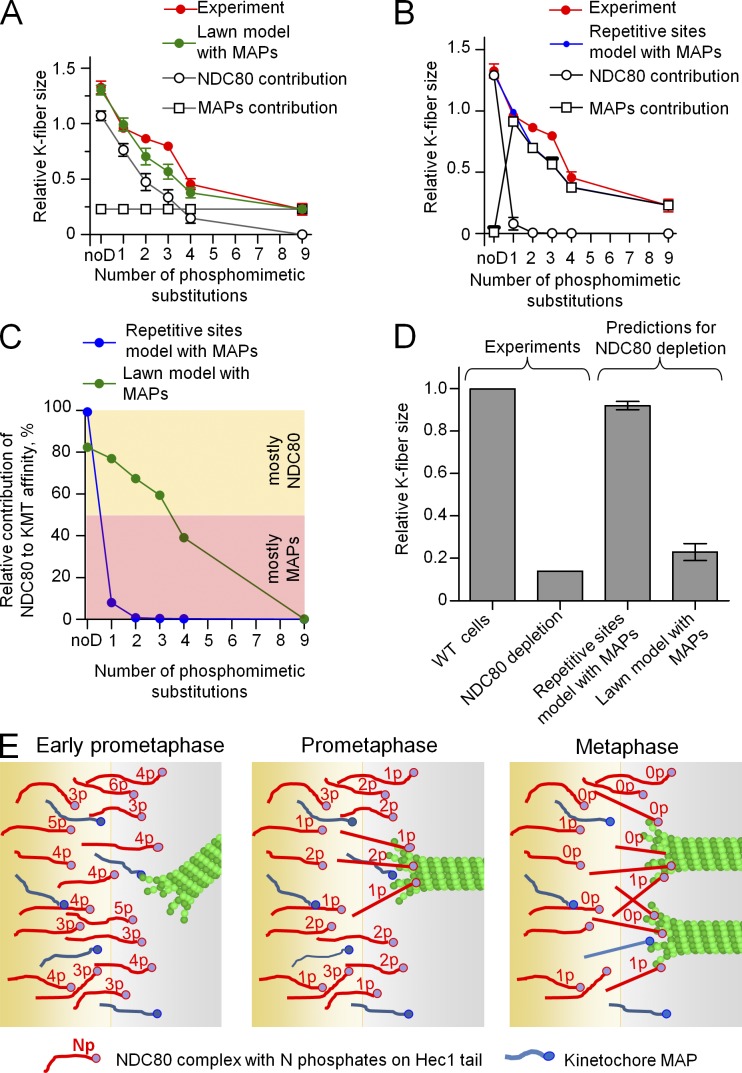
Quantitative analysis of the phosphoregulation of K fibers in metaphase cells. (A) Example metaphase PtK1 cell depleted of endogenous Hec1 and rescued with 1D Hec1, fixed, and stained with antitubulin and anti-Hec1 antibodies. The white box shows the area in which the mean intensity of tubulin staining was measured to compare relative sizes of K fibers in different cells. Bar, 1 µm. (B) Interkinetochore distances in metaphase cells expressing Hec1 with different numbers of phosphomimetic substitutions. Results for 1D Hec1 are averaged between three mutants (Fig. S2 D). Shaded area shows the range for interkinetochore distances in WT Hec1 cells. The broken line shows interkinetochore distance in the absence of MTs: 1.0 µm (DeLuca et al., 2006, 2011). At least 127 kinetochores were analyzed in ≥30 cells for each Hec1 mutant. (C) Relative K-fiber intensities in cells expressing Hec1 with different numbers of phosphomimetic substitutions. Results for 1D Hec1 are averaged between three Hec1 mutants (Fig. S2 E). Mean intensity of K fibers in WT cells was chosen as 1; shaded area shows the corresponding range. The value for 9D Hec1 is significantly different from 0 (P < 0.05), which is predicted from the linear relationship. The black line is a best linear fit of experimental points for zero to four phosphomimetic substitutions. At least five K fibers were measured per cell in ≥22 cells for each Hec1 mutant. Error bars are SEMs. a.u., arbitrary unit.
This approach allowed us to determine the exact relationship between the level of Hec1 tail phosphorylation and K-fiber size in metaphase cells. As the number of phosphomimetic substitutions increased from zero to four, the MT binding affinity of kinetochores decreased gradually (Fig. 3 C, black line). Consistent with previous reports, cells expressing 9D Hec1 exhibited a severe decrease in mean K-fiber size; however, this loss was significantly less than what is expected from a linear dependence (Fig. 3 C; Guimaraes et al., 2008). Thus, the overall affinity of kinetochores for MTs at the KMT interface responds to Hec1 phosphorylation in a complex manner.
A molecular model with repetitive binding sites can accurately describe KMT properties observed in cells expressing 1D Hec1
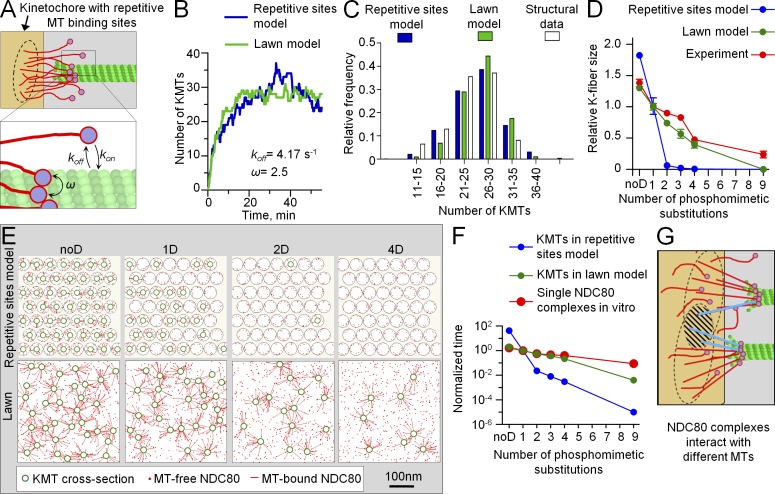
KMT interactions are highly complex, and multiple kinetochore proteins are involved in regulating the overall MT binding affinity during mitosis. Because Hec1-containing NDC80 complexes are essential for these attachments, we next examined how much of this cellular phenomenology could be explained by the molecular properties of this one component. We initially used a simple mathematical model of the KMT interface, which was composed of repetitive MT binding sites, each containing multiple copies of NDC80 complexes (Fig. 1 A; Zaytsev et al., 2013). All NDC80 complexes that belonged to one site could bind and unbind MTs stochastically, but these molecular interactions were constrained: only one MT interacted with NDC80 complexes that made up one site. As in our cell experiments, the NDC80 complexes in silico had a specific number of phosphomimetic mutations that did not change during the course of the simulations. The K-fiber size was calculated as a mean number of MTs attached to such a kinetochore interface at steady state (Video 1).
The values of model parameters that described the composition of a KMT interface (i.e., number of MT binding sites, number of NDC80 complexes per site, etc.) were taken from the literature or selected as described in the Materials and methods. Three kinetic parameters were used to describe stochastic molecular binding between NDC80 complexes and MTs: the MT association and dissociation rates for single NDC80 complexes and the cooperativity factor, which took into account NDC80–NDC80 interactions when these molecules were bound on an MT wall (Fig. 4 A). The values of the dissociation rate and cooperativity factor for NDC80 complexes were taken from our in vitro affinity measurements of recombinant NDC80 complexes containing 1D Hec1 (Table 1). The single unconstrained parameter, which corresponded to the MT association rate of the NDC80 complex, was varied to achieve normal K-fiber size at steady state (Fig. S3 A). This resulted in a fully defined molecular-kinetic model, which reproduced not only the final K-fiber size but also the kinetics of its formation (Fig. 4 B). The distribution of KMT numbers at steady state also matched the structural data for metaphase PtK1 cells (Fig. 4 C). Moreover, these KMTs turned over with a half-life of 8–10 min, similar to the rates reported in vivo (DeLuca et al., 2006). Thus, a model with repetitive, constrained NDC80-containing binding sites can describe many features of KMT interactions at metaphase, when the phosphorylation status of Hec1 is constant.
Figure 4.
Mathematical model of the KMT interface. (A) NDC80–MT binding is characterized by three molecular parameters: association rate (kon), dissociation rate (koff), and cooperativity parameter (ω). (B) Calculated kinetics of the increase in K-fiber size for the kinetochore with 1D NDC80 complexes for the repetitive sites model and lawn model. Values of other model parameters are provided in Table 1 and Table 2. Both models correctly predict that formation of a K fiber with a mean of 27 KMTs takes ∼20 min in PtK1 cells (McEwen et al., 1997). (C) Histogram distribution of the number of KMTs at a steady state in the repetitive sites model and lawn model relative to metaphase cells (experimental data are from McEwen et al., 1997). (D) K-fiber size for different numbers of phosphomimetic substitutions predicted in the two models normalized relative to the normal K-fiber size (27 KMTs in PtK1 cells). Error bars here and for all theoretical plots are SDs. Here and in Fig. 6 (A and B), experimental data for K-fiber size are the same as in Fig. 3 C. (E) Snapshots from theoretical videos for kinetochores with different NDC80 complexes (Video 1 for 1D mutant in the repetitive sites model and Video 2 in the lawn model). Each image is a fragment of a kinetochore cross section at steady state. The exact positions of attached MTs change with time, but the mean number of MTs bound to the kinetochores stays roughly the same at steady state. (F) Mean KMT half-life calculated for kinetochores with different phosphomutants and normalized relative to KMT half-life for 1D NDC80 complexes, which was 9 min in both models (owing to calibration) and was taken as 1 on this graph. Experimental data for single NDC80 complexes are mean residency times in vitro, which is an inverse of the dissociation rate (Table 1). Residency time for 1D NDC80 is 240 ms (taken as 1 on this graph). Note that time is plotted on a logarithmic scale. (G) Schematic of the KMT interface in the lawn model, in which the sites of MT attachment to the kinetochore do not have a predetermined location or molecular makeup, and they form via stochastic contacts between MTs and the NDC80 complexes that can reach them (black circles). The hatched area shows an overlap between two such circles; molecules from this area (in blue) can engage in binding to either of the two MTs.
A repetitive binding site model cannot explain the observed phosphoregulation of KMT affinity
We next asked whether this model could also match our observed changes in K-fiber size in response to Hec1 phosphorylation. These calculations were performed using the same values for all model parameters as described for 1D Hec1, but the dissociation rates for NDC80–MT binding were varied to match the experimental measurements in vitro with NDC80 complexes containing different numbers of phosphomimetic substitutions (Table 1). Fig. 4 D shows that in the repetitive sites model, the K-fiber size is predicted to be highly sensitive to phosphorylation: KMTs in silico were densely packed at kinetochores with unphosphorylated Hec1 (Fig. 4 D, noD, which is analogous to 9A Hec1 in cells), whereas a good match was obtained with 1D Hec1. However, there were virtually no KMTs when Hec1 was phosphorylated on more than one residue (Fig. 4 E). Consequently, the number of KMTs in silico decreased with phosphorylation much more steeply than what we measured in cells (Fig. 4 D). To try to improve the fit, we systematically varied relevant model parameters and model features, but none of these changes improved the match to our experimental data (see Materials and methods, Mathematical modeling of the kinetochore–MT interface). For example, the response to phosphorylation remained very steep in the repetitive sites model with different numbers of NDC80 complexes per MT binding site (Fig. S3 B), in the model in which the position of these sites within the kinetochore was randomized (Fig. S3 C) or when the cooperativity factor for NDC80–MT binding was varied (Fig. S3 D). Next, we hypothesized that the ratio of dissociation rates of differentially phosphorylated NDC80 complexes in cells was different from that measured in vitro. Such a difference could arise from kinetochore tension (Akiyoshi et al., 2010), which likely changes in response to K-fiber size, as seen from changes in interkinetochore stretch for different phosphomutants (Fig. 3 B and Fig. S3 E). However, when the dissociation rates for NDC80 complexes in silico were adjusted to take this effect into account, the phosphorylation response was still steeper than in the experiment (Materials and methods, Mathematical modeling of the kinetochore–MT interface; Fig. S3 F). Finally, we used two additional strategies to calibrate the repetitive sites model, one based on the 2D Hec1 data instead of 1D and another that used the value of kon estimated from the known kinetochore size and NDC80 density (Materials and methods section Model calibration). With these modifications, the phosphoregulation in the repetitive sites model was also toggle-like (Fig. S3, F and G), demonstrating that the details of calibration do not affect this behavior.
Based on these results, we concluded that the abrupt response to phosphorylation is a robust feature of the repetitive sites model, and it cannot be improved by changing parameter values or other model features. The root of the problem appears to be in the overall design of a KMT interface that contains clusters of NDC80 complexes in each MT binding site. This is illustrated in Fig. 4 F by comparing the relative changes in mean interaction time for NDC80 phosphoproteins and MTs in vitro and the relative changes in mean KMT half-life predicted for the corresponding kinetochores. Each phosphomimetic substitution decreases the molecular interaction time gradually by ∼1.7-fold per substitution (Table 1). In contrast, the interface with repetitive sites composed of such molecules strongly amplifies this relatively small phosphorylation-dependent tuning of individual NDC80 complexes (Fig. 4 F). Such amplification is a straightforward consequence of the combinatorial action of multiple NDC80 complexes within each site and the fact that complexes from one site are restrained to bind only one MT (see Discussion).
A model in which NDC80 complexes have unrestrained interactions with KMTs provides a good fit to all experimental data
We then examined an alternative model in which the KMT interface did not have defined binding sites. Instead, the same number of NDC80 complexes used in the repetitive sites model was distributed randomly on the kinetochore surface (Fig. 4 G and Video 2). Importantly, the interactions between different NDC80 complexes and MTs were constrained only by their distance to an MT, not by their association with a specific site. Thus, in the course of a simulation, the same NDC80 complex could switch randomly between several MTs (Video 3). Unlike in the repetitive sites model, this forced the incoming MTs to compete for NDC80 complexes, interacting with any adjacent and available NDC80 complex within a molecular lawn. As a result, molecular interactions are not restricted and can detect and respond to what is happening in nearby areas, not just in one site. Our calculations demonstrate that the lawn model retained all positive features of the repetitive sites model with a constant level of Hec1 phosphorylation. Such an interface could support the formation of WT-sized K fibers, and the model described well both the dynamic steady-state distribution of KMTs and their turnover rate (Fig. 4, B and C). Unlike in the repetitive sites model, however, the “tuning” of the lawn KMT interface corresponded directly to the phosphorylation-induced changes at the single molecule level with little additional amplification in the zero to four range of phosphomimetic substitutions (Fig. 4 F). In this range, the model showed a graded response to phosphorylation, which fit well to the phosphomimetic-dependent changes of the K fiber in cells (Fig. 4, D and E). This behavior of the lawn model was robust, as it did not depend on how the model was calibrated (Fig. S3 G) or the residual amount of endogenous NDC80 complexes (Fig. S3 H), which we included in the kinetochore models to mimic incomplete siRNA depletion (Fig. S1 D).
KMT turnover in early mitosis requires three to four phosphomimetic substitutions on the Hec1 tail, consistent with the lawn model prediction
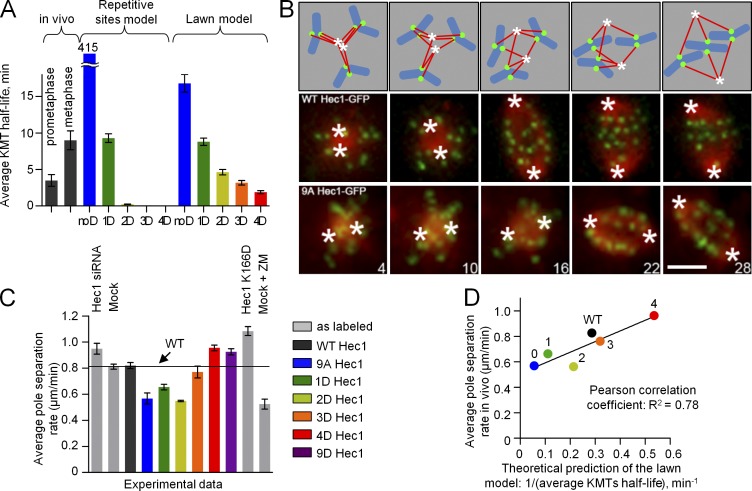
To further discriminate between the models, we examined their predictions regarding KMT interactions in prometaphase cells, when KMTs are less stable. We calculated the mean KMT half-life in these models for different levels of Hec1 phosphorylation. The experimental mean KMT half-life in metaphase cells (9 min; DeLuca et al., 2006) was matched well in both models by using the dissociation rate of 1D Hec1 complexes (Fig. 5 A). The experimental mean KMT half-life in prometaphase cells (3.5 min; Cimini et al., 2006) was best matched by the repetitive sites model when the interface was composed of 1.4D Hec1 proteins. Therefore, this model predicted that cells expressing Hec1 with one or two phosphomimetic substitutions should behave in prometaphase most similarly to WT Hec1. The lawn model, however, predicted that the best match to prometaphase should be seen in 3D Hec1-expressing cells. To test these predictions, we used an established assay with monastrol, which blocks cells in prometaphase (Mayer et al., 1999). Upon mitotic entry, the monastrol-treated cells become monopolar and accumulate numerous incorrect KMT attachments (Fig. 5 B; Kapoor et al., 2000). Upon monastrol washout, the spindle poles separate at a rate limited by the kinetics of the release of MTs from kinetochores (Vader et al., 2007). We therefore used the rate of pole separation as a readout for the efficiency of KMT release in prometaphase cells (Fig. S4). As the number of phosphomimetic substitutions increased, the pole separation rate also increased, as expected (Fig. 5 C). Importantly, cells with three to four substitutions had the rate most similar to WT Hec1-expressing cells. Moreover, the entire dataset with phosphomutants in vivo showed a significant correlation with predictions for the lawn model but not the repetitive sites model (Fig. 5 D). Thus, consistent with the lawn model, three to four phosphomimetic substitutions in the Hec1 tail are required to support prometaphase KMT dynamics.
Figure 5.
Phosphoregulation of the KMT interface in prometaphase cells. (A) Mean KMT half-life calculated with the two models relative to these values measured previously in PtK1 cells in prometaphase and metaphase (3.5 and 9 min, respectively; Cimini et al., 2006; DeLuca et al., 2006). Both models were calibrated such that the interfaces with 1D NDC80 complexes matched the metaphase half-life. The repetitive sites model predicts very unstable KMT attachments for 2D, 3D, and 4D proteins, so these bars are virtually invisible, whereas the large half-life for noD NDC80 kinetochores is plotted with an interrupted axis. (B) Schematics and representative still images from time-lapse videos of PtK1 cells subjected to a monastrol washout assay. Numbers are times (in minutes) from monastrol washout. Hec1-GFP is shown in green (kinetochores); mCherry-tubulin is shown in red (MTs); cartoon chromosomes are shown in blue; asterisks show position of poles. Bar, 5 µm. (C) Rate of pole separation after monastrol washout. ZM, Aurora B inhibitor ZM447439. The bar labeled Hec1 K166D corresponds to a non-MT–binding Hec1K166D mutant (Ciferri et al., 2008; Sundin et al., 2011). n = 10 cells were analyzed for each Hec1 mutant. (D) Correlation plot for in silico and in vivo data from A and C, respectively. Pearson correlation analysis shows that the correlation is significant for the lawn model but not the repetitive sites model (R2 is 0.78 and 0.51, respectively, with threshold value of 0.05). Error bars are SEMs.
Incorporation of other MT-binding proteins in the lawn model further improves the fit to experimental data
The lawn model most accurately predicted KMT phosphoregulation in cells; however, the MT-binding capacity of 9D Hec1 kinetochores in cells was higher than predicted by the model (Fig. 4 D). This suggests that at higher levels of Hec1 phosphorylation, the contribution from other kinetochore MAPs is significant. There are many known kinetochore MAPs, but their molecular constants are not yet known. For simplicity, we have supplemented the NDC80 lawn with only one type of MAP; its affinity was chosen such that this “generic” MAP compensated the K-fiber size in the model with 9D NDC80. The contribution of this MAP during other mitotic stages (corresponding to the different levels of Hec1 phosphorylation) was set as constant. This combined NDC80-MAP lawn improved the model fit relative to a model with only NDC80 complexes (Fig. 6 A vs. Fig. 4 D).
Figure 6.
Analysis of contribution of other kinetochore MAPs to KMT affinity. (A) Relative K-fiber size predicted for different molecular lawns. The green curve is the total kinetochore affinity, and it is a sum of the contributions from NDC80 complexes with different degrees of phosphorylation and MAPs. (B) Relative K-fiber size predicted for different phosphomutants in the repetitive sites model. Contribution from NDC80 complexes was renormalized (compare with Fig. 4 D), such that the predicted number of KMTs did not exceed the experimentally measured K-fiber size. The MAP contribution is a difference between the best fit with the lawn model (A) and the contribution from NDC80 complexes. The combined affinity in the repetitive sites model was thus matched to the combined affinity in the lawn model (A). (C) Percentage of contribution of NDC80 complexes to the total KMT affinity calculated for the two models with MAPs. Both models provide a match to K-fiber size in cells (see A and B), but they predict different relative NDC80 versus MAP contribution. (D) Relative K-fiber size for kinetochores with depleted Nuf2 (a subunit of the NDC80 complex) in vivo (data from DeLuca et al., 2005) and predictions of the two models in which only MAPs are present. Experimental and theoretical data were normalized to the number of KMTs for WT cells. (E) Summary diagram of the tunable KMT interface. NDC80 complexes and other MAPs comprise a molecular lawn that interacts dynamically with competing MTs. Early in mitosis, NDC80 is likely to be hyperphosphorylated, so other MAPs provide a major contribution to the overall weak KMT binding. As MTs begin to attach, the mean phosphorylation on the Hec1 tail decreases to a range of one to two phosphates, and the contribution of NDC80 becomes significant. Further dephosphorylation in metaphase reduces the mean number of phosphates to a range of zero to one, and the contribution of NDC80 complexes to MT binding dominates. p indicates the number of phosphates. This drawing underrepresents the number of MT-bound NDC80 complexes, which is estimated to be 10–12 (Fig. S5 B). Error bars are SEMs.
Similar modifications were then applied to the repetitive sites model, but a good match to the KMT phosphoregulation in cells was still not possible (Fig. S3 J). This combined sites model produced the same fit as the combined lawn model only if the contributions from MAPs were adjusted for every level of Hec1 phosphorylation to compensate for the differences (Fig. 6 B). However, even with such arbitrary adjustments, which allowed fitting to the phosphoregulation curve, the repetitive sites model still conflicted with other experimental findings because it predicted a yin–yang relationship for NDC80 complexes and MAPs. Indeed, in such a combined repetitive sites model, the toggle-like contribution from NDC80 was complemented by the toggle-like contribution from MAPs, such that the overall KMT affinity was provided either by MAPs or by NDC80, when it was fully dephosphorylated (Fig. 6 C). Clearly, this behavior contradicts the established essential role of the NDC80 complex as a core KMT attachment factor. When NDC80 complexes were depleted in silico, the combined sites model predicted only a 5% reduction in K-fiber size, in contrast to our measurements in cells (DeLuca et al., 2005). In the combined lawn model, however, the metaphase K-fiber size was reduced approximately fivefold, consistent with experimental findings (Fig. 6 D). Thus, the presence of other compensatory MAPs within the repetitive sites cannot explain the severe lack of consistency between this model and the experimental data.
Discussion
To create a comprehensive view of MT binding affinity at a dynamic kinetochore interface, we have performed an integrated study of the impact of phosphorylation of the NDC80 complex, the major kinetochore MT-binding component. Phosphomimetic substitutions in the N-terminal tail of its Hec1 subunit were systematically and quantitatively examined at the cellular level and with advanced computational approaches. These experiments have led to new insights into the regulation of KMT affinity by Hec1 tail phosphorylation, the respective contributions of the NDC80 complex and other MAPs to overall KMT affinity, and the general design of the KMT interface.
Phosphorylation of Hec1 tunes KMT affinity from prometaphase to metaphase
The Hec1 tail is hyperphosphorylated early in mitosis, and phosphorylation decreases from prometaphase to metaphase (DeLuca et al., 2011). However, of the nine Aurora B sites on the Hec1 tail, the physiological range of sites phosphorylated during these stages was previously unknown. Here, we show that during prometaphase and metaphase, the range of Hec1 phosphorylation is surprisingly narrow: from three or four to one phosphomimetic substitutions. The charge of one phospho-Ser is 1.9× higher than that of Asp (Zachariou et al., 1996), so these data suggest that Hec1 phosphorylation varies from not more than two phosphoresidues in prometaphase down to zero or one in metaphase. Because the dynamicity of this posttranslational modification is necessary for accurate chromosome segregation (Fig. 1), these values are likely to correspond to the mean level of Hec1 tail phosphorylation during these mitotic stages. The actual range of phosphorylation at single NDC80 complexes may be higher. It may also vary more significantly at shorter time scales or differ within the kinetochore locally, depending on the number and accuracy of KMT connections in different kinetochore areas. It also remains to be seen whether the exact molecular location of the added phosphate in the Hec1 tail is important. Although all our assays showed highly similar results with mutants that have the same number of phosphomimetic substitutions regardless of their exact locations, this collection of mutants is far from being exhaustive (512 different combinations are possible). In cells, phosphorylation at different positions may have additional physiological consequences, affecting Hec1 tail interactions with other kinetochore components or processes.
We also show that in metaphase spindles, the incremental changes in Hec1 tail phosphorylation are translated into small, gradual changes in overall kinetochore–MT affinity (Fig. 3, B and C). Such tuning ability is likely to be important for accurate adjustment of KMT attachments, which take place dynamically at the kinetochores of each separate chromosome. The variability in different system parameters during mitosis (chromosome and kinetochore size, chromosome position within the metaphase plate, etc.) is likely to be significant, so accurate and precise local tuning may be required. Importantly, our study reveals that the role of the NDC80 complex in KMT affinity increases gradually during mitotic progression (Fig. 6 C). Early in mitosis, when the Hec1 tail is hyperphosphorylated (DeLuca et al., 2011), its contribution to kinetochore–MT binding appears to be too low to explain the formation of stable KMT attachments. At this stage, other kinetochore-associated MAPs, such as CENP-E, CENP-F, KNL1, dynein, or the Ska complex (Santaguida and Musacchio, 2009), may play a crucial role in engaging the MTs. As Hec1 phosphorylation decreases, the NDC80 contribution to overall KMT affinity begins to dominate (Fig. 6 C). As new quantitative details about other MAPs and regulation of their MT affinity begin to emerge, our modeling approaches will be valuable in dissecting specific contributions of different MAPs and in revealing how their complex ensemble tunes the kinetochore for diverse mitotic functions. We emphasize, however, that the current lawn model is already significantly constrained by numerous experimental juxtapositions. This suggests that although the exact ratio and percentage of contributions for different MAPs may change, the major conclusions about the rules of engagement for MAPs (NDC80 and others) with KMTs and the overall design of the KMT interface are likely to hold true.
A dynamic lawn model describes the major features of KMT affinity of mammalian kinetochores
Although molecular interactions between single NDC80 complexes and MTs in vitro are very short lived (Powers et al., 2009; Umbreit et al., 2012; Tien et al., 2013), our modeling shows that a kinetochore composed of multiple such molecules can support lasting KMT attachments (Fig. 4; Zaytsev et al., 2013). A good match to WT metaphase kinetochores can be obtained with a stochastic molecular model regardless of the overall design of the MT-binding interface. However, the rules of engagement between kinetochore proteins and MTs have a major impact on how this interface responds to NDC80 complex phosphorylation. In vitro, single phosphomimetic substitutions decrease the interaction time between individual NDC80 complexes and MTs gradually (Fig. 4 F), but the repetitive sites design dramatically amplifies this single molecule tuning, leading to the major conflicts between this model’s predictions and observations in cells (Fig. 4 D, Fig. 5 A, and Fig. 6 D). In contrast, the lawn model with unrestricted molecular interactions was successful in describing the experimental observations for different mitotic stages, including (a) size of a metaphase K fiber and kinetics of its acquisition (Fig. 4 B), (b) distribution of the number of MTs in metaphase K fibers (Fig. 4 C), (c) mean KMT half-life during both metaphase and prometaphase (Fig. 5 A), (d) aspects of KMT stability in monastrol washout cells for a panel of Hec1 phosphomutants (Fig. 5 D), (e) dependence of K-fiber size on molecular characteristics of NDC80 complexes with different degrees of phosphorylation (Fig. 6 A), and (f) results of NDC80 complex depletion experiments in cells (Fig. 6 D). These findings represent an important step toward creating a realistic theoretical model of the KMT interface, which should ultimately link the specific molecular and structural features of this interface with its physiological behavior.
Molecular interactions between MTs and kinetochore MAPs are predicted to be unconstrained
The current view of the molecular design of the KMT interface is firmly rooted in the concept of repetitive units, and KMT interactions are thought to occur via specialized MT attachment sites (Alberts et al., 2008). Our study argues strongly against a key feature of such a design: that the MAPs forming one site are constrained in their binding to a single MT. Although this feature is not usually emphasized, this assumption was present in previous theoretical studies of the KMT interface (Hill, 1985; Joglekar and Hunt, 2002; Liu and Onuchic, 2006; Efremov et al., 2007; Powers et al., 2009; Armond and Turner, 2010; Shtylla and Keener, 2010; Civelekoglu-Scholey et al., 2013; Keener and Shtylla, 2014). Abandoning this assumption and modeling the KMT interface as a molecular lawn has a surprisingly large impact on the regulatory characteristics of the kinetochore. Computer modeling helps to understand the molecular mechanism that is responsible for this nonintuitive result. With increasing phosphorylation, the mean time that one NDC80 complex remains bound to the KMT decreases (Fig. 4 F). Such weakening of the individual molecular bonds takes place regardless of the design of kinetochore interface. In the repetitive sites model, in accordance with the mass action law, this leads to a decrease in the number of NDC80 complexes bound to the KMT at each site (Fig. S5 A); KMTs begin to detach, and the K-fiber size drops precipitously (Fig. 4, D and E). Even small variations in the duration of molecular binding events in the repetitive sites model lead to a dramatic change in the number of KMTs for a wide range of model parameters and regardless of the calibration procedure (Fig. S3, F and G; Zaytsev et al., 2013). Such combinatorial amplification is a robust feature of any system in which multiple molecules are involved, consistent with our finding that the steepness of the response increases with an increasing number of NDC80 complexes per site (Fig. S3, B and F; and Fig. S5 C).
In the lawn interface, the weakening of the individual NDC80 bonds to KMTs also initially leads to a fast decrease in the number of KMT-bound NDC80 complexes (Fig. S5 F). But this decrease is transient because the loss of some KMTs frees the NDC80 complexes, thereby increasing the pool of complexes that are available for binding to the remaining KMT (Fig. S5 B and Video 3). Such binding reorganization is not possible at the interface built from repetitive sites, where by definition the NDC80 complexes in one site are dedicated to binding only the KMT at this site, such that the pool of NDC80 complexes that are available for interaction with one KMT is always constant (Fig. S5 A). Reorganization of the molecular bonds at the lawn interface provides the inherent compensatory effect that buffers the number of complexes bound to the remaining KMTs (Fig. S5, B and G), thereby dampening the systems’ response to molecular variations (Fig. 4, D and F). Such buffering explains why changes at the single molecule level of the NDC80 lawn are matched by the changes in KMT attachments without significant amplification (Fig. 4 F).
We emphasize that the crucial distinction between these models is not structural because, even in the lawn interface, the underlying molecular interactions could in principle become constrained. When we impose the constraining rule in our lawn model, it leads to the same results as the repetitive sites model, demonstrating that the rules of molecular engagement constitute the key difference between the two interfaces. Our work also does not argue against using the word “site” to describe the organization and molecular makeup of proteins that contribute to linking MT ends to kinetochores. The important implication of our study is that the local concentration of proteins that form the attachment site for an MT end at the mammalian kinetochore is variable, and it is sensitive to the number and stability of KMTs. The binding proteins can switch between several adjacent KMTs, so some proteins can be shared (Fig. S5, D and E). We propose that such variability in the number of molecular bonds to the KMT end allows the interface to monitor and respond accurately and expediently to the number and stability of KMT attachments during mitotic progression.
Materials and methods
Experimental methods
Cell culture and transfection of siRNA and DNA.
PtK1 cells were cultured in Ham’s F-12 medium, and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) at 37°C in 5% CO2. Media were supplemented with 10% FBS (Atlanta Biologicals) and 1% antibiotic/antimycotic solution. siRNAs directed to PtK1 Hec1 (5′-AATGAGCCGAATCGTCTAATA-3′) or human Hec1 (5′-AACCCTGGGTCGTGTCAGGAA-3′; catalog No. 1027424) were purchased from QIAGEN. 9A Hec1-GFP template (Guimaraes et al., 2008) was used to make specific serine (S) to aspartic acid (D) mutations using site-directed mutagenesis. PtK1 cells were harvested and counted to ensure that ≥106 cells were used for each reaction and then cotransfected with both Hec1 siRNA and plasmids encoding for Hec1-GFP fusion proteins using Solution R (Lonza) and Nucleofector program T-020 (Lonza). Cells were plated on acid-washed coverslips or in glass-bottomed dishes (MatTek Corporation) and analyzed 48 h after electroporation. All transfections of siRNA and DNA in HeLa cells were performed as described in Sundin et al. (2011). In brief, cells were transfected with siRNA using Oligofectamine (Invitrogen) and were subsequently transfected with plasmid DNA using FuGENE 6 (Roche) 24 h later. Cells were assayed 24 h after DNA transfection.
Immunofluorescence and microscopy imaging.
For PtK1 fixed cell immunofluorescence experiments, cells were extracted for 5 min on ice with PHEM (60 mM Pipes, 25 mM Hepes, pH 6.9, 10 mM EGTA, and 4 mM MgSO4) + 0.5% Triton X-100 and fixed for 3 min at room temperature with ice-cold methanol containing 5 mM EGTA. Cells were then transferred to a −20°C freezer for 20 min. Coverslips were blocked for 1 h at room temperature in 10% boiled donkey serum (BDS). Primary antibodies were prepared in 5% BDS: mouse α–α-tubulin (Sigma-Aldrich) at 1:200 and human α–anticentromere antibody (Antibodies, Inc.) at 1:300. Primary antibodies were incubated for 1 h at room temperature followed by an overnight incubation at 4°C. Secondary antibodies were incubated for 45 min at room temperature; secondary antibodies conjugated to Alexa Fluor 647, Alexa Fluor 488, or Rhodamine red-X (Jackson ImmunoResearch Laboratories, Inc.) were used at a dilution of 1:300. Coverslips were counterstained with DAPI and mounted in an antifade solution containing 90% glycerol and 0.5% N-propyl gallate. Fixed cell immunofluorescence experiments using HeLa cells were performed as described in Sundin et al. (2011), except that 100 nM microcystin (Sigma-Aldrich) was added to PHEM + 0.5% Triton X-100 during extraction, and cells were fixed with 2% paraformaldehyde in PHEM buffer. Primary antibodies were prepared in 5% BDS and used at the following concentrations: α-phospho-Dsn1 at 1:1,000, α–anticentromere antibody at 1:300, and α-phospho-KNL1 (a gift from I. Cheeseman, Whitehead Institute for Biomedical Research, Cambridge, MA) at 1:1,000. Secondary antibodies conjugated to Alexa Fluor 647, Alexa Fluor 488, or Rhodamine red-X were used at a dilution of 1:300. Affinity-purified antibodies against phosphorylated Dsn1 were generated at 21st Century Biochemicals, rabbits were immunized with the phosphorylated Dsn1 peptide QSWRRA[pS]MKETN, and serum was affinity purified. For fixed cell immunofluorescence, imaging was performed at room temperature. For live-cell imaging, experiments were performed at 37°C in Leibovitz’s L-15 media (Invitrogen) supplemented with 10% FBS, 7 mM Hepes, pH 7.0, and 4.5 g/liter glucose. Cells were chosen for analysis if they were both Cy5 (siRNA) and GFP (Hec1-GFP fusion protein) positive. All microscopy was performed using an imaging system (DeltaVision Personal DV; Applied Precision) with a 60×, 1.42 NA Plan Apochromat differential interference contrast oil immersion lens (Olympus) and a camera (CoolSNAP HQ2; Photometrics/Roper Scientific).
Data analyses.
To ensure that our analysis was not affected by different expression levels, the fluorescence intensity of Hec1-GFP at kinetochores was quantified, and only data from cells that expressed GFP within a defined range were collected (Fig. S1). For chromosome alignment assays, cells were scored as follows: aligned = all chromosomes contained within a metaphase plate at the spindle equator; unaligned = no discernable metaphase plate; and partially aligned = a visible metaphase plate with one or more chromosomes not aligned at the spindle equator. For live-cell imaging of kinetochore oscillations, cells were imaged 24 h after DNA transfection in Leibovitz’s L-15 media supplemented with 10% FBS, 7 mM Hepes, pH 7.0, and 4.5 g/liter glucose. Stage temperature was maintained at 37°C with an environmental chamber (Precision Control). Fluorescence images of GFP-Hec1–expressing cells were acquired every 3 s for 10 min. At each time point, five images were collected in a z stack, using a 0.5-µm step size. Cells chosen for analysis were in late prometaphase or metaphase with primarily bioriented chromosomes. Sister kinetochore pairs chosen for analysis were located within the middle of the spindle. Kinetochore movements were tracked using GFP-Hec1 fluorescence on maximum projection time-lapse sequences using the Track Points function in MetaMorph software (Molecular Devices). Tracking data were analyzed in Excel (Microsoft). A pause event was recorded when a kinetochore did not move for two sequential time frames. Velocity was calculated by linear regression analysis of kinetochore distance versus time plots, and deviation from average position was determined by subtracting the position of the kinetochore in the regression line from the original kinetochore position (Stumpff et al., 2008; DeLuca et al., 2011). For quantification of spindle pole separation rates, PtK1 cells were imaged every 60 s for 1 h. The distance between the spindle poles was measured at each time point using softWoRx software (Applied Precision). Distance and time were plotted in x and y, and the slope of the line generated from the first 6–10 time points was used as the initial rate of spindle pole separation. For fixed cell experiments, images were acquired at room temperature as z stacks at 0.2-µm intervals. Kinetochore fluorescence intensity measurements were performed using MetaMorph software: the integrated fluorescence intensity minus the calculated background was determined for each kinetochore in control and treated samples (Hoffman et al., 2001) and normalized to the mean value obtained from control cells. Quantification of K-fiber intensities in PtK1 cells was performed as in Cimini et al. (2003). In brief, a 5 × 5–pixel region was centered on an image of a fiber at the kinetochore–MT contact point. The integrated intensity of the K fiber and two background region intensities were measured. The mean of the two background intensity measurements was calculated and subtracted from the measured integrated intensity of the K fiber.
Mathematical modeling of the kinetochore–MT interface
General description.
Our approach for mathematical modeling of the KMT interface is based on the probabilistic model of single-molecule interactions between kinetochore-associated molecular complexes and MTs (Zaytsev et al., 2013). The kinetochore was represented as a planar square surface with a linear size Lkin (Table 2). Binding events between this surface and new MTs were treated as stochastic events that occurred with rate katt. MT binding to the kinetochore was mediated by Ntotal molecules of the NDC80 complex, located at the kinetochore surface. Stochastic interactions between individual NDC80 complexes and MTs in silico were described with three parameters: association rate (kon), dissociation rate (koff), and NDC80–MT binding cooperativity parameter (ω) as previously described (Zaytsev et al., 2013). An MT was considered to be attached to the kinetochore if it had at least one attached NDC80 complex. An MT that lost all attached NDC80 complexes was referred to as a detached MT. To examine phosphoregulation of the KMT interface, the molecular characteristics of NDC80 complexes were modified to match their behavior in vitro. Specifically, calculations for different interfaces were performed using different values of koff, whereas all other model parameters were fixed (see Choice of model parameters). The same model parameters were used in the repetitive sites and lawn models, except for the value of kon, which was slightly different in these models (see Model calibration).
Table 2.
List of general model parameters
| Symbol | Model parameter | Value | Reference |
| Parameters used in both models | |||
| Lkin | Kinetochore linear size | 430 nm | Estimated based on McDonald et al., 1992 |
| katt | Rate of binding events between new MTs and the kinetochore | 4 min−1 | Materials and methods part 2 section “Choice of model parameters” |
| Ntotal | Total number of NDC80 complexes per kinetochore | Nsites × N0 = 600 | By definition |
| Δt | Time step in computational algorithm | 1 ms | Materials and methods |
| Additional parameters in the model with repetitive sites | |||
| Nsites | Number of MT binding sites per kinetochore | 50 | Estimated based on McEwen et al., 1997 |
| N0 | Number of NDC80 complexes per MT binding site | 12 | Johnston et al., 2010; Lawrimore et al., 2011; Aravamudhan et al., 2013 |
| Additional parameters in the lawn model | |||
| LNDC80 | Length of NDC80 complex | 60 nm | Ciferri et al., 2008 |
| Lmin | Minimal distance between KMTs | 35 nm | McDonald et al., 1992 |
These values are based on published data for PtK1 cells (for details see Materials and methods). All calculations in this work were carried out using these parameter values unless stated otherwise.
In the repetitive sites model, all NDC80 complexes were divided into “sites,” each containing N0 molecules (Fig. 4, A and E). Sites were numbered (denoted with i) and positioned regularly at the kinetochore surface using the two-dimensional coordinates specified as follows (nm): for i = 1–42, x coordinate of site i = 28.9 + 57.8(i mod 7) + 22.2((i div 7) mod 2) and y coordinate of site i = 33.3 + 60(i div 7); for i = 43–50, x coordinate of site i = 28.9 + 53.3(i mod 7) and y coordinate of site i = 393.2, in which (i mod n) is the remainder of the division of i by n, and (i div n) is the largest integer number less or equal to i/n. All calculations reported here for the repetitive sites model were performed using this spatial arrangement. However, the main model conclusions remain true for the interface with random site organization (Fig. S3 C), so the exact positioning of the sites is not an essential feature of this model.
A new MT contacts the kinetochore surface at a random position. When the nearest binding site to this position is already occupied by another MT, the new MT does not form any molecular bonds and detaches. If the nearest site is unoccupied, the new MT immediately engages in stochastic molecular interactions with NDC80 complexes that belong to that site, now associated with this MT. The key feature of the repetitive sites model, which distinguishes it from the lawn model, is that each NDC80 complex belongs to a specific site, and it can form a bond to the MT that is associated with this but not other sites. This model assumption is analogous to that in the model of the kinetochore interface described by Joglekar and Hunt (2002), which is based on the Hill’s sleeve model (Hill, 1985). However, the sites in our model are significantly different from those in Hill’s sleeve model because the NDC80 complexes within a single site in our model interact with the MT independently from each other, whereas the MAPs in the sleeve were coordinated (for additional discussion, see Zaytsev et al., 2013).
In the lawn model, the NDC80 complexes are distributed randomly on the kinetochore surface (Fig. 4 G). New MTs arrive at random positions within the kinetochore, but they do not attach if one or more MTs are found bound to the kinetochore at a distance Lmin or less from the new MT. This rule was introduced to ensure that KMTs did not pack more densely than at the PtK1 kinetochores (McDonald et al., 1992). If there are no other MTs found within the radius Lmin, the new MT forms a bond with one randomly selected, MT-free NDC80 complex that is located within radius LNDC80 from this MT (Fig. 4 G, two such areas are indicated with circles). The maximum number of NDC80 complexes that can reach a single MT can be calculated as π × Ntotal × LNDC802/Lkin2, which is equal to 37 for the parameters used (Table 2). All subsequent interactions between the MT and NDC80 complexes within this circle are described with the kinetic constants kon, koff, and ω, as in the repetitive sites model. Unlike in the repetitive sites model, however, each kinetochore-associated NDC80 complex in the molecular lawn can interact with any MT within a radius LNDC80 from this complex, so one NDC80 complex can alternate its binding between several KMTs, allowing it to kinetically monitor the density of KMTs and their stability. If this rule of engagement with MTs is abandoned (for example, by setting LNDC80 < Lmin), all “shared” areas in the lawn model are eliminated. Consequently, such a lawn is no longer able to “sense” the KMT density, and the response to phosphorylation of such a lawn interface is the same as in the repetitive sites model. Thus, sharing of NDC80 complexes by different MTs and the resulting competition between MTs for these complexes, which takes place in the lawn but not in the repetitive sites model, significantly alters the overall behavior of the interface and is the key difference between these designs.
Choice of model parameters.
Values of model parameters were selected based on published results for PtK1 cells, when possible (Table 1 and Table 2). The number of sites, Nsites = 50, was chosen to match the observed maximum number of KMTs per kinetochore in metaphase PtK1 cells (McEwen et al., 1997). The rate of binding events between new MTs and the kinetochore surface, katt, was proportional to the probability of a new MT to encounter one site (Pat), the association rate with any of N0 NDC80 complexes at one binding site (kon × N0), and the total number of MT binding sites at the kinetochore (Nsites). Therefore, katt = Pat × Nsites × kon × N0. Using this formula and Eq. 13 for Pat from Zaytsev et al. (2013), we obtained the following expression for katt: katt = NMT × ln2/τ × Lkin/(Lkin2 − NMT × π × Lmin2), in which NMT is the mean number of KMTs per kinetochore in metaphase cells, and τ is the mean KMT half-life in metaphase. Using the values listed in Table 2, NMT = 27 (McDonald et al., 1992) and the mean KMT half-life in metaphase for PtK1 cells τ = 9 min (Cimini et al., 2006; DeLuca et al., 2006), we find that katt = 4 min−1. Other parameters for the repetitive sites model were described in Zaytsev et al. (2013). Number of NDC80 complexes per site (N0) was 12, which is the mean of estimates from different studies (Johnston et al., 2010; Lawrimore et al., 2011; Aravamudhan et al., 2013).
The value of kinetic constants koff and ω were based on our in vitro study (Table 1), whereas kon was determined using the calibration procedure described in Model calibration. The value of parameter ω was similar for different NDC80 phosphomutants in vitro (Table 1), so for all NDC80 complexes in silico, we used their experimental mean (ω = 2.5; Table 1), but the same results were obtained with the repetitive sites model for other values of ω (Fig. S3 D). The resulting density of KMTs for 1D NDC80 interfaces in both models was 146 KMTs/µm2, similar to that measured with electron microscopy at PtK1 kinetochores (McDonald et al., 1992). The resulting initial rate of KMT acquisition in both models was 2.2 KMTs/min (Fig. 4 B), similar to that reported in McEwen et al. (1997).
The time step for iterations (Δt) was chosen at least two orders of magnitude smaller that the fastest characteristic time parameters in the model. For most of the simulations, Δt was 1 ms.
Model calibration.
By measuring the K-fiber size and kinetochore oscillations in metaphase cells with Hec1 mutant proteins, we found that the MT-binding properties of 1D NDC80 corresponded well to the WT KMT affinity in metaphase (Fig. 2, C–E; and Fig. 3 C). This finding provided a strategy to calibrate our mathematical model of the phosphoregulation of the KMT interface: the normal K-fiber size was fitted based on the in vitro molecular parameters of NDC80 complexes with a single phosphomimetic substitution on the Hec1 tail (1D NDC80). A similar calibration procedure was used to calibrate both the repetitive sites and lawn models. The in vitro values of kinetic parameters koff and cooperativity factor ω for 1D NDC80 complexes were used directly, and the remaining free parameter kon, the MT association rate for kinetochore-bound NDC80 complexes, was then varied to fit the K-fiber size (Fig. S3 A). After the fit was achieved for 1D NDC80, the values of kon and all other parameters (except koff) were fixed, so the same parameters were used for all model calculations unless stated otherwise. To change the level of phosphorylation of the kinetochore interface in silico, the koff measured with corresponding NDC80 mutant proteins in vitro was used (Table 1). NoD NDC80 in the model corresponds to 9A Hec1 in cell experiments. The use of the same values of kon and ω for all NDC80 complex mutants is justified by our findings in vitro that changing the number of phosphomimetic substitutions in the Hec1 tail changes the koff for NDC80–MT interactions but does not affect kon or ω (Table 1). Furthermore, proteins with the same number of substitutions behaved similarly in vitro regardless of their exact location, consistent with our data in cells (Fig. S2). Therefore, we based our modeling on variations in the level of NDC80 complex phosphorylation and did not vary the location of phosphorylated residues. Also, because the goal of this model was to examine the consequences of NDC80 phosphorylation, the upstream factors that cause changes in NDC80 phosphorylation during mitotic progression were not included.
To verify that this calibration procedure did not affect the main model conclusions, we examined results from a repetitive sites model that was calibrated differently. In the first test, we capitalized on the previously estimated value of kon for NDC80–MT interactions at the kinetochore (1–100 s−1; Zaytsev et al., 2013). kon = 50 s−1, which is in the middle of this range, was used. The value of koff was then varied to obtain a model match with WT K-fiber size (Fig. S3 G), and the resulting value of koff was assigned to the 1D NDC80 interface in silico. All other phosphomutants were modeled by changing the koff using the ratios derived from the in vitro differences for corresponding proteins. The value of ω was kept constant because it does not affect the steepness of the phosphoregulation response (Fig. S3 D). With this calibration strategy, the repetitive sites model also predicted a binary response (Fig. S3 G), illustrating that the use of kon versus koff for calibration does not affect its results. In the second test, we performed calibrations based on the experimental data for 2D NDC80 complexes (Fig. S3 F). The values of koff and ω measured previously in vitro for 2D NDC80 complexes were used when fitting the kon value to achieve the K-fiber size in 2D Hec1 cells, which was 86% of the K-fiber size in WT Hec1 cells (Fig. 3 C). Thus, this calibration assured that the interface with 2D NDC80 complexes could maintain on average 23 KMT attachments at steady state in silico. Fig. S3 F shows predictions of the repetitive sites model based on this calibration strategy. It is clear that the predicted phosphoregulation of K-fiber size is very similar to the one obtained with the main calibration procedure, based on data for 1D NDC80 complexes. Additionally, we performed calculations in both models in which 10% of the NDC80 complexes were considered to be endogenous, which were randomly selected from Ntotal NDC80 complexes at the kinetochore. Based on data in Fig. 3 C, we assume that endogenous NDC80 is in a form of 1D. For simulations of different phosphomutants, koff for 90% of the total number of NDC80 complexes (corresponding to the exogenously expressed protein) was varied as in Table 1, and the remaining 10% of the NDC80 complexes (corresponding to the endogenous complexes) were in a 1D form with the corresponding constant koff. The obtained results were highly similar to the theoretical control (no endogenous NDC80; Fig. S3 H). Thus, our conclusion that the repetitive sites model strongly amplifies the single molecule phosphotuning is robust and does not depend on the exact calibration procedure. The procedure is relatively unimportant because it simply provides a match of one point on the phosphoregulation curve, and it does not affect the model’s behavior when other points are calculated.
Stochastic simulation algorithm.
After specifying all model parameters, the stochastic simulation algorithm was performed as follows.
Encounters between new MTs and the kinetochore surface.
The probability Ψat for an MT end to encounter the kinetochore surface during time Δt was calculated as follows: Ψat = 1 − Exp(−Δt × katt). Then, the random number p from the range [0, 1] was generated. If p was larger than Ψat, no contact was made, and this MT was not followed. If p was smaller than Ψat, the position of the MT end was defined with coordinates (xMT, yMT), which were generated in the following range: 0 < xMT < Lkin and 0 < yMT < Lkin. In the repetitive sites model, this new MT became immediately associated with the nearest MT binding site. If this site was already occupied, the MT end did not associate with any sites, and this MT was not followed. In the dynamic lawn model, the circular area with radius Lmin centered at position (xMT, yMT) was searched. If no other attached MTs were found, this MT end formed a bond with a randomly selected NDC80 complex located within distance LNDC80 from this MT. If other MTs were found in the vicinity of this position, the MT end did not bind any NDC80 complexes, and this MT was not followed.
Binding of new NDC80 complexes to KMTs.
Dynamic lawn model. The probability Ψon for a free NDC80 complex to attach to an MT located within distance LNDC80 from this molecule was calculated as follows: Ψon = 1 − Exp(−Δt × kon × M), in which M is the number of MTs within distance LNDC80 from this molecule. Then, the random number p from the range [0, 1] was generated. If p was smaller than Ψon, the NDC80 complex became associated with one of these MTs, selected randomly. If p was larger than Ψon, the NDC80 complex continued to be unbound. All other molecular interactions in the lawn model were calculated identically to those in the repetitive sites model. The entire algorithm was repeated until a programmed number of iterations was achieved. The typical simulated time to calculate mean values for one steady-state configuration was 6 h.
Repetitive sites model.
Binding of new NDC80 complexes to KMTs in the repetitive sites model was calculated as in Zaytsev et al. (2013). In brief, the probability Ψon for an NDC80 complex to bind to the KMT that was already bound to at least one NDC80 at this site during Δt was Ψon = 1 − Exp(−Δt × kon). For each unattached NDC80 complex within the occupied sites, the random number p from the range [0, 1] was generated. If p was smaller than Ψon, this NDC80 was called bound to the MT. If p was larger than Ψon, this NDC80 complex remained unbound.
Detachment of NDC80 complexes from KMTs.
The probability Ψoff for an NDC80 complex to dissociate from the KMT during Δt was calculated as follows (Zaytsev et al., 2013):
in which N denoted the number of NDC80 complexes attached to this KMT. For each attached NDC80 complex, the random number p from the range [0, 1] was generated. If p was larger than Ψoff, this NDC80 complex remained attached to the MT. If p was smaller than Ψoff, the NDC80 complex dissociated from the MT.
Detachment of the MTs from the kinetochore.
The calculations were stopped for the KMTs that lost all attachments with NDC80 complexes. In the repetitive sites model, the unoccupied sites became available immediately for interactions with the incoming MTs (see Encounters between new MTs and the kinetochore surface).
Sensitivity analysis of the repetitive sites model.
The repetitive sites model predicts that kinetochores containing noD NDC80 complexes bind MTs so strongly that all MT binding sites become occupied, whereas the 2D NDC80 complex-containing kinetochores have almost no attached MTs (Fig. 4, D and E). Thus, this model is hypersensitive to NDC80 complex phosphorylation and produces a steep response, contrary to a gradual response observed in cell-based experiments (Fig. S3 B, first three bars). To investigate the robustness of this conclusion, we varied different model parameters and features.
Number of NDC80 complexes per site.
Number of NDC80 complexes per site (parameter N0) was varied from 3 to 16. Steepness of the model response was quantified by measuring the slope of the phosphorylation response curve versus koff at the point corresponding to 1D NDC80 complexes. The steepness decreased for smaller N0, but even for N0 = 3 (much less than expected based on Lawrimore et al., 2011), the steepness was threefold higher than in vivo (Fig. S3 B). For N0 > 12, the steepness increased, leading to an even stronger discrepancy with the experimental data. Additionally, we performed a simulation with N0 = 37, which is the maximum number of available NDC80 complexes per KMT in the lawn model, and the steepness of the model response increased even further, as expected (Fig. S3 F).
Cooperativity parameter ω.
Cooperativity parameter ω for NDC80–MT binding at the kinetochore is not known, but our previous theoretical study demonstrates that its value is unlikely to be high (Zaytsev et al., 2013). Varying ω from 1 (noncooperative binding) to 10 (corresponds to Hill coefficient 2.6) changed the steepness of the response by only 5% (Fig. S3 D).
Interkinetochore tension.
We have also investigated the possibility that KMT binding affinity is modulated by interkinetochore tension (Nicklas, 1997). Because the distance between sister kinetochores is larger in cells with fewer D substitutions (Fig. 3 B), the KMTs at such kinetochores may experience a larger tension, which in turn may weaken the NDC80–MT molecular bonds (Powers et al., 2009). To examine this hypothesis quantitatively, we estimated the interkinetochore tension in these cells, assuming that sister kinetochores are connected via a linear spring with a resting length of 1 µm (DeLuca et al., 2006, 2011) and a spring constant of 0.1 pN/nm (Fig. S3 E; Joglekar and Hunt, 2002). Next, we calculated the mean force per KMT at these kinetochores as the ratio of interkinetochore tension and the number of KMTs, which was estimated based on Fig. 3 C, assuming that WT PtK1 cells have on average 27 KMTs per kinetochore (McEwen et al., 1997). The force per single NDC80 complex attached to a KMT was estimated as the mean force per KMT divided by the number of attached NDC80 complexes, as calculated with our model. This led to the effective dissociation rate keffoff for NDC80 complexes under force F: keffoff = koff Exp(F/F0), in which koff is the dissociation rate without an applied external force, F0 = 3 pN (Civelekoglu-Scholey et al., 2013). Although the estimated total tension at kinetochores with less phosphorylated NDC80 complexes was larger, such kinetochores had more KMTs and MT-bound NDC80 complexes, so the resulting force per NDC80 complex ended up being similar for kinetochores with different phosphomutants. The effective dissociation constants for all interfaces were therefore similar, explaining a weak effect of interkinetochore tension on the steepness of the response to NDC80 phosphoregulation in the repetitive sites model (Fig. S3 F).
Modeling the kinetochore with a mixture of NDC80 complexes and generic MAP molecules.
Calculations for interfaces supplemented with non-NDC80 MAPs were performed analogously to the aforementioned kinetochore surfaces, but the number of KMTs in a K fiber was assumed to result from two affinities: from the NDC80 molecular binding events and from MAPs. In the lawn model, the MAP component was assumed to support attachment of six KMTs on average, regardless of the phosphorylation level of the NDC80 complex. This number of KMTs corresponded to the K fiber observed in 9D Hec1-expressing cells (Fig. 3 C and Fig. 4 D). This MT-binding MAP component was combined with the NDC80 lawn model, which was calibrated as described in Model calibration, except 19 KMTs were attached on average to the interfaces with 1D NDC80 complexes; all other model parameters were the same as in the corresponding models with no MAPs. This combined model produced a better fit to the experimental curve than the interface with no MAPs (Fig. 6 A).
Contribution of non-NDC80 MAPs in the repetitive sites model was considered using two approaches. First, we assumed constant contribution from MAPs analogously to the aforementioned approach with the lawn model. To build such a curve, we varied the magnitude of the MAP contribution and quantified the resulting fit to the experimental curve using the sum of the residual squares. The minimal sum of the squared residuals was achieved when non-NDC80 MAPs provided 46% of the affinity (11.5 KMTs); however, even this curve provided a poor match to the experimental data (Fig. S3 J). Next, we assumed that the MAPs contribution was different for kinetochores with different NDC80 proteins. Because, in the repetitive sites model, the kinetochores with noD NDC80 are at maximum KMT occupancy (Fig. 4 D), the contribution of MAPs for noD NDC80 was set to 0. The repetitive sites model was then calibrated such that, with noD NDC80, this model had the same number of KMTs as in the lawn model with noD NDC80 complexes (Fig. 6 B), so that these two models could be compared under similar conditions. For all other levels of NDC80 complex phosphorylation, the contribution of non-NDC80 MAPs was simply a difference between the predictions based on NDC80 alone and the lawn model, so the full match was achieved. This model was then used to calculate KMT binding in the absence of NDC80 complexes (Fig. 6 D).
Visualization of model simulations.
3D visualization of the numerical results was performed with Mathematica software (Wolfram Research; Video 1 and Video 2). Fluctuations in the orientation of NDC80 complexes were achieved by randomizing the polar and azimuthal angles from 0 to 15° and 0 to 360°. This video feature was used to illustrate the transient and highly dynamic nature of binding/unbinding between NDC80 complexes and KMTs. Model visualization for Video 3 was obtained using a Delphi programming environment (Borland).
Online supplemental material
Fig. S1 shows results of the immunostaining of mitotic cells with antibodies to phospho-Dsn1 or phospho-KNL1 and quantification of Hec1-GFP fusion protein levels at the kinetochores. Fig. S2 shows how parameters of chromosome oscillations (such as mean oscillation velocity, percentage of time spent in pause, and deviation from average position), interkinetochore distance, and relative K-fiber intensity depend on the number and position of phosphomimetic substitutions in the Hec1 tail. Fig. S3 reports additional theoretical results that illustrate and/or justify model conclusions. Fig. S4 contains data for the spindle pole separation assay. Fig. S5 illustrates differences in molecular interactions in the repetitive sites and lawn models of the KMT interface and reports additional investigations of the repetitive sites model. Video 1 shows the behavior of the KMT interface in the repetitive sites model, and Video 2 shows analogous data for the dynamic lawn model. Video 3 illustrates that phosphorylation of NDC80 complexes in the dynamic lawn model leads to a smooth, graded adjustment of K-fiber size. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201312107/DC1.
Supplementary Material
Acknowledgments
We are grateful to F.I. Ataullakhanov for help with mathematical modeling. We also thank members of the DeLuca and Grishchuk laboratories, T. Salmon, J.R. McIntosh, M. Lampson, and I. Cheeseman for stimulating discussions, J.R. McIntosh for supporting the initial stages of this project (GM033787), and I. Cheeseman for pKNL1 antibodies.
This work was supported by National Institutes of Health grant R01GM088371 to J.G. DeLuca and grant R01GM098389 to E.L. Grishchuk, grants 13-04-40188-H and 13-04-40190-H from the Russian Fund for Basic Research, and grants from the Presidium of Russian Academy of Sciences Mechanisms of the Molecular Systems Integration and Molecular and Cell Biology program to F.I. Ataullakhanov. E.L. Grishchuk is a Kimmel Scholar. J.G. DeLuca is a Pew Scholar in the Biomedical Sciences.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BDS
- boiled donkey serum
- K fiber
- kinetochore MT fiber
- KMT
- kinetochore-bound MT
- MAP
- MT-associated protein
- MT
- microtubule
- WT
- wild type
References
- Akiyoshi, B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., and Biggins S.. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, B., Johnson A., Lewis J., Raff M., Roberts K., and Walter P.. 2008. Molecular Biology of the Cell. Fifth edition Garland Science, New York: 1392 pp [Google Scholar]
- Alushin, G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., and Nogales E.. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudhan, P., Felzer-Kim I., and Joglekar A.P.. 2013. The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr. Biol. 23:770–774 10.1016/j.cub.2013.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armond, J.W., and Turner M.S.. 2010. Force transduction by the microtubule-bound Dam1 ring. Biophys. J. 98:1598–1607 10.1016/j.bpj.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R. III, Chan C.S., Drubin D.G., and Barnes G.. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Ciferri, C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini, D., Moree B., Canman J.C., and Salmon E.D.. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 116:4213–4225 10.1242/jcs.00716 [DOI] [PubMed] [Google Scholar]
- Cimini, D., Wan X., Hirel C.B., and Salmon E.D.. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16:1711–1718 10.1016/j.cub.2006.07.022 [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey, G., He B., Shen M., Wan X., Roscioli E., Bowden B., and Cimini D.. 2013. Dynamic bonds and polar ejection force distribution explain kinetochore oscillations in PtK1 cells. J. Cell Biol. 201:577–593 10.1083/jcb.201301022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., and McEwen B.F.. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- DeLuca, K.F., Lens S.M., and DeLuca J.G.. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., Vanden Beldt K.J., Meng X., Khodjakov A., and McEwen B.F.. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522 10.1038/ncb1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov, A., Grishchuk E.L., McIntosh J.R., and Ataullakhanov F.I.. 2007. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc. Natl. Acad. Sci. USA. 104:19017–19022 10.1073/pnas.0709524104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen, S., Akiyoshi B., Iadanza M.G., Shi D., Duggan N., Biggins S., and Gonen T.. 2012. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 19:925–929 10.1038/nsmb.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes, G.J., Dong Y., McEwen B.F., and Deluca J.G.. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, T.L.1985. Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA. 82:4404–4408 10.1073/pnas.82.13.4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P., and Hunt A.J.. 2002. A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys. J. 83:42–58 10.1016/S0006-3495(02)75148-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., and Bloom K.S.. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594 10.1083/jcb.200803027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P., Bloom K., and Salmon E.D.. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, K., Joglekar A., Hori T., Suzuki A., Fukagawa T., and Salmon E.D.. 2010. Vertebrate kinetochore protein architecture: protein copy number. J. Cell Biol. 189:937–943 10.1083/jcb.200912022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T.M., Mayer T.U., Coughlin M.L., and Mitchison T.J.. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener, J.P., and Shtylla B.. 2014. A mathematical model of force generation by flexible kinetochore-microtubule attachments. Biophys. J. 106:998–1007 10.1016/j.bpj.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore, J., Bloom K.S., and Salmon E.D.. 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195:573–582 10.1083/jcb.201106036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Onuchic J.N.. 2006. A driving and coupling “Pac-Man” mechanism for chromosome poleward translocation in anaphase A. Proc. Natl. Acad. Sci. USA. 103:18432–18437 10.1073/pnas.0608962103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, R., Lenobel R., Santamaria A., Ries A., Nigg E.A., and Körner R.. 2009. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome Res. 8:4553–4563 10.1021/pr9003773 [DOI] [PubMed] [Google Scholar]
- Mayer, T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., and Mitchison T.J.. 1999. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286:971–974 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- McDonald, K.L., O’Toole E.T., Mastronarde D.N., and McIntosh J.R.. 1992. Kinetochore microtubules in PTK cells. J. Cell Biol. 118:369–383 10.1083/jcb.118.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B.F., Heagle A.B., Cassels G.O., Buttle K.F., and Rieder C.L.. 1997. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 137:1567–1580 10.1083/jcb.137.7.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, J.R., O’Toole E., Zhudenkov K., Morphew M., Schwartz C., Ataullakhanov F.I., and Grishchuk E.L.. 2013. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J. Cell Biol. 200:459–474 10.1083/jcb.201209154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.A., Johnson M.L., and Stukenberg P.T.. 2008. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr. Biol. 18:1785–1791 10.1016/j.cub.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas, R.B.1997. How cells get the right chromosomes. Science. 275:632–637 10.1126/science.275.5300.632 [DOI] [PubMed] [Google Scholar]
- Nousiainen, M., Silljé H.H., Sauer G., Nigg E.A., and Körner R.. 2006. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. USA. 103:5391–5396 10.1073/pnas.0507066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., and Asbury C.L.. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida, S., and Musacchio A.. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtylla, B., and Keener J.P.. 2010. A mechanomolecular model for the movement of chromosomes during mitosis driven by a minimal kinetochore bicyclic cascade. J. Theor. Biol. 263:455–470 10.1016/j.jtbi.2009.12.023 [DOI] [PubMed] [Google Scholar]
- Stumpff, J., von Dassow G., Wagenbach M., Asbury C., and Wordeman L.. 2008. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 14:252–262 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin, L.J., Guimaraes G.J., and Deluca J.G.. 2011. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell. 22:759–768 10.1091/mbc.E10-08-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien, J.F., Fong K.K., Umbreit N.T., Payen C., Zelter A., Asbury C.L., Dunham M.J., and Davis T.N.. 2013. Coupling unbiased mutagenesis to high-throughput DNA sequencing uncovers functional domains in the Ndc80 kinetochore protein of Saccharomyces cerevisiae. Genetics. 195:159–170 10.1534/genetics.113.152728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley, J., and Stukenberg P.T.. 2011. The Ndc80 complex: integrating the kinetochore’s many movements. Chromosome Res. 19:377–391 10.1007/s10577-010-9180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit, N.T., Gestaut D.R., Tien J.F., Vollmar B.S., Gonen T., Asbury C.L., and Davis T.N.. 2012. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. USA. 109:16113–16118 10.1073/pnas.1209615109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader, G., Cruijsen C.W., van Harn T., Vromans M.J., Medema R.H., and Lens S.M.. 2007. The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule kinetochore interactions. Mol. Biol. Cell. 18:4553–4564 10.1091/mbc.E07-04-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R.R., Al-Bassam J., and Harrison S.C.. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59 10.1038/nsmb1186 [DOI] [PubMed] [Google Scholar]
- Zachariou, M., Traverso I., Spiccia L., and Hearn M.T.W.. 1996. Potentiometric investigations into the acid-base and metal ion binding properties of immobilized metal ion affinity chromatographic (IMAC) adsorbents. J. Phys. Chem. 100:12680–12690 10.1021/jp9601476 [DOI] [Google Scholar]
- Zaytsev, A.V., Ataullakhanov F.I., and Grishchuk E.L.. 2013. Highly transient molecular interactions underlie the stability of kinetochore-microtubule attachment during cell division. Cell. Mol. Bioeng. 6:393–405 10.1007/s12195-013-0309-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski, R.P., Meyne J., and Brinkley B.R.. 1991. The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 113:1091–1110 10.1083/jcb.113.5.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.