Abstract
Mitophagy, or mitochondria autophagy, plays a critical role in selective removal of damaged or unwanted mitochondria. Several protein receptors, including Atg32 in yeast, NIX/BNIP3L, BNIP3 and FUNDC1 in mammalian systems, directly act in mitophagy. Atg32 interacts with Atg8 and Atg11 on the surface of mitochondria, promoting core Atg protein assembly for mitophagy. NIX/BNIP3L, BNIP3 and FUNDC1 also have a classic motif to directly bind LC3 (Atg8 homolog in mammals) for activation of mitophagy. Recent studies have shown that receptor-mediated mitophagy is regulated by reversible protein phosphorylation. Casein kinase 2 (CK2) phosphorylates Atg32 and activates mitophagy in yeast. In contrast, in mammalian cells Src kinase and CK2 phosphorylate FUNDC1 to prevent mitophagy. Notably, in response to hypoxia and FCCP treatment, the mitochondrial phosphatase PGAM5 dephosphorylates FUNDC1 to activate mitophagy. Here, we mainly focus on recent advances in our understanding of the molecular mechanisms underlying the activation of receptor-mediated mitophagy and the implications of this catabolic process in health and disease.
Keywords: mitochondria, quality control, mitophagy receptor, protein phosphorylation, mitochondrial stress, selective autophagy
Introduction
Mitochondria determine both cell life and death1,2,3,4. Healthy mitochondria function as the powerhouses for energy conversion through the TCA cycle and oxidative phosphorylation. In response to death stimuli, mitochondrial outer membrane becomes permeabilized to release cytochrome c that could bind its cytoplasmic receptor Apaf1 to form the apoptosome and to activate caspase cascades for apoptosis, a form of programmed cell death5. Mitochondria are also the major sites for producing superoxide as an inevitable byproduct during oxidative phosphorylation6,7,8,9,10. Moreover, mitochondria are the center for iron metabolism and lipid oxidation9,11. Given these key roles, damaged mitochondria could be detrimental to the cell. Accumulation of dysfunctional mitochondria is the characteristic of multiple types of diseases including heart failure, Alzheimer's disease, Parkinson's disease and cancers12,13,14,15.
To maintain the well-being of the cell, eukaryotes have evolved a mechanism to segregate and remove damaged or unwanted mitochondria through mitophagy, an autophagy-dependent process specific to the energy-converting organelles. One of the relevant aspects about mitophagy is its evolutionary conservation. In the budding yeast Saccharomyces cerevisiae, transport of mitochondria to the vacuole, a lytic compartment, has been suggested as a possible mechanism by which mitochondrial DNA escapes to the nucleus16. Subsequent genetic approaches unambiguously reveal that mitochondria degradation in this unicellular eukaryote fully depends on a set of “core” autophagy-related (Atg) proteins essential for formation of autophagosomes, double membrane-bound vesicles enclosing disposable cargoes17,18,19,20. In addition to core Atg proteins and other factors required for autophagosome-vacuole fusion and the breakdown of autophagic bodies in the vacuolar lumen, a dozen molecules have been reported as important for mitophagy in yeast (see Figure 1A and 1B). Among those, Atg32 is the key molecule that has been identified through two independent genome-wide screens for non-essential gene deletion mutants defective in degradation of mitochondria19,20. Importantly, Atg32 serves as a protein receptor for recruiting Atg8, a conserved ubiquitin-like protein essential for all autophagy-related processes that is conjugated to the phospholipid phosphatidylethanolamine (PE) and localized to autophagosomes, and Atg11, a scaffold protein acting as a platform for core Atg protein assembly during selective autophagy-related processes19,21,22.
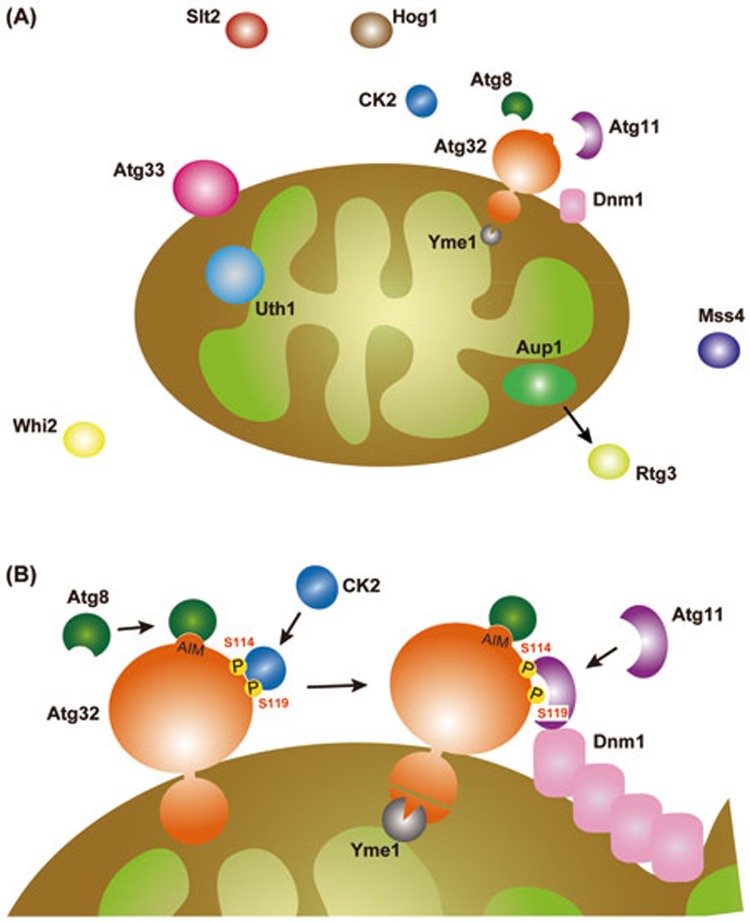
Figure 1.
(A) Mitophagy-related proteins in yeast. Aup1 is a protein phosphatase in the mitochondrial IMS that may be linked to Rtg3, a transcription factor responsible for signaling from mitochondria to the nucleus83,84. Uth1 is a mitochondrial inner membrane protein of unknown function26,85. Atg33 is a mitochondrial outer membrane protein of unknown function18,26. Whi2 is a stress response protein in the Ras-PKA signaling pathway41,86. Mss4 is a phosphatidylinositol-4-phosphate 5-kinase87. Slt2 and Hog1 are MAP kinases acting in the Wsc1-Pkc1-Bck1-Mkk1/2-Slt2 and Ssk1-Pbs2-Hog1 signaling cascades, respectively21,34. (B) Atg32 and its interacting partners. See text for details.
In mammalian cells, previous studies have suggested that both NIX/BNIP3L and BNIP3 have a putative LC3-interacting region (LIR) that interacts with LC3, an Atg8 homolog, and defines NIX/BNIP3L as a mitophagy receptor. Another important mitophagy receptor is FUNDC1 containing a typical LIR motif Y(18)xxL23. In this review, recent progress toward understanding the regulation of receptor-mediated mitophagy will be focused and described.
Mitophagy in yeast
Mitophagy in yeast absolutely relies on Atg32, a single-pass membrane protein of 59 kDa localized on the outer membrane of mitochondria with its N and C termini facing the cytosol and mitochondrial intermembrane space (IMS), respectively19,20. When cells are grown in non-fermentable medium, a condition in which mitochondrial respiration is indispensable for viability, Atg32 is strongly induced and accumulates on the surface of mitochondria19. Suppression of Atg32 expression reduces mitophagy efficiency19. Conversely, overexpression of Atg32 has the opposite effect24. Thus, it seems likely that Atg32 is a rate-limiting factor to control the amount of mitochondria to be degraded. Yeast cells lacking Atg32 exhibit no obvious defects in general autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway, an autophagy-related process for selective transport of several proteins to the vacuole19,20, supporting the idea that Atg32 is a bona fide mitophagy-specific factor. Although mitochondria autophagy is an evolutionarily conserved process, Atg32 homologs have so far been identified only in yeast species.
Atg32 domain features
The key mitophagy protein Atg32 consists of three major modules, an N-terminal 43 kDa cytosolic domain, a predicted single-helical transmembrane (TM) domain and a C-terminal 13 kDa mitochondrial IMS domain19. The TM domain functions in targeting to mitochondria and insertion into the outer membrane19,21. The cytosolic domain contains two consensus motifs critical for interaction with Atg8 and Atg1119,21,22 (see below for details). Strikingly, a variant of this module anchored to peroxisomes can promote peroxisome autophagy (pexophagy)22, suggesting that the Atg32 cytosolic domain is necessary and sufficient for recruiting autophagic machineries. The IMS domain, which is dispensable for mitophagy21,22, seems to be processed by Yme1, a mitochondrial inner membrane AAA (ATPases associated with diverse cellular activities) protease facing the IMS25. The role of Yme1 in mitophagy is, however, controversial16,25,26. Nevertheless, Yme1-dependent processing has been proposed to regulate Atg32-Atg11 interaction25.
Atg32 induction
Although how yeast cells trigger mitophagy is not fully understood, oxidative stress is likely to be a signal to induce Atg32 expression. Supporting this idea, the Atg32 protein level drastically increases in cells during respiratory growth (10-20 fold higher than that in cells during fermentable growth)19. In addition, the antioxidant N-acetylcysteine (NAC) can suppress Atg32 induction and mitophagy, possibly by increasing the glutathione pool19,27. Since glutathione is a major component that maintains cellular redox homeostasis, one scenario could be that an increase in mitochondrial and/or cytosolic oxidative stress results in a shift of cellular redox balance to oxidized state, which in turn activates redox-sensing transcription factors responsible for Atg32 expression.
It has been thought that a shift from respiration to starvation, or prolonged respiratory growth is needed to induce Atg32 and mitochondria degradation. Surprisingly, a recent study reveals that mitophagy can also be promoted independently of mitochondrial respiration24. When cells grown in fermentable medium are challenged with nitrogen starvation, bulk autophagy is rapidly activated. Under the same conditions, mitophagy also occurs without strong Atg32 induction, but much later than bulk autophagy24. Strikingly, respiration-deficient mutants are competent for this starvation-induced mitophagy24, suggesting that oxidative stress is not a general prerequisite for degradation of mitochondria.
Atg32-Atg8 and -Atg11 interactions
The Atg32 43 kDa cytosolic domain contains a motif called Atg8-family interacting motif (AIM, equivalent to LIR) crucial for binding to Atg819,22. Mutations in Trp86 and Ile89 of the W/YXXI/L/V consensus sequence weaken Atg32 binding to Atg8, resulting in a partial mitophagy defect19. Crystal structure of the Atg8-Atg32 AIM peptide complex reveals an interaction mode very similar to those of the Atg8-Atg19 (Cvt receptor) and Atg8-Atg3 (E2 enzyme for Atg8 lipidation) complexes in yeast, and the LC3-p62 (selective autophagy adaptor), LC3-NIX (mitophagy receptor in erythrocyte maturation) and LC3-Atg4B (cysteine protease for Atg8 lipidation) complexes in mammalian cells22,28,29,30,31,32. Atg32 binds both soluble and PE-conjugated Atg8 independently of other core Atg proteins22. The AIM is not absolutely required for mitophagy19,21,22, suggesting that Atg32-Atg8 interaction serves as an auxiliary to facilitate formation of autophagosomes surrounding mitochondria. Interestingly, in the methylotrophic yeast Pichia pastoris, a threonine residue near the AIM of PpAtg32 seems to be a potential phosphorylation site that contributes to Atg8 binding and mitophagy33.
Atg32 also has I/VLS, a consensus motif important for binding to Atg1121. An alanine substitution in Ser114 of the I/VLS motif affects Atg32 phosphorylation and strongly impairs mitophagy21. PpAtg32 Ser159 is the corresponding amino acid residue in its I/VLS motif that appears to function in a manner similar to that of Atg32 Ser11433. The Atg32 I/VLS motif-containing region physically associates with the Atg11 C-terminal coiled-coil domain that is also crucial for Atg19 binding21,22. Atg32 interacts with Atg11 at the early stage of mitophagy, and even in the absence of Atg8 and other core Atg proteins22. Hence, it seems likely that Atg32-Atg11 interaction is an initial event distinct from autophagosome formation. Mitophagy absolutely requires Atg11, whereas the Cvt pathway in cells lacking Atg11 can still partially operate in a manner dependent on Atg17, another scaffold protein crucial for bulk autophagy19. The reason for this different Atg11 dependency remains uncertain. In conclusion, the primary function of Atg32 is to recruit Atg11 to the surface of mitochondria and localize the site of core Atg protein assembly for mitophagy.
Protein kinases and Atg32 phosphorylation
Recent studies demonstrate that two mitogen-activated protein kinase (MAPK) signal transduction pathways, Wsc1-Pkc1-Bck1-Mkk1/2-Slt2 and Ssk1-Pbs2-Hog1, are important for mitophagy in yeast21,34. In contrast, Slt2 and Hog1, MAP kinases known to be involved in cell wall integrity signaling and hyperosmotic stress response, respectively, are dispensable for bulk autophagy and the Cvt pathway21,34. Notably, Atg32 is phosphorylated under mitophagy conditions21,22. This posttranslational modification depends, at least in part, on Hog1, but not Slt2, although Atg32 is not a direct substrate for Hog121. Whether and how signals specific for mitophagy are transduced into these pathways remains to be investigated. Atg1, a serine/threonine protein kinase essential for all autophagy-related processes, also plays a direct or indirect role in Atg32 phosphorylation22, although the underlying molecular mechanism is currently unknown.
In addition to the induction of its protein synthesis, phosphorylation is a critical posttranslational modification for Atg32 to become active for mitophagy. In particular, Ser114 and Ser119 are key amino acid residues for Atg32 phosphorylation by casein kinase-2 (CK2), an extremely multitasking protein kinase conserved during evolution35. CK2-dependent phosphorylation of Atg32 stabilizes Atg32-Atg11 interaction, which then leads to core Atg protein assembly, subsequent autophagosome formation and ultimately mitochondria degradation35. CK2 is not important for bulk autophagy, pexophagy and the Cvt pathway35,36, suggesting its specific role in mitophagy. CK2 in yeast also phosphorylates protein import factors in the outer membrane of mitochondria, thereby positively regulating the biogenesis of this organelle36. In mammalian cells, p62 is a target for CK2: its specific phosphorylation promotes selective autophagic turnover of polyubiquitinated cargo proteins37. How CK2-dependent phosphorylation of Atg32 is regulated upon mitophagy is not clear. Moreover, whether there is a direct link between CK2 and the HOG signaling pathway remains to be explored.
Atg32-Atg8 interaction may also be regulated through phosphorylation. In P. pastoris, PpAtg32 contains a potential phosphorylation site near its AIM, Thr119, that is required for full mitophagy activities in this methylotrophic yeast33. Whether phosphorylation regulates Atg32-Atg8 interaction in S. cerevisiae has not yet been clarified.
Mitochondrial fission and mitophagy
It is quite conceivable that fragmented mitochondria would be easier targets for mitophagy than tubular mitochondria, since the size of autophagosomes containing mitochondria in yeast mitophagy under prolonged respiratory growth is limited to 200-300 nm in diameter19. In addition, autophagosome formation per se is unlikely to mediate mitochondrial fragmentation. Consistent with this idea, studies in mammalian cells demonstrate that fragmentation is a critical step for mitochondria to be efficiently sequestered into autophagosomes38,39,40.
Recently, it has been reported that Atg11 interacts with Dnm1, a dynamin-related GTPase required for mitochondrial fission in yeast41. A single mutation, E728R or D729R, in the Dnm1 C-terminal GTPase effector domain does not affect mitochondrial shape, but impairs Atg11 binding and partially suppresses mitophagy41. It remains uncertain if Dnm1 contributes to stabilizing Atg32-Atg11 interaction, and/or assists in any other events during degradation of mitochondria. Whether Dnm1 foci associated with the Atg32-Atg11 complex are indeed active fission sites to generate small mitochondrial fragments is also an intriguing issue for future studies. Nonetheless, there may be other factor(s) and mechanism(s) mediating mitophagy-specific mitochondrial fission, as loss of Dnm1 does not completely block degradation of mitochondria.
Physiological significance of mitophagy
Although cells lacking Atg32 exhibit no obvious defects in respiratory growth19,20, mitophagy seems to become important under stress conditions. In particular, mitochondrial DNA deletion frequently occurs in the atg32-null mutant during prolonged nitrogen starvation42. This phenotype is due to oxidative stress derived from excess reactive oxygen species (ROS), which can be suppressed by NAC treatment42. Similarly, the elevated ROS levels in mitophagy-deficient cells under caloric restrictions cause mitochondrial dysfunctions such as reduced respiration, lowered membrane potential and aberrant morphologies, ultimately leading to shortened chronological life span43.
A closely related study in the fission yeast Schizosaccharomyces pombe reveals that transport of mitochondria to the vacuole is drastically promoted in proteasome-deficient cells at G0 phase (quiescent state)44. Under the same conditions, ROS accumulate in mitochondria and the nucleus44. Disruption of the ATG8 gene causes a strong increase in the ROS levels and loss of the mutant viability44, suggesting a critical role of autophagy-dependent mitochondria degradation in cell homeostasis. Strikingly, NAC treatment prevents ROS accumulation and restores cell survival44. It should be noted that mitochondria degradation is neither facilitated in vegetatively growing proteasome-deficient mutants, nor in wild-type cells at G0 phase44. Hence, both autophagy and the proteasome may synergistically contribute to mitochondrial quality control in the quiescent state.
In conclusion, mitophagy in yeast serves as one of the multilayered systems for the management of mitochondrial fitness. When non-dividing cells are exposed to severe stresses, mitophagy becomes essential for the maintenance of healthy mitochondria.
Mitophagy receptors in mammalian systems
Like Atg32 in yeast, mitophagy receptors in mammalian systems are localized at the outer membrane of mitochondria and have the classic tetrapeptide sequence W/F/YxxL/I that mediates the interaction with LC3 for selective autophagy. Currently, there are two types of mitophagy receptors identified in mammalian cells. One family includes NIX/BNIP3L and BNIP345, and the other mitophagy receptor is FUNDC123.
NIX/BNIP3L and BNIP3
BNIP3 was first identified as a Bcl-2 interacting molecule, and NIX/BNIP3L was identified based on its homology (56% identity) to BNIP346,47. NIX/BNIP3L and BNIP3 are localized to mitochondria and the ER, and are involved in regulating apoptotic death or programmed necrosis by affecting mitochondrial respiration or ROS production48,49. Studies from Ivan Dikic and colleagues showed that NIX/BNIP3L has a putative LIR (equivalent to AIM in yeast) that interacts with LC3 and thus defines NIX/BNIP3L as a mitophagy receptor30,50. BNIP3 also contains a typical LIR motif and functions in mitophagy51,52. Mutations in the conserved LIR motif of these proteins abolish their interactions with LC3/GABARAP. Genetic ablation analysis revealed that NIX/BNIP3L functions in autophagic degradation of mitochondria in reticulocytes, a process essential for red blood cell maturation53. NIX/BNIP3L is highly induced at the late stage of erythrocyte maturation and anchored to the surface of mitochondria. It should be noted that mitochondrial clearance in reticulocytes also occurs, at least to some extent, independently of NIX and some core Atg proteins54,55.
NIX/BNIP3L and BNIP3 are also involved in hypoxia-induced mitophagy. One would expect that these mitophagy receptors are induced by hypoxia. Indeed, NIX/BNIP3L and BNIP3 are transcriptionally regulated through HIF or FOXO348,56. In addition, it has been shown that phosphorylation of BNIP3 at Ser17 and Ser24 promotes its binding to LC3-B and GATE-16, and facilitates subsequent mitophagy52. Kinases and phosphatases for BNIP3 are yet to be discovered. Recently, NIX/BNIP3L has been suggested to interact with Rheb, a small GTPase of the Ras superfamily, to promote mitophagy in cells highly active for oxidative phosphorylation57.
FUNDC1 and its posttranslational modifications
FUNDC1 is a mitochondrial outer membrane protein with three TM domains23. The N-terminal region (aa 1-50 of FUNDC1) is exposed to the cytosol and it contains a typical LIR, Y(18)xxL23. The conserved Y18 and L21 are essential for the interaction between FUNDC1 and LC323. Mutations in Y18 and L21, or deletion of the LIR abolish FUNDC1 interaction with LC3 and its function to mediate mitophagy23. Although FUNDC1 is involved in hypoxia-induced mitophagy, the mRNA level of FUNDC1 decreases under hypoxic conditions and its protein levels were significantly reduced due to mitophagy23. The exact mechanism by which hypoxia suppresses FUNDC1 expression was not yet clear. Promoter analysis does not reveal any conserved HIF-1 recognition site (unpublished observation).
The activity of FUNDC1-mediated mitophagy is regulated by reversible phosphorylation at the posttranslational level. We have found that dephosphorylation activates FUNDC1-mediated mitophagy in mammalian systems23. Both Ser13 and Try18 of FUNDC1 are dephosphorylated in response to hypoxic stress and the loss of mitochondrial membrane potential58. Specifically, we found that under normal physiological conditions Tyr18 of FUNDC1 is phosphorylated by Src kinase and Ser13 is phosphorylated by CK258. These two kinases are highly abundant and constitutively active, ensuring the inhibition of mitophagy under unstressed conditions. Inactivation of either Src kinase or CK2 alone by knockdown approach or by pharmacological inhibitors was not sufficient to activate mitophagy, while inhibition of both kinases strongly activates mitophagy58. Furthermore, we have identified that the mitochondria-localized phosphatase PGAM5 is responsible for dephosphorylation of FUNDC1 at Ser1358. Dephosphorylated FUNDC1 has a significantly higher affinity to LC3 and thus this results in an increased interaction between FUNDC1 and LC3, leading to selective autophagosome incorporation and subsequent autophagic removal of the affected mitochondria23,58 (see Figure 2).
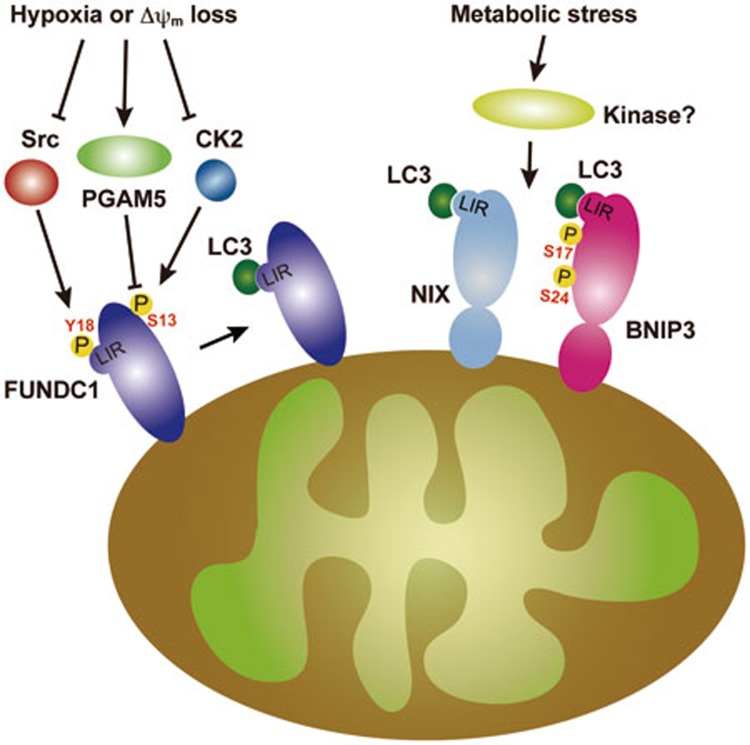
Figure 2.
Mitophagy receptors in mammalian systems. FUNDC1, NIX/BNIP3L and BNIP3 interact with LC3 through LIRs in their N-terminal region. The interaction between FUNDC1 and LC3 is regulated by reversible phosphorylation. In normal conditions, FUNDC1 is phosphorylated at Try18 and Ser13 by Src kinase and CK2, respectively. Hypoxia and FCCP promote FUNDC1 dephosphorylation by inactivating Src kinase and CK2, and by activating PGAM5. Dephosphorylated FUNDC1 has a significantly higher affinity to LC3. The interaction between BNIP3 and LC3 is positively regulated by phosphorylation of Ser17 and Ser24 in BNIP3. The kinase for BNIP3 phosphorylation remains unknown.
Mitochondrial segregation and mitophagy
It is well documented that mitochondrial fission or fragmentation precedes mitophagy. Mitochondrial fission could be part of the sorting mechanism that distinguishes the mitochondria to be engulfed by mitophagy. It was found that the fragmented mitochondria either fuse back to the network when their membrane potential is sustained, or undergo mitophagy when the membrane potential is dissipated40. This sorting mechanism is well illustrated in PINK1/Parkin-mediated mitophagy. Loss of mitochondrial membrane potential will stabilize PINK1 at the outer membrane that is able to recruit Parkin onto the surface of mitochondria59,60. Parkin as a potent E3 ligase then ubiquitinates mitofusin 1 and mitofusin 261, two key mitochondrial fusion mediators, and a number of other mitochondrial proteins62,63. Proteasome-dependent degradation of ubiquitinated mitofusins leads to mitochondrial fragmentation and subsequent mitophagy. Conversely, excessive fusion, such as in the cases of overexpression of dominant-negative Drp1 or wild-type OPA1, retards autophagic degradation of mitochondria in mammalian cells64,65. It should, however, be noted that mitochondrial fragmentation without membrane potential dissipation does not induce Parkin-dependent mitochondrial turnover, suggesting that mitochondrial fission is necessary but not sufficient for mitophagy66. Parkin-labeled mitochondria are bit-by-bit engulfed by autophagic membranes one at a time67. Using a light-activation scheme to impair long mitochondrial tubules, it was shown that sites undergoing bit-by-bit mitophagy display preferential ubiquitination67, and are situated where Parkin-labeled mitochondrial tubules and the endoplasmic reticulum intersect.
The exact contribution of mitochondrial fission in receptor-mediated mitophagy is a subject that is of substantial interest. Overexpression of FUNDC1 induces massive mitochondrial fragmentation prior to mitophagy and knockdown of FUNDC1 enhances mitochondrial fusion, suggesting that mitochondrial fission is required for mitophagy23. Interestingly, deletion of the LIR motif of FUNDC1 almost completely abolished its mitophagy activity, while the mutant protein still strongly induced mitochondrial fragmentation, suggesting that FUNDC1-mediated mitochondrial fission and degradation can be uncoupled23.
Early studies have shown that PGAM5 recruits the mitochondrial fission factor Drp1 and activates its GTPase activity by dephosphorylating Ser637 of Drp168. Since FUNDC1 is also a substrate of PGAM5, it is possible that mitochondrial fission and mitophagy can be highly orchestrated by PGAM5. The molecular details of this orchestration for mitochondrial fission and mitophagy warrant further investigations.
Signaling toward receptor-mediated mitophagy
Current advances in the field have shown that mitophagy activity is highly sensitive to a wide array of cellular and mitochondrial cues. Indeed, perturbation of the cellular bioenergetic status, changes of oxygen tension, loss of mitochondrial membrane potential, an increase in cellular ROS69 (either derived from the cytosol or mitochondria), a disturbance of Ca2+ signaling, defects in mitochondrial protein import or export70, mtDNA damages71, and perturbation of the mitochondrial protein quality control system or accumulation of protein aggregates in mitochondria can all activate mitophagy72.
Whether receptor-mediated mitophagy is involved in some or all of these above mentioned mitochondrial or cellular perturbations to activate mitophagy remains to be examined. It would also be interesting to investigate if these distinct damage or stress signals converge at the mitochondrial receptor level for cross talk with the autophagy machinery. Given the critical role of reversible phosphorylation in the regulation of mitophagy, we propose that distinct kinases such as Src kinase and CK2 or phosphatases such as PGAM5 function as the sentinel for mitochondrial stress signals responsible for initiation of mitophagy. Perturbation of mitochondrial physiology is likely to either activate or inactivate kinases or phosphatases, modulating the interation between mitophagy receptors and the core autophagy-related proteins, thereby leading to activation or prevention of mitophagy. Indeed, both FCCP and hypoxia inactivate Src kinase as well as CK2, and activate PGAM5 to promote FUNDC1-mediated mitophagy in mammalian cells23,58.
Early studies have clearly shown that ROS can activate mitophagy in mammals73. The antioxidant NAC prevents mitophagy induction in yeast27,74, suggesting that the mitochondrial redox status or the ROS production level is one of the common factors contributing to the regulation of mitophagy. Acute ROS production could lead to the opening of the mitochondrial permeability transition pore and loss of mitochondrial membrane potential, which ultimately activates PINK1/Parkin-mediated mitophagy. Indeed, ROS production induced by a mitochondria-targeted photosensitizer resulted in a loss of membrane potential and subsequent activation of Parkin-dependent mitophagy75. One could speculate that slight, but neither acute nor severe, increases in ROS levels may be sufficient to affect kinases or phosphatases to activate receptor-mediated mitophagy. In yeast, the MAPK Hog1 pathway, which can be modulated by oxidative stress, is known to regulate CK2- and Atg32-dependent processes in mitophagy76. We also found that hypoxia can inactivate Src kinase to activate FUNDC1-mediated mitophagy in mammalian cells23. NAC can also prevent hypoxia-induced mitophagy, suggesting that ROS are involved (unpublished observations). Interestingly, it has been shown that ROS and the mitophagy receptor NIX/BNIP3L promote the induction and initiation of mitophagy by enhancing the translocation of Parkin onto damaged mitochondria77, suggesting that PINK1/Parkin-mediated mitophagy and the receptor-mediated pathways could be interdependent.
Perspectives
Receptor-mediated mitophagy is a highly conserved mechanism to selectively remove unwanted or damaged mitochondria. It plays a critical role in controlling mitochondrial quality and quantity in response to the energy needs and other mitochondrial and cellular cues. Mitophagy receptors are highly regulated by reversible phosphorylation by distinct kinases such as CK2 and Src kinase, which can be counterbalanced by phosphatases such as PGAM5 in mammalian cells. Regulation of mitophagy could be more complex and additional players are likely to be discovered as the field is attracting greater attention. For example, cardiolipin can also bind to LC3 to mediate mitophagy78. Selective mitophagy induced by 6-OHDA, rotenone and staurosporine in neuronal cells involves the externalization of cardiolipin to the outer mitochondrial membrane, which acts as a signal for subsequent mitophagy78. Moreover, iron chelation can induce mitophagy in a BNIP3- and Parkin-independent manner79. Although the PINK1/Parkin-mediated pathway has been extensively explored in Drosophila80, whether specific receptors also regulate mitophagy in this fly model remains uncertain. Notably, paternal mitochondria are eliminated via fertilization-triggered autophagy in Caenorhabditis elegans embryos81,82, yet the underlying molecular mechanisms are not understood. Future studies will reveal how receptor-mediated mitophagy coordinates with sorting mechanisms for selective removal of unwanted or damaged mitochondria to regulate mitochondrial quality and quantity.
Defects in mitophagy are proposed to cause accumulation of dysfunctional mitochondria, which is widely observed in aging-related diseases. However, this idea has not been rigorously tested, due to the lack of suitable model systems and reliable quantitative measurements of mitophagy in vivo. Utilization of appropriate model systems for extensive molecular studies will pave new avenues for treatments of diseases involving dysfunctional mitochondria.
References
- Wallace DC, Brown MD, Melov S, Graham B, Lott M. Mitochondrial biology, degenerative diseases and aging. Biofactors. 1998;7:187–190. doi: 10.1002/biof.5520070303. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ Res. 2012;111:1198–1207. doi: 10.1161/CIRCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezek P, Hlavata L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol. 2005;37:2478–2503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Bartosz G. Reactive oxygen species: destroyers or messengers. Biochem Pharmacol. 2009;77:1303–1315. doi: 10.1016/j.bcp.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Nohl H, Gille L, Staniek K. Intracellular generation of reactive oxygen species by mitochondria. Biochem Pharmacol. 2005;69:719–723. doi: 10.1016/j.bcp.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Cheng VW, Ma E, Zhao Z, Rothery RA, Weiner JH. The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. J Biol Chem. 2006;281:27662–27668. doi: 10.1074/jbc.M604900200. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Tavernarakis N. Mitophagy in neurodegeneration and aging. Front Genet. 2012;3:297. doi: 10.3389/fgene.2012.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R, Goldman SJ. Mitophagy and disease: new avenues for pharmacological intervention. Curr Pharm Des. 2011;17:2056–2073. doi: 10.2174/138161211796904768. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci. 1998;111(Pt 16):2455–2464. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Baba M, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Kanki T, Hirota Y, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Noda NN, Suzuki SW, et al. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J Biol Chem. 2012;287:10631–10638. doi: 10.1074/jbc.M111.299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- Eiyama A, Kondo-Okamoto N, Okamoto K. Mitochondrial degradation during starvation is selective and temporally distinct from bulk autophagy in yeast. FEBS Lett. 2013;587:1787–1792. doi: 10.1016/j.febslet.2013.04.030. [DOI] [PubMed] [Google Scholar]
- Wang K, Jin M, Liu X, Klionsky DJ. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy. 2013;9:1828–1836. doi: 10.4161/auto.26281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter E, Montino M, Reinhold R, et al. Uth1 is a mitochondrial inner membrane protein dispensable for post-log-phase and rapamycin-induced mitophagy. FEBS J. 2013;280:4970–4982. doi: 10.1111/febs.12468. [DOI] [PubMed] [Google Scholar]
- Deffieu M, Bhatia-Kissova I, Salin B, Galinier A, Manon S, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. J Biol Chem. 2009;284:14828–14837. doi: 10.1074/jbc.M109.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoo K, Noda NN, Kumeta H, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Noda NN, Nakatogawa H, Kumeta H, Ohsumi Y, Inagaki F. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J Biol Chem. 2010;285:29599–29607. doi: 10.1074/jbc.M110.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14:441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193:755–767. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Kurihara Y, Jin X, et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013;14:788–794. doi: 10.1038/embor.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, et al. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Kanki T, Aoki Y, et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem. 2012;287:3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard VR, Leonov A, Beach A, et al. Macromitophagy is a longevity assurance process that in chronologically aging yeast limited in calorie supply sustains functional mitochondria and maintains cellular lipid homeostasis. Aging. 2013;5:234–269. doi: 10.18632/aging.100547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Yoshida T, Kikuchi S, et al. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc Natl Acad Sci USA. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu L, Zhu Y, Chen Q. Molecular signaling toward mitophagy and its physiological significance. Exp Cell Res. 2013;319:1697–1705. doi: 10.1016/j.yexcr.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Chen G, Ray R, Dubik D, et al. The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J Exp Med. 1997;186:1975–1983. doi: 10.1084/jem.186.12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima M, Fujiwara T, Takahashi E, et al. Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosomes Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- Bursch W, Karwan A, Mayer M, et al. Cell death and autophagy: cytokines, drugs, and nutritional factors. Toxicology. 2008;254:147–157. doi: 10.1016/j.tox.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Novak I, Dikic I. Autophagy receptors in developmental clearance of mitochondria. Autophagy. 2011;7:301–303. doi: 10.4161/auto.7.3.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Massen S, Terenzio M, et al. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Yamamoto A, Kuma A, Ohsumi Y, Mizushima N. Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem Biophys Res Commun. 2006;339:485–489. doi: 10.1016/j.bbrc.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G, Vijayalingam S, Gibson SB. BNIP3 subfamily BH3-only proteins: mitochondrial stress sensors in normal and pathological functions. Oncogene. 2008;27(Suppl 1):S114–127. doi: 10.1038/onc.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser S, Chatelain EH, Lavie J, et al. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 2013;17:719–730. doi: 10.1016/j.cmet.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Chen G, Han Z, Feng D, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125:795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson's disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Reichert AS. Mitophagy, mitochondrial dynamics and the general stress response in yeast. Biochem Soc Trans. 2011;39:1514–1519. doi: 10.1042/BST0391514. [DOI] [PubMed] [Google Scholar]
- Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Yang WY. Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nat Commun. 2013;4:2428. doi: 10.1038/ncomms3428. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Byrne AM, Lemasters JJ, Nieminen AL. Contribution of increased mitochondrial free Ca2+ to the mitochondrial permeability transition induced by tert-butylhydroperoxide in rat hepatocytes. Hepatology. 1999;29:1523–1531. doi: 10.1002/hep.510290521. [DOI] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;12:1613–1621. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- Kim I, Lemasters JJ. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid Redox Signal. 2011;14:1919–1928. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter DH. Liver injury in alpha1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J Clin Invest. 2002;110:1579–1583. doi: 10.1172/JCI16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Kissova IB, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. Autophagy. 2009;5:872–873. doi: 10.4161/auto.9065. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012;8:1462–1476. doi: 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- Prick T, Thumm M, Kohrer K, Haussinger D, Vom Dahl S. In yeast, loss of Hog1 leads to osmosensitivity of autophagy. Biochem J. 2006;394:153–161. doi: 10.1042/BJ20051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Li M, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. Drosophila as a model to study mitochondrial dysfunction in Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2.pii:a009944. doi: 10.1101/cshperspect.a009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rawi S, Louvet-Vallee S, Djeddi A, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- Journo D, Mor A, Abeliovich H. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem. 2009;284:35885–35895. doi: 10.1074/jbc.M109.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Mendl N, Occhipinti A, Muller M, Wild P, Dikic I, Reichert AS. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci. 2011;124:1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- Wang K, Yang Z, Liu X, Mao K, Nair U, Klionsky DJ. Phosphatidylinositol 4-kinases are required for autophagic membrane trafficking. J Biol Chem. 2012;287:37964–37972. doi: 10.1074/jbc.M112.371591. [DOI] [PMC free article] [PubMed] [Google Scholar]