Abstract
The enteric pathogen, Salmonella enterica serovar Typhimurium, causes food poisoning resulting in gastroenteritis. The S. Typhimurium effector protein, SipA, promotes gastroenteritis by functional motifs that either trigger mechanisms of inflammation or bacterial entry. During infection of intestinal epithelial cells, SipA was found to be responsible for the early activation of caspase-3, an enzyme that is required for SipA cleavage at a specific recognition motif that divided the protein into its two functional domains, and activated SipA in a manner necessary for pathogenicity. Other caspase-3 cleavage sites identified in S. Typhimurium appeared to be restricted to secreted effector proteins, indicating this may be a general strategy employed by this pathogen for processing of its secreted effectors.
Salmonella enterica serovar Typhimurium acquires virulence via a 40-kb segment of the bacterial chromosome designated Salmonella Pathogenicity Island 1 (SPI-1) (1). SPI-1 contains more than 25 genes encoding structural components and substrates of a type III protein-secretion system that mediates the translocation of effector proteins from Salmonella into mammalian cells (1). One of these, Salmonella invasion protein A (SipA), is a bifunctional molecule responsible for promoting actin polymerization, a process that facilitates bacterial entry into epithelial cells (2), and is required to trigger signal transduction cascades that promote polymorphonuclear leukocyte (PMN) migration across the intestinal epithelium (3). The actin binding function of SipA is known to be localized to a C-terminal fragment (amino acids 426–684), termed SipAb (4). We have reported that the N-terminal fragment of the SipA effector protein (amino acids 2-425), harbors the functional domain that induces PMN transepithelial migration (5), which underlies the clinical manifestations of salmonellosis.
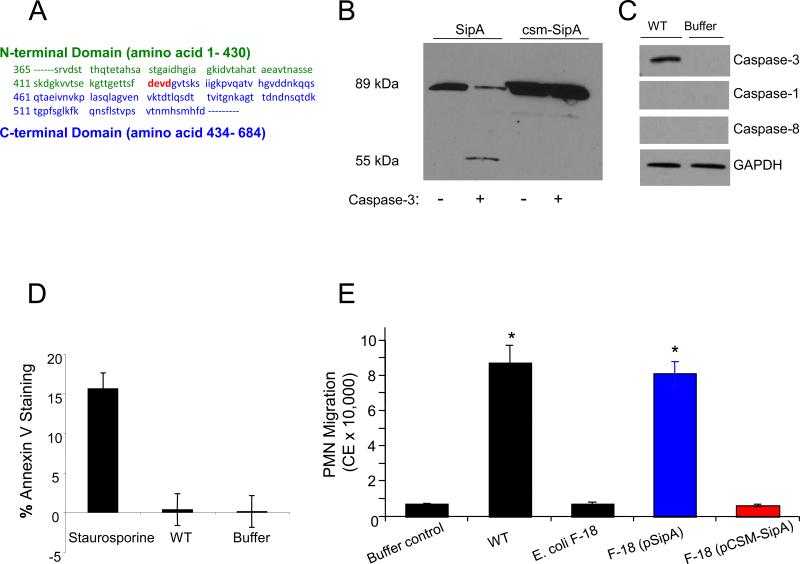
We discovered a caspase-3 motif, Asp-Glu-Val-Asp (DEVD), at amino acid position 431-434 (Fig. 1A) of SipA at the junction between the two functional domains. Treatment of a purified fraction of SipA to activated caspase-3 enzyme generated a predicted fragment (55 kDa; Fig. 1B) and confirmed that the SipA DEVD cleavage motif was functionally active. The expected lower molecular weight band was not seen most likely due to lack of antibody recognition; however, mutation of the caspase-3 site by changing aspartic acid at position four to alanine (A) (DEVD →DEVA; termed caspase site mutant: csm-SipA) rendered SipA insensitive to caspase-3 cleavage (Fig. 1B).
Fig. 1.
The effector protein SipA harbors a functional caspase-3 cleavage site. (A) Primary amino acid sequence of SipA showing the caspase-3 cleavage site (highlighted in red) between the N-terminal (green) and C-terminal (blue) domains. (B) Immunoblot of SipA in the absence and presence of caspase-3. The full length 89 kDa SipA effector is cleaved resulting in the 55 kDa truncated N-terminal domain being produced. The caspase site mutant SipA (csm-SipA) is not cleaved by caspase-3. (C) Whole cell extracts from T84 cells that were uninfected or infected with wild-type S. Typhimurium (WT) were probed with anti-caspase-3, -1, -8 and a GAPDH control showing equal loading of lanes. (D) Annexin V staining as a measurement of apoptosis in T84 cells 2 hours post infection with wild-type S. Typhimurium (WT). Staurosporine treatment served as the positive control for apoptosis. (*) P < 0.01. (E) Alteration of the SipA caspase-3 motif rendered SipA resistant to cleavage and attenuated the ability of S. Typhimurium to induce neutrophil (PMN) movement. PMN migration induced by SipA (blue bars) was far greater than with the pCSM-SipA (red bars) and was of the same extent as the wild-type S. Typhimurium strain (WT). (*), P < 0.01; (NS) not significant.
Although caspase-3 is a frequently activated death protease, this enzyme also catalyzes specific cleavage of many cellular proteins (6), and promotes cell proliferation and inflammation without inducing apoptosis (7). We found that S. Typhimurium infection of model intestinal epithelia induced the activated form of the caspase-3 enzyme within 2 hour and persisted for at least 4 hour (Fig. 1C and Fig. S1A and B). Apoptosis occurred 24 hours following infection of epithelial cells (8). No activation of other initiator cysteine proteases known to play essential roles in apoptosis (i.e., caspase-1 or caspase-8) (Fig. 1C) was observed. Staurosporine treatment, which induces apoptosis and was used as a positive control, showed significant Annexin V staining, whereas the S. Typhimurium treated T84 cells showed minimal Annexin V binding and no propidium iodide staining (Fig. 1D), confirming that early after infection this pathogen can activate caspase-3 without inducing apoptosis or necrosis.
A plasmid bearing the mutant clone, pCSM-SipA (for plasmid caspase site mutant), was transformed into both Escherichia coli F-18, an intestinal commensal isolate (9), and Salmonella. Since E. coli F-18 secretes SipA through the flagella basal body (5), we used this strain to assess the ability of the caspase-3 site mutant to drive PMN transepithelial migration in the absence of other S. Typhimurium virulence determinants. Using our in vitro inflammatory model system (10), we found that the mutant had a reduced capacity to induce PMN transepithelial migration (>95%; P<0.01) compared with the E. coli F-18 strain expressing the wild-type SipA protein (pSipA) (Fig. 1E).
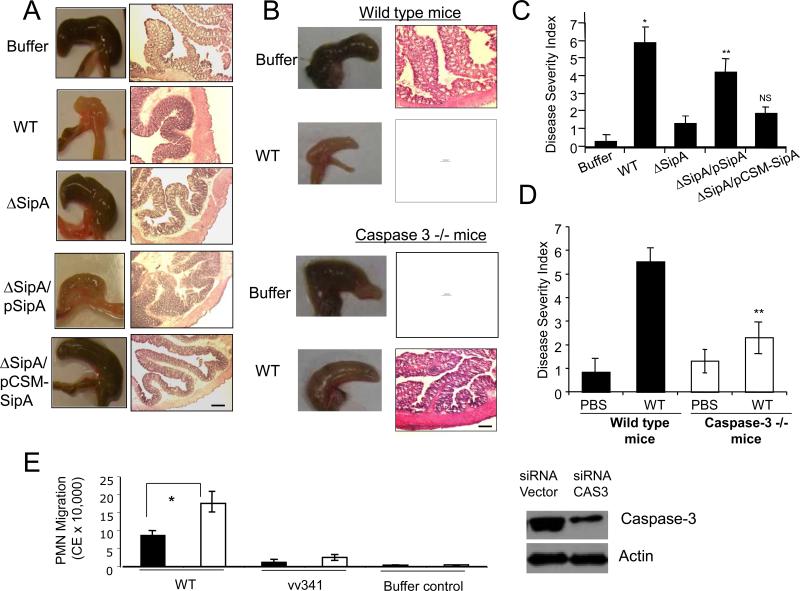
Salmonella enterica serovar Dublin was used in the in vivo enteritis model because this strain constitutively expresses proteins from the pBH plasmid and allows the expression of proteins of interest in animal infections (5,11, 12). Macroscopic inspection of the intestines revealed inflammation of the cecum in infections with both wild-type Salmonella and the ΔSipA mutant strain expressing SipA (ΔSipA/pSipA) (Fig. 2A). By contrast, no overt signs of intestinal inflammation were observed in mice infected with the ΔSipA mutant strain expressing the caspase-3 site mutant (ΔSipA/pCSM-SipA), the ΔSipA mutant strain, or the buffer negative control (Fig. 2A). Histological evaluation of hematoxylin and eosin (H&E) stained tissue sections and the pathology scores for disease severity showed significant inflammation of the proximal colons of mice infected with either wild-type Salmonella or ΔSipA/pSipA, as determined by colonic wall thickening, crypt elongation, PMN infiltration, epithelial erosion, and edema (Fig. 2A and 2C). By contrast, infections with both the ΔSipA/pCSM-SipA and ΔSipA mutant strains did not cause pronounced intestinal inflammation (Fig. 2A and 2C). Quantification of infiltrating PMNs, as determined by myeloperoxidase (MPO) activity, supported these observations (Fig. S2).
Fig. 2.
Mutation of the caspase-3 cleavage site attenuates the ability of the SipA effector to promote inflammation in an in vivo mouse model. (A) Disease pathology following a 48 hr infection of mice with different strains of Salmonella. The left panel shows gross morphology of the cecum while the right panels shows the histopathology of the sections stained with hematoxylin and eosin (H&E). The magnification bar is 100 μM. The ΔSipA mutant strain was complemented with either SipA (pSipA) or caspase-site-mutant SipA (pCSM-SipA). Phosphate buffered saline (PBS) treatment was the negative control. (B) Disease pathology following a 48 hour infection of wild-type mice and caspase-3 knock out mice (both on C57Bl/6 background). The left panel shows the gross morphology of the cecum, and the H&E stained sections of the proximal colon are shown on the right. The magnification bar is 100 μM. WT refers to the wild-type Salmonella strain used for infection, and buffer refers to the PBS treated negative control. (C) The colonic histopathology for the sections shown in part A were quantified using a disease severity index (scale 0-8), with “0” scored normal and a score of “8” showing the most significant level of disease pathology (n=5). Sections were scored blinded by a trained pathologist. Data represent ± SD. (*), P < 0.01; (**), P < 0.05; (NS) not significant. (D) The colonic histopathology for sections in part B was quantified using the same disease severity index (scale 0-8, n=5). Data represent ± SD. (**), P < 0.05. (E) Neutrophil (PMN) transepithelial migration following small interfering RNA (siRNA) knockdown of caspase-3 in HCT8 cells expressing siRNA against caspase-3 (black bars) or containing a vector control (open bars). Cells were infected with either wild-type S. Typhimurium (WT) or a ΔHilA isogenic negative mutant (vv341), which is incapable of inducing migration (*), P<0.05. Immunoblot shows knockdown of caspase-3 expression by siRNA directed against caspase-3, actin was used a loading control.
Cleavage of the SipA DEVD recognition site appears to be important for promoting proinflammatory responses and suggests a role for caspase-3. Consistent with this notion, Salmonella is less virulent in caspase-3 knockout (caspase-3−/−) mice, as judged by gross cecal and histopathologic examination of the proximal colons of Salmonella-infected caspase-3−/− mice, which reveals significantly less intestinal pathology compared to Salmonella-infected wild-type mice (Fig. 2B and 2D). In addition, reduced S. Typhimurium invasiveness in bone-marrow-derived macrophages from caspase-3−/− mice, compared with wild-type macrophages further indicates that a lack of caspase-3 has an impact on the infection outcome (Fig. S2C). Likewise, in vitro inhibition of epithelial derived caspase-3 by siRNA significantly reduced S. Typhimurium-induced PMN transepithelial migration (Fig. 2E), as did pharmacologic inhibition of caspase-3 (Fig. S3).
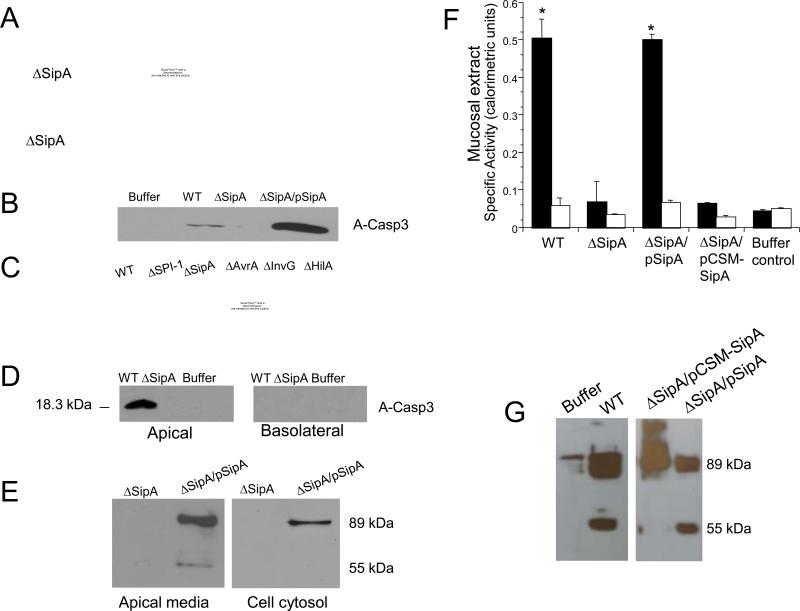
Remarkably, the SipA effector itself stimulated activation of caspase-3, whereas the ΔSipA mutant strain was not able to (Fig. 3A and 3B), indicating that the effector is necessary and sufficient to promote activation of caspase-3. Moreover, S. Typhimurium non-polar, isogenic mutant strains defective in SipA expression and secretion (ΔSPI-1 and ΔInvG, respectively), or lacking transcription factors controlling SipA expression (ΔHilA) also failed to induce caspase-3 activation (Fig. 3C). Mutations in genes encoding other effector proteins not associated with SipA expression, such as avrA, did not adversely affect the expression of activated caspase-3 (Fig. 3C). Levels of caspase-3 expression measured during S. Typhimurium infection of epithelial cells by quantitative PCR showed no new procaspase-3 was being produced (Fig. S4). However, activated caspase-3 does appear in the membrane insoluble fraction (Fig. 2D), suggesting that S. Typhimurium infection modifies existing pools of caspase-3.
Fig. 3.
SipA causes activation of caspase-3 and its release at the apical surface of the epithelium. (A) Time course of caspase-3 activation during S. Typhimurium infection of T84 cell monolayers. Cells were infected with either wild-type S. Typhimurium (WT) or the ΔSipA mutant. Casp3 refers to pro-caspase-3 while A-Casp3 represents the activated form of the enzyme; Soluble and insoluble refers to the respective membrane fractions. (B) The ΔSipA mutant strain complemented with a vector expressing SipA (ΔSipA/pSipA) rescued the ability to induce the activation of caspase-3 following a 2 hour infection. Buffer refers to the buffer control. (C) Infection of T84 cells with different isogenic mutant strains of S. Typhimurium. Immunoblots were performed on cell extracts 3 hour post infection with anti-caspase-3 antibody. Casp3 refers to pro-caspase-3; A-Casp3 represents the activated form of the enzyme. (D) Activated caspase-3 was released at the apical but not basolateral surface during in vitro infection of T84 cells with wild-type S. Typhimurium (WT). No caspase-3 was detected in ΔSipA mutant infected wells or in the buffer control, uninfected wells. (E) Cleaved SipA was detected in apical media but not in the cell cytosol following infection with the ΔSipA mutant complemented with SipA (ΔSipA/pSipA). The arrow shows the N-terminal fragment generated by cleavage. No SipA is detected in the SipA negative mutant infected cells. (F) Caspase-3 (black bars) or caspase-1 (open bars) activity was measured in mucosal extracts post in vivo infection of wild-type mice. Infection was carried out with wild-type Salmonella (WT), a ΔSipA mutant, and ΔSipA complemented with either SipA (ΔSipA/pSipA) or pCSM-SipA (ΔSipA/pCSM-SipA). Buffer refers to the uninfected buffer control. (G) Immunoblots showing cleavage of SipA after addition of mucosal extract isolated from mice infected with various indicated strains. WT refers to wild-type Salmonella and ΔSipA is the ΔSipA strain complemented with either wild-type SipA (SipA/pSipA) or the pCSM-SipA (ΔSipA/pCSMSipA). Buffer refers to the negative control.
In the intestinal lumen several environmental cues trigger the upregulation of the Salmonella SPI-1 type III secretion system (i.e., osmolarity, oxygen tension) (13, 14). Therefore, upon colonization, it is likely that Salmonella has begun to secrete effector proteins into the intestinal milieu. Although SipA is a type III secreted protein it is unique in its ability to act extracellularly and elicit a proinflammatory response (5). We hypothesized that processing of SipA by caspase-3 cleavage occurs at the apical epithelial surface, liberating the N-terminal SipA domain for its extracellular function, which presumably requires binding to a surface receptor (3). This offers an explanation of why the N-terminal domain, when expressed independently of the C-terminal domain, is a more potent inflammatory stimulus than the full-length protein (5).
In support of this hypothesis, S. Typhimurium infection of polarized T84 monolayers resulted in the preferential secretion of activated caspase-3, in a SipA dependent manner, into the apical epithelial compartment, but not the basolateral compartment (Fig. 3D). SipA processing also occurs extracellularly at the apical surface, as SipA cleavage products were recovered within apical supernatants following S. Typhimurium infection, but were not observed within the epithelial cell (Fig. 3E). Colonic mucus extracts were analyzed following Salmonella-induced colitis for the functional activity of caspase-3 and caspase-1 (a non-specific control) (9). Consistent with our in vitro findings, only caspase-3 activity could be isolated from colonic mucus infected with Salmonella bearing a functional SipA (Fig 3F), and Salmonella infected mice were only capable of cleaving SipA possessing an intact caspase-3 recognition motif (Fig. 3G). Because these data demonstrate a novel mode of caspase-3 action we attempted to model these interactions in vitro. Exogenous addition of the caspase-3 enzyme to S. Typhimurium infected T84 monolayers significantly and specifically enhanced PMN transepithelial migration (Fig. S5). While these results validate the concept that caspase-3 dependent processing plays an important role in bacterial-induced inflammation, the underlying molecular mechanism of how caspase-3 becomes activated remains to be defined. However, during infection, caspase-3 secretion does not appear to correlate with cell lysis (Fig. S6).
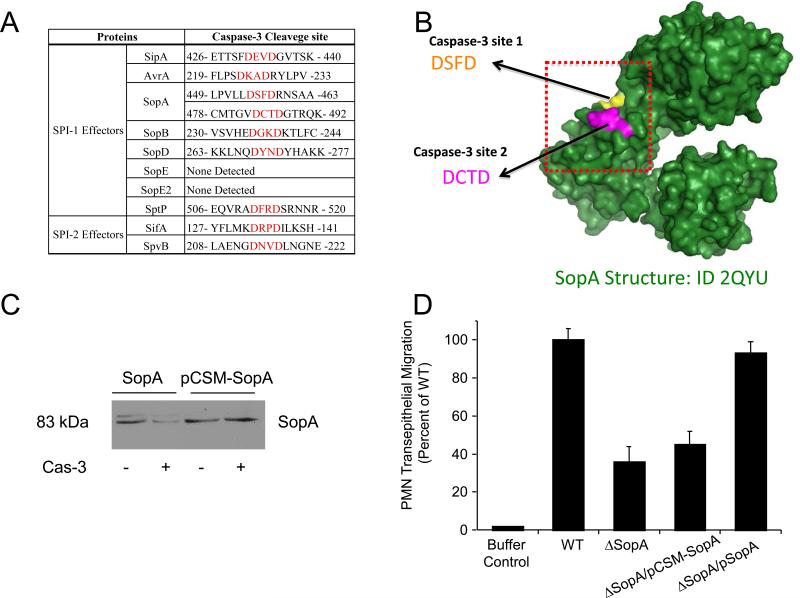
A detailed analysis of S. Typhimurium SPI-1 proteins revealed the existence of caspase-3 recognition and cleavage sites in other secreted S. Typhimurium effectors (i.e., AvrA, SopB, SifA, SipB and SopA), but not type III secretion system structural proteins, chaperones or transcriptional regulators (Table 1). Like SipA, many of the caspase-3 cleavage sites in these other effector proteins are centrally located within the protein sequence, dividing the effectors into what, at least in the case of SifA, have been identified as two functional domains (15). These observations are further supported by studies with the effector protein, SopA, a HECT-like E3 ubiquitin ligase, which is also involved in inducing transepithelial migration of PMNs (16, 17). SopA harbors two caspase-3 recognition motifs (449-463 and 478-492) that not only are positioned close to each other (Fig. 4A), but also are located in an exposed domain of the effector molecule (Fig. 4B). These SopA caspase-3 cleavage sites are functional, as exposure of SopA to the activated caspase-3 enzyme led to nearly complete digestion of the effector (Fig. 4C). By contrast, single amino acid substitution at position four of the caspase-3 recognition site to a motif not recognized by the enzyme (aspartic acid (D) to alanine (A): DSFD to DSFA and DCTD to DCTA), was refractory to capase-3 digestion (Fig. 4C). The inability to visualize the digested fragments is most likely due to instability of the resulting fragments or poor antibody recognition. Nonetheless, the SopA caspase-3 site double mutant had significantly less capacity to induce PMN transepithelial migration than the S. Typhimurium wild-type strain (> 50%; P < 0.01).
Fig. 4.
Cleavage sites have been identified in numerous other S. Typhimurium effectors. (A) Functional caspase-3 cleavage sites in SipA, SopA, and putative caspase-3 cleavage sites in other S. Typhimurium effectors. (B) Three dimensional image of the wild-type SopA protein showing the position of the two caspase-3 cleavage sties that are surface exposed. (C) Immunoblot showing cleavage of SopA with caspase-3 and the absence of cleavage in the SopA caspase site double mutant. (D) Neutrophil (PMN) transepithelial migration induced in vitro by infection with wild-type S. Typhimurium and a SopA mutant strain (ΔSopA). ΔSopA/pCSM-SopA and ΔSopA/pSopA are the ΔSopA strain complemented with the caspase site mutant of SopA (csm-SopA) and wild-type SopA respectively. (*) P < 0.01. Migration levels shown are presented as the percent of the wild-type S. Typhimurium strain.
Secreted effector proteins from other enteric pathogens, such as Shigella flexneri and Enteropathogenic Escherichia coli (EPEC), also harbor potential caspase-3 recognition sites (Tables S1 and S2). While some protein sequences were found to contain multiple caspase-3 cleavage sites (i.e., IpaA and EspC), other sequences such as EPEC EspB protein, which is involved in the translocation of effector proteins during EPEC infection (18), exhibit discrete localization of the caspase-3 sites, depending on whether the strain is isolated from humans or other mammals (rabbits or mice). Of potential significance, most of the proteins having a putative caspase-3 recognition and cleavage site exhibit a dual function, like SipA, with distinct activities of the N- or C-terminus of the protein (19-22). Future studies will determine whether these secreted effectors are also processed post-transcriptionally by caspase-3.
Our work reveals that S. Typhimurium has evolved a mechanism to deliver effector proteins in a precursor form to the host cell where they are subsequently processed into independent functionally active domains. Alteration of the caspase-3 motif or modulation of caspase-3 expression attenuates the ability of S. Typhimurium to induce gastroenteritis. Many effector proteins secreted by bacteria share caspase-3 cleavage motifs, inviting speculation that caspase-3 processing may be a common host-pathogen interaction. The intestinal epithelium is a barrier against invading pathogens, and caspase-3 induction/release may have evolved as a rapid innate immune response to pathogens, perhaps in an attempt to kill or disarm the intruder (i.e., defensins). However, some pathogens such as S. Typhimurium appear to be able to subvert such defenses.
Supplementary Material
Acknowledgements
The research was supported by grants from the National Institutes of Health (DK56754 and DK33506), and the Crohn's and Colitis Foundation of America to B.A.M. Additional grant support was provided by Tenovus Scotland, and a Society for General Microbiology Travel Award to D.M.W. The use of human volunteers in this study was in accordance with appropriate guidelines and was approved by the University of Massachusetts Medical School Review Board for the Protection of Human Subjects (approval #13006).
References and Notes
- 1.Hueck CJ. Microbiol Mol Biol Rev. 1998 Jun;62:379. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou D, Mooseker MS, Galan JE. Proc Natl Acad Sci U S A. 1999 Aug 31;96:10176. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CA, et al. Proc Natl Acad Sci U S A. 2000 Oct 24;97:12283. doi: 10.1073/pnas.97.22.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lilic M, et al. Science. 2003 Sep 26;301:1918. doi: 10.1126/science.1088433. [DOI] [PubMed] [Google Scholar]
- 5.Wall DM, et al. Cell Microbiol. 2007 Sep;9:2299. doi: 10.1111/j.1462-5822.2007.00960.x. [DOI] [PubMed] [Google Scholar]
- 6.Zakeri Z, Lockshin RA. Adv Exp Med Biol. 2008;615:1. doi: 10.1007/978-1-4020-6554-5_1. [DOI] [PubMed] [Google Scholar]
- 7.Nhan TQ, Liles WC, Schwartz SM. Am J Pathol. 2006 Sep;169:729. doi: 10.2353/ajpath.2006.060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JM, et al. J Clin Invest. 1998 Nov 15;102:1815. doi: 10.1172/JCI2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormick BA, Stocker BA, Laux DC, Cohen PS. Infect Immun. 1988 Sep;56:2209. doi: 10.1128/iai.56.9.2209-2217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL. J Cell Biol. 1993 Nov;123:895. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barthel M, et al. Infect Immun. 2003 May;71:2839. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherayil BJ, McCormick BA, Bosley J. Infect Immun. 2000 Oct;68:5567. doi: 10.1128/iai.68.10.5567-5574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CA, Falkow S. Proc Natl Acad Sci U S A. 1990 Jun;87:4304. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CA, Jones BD, Falkow S. Proc Natl Acad Sci U S A. 1992 Mar 1;89:1847. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohlson MB, et al. Cell Host Microbe. 2008 Nov 13;4:434. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood MW, et al. Cell Microbiol. 2000 Aug;2:293. doi: 10.1046/j.1462-5822.2000.00054.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. Mol Microbiol. 2006 Nov;62:786. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 18.Deng W, et al. Proc Natl Acad Sci U S A. 2004 Mar 9;101:3597. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu HJ, Syu WJ. Microbiology. 2005 Oct;151:3277. doi: 10.1099/mic.0.28115-0. [DOI] [PubMed] [Google Scholar]
- 20.Deane JE, Roversi P, King C, Johnson S, Lea SM. J Mol Biol. 2008 Apr 4;377:985. doi: 10.1016/j.jmb.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramarao N, et al. FEBS Lett. 2007 Mar 6;581:853. doi: 10.1016/j.febslet.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 22.Tran Van Nhieu G, Caron E, Hall A, Sansonetti PJ. Embo J. 1999 Jun 15;18:3249. doi: 10.1093/emboj/18.12.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.