Dear Editor,
Type II clustered, regularly interspaced, short palindromic repeat (CRISPR)-associated (Cas) system is a novel genome-editing tool for targeted mutagenesis in cultured cells and whole organisms1,2,3. Recently, the CRISPR-Cas9 system has been applied in C. elegans using various delivery approaches including the transgene method and the direct injection of Cas9 mRNA/single guide RNA (sgRNA) or Cas9-sgRNA ribonucleoproteins4,5,6. In the present study, we have developed a CRISPR-Cas9-based genome-editing tool for C. elegans by delivering gene-specific guide RNA (gRNA) through bacterial feeding to achieve gene disruptions in a time- and labor-saving manner.
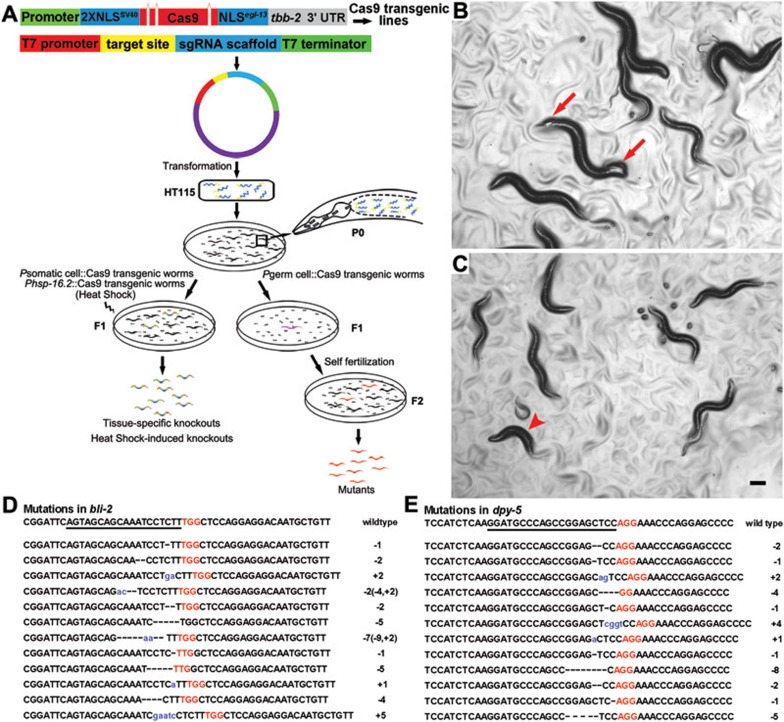
The RNAi feeding is a powerful technique for silencing gene function by using bacteria to deliver gene-specific double-stranded RNAs to major tissues in C. elegans7. This technique has been widely used to systematically study gene function in the worm8. We sought to determine whether feeding Cas9-expressing worms with bacteria expressing single-stranded target-specific gRNA (ts-gRNA) would produce preferred genome editing in C. elegans. First we created an RNAi-like cassette using the T7 promoter to ensure efficient transcription of ts-gRNAs in HT115 bacteria. Two BbsI restriction enzyme sites were added between the T7 promoter and the gRNA scaffold to allow the insertion of each target-specific sequence in the same fashion (Figure 1A). To ensure the correct termination of ts-gRNA transcription, a 46 bp or 129 bp of T7 RNA polymerase terminator was added to the 3′ end of the gRNA scaffold, generating two RNAi-like cassettes termed gRNA-46 and gRNA-129, respectively (Supplementary information, Figure S1Aa). We selected dpy-5 and bli-2 as targets because of their easy-to-score phenotypes. Mutations in dpy-5 are semi-dominant and usually produce a progressively severe short body and dumpy phenotype, known as Dpy9, while mutations in bli-2 are recessive and exhibit a blistered phenotype (large fluid-filled blisters on the body surface), known as Bli10 (Figure 1B and 1C). An 18∼20 bp DNA fragment specific to the target gene was inserted into the gRNA-46 or gRNA-129 cassette, generating dpy-5ts-gRNA-46 or bli-2ts-gRNA-46, and dpy-5ts-gRNA-129 or bli-2ts-gRNA-129 plasmids. As both constructs were able to be actively transcribed in bacteria (Supplementary information, Figure S1Ab), they were eventually used in gene knockout experiments (Figure 1B-1E and Supplementary information, Table S1A).
Figure 1.
Inheritable germline mutagenesis and spatial-temporal somatic knockout using the CRISPR-Cas9 feeding system in C. elegans. (A) Schematic description of the CRISPR-Cas9 feeding system. Germline-specific or tissue-specific Cas9 expression vectors were used to generate transgenic lines. Three artificial introns are engineered into the Cas9 coding sequence that is flanked with SV40 and egl-13 NLSs. The resulted cassette is under control of a germline-specific or tissue-specific promoter. To perform the germline or somatic mutagenesis, L4 larval staged Pgerm cell::Cas9 or young adult staged Psomatic cell::Cas9 (P0) worms were fed with HT115 bacteria transformed by the modified pMD18-T vector containing ts-gRNA-46. Individual progeny of P0 (F1) is picked up and seeded onto a culture dish with normal HT115 bacteria. Worms with desired phenotypes could be observed in either F1 (Psomatic cell::Cas9) or F2 (Pgerm cell::Cas9) worms, and are subjected to genotyping or PCR/restriction confirmation. Conditional and direct knockout in somatic lineage of C. elegans can be achieved by feeding transgenic animals, in which the Cas9 gene is under the control of inducible (promoter of heat shock proteins, such as Phsp-16.2) or tissue-specific promoter, with HT115 bacteria containing the plasmid encoding ts-gRNA-46. More details are described in Supplementary information, Figure S1G-S1I, Table S1E-S1F, and Data S1. (B, C) Germline mutagenesis of bli-2 (B) or dpy-5 (C) mediated by the CRISPR-Cas9 feeding system. Worms with strong Bli (arrows) or Dpy (arrowhead) phenotype were observed in F2 worms derived from Ppie-1::Cas9 (P0) fed with (bli-2 or dpy-5)ts-gRNA-46 plasmid-transformed bacteria. Scale bar, 100 μm. (D) Sequences of 12 indels of bli-2 mutants obtained by the CRISPR-Cas9 feeding system. (E) Sequences of 12 indels of dpy-5 mutants obtained by the CRISPR-Cas9 feeding system. The underlined letters are the target sequence (bli-2 or dpy-5). The short lines represent deletions and blue lowercase letters are the insertions. Red capital letters represent the PAM (protospacer adjacent motif), NGG, which must be adjacent to the 3′ end of the target sequence. The nature of the mutations is indicated as the number of base pairs inserted (+) or deleted (−). In cases where both deletion and insertion were observed, details are then explained in parentheses (D).
We monitored the production of heritable mutations by directly injecting in vitro-synthesized ts-gRNA and Cas9 mRNA into the gonads of C. elegans hermaphrodite. We successfully obtained heritable mutants and observed that the majority of mutant progeny were produced in a relatively narrow time window (8-16 h post gonad injection) (Supplementary information, Figure S1B, S1C and Table S1B). To generate ideal transgenic lines that express Cas9 in the germline, we optimized and reengineered the plasmids. We modified the flanking sequences of the cas9 gene as shown in Figure 1A. The newly optimized Cas9 was driven by the pie-1 promoter, which worked with the tbb-2 3′ UTR to effectively express exogenous cas9 or gfp in the germline11,12 (Supplementary information, Figure S1Da and S1Db). By the direct gonad microinjection of a high concentration of Ppie-1::Cas9 and U6::dpy-5ts-gRNA plasmid DNA into P0 hermaphrodites, we obtained 37 and 43 F1 worms in two separate experiments. In each experiment, 3 F1 worms produced F2 Dpy progeny (Supplementary information, Table S1C). Using a low concentration of Ppie-1::Cas9 and U6::dpy-5ts-gRNA plasmid DNA, two (out of 91) and one (out of 42) F1 animals produced F2 Dpy progeny. Thus, injection of high and low concentrations of DNA resulted in mean mutation rates of 7.54% and 2.29%, respectively (Supplementary information, Table S1C). All mutations were confirmed by sequencing the dpy-5 locus of the F2 Dpy worms (Supplementary information, Figure S1E and Table S1D).
We next generated Ppie-1::Cas9 transgenic worms through direct gonad DNA injection using a low concentration regime (Supplementary information, Data S1). Ppie-1::Cas9 worms were then fed the HT115 bacteria transformed with (dpy-5 or bli-2) ts-gRNA-46 or (dpy-5 or bli-2) ts-gRNA-129 vectors starting at the L4 larval stage (P0). We singled out all F1 progeny from the continuously fed P0 dish, seeded each into a new dish and evaluated the F2 worms for the Dpy or Bli phenotype (Figure 1A). In three independent experiments in which P0 worms were fed bacteria containing bli-2ts-gRNA-46, 213, 492 and 227 F1 worm dishes were obtained. In these experiments, we found three, five and four dishes, respectively, produced F2 worms with a blistered cuticle, representing an average bli mutation rate of 1.40%. Following the same procedure, F2 worms from 4/487, 4/330 and 4/542 F1 dishes showed the Dpy phenotype when dpy-5ts-gRNA-46 was used, representing an average mutation rate of 0.92% (Supplementary information, Table S1A). The Dpy or Bli F2 progeny were collected (10-15 F2 animals were randomly selected from each positive F1 dish), and their genomic DNA fragments spanning the specific target sequence were PCR-amplified and sequenced. A total of 12 independent indels were obtained in the Bli F2 progeny, with seven deletion, three insertion and two insertion/deletion mutations; 12 independent indels comprising nine deletion and three insertion mutations were obtained in the Dpy F2 progeny (Figure 1D-1E). The sequencing results indicated that F2 progeny from the same F1 dish contained identical indels, and the lesion type in the sequenced Dpy or Bli worms was frame-shift mutations (Supplementary information, Figure S1F and Table S1D). No mutants were identified in worms that were fed bacteria containing ts-gRNA-129 (Supplementary information, Table S1A), which may be due to distinct secondary or tertiary structures between bacterially transcribed ts-gRNA-46 and ts-gRNA-129. Notably, because we identified candidate mutants by determining the Dpy or Bli phenotype before sequencing, the actual genomic mutation rate may be underestimated, considering both the occurrence of in-frame and silent mutations and the penetrance of the Dpy and Bli phenotypes.
To generate somatic knockout worms, we replaced the tbb-2 3′ UTR with the unc-54 3′ UTR (Supplementary information, Figure S1Ga), which is known to be permissive for ubiquitous expression. We also replaced the pie-1 promoter with either a ubiquitous (dpy-30) promoter or a tissue-specific promoter such as ges-1 (which is predominantly expressed in the intestine), unc-119 (neuron), unc-54 (muscle) and dpy-7/dpy-5 (hypodermis) (Supplementary information, Figure S1G). Because dpy-5 functions autonomously in the hypodermis to affect body length9,13, the Pdpy-5::Cas9 animals served as controls. In theory, when fed with bacteria containing the dpy-5ts-gRNA-46 plasmid, Cas9 could disrupt the dpy-5 locus in specific tissues according to the tissue-specificity of the employed promoter (Supplementary information, Figure S1Hb and Figure S1Hc). P0 worms with Cas9 driven by the dpy-5, dpy-7 or dpy-30 promoter were expected to generate F1 Dpy worms (Supplementary information, Figure S1Ha), since all three promoters can drive Cas9 expression in hypodermal cells. Indeed, when fed on bacteria containing the dpy-5ts-gRNA-46 plasmid starting at the young adult stage, an average of 43.30% and 23.32% of F1 Pdpy-30::Cas9 and Pdpy-7::Cas9 worms showed the Dpy phenotype, respectively, higher than that of Pdpy-5::Cas9 worms (15.71%). Very few (1.08%, 0.22% and 2.09%, respectively) F1 Dpy worms with Pges-1::Cas9, Punc-119::Cas9 and Punc-54::Cas9 were observed (Supplementary information, Table S1E and Figure S1H). The molecular lesions in the tissue-specific knockout mutants were confirmed by SacI restriction of the PCR product amplified from pooled Dpy F1 (10 or 15 worms per dish) genomic DNA (Supplementary information, Figure S1Hb); and sequencing analyses of some SacI-resistant fragments revealed indels (Supplementary information, Figure S1Hc).
To test inducible gene knockouts, we generated Cas9 transgenic worms driven by the hsp-16.2 promoter. Phsp-16.2::Cas9 P0 worms were fed with dpy-5 ts-gRNA-46-expressing bacteria, and their F1 progeny were heat shocked at the larval L1 stage. We observed that ∼28.3% of the F1 worms had the Dpy phenotype; from these animals (n = 143), 10 or 15 F1 Dpy worms were randomly selected and their genomic DNA was pooled together for PCR analysis, which yielded a SacI-resistant DNA band (∼457 bp). Neither the Dpy worms nor the undigested DNA band was observed in the F1 worms without heat shocking (n = 86) (Supplementary information, Figure S1I and Table S1F). No F2 Dpy worms were produced by crossing F1 Dpy worms because the hsp-16.2 promoter is only active in somatic cells. Altogether, using our CRISPR-Cas9 feeding system, not only heritable mutants but also conditional gene disruptions (in a spatial or temporal manner) could be quickly generated on a large-scale.
To assess the potential off-target effects of CRISPR-Cas9 cleavage in our mutant strains, we sequenced the potential off-target sites in mutant genomes3,4,5. In all examined cases, i.e., four gfp mutants (obtained from Cas9 mRNA/gfpts-gRNA gonad injection), eight dpy-5 mutants (three mutants obtained from Cas9 mRNA/dpy-5ts-gRNA gonad injection, two mutants obtained from the Ppie-1::Cas9+U6::dpy-5ts-gRNA transgene and three mutants obtained from the CRISPR-Cas9 feeding system) and 12 bli-2 mutants (obtained from the CRISPR-Cas9 feeding system), no indels were detected at potential off-target loci (Supplementary information, Table S1D and S1G).
Despite the low mutation rate, the novel CRISPR/Cas9 feeding technique has particular advantages: the continuous feeding of ts-gRNA bacteria facilitates experimental management, and the method can be modified for large-scale studies, with a worm ts-gRNA library. To improve this CRISPR/Cas9 feeding system, three strategies could be employed to address the following aspects. First, no mutants were identified in worms that were fed bacteria containing ts-gRNA-129 (Supplementary information, Table S1A), suggesting that the nucleotide composition and/or folding of the ts-gRNAs may play a critical role in the function of the Cas9/gRNA complex. Therefore, the CRISPR-Cas9 feeding system can be optimized by modifying the T7 RNA polymerase terminator to create ts-gRNAs that are more stable, properly folded and efficiently transcribed in HT115 bacteria. Second, to overcome germline silencing, we would select a low- or single-copy-transgene carrier worm line with chromosomally integrated Cas9 (such as the Ppie-1::Cas9 line) as the host for feeding with the bacteria. Finally, the appropriate genetic background could be selected to enhance the intake of ts-gRNA. SID-1 and SID-2 transgenic worms may be appropriate candidates to uptake ts-gRNA with improved efficiency14,15. Despite the need for further work, this newly developed feeding system will facilitate reverse genetics studies in C. elegans.
Acknowledgments
We are grateful to Drs M Dong and M Ding for their critical comments. We thank the Caenorhabditis elegans Genetic Center for providing additional worm strains and greatly appreciate comments from and proof-reading by Drs R Peterson and L Tao. This project has been supported by the National Basic Research Program of China (973 Program; 2011CBA01102 and 2012CB944503 to DL, 2012CB944501 to J-W X), the National Natural Science Foundation of China (90919034 to DL) and the Peking-Tsinghua Center for Life Sciences (to DL). PL was a recipient of the PKU President Graduate Scholarship.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
The ts-gRNA encoding plasmids, used for the CRISPR-Cas9 feeding system and, are transcribed normally in bacteria.
A Mutagenizing dpy-5 or bli-2 by the CRISPR-Cas9 feeding system using two types of T7 RNA polymerase terminator
Materials and Methods
References
- Cong L, Ran FA, Cox D, et al. Science. 2013. pp. 819–823. [DOI] [PMC free article] [PubMed]
- Mali P, Yang L, Esvelt KM, et al. Science. 2013. pp. 823–826. [DOI] [PMC free article] [PubMed]
- Chang N, Sun C, Gao L, et al. Cell Res. 2013. pp. 465–472. [DOI] [PMC free article] [PubMed]
- Friedland AE, Tzur YB, Esvelt KM, et al. Nat Methods. 2013. pp. 741–743. [DOI] [PMC free article] [PubMed]
- Chiu H, Schwartz HT, Antoshechkin I, et al. Genetics. 2013. pp. 1167–1171. [DOI] [PMC free article] [PubMed]
- Cho SW, Lee J, Carroll D, et al. Genetics. 2013. pp. 1177–1180. [DOI] [PMC free article] [PubMed]
- Timmons L, Court DL, Fire A. Gene. 2001. pp. 103–112. [DOI] [PubMed]
- Kamath RS, Ahringer J. Methods. 2003. pp. 313–321. [DOI] [PubMed]
- Thacker C, Sheps JA, Rose AM. Cell Mol Life Sci. 2006. pp. 1193–1204. [DOI] [PMC free article] [PubMed]
- Hodgkin J. Genetics. 1983. pp. 43–64. [DOI] [PMC free article] [PubMed]
- Merritt C, Rasoloson D, Ko D, et al. Curr Biol. 2008. pp. 1476–1482. [DOI] [PMC free article] [PubMed]
- Merritt C, Seydoux G. WormBook. 2010 Feb 8. pp. 1–21. [DOI] [PMC free article] [PubMed]
- Cheng Z, Yi P, Wang X, et al. Nat Biotechnol. 2013. pp. 934–937. [DOI] [PubMed]
- Feinberg EH, Hunter CP. Science. 2003. pp. 1545–1547. [DOI] [PubMed]
- McEwan DL, Weisman AS, Hunter CP. Mol Cell. 2012. pp. 746–754. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ts-gRNA encoding plasmids, used for the CRISPR-Cas9 feeding system and, are transcribed normally in bacteria.
A Mutagenizing dpy-5 or bli-2 by the CRISPR-Cas9 feeding system using two types of T7 RNA polymerase terminator
Materials and Methods