Abstract
Macroautophagy has been implicated in numerous diseases, yet our understanding of the proteins responsible for the turnover of specific cargo by autophagy is limited. In a recent paper published in Nature, Mancias et al. used quantitative proteomics to identify a cohort of autophagosome-enriched proteins, one of which, nuclear receptor coactivator 4 (NCOA4) was shown to be required for the selective delivery of ferritin to the lysosome, ultimately regulating intracellular iron by autophagic turnover of ferritin, or ferritinophagy.
Autophagy is a cell survival process whereby double-membrane structures, autophagosomes, sequester bulk cytoplasm, damaged proteins, and organelles for delivery to the lysosome and turnover to maintain homeostasis. Autophagosomes are identified by ATG8 proteins (in mammalian cells these are LC3 and its family members) that have been shown to recruit selective receptors that deliver specific cargos for degradation; however, the full range of cargo proteins and their related receptor proteins are still largely unknown1. Understanding which proteins are responsible for specific cargo degradation is needed to clarify the complicated roles of autophagy in human diseases, for example, to explain the dual roles that autophagy is thought to play in tumor suppression or in the survival and growth of tumors2.
A recent study from the Kimmelman and Harper labs has taken a crucial first step in robustly identifying proteins that are associated with autophagosomes and their turnover3. Although previous studies have identified proteins in autophagosome preparations, these studies had limitations and low protein overlap with one another4,5,6. Most importantly, because autophagy non-selectively degrades cytosolic proteins as well as selectively targets specific cargos, the previous studies were bedeviled by the problem that many proteins targeted to autophagosomes might simply have been material whose likelihood of being in the autophagosome is not regulated but instead related to their overall abundance. In this paper, proteomics using stable isotope labeling by amino acids in cell culture (SILAC) was paired with a density gradient separation protocol for autophagosomes and a clever refinement where “hits” were filtered based on their abundance in the total proteome. This allows for a powerful association of proteins that are specific to autophagosomes. In addition, use of multiple human cell lines with differing reliance on autophagy (PANC-1, PA-TU-8988T, and MCF7) helped to strengthen the protein associations in regards to autophagy.
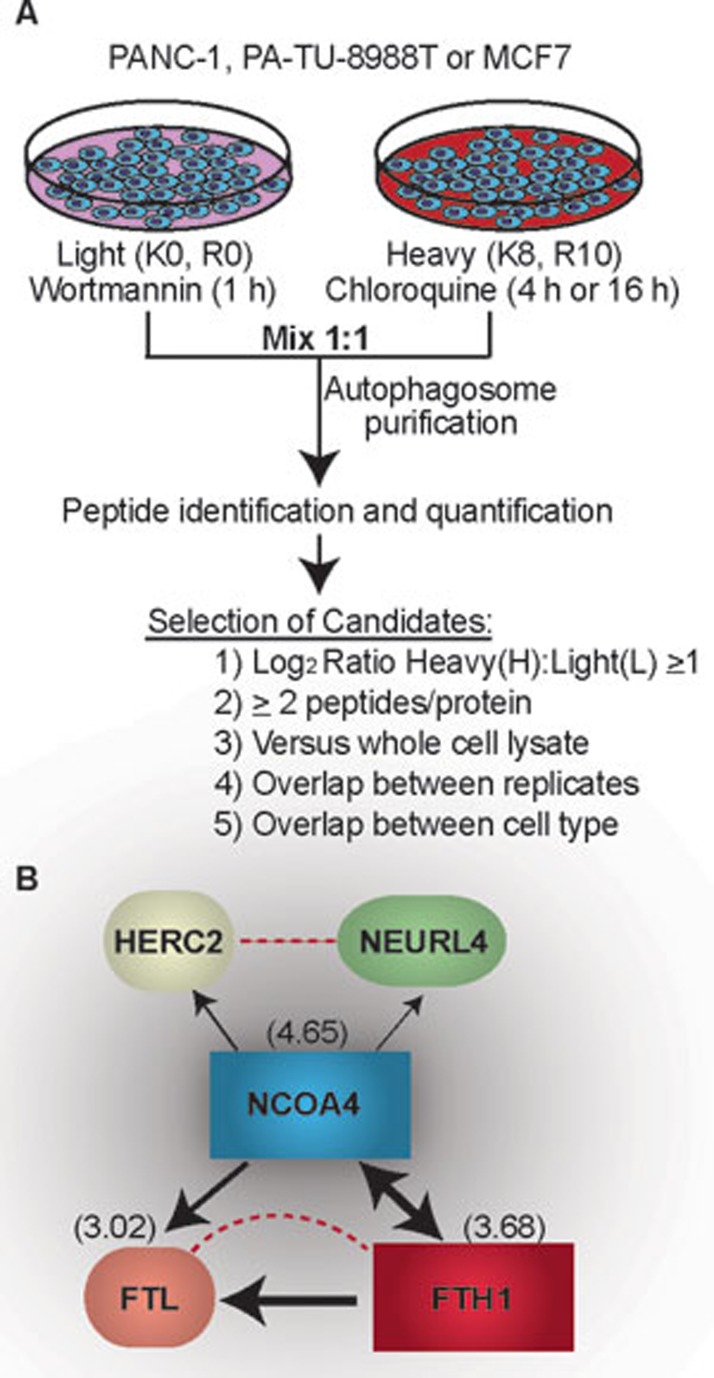
Identification of specific autophagy-related proteins was achieved through the treatment of light isotope-labeled cells with wortmannin, a phosphoinositide 3-kinase inhibitor that prevents autophagosome formation, and heavy isotope-labeled cells treated with chloroquine, a lysosomal inhibitor to prevent autophagosome degradation and increase numbers. Autophagosomes were then isolated from the differentially labeled isotope samples which allowed for the identification of specific autophagy proteins (heavy) from those proteins that were isolated at the same density upon gradient centrifugation (light) (Figure 1A). A total of 50 high-stringency proteins were selected based on an equal or greater than one log2 increase in the heavy:light ratio, protein overlaps between cell replicates, and protein overlap between the different cell types utilized. Several known autophagy proteins and cargo receptor proteins were identified, as well as plasma membrane and endocytosis-related proteins, which is consistent with previous findings and the intermixing of membranes during autophagosome maturation. Of the proteins not previously shown to associate with autophagy, NCOA4 was the highest and most consistently enriched protein identified. NCOA4 had previously been suggested to interact with androgen receptor7,8; however, the new study describes an unrecognized role of NCOA4 as a specific cargo receptor for autophagy, which interacts with LC3 proteins to deliver selective cargo to the autophagosome. For example, characterization of GFP-labeled NCOA4 showed puncta accumulation that tended to localize with LC3B-positive puncta in response to chloroquine treatment.
Figure 1.
Identification of autophagy-associated proteins and protein interaction of top HCIP: NCOA4. (A) Workflow of autophagy-associated protein identification. (B) Different complex interactions of NCOA4 with HERC2, NEURL4, and the ferritin complex. Arrows depict the directionality of interaction with line weight indicating the weighted and normalized D score (WDN). Dotted lines represent data from the STRING database. Numbers in parentheses are the log2(H:L) ratio obtained from A.
Further experiments were performed to understand how NCOA4 functions as a selective autophagy cargo receptor. Affinity purification-mass spectrometry was used to identify high-confidence interacting proteins (HCIP) associating with NCOA4. Among the HCIPs identified, ferritin heavy chain (FTH1), ferritin light chain (FTL), HERC2, and NEURL4 were verified to associate with NCOA4 by immunopreciptation followed by immunoblotting. As HERC2 and NEURL4 are not found in the autophagosome fraction and do not associate with ferritin immune complexes, it is believed that NCOA4 creates separate distinct complexes with ferritin and HERC2-NEURL4 (Figure 1B). This result indicates a further level of control whereby NCOA4 does not just deliver everything that it binds to the autophagosome. Instead, NCOA4 must have some mechanism by which it “knows” to only deliver cargo such as ferritin to the autophagosome.
In previous studies, it was shown that ferritin can concentrate in the lysosome and upon iron chelation, ferritin is transported to the lysosome for degradation9,10, thus allowing release of iron to the cell. Here co-localization of NCOA4, LC3B, and ferritin into puncta was shown to occur upon stimulation of ferritin expression with ferric ammonium citrate, reflecting the targeting of ferritin into autophagosomes for degradation through the lysosome. This process has been termed “ferritinophagy” by the authors. It was confirmed that NCOA4 and autophagy play a central role in ferritin degradation by the prevention of ferritin turnover through genetic inhibition of either ATG5 or NCOA4 by RNA interference (RNAi). On the other hand, RNAi against HERC2 did not prevent ferritin turnover, further confirming that the separate, apparently autophagy-independent complex NCOA4 forms with HERC2 that has no relationship to the turnover of ferritin. This provides a molecular explanation for how bioavailability of iron is controlled – when iron is needed, ferritin is shuttled to the autophagosome by NCOA4 and degraded, allowing release of iron to the cytoplasm.
In summary, this study describes not only a comprehensive technique to identify autophagy-specific cargo proteins and a valuable list of autophagosome-associated proteins that the autophagy research community can start to mine, but also the first mechanistic understanding of ferritin degradation through autophagy. More generally, the study reveals an example of how sophisticated proteomic analysis can provide a much needed understanding of how particular proteins, organelles, and nutrients are turned over through autophagy, ultimately identifying targets for therapeutically directed strategies designed to manipulate these mechanisms in disease processes.
References
- Green DR, Levine B. Cell. 2014. pp. 65–75. [DOI] [PMC free article] [PubMed]
- Wu WK, Coffelt SB, Cho CH, et al. Oncogene. 2012. pp. 939–953. [DOI] [PubMed]
- Mancias JD, Wang X, Gygi SP, et al. Nature. 2014. pp. 105–109. [DOI] [PMC free article] [PubMed]
- Dengjel J, Høyer-Hansen M, Nielsen MO, et al. Mol Cell Proteomics. 2012. p. M111.014035. [DOI] [PMC free article] [PubMed]
- Gao W, Kang JH, Liao Y, et al. J Biol Chem. 2010. pp. 1371–1383. [DOI] [PMC free article] [PubMed]
- Overbye A, Fengsrud M, Seglen PO. Autophagy. 2007. pp. 300–322. [DOI] [PubMed]
- Gao T, Brantley K, Bolu E, et al. Mol Endocrinol. 1999. pp. 1645–1656. [DOI] [PubMed]
- Yeh S, Chang C. Proc Natl Acad Sci USA. 1996. pp. 5517–5521. [DOI] [PMC free article] [PubMed]
- Asano T, Komatsu M, Yamaguchi-Iwai Y, et al. Mol Cell Biol. 2011. pp. 2040–2052. [DOI] [PMC free article] [PubMed]
- Kidane TZ, Sauble E, Linder MC. Am J Physiol Cell Physiol. 2006. pp. C445–C455. [DOI] [PubMed]