Abstract
The retinoic acid receptors α, β and γ (RARα, RARβ and RARγ) are nuclear hormone receptors that regulate fundamental processes during embryogenesis, but their roles in skeletal development and growth remain unclear. To study skeletal-specific RAR function, we created conditional mouse mutants deficient in RAR expression in cartilage. We find that mice deficient in RARα and RARγ (or RARβ and RARγ) exhibit severe growth retardation obvious by about 3 weeks postnatally. Their growth plates are defective and, importantly, display a major drop in aggrecan expression and content. Mice deficient in RARα and RARβ, however, are virtually normal, suggesting that RARγ is essential. In good correlation, we find that RARγ is the most strongly expressed RAR in mouse growth plate and its expression characterizes the proliferative and pre-hypertrophic zones where aggrecan is strongly expressed also. By being avascular, those zones lack endogenous retinoids and thus RARγ is likely to exert ligand-less repressor function. Indeed, our data indicate that: aggrecan production is enhanced by RARγ over-expression in chondrocytes under retinoid-free culture conditions; production is further boosted by corepressor Zac1 or pharmacologic agents that enhance RAR repressor function; and RAR/Zac1 function on aggrecan expression may involve Sox proteins. In sum, our data reveal that RARs, and RARγ in particular, exert previously unappreciated roles in growth plate function and skeletal growth and regulate aggrecan expression and content. Since aggrecan is critical for growth plate function, its deficiency in RAR-mutant mice is likely to have contributed directly to their growth retardation.
Keywords: Retinoic acid receptors, Skeletal growth, Aggrecan, Growth plate, Zac1, Transcriptional repressors
Introduction
Prenatal and postnatal growth of the skeleton is a complex and tightly-regulated spatio-temporal process. In most skeletal elements, growth is attributable to the activity of the growth plates where chondrocytes proliferate, enlarge into pre-hypertrophic and hypertrophic cells, and are ultimately replaced by endochondral bone and marrow (Wilsman et al., 1996). Growth plate function is directed and orchestrated by multiple systemic and local factors that include growth hormone, hedgehog proteins, and Runx and Sox transcription factors (Iwamoto et al., 1999; Lefebvre and Smits, 2005; Nilsson et al., 2005). In addition, functioning of growth plates depends on their extracellular matrix composed of aggrecan, collagen II and other macromolecules (Schwartz and Domowicz, 2002; van der Eerden et al., 2003). Indeed, abnormalities in matrix gene expression and structure can cause growth plate malfunction and chondrodysplasias including dwarfism (ul Haque et al., 1998; Watanabe et al., 1994). For example, the lethal mouse chondrodystrophy Cartilage Matrix Deficiency (cmd) is caused by stop codon mutations in aggrecan gene; the mutant mice exhibit short limbs, an enlarged abdomen and cleft palate, and their growth plates are disorganized (Watanabe et al., 1994). In addition, aggrecan expression levels and turn-over rates are tightly regulated and important for normal growth plate function (Shapses et al., 1994; Wang et al., 2004). Clearly, a normal complement of matrix components is critical for growth plate function and skeletal growth, but it is far from clear particularly at the molecular level how matrix homeostasis is established and maintained.
The retinoic acid receptors α, β and γ (RARα, RARβ and RARγ) are nuclear hormone receptors that regulate fundamental processes during development (Chambon, 1996; Sucov and Evans, 1995). The RARs form heterodimers with RXRα, RXRβ and RXRγ, interact with specific DNA response elements (RAREs) and can act as ligand-dependent transcriptional activators or ligand-less transcriptional repressors. When ligands are present, a conformational change allows interaction with co-activators and proteins with histone acetyltransferase (HAT) activity (Glass and Rosenfeld, 2000; Moras and Gronemeyer, 1998). When ligands are absent, the RAR-RXR dimers bound to RAREs recruit co-repressors, including nuclear receptor corepressor-1 (NCoR-1) and silencing mediator for retinoic acid and thyroid hormone receptors (SMRT/NCoR-2) (Chen et al., 1996; Horlein et al., 2002), and additional factors such as mSin3A and histone deacetylases (HDACs), thus promoting chromatin compaction and transcriptional repression. Studies by Chambon and coworkers first established that RARs are differentially expressed during early steps of limb skeletogenesis and required for skeletal development (Mendelsohn et al., 1992). While single RAR-null mice were found to be viable and displayed few defects, double-null mice lacking RARα/RARγ or RARβ/RARγ died before or immediately after birth and displayed severe skeletal defects including homeotic transformations, growth retardation and malformation of limb bones (Lohnes et al., 1994). It has remained largely unclear ever since, however, what specific roles the RARs have in mammalian skeletal development and growth plate functioning. To address this important issue, we created conditional double mutant mice deficient in RARα/RARβ, RARα/RARγ or RARβ/RARγ in cartilage. We find that mice deficient in RARα/RARγ or RARβ/RARγ exhibit significant skeletal growth retardation and growth plate abnormalities, whereas mice deficient in RARα/RARβ are essentially normal. The apparent centrality of RARγ is reaffirmed by our extensive in vitro experiments with wild type and RAR-deficient chondrocytes in which we have also identified Zac1 as a likely RAR functional partner. The data provide novel insights into the molecular regulation of growth plate function and skeletal growth.
Materials and methods
RAR-deficient mice and genotyping
RAR-deficient mice were generated by mating floxed RAR mice (RARαf/f, RARβf/f or RARγf/f) (Chapellier et al., 2002a; Chapellier et al., 2002b; Chapellier et al., 2002c) with Col2a1-Cre mice (Ovchinnikov et al., 2000) to eventually create homozygous double RAR-deficient mice in cartilage. Briefly, Col2a1-Cre mice were first mated with a single RAR floxed homozygous mouse to create heterozygous Col2a1-Cre+/RARf/+ mice that were in turn mated to produce Col2a1-Cre+/RARf/f mice deficient in one RAR in cartilage. To generate double mutant mice, single RAR deficient mice were mated to each other to generate double heterozygous mice (Col2a1-Cre+/RARf/+/RARf/+). Resulting mice, usually Col2a1-Cre+/RARf/f/RARf/+, were mated to obtain double RAR-deficient mice (Col2a1-Cre+/RARf/f/RARf/f) in all receptor combinations (α/β, α/γ, β/γ). Cre-mediated excision deletes exon 8 in RARα and RARγ that encodes most of DNA binding domain, and deletes exons 9 and 10 in RARβ that encode much of the ligand-binding domain. In each case, deletions cause also a frameshift mutation, thus resulting in functional ablation of all spice forms. Genotyping procedures and results are shown as supplemental Fig. S1 and S2.
In situ hybridization, anatomical measurements and RT-PCR
In situ hybridization was carried out with radiolabeled riboprobes (Iwamoto et al., 2000; Koyama et al., 1999). Plasmids were: pGEM-T encoding 548-1627 of NM_009024 for RARa; pGEM-T encoding 1480-2116 of XM_127583 for RARb; pGEM-T encoding 1673-2628 of NM_011244 for RARg that recognize all RARg splice forms. Col9a1, Sox 9, Aggrecan and H4C probes were described previously (Tamamura et al., 2005). Dark and bright field images were captured using a digital camera. In some experiments, dark field images were pseudo-colored using Adobe Photoshop software.
Mice sedated by brief isofluorane exposure were measured from nose to tail base to nearest 1/10 centimeter. Femurs were dissected out and distance from greater trochanter to the medial condyle was measured with digital calipers to nearest 1/100 millimeter.
Tibias and vertebrae at L3 level collected from 3- and 5-week-old double RARβ/RARγ- deficient and double floxed littermates were fixed in 4% paraformaldehyde, decalcified in 10% EDTA (pH 7.4) for three weeks at 4°C, and embedded in paraffin. Control and mutant sections mounted on same slides were stained 0.01-0.1%Safranin-O/2% fast green or 0.1% alcian blue pH 1.0. Staining was as brief as possible to avoid dye saturation. Real-time PCR was used to compare levels of transcriptional co-factors using RNA isolated from newborn tibial and femoral epiphyseal cartilage. Primer pairs are listed in Table S2 and genes tested were the following: NCoR1 (nuclear receptor co-repressor 1) (Jepsen et al., 2000); NCoR2 (also called SNRT) (Hauksdottir et al., 2002); Lcor (ligand-dependent nuclear receptor corepressor) (Fernandes et al., 2003); PRAME (preferentially expressed antigen in melanoma) (Epping et al., 2005); NSD1 (nuclear receptor binding SET domain protein 1) (Huang et al., 1998); Sorbs3 (sorbin and SH3 domain containing protein 3) (Bour et al., 2005); Msx1 (msh homeobox 1) (Weston et al., 2003); and Mta1 (metastasis-associated 1) (Mazumdar et al., 2001).
To analyze chondrocyte proliferation, serial sections were prepared from the proximal tibial growth plates of 3- and 5-week-old double RARβ/RARγ-deficient and double floxed littermates. Sections were hybridized with radiolabeled H4C riboprobes and then stained with eosin. Bright and dark field pictures were taken from a minimum of 6 slides from each genotype and age and printed. Each picture was used to manually count every cell present in the proliferative zone, defined as the tissue area above the Indian hedgehog-expressing prehypertrophic zone. The same process was repeated by determining the number of H4C-positive cells; positive signal was defined as that given by a minimum 5 hybridization grains per cell. Statistical significance was determined by two-tailed unpaired student t-test.
Microcomputed tomography (μCT) analysis
Tibia and femurs were analyzed by μCT at 45 kVp. Trabecular bone volume, connectivity, trabecular thickness and mineral density were analyzed and quantified with a Scanco μCT 40 apparatus using proprietary software.
Chondrocyte cultures
Cartilage fragments from neonatal epiphyses and 3 to 5-week old ribs were washed with Ca/Mg-free HBSS plus antibiotics and with 0.05% trypsin/0.01% EDTA. Fragments were incubated in fresh trypsin/EDTA mixture at 37°C for 20 minutes, washed and incubated with 0.1% testicular collagenase in αMEM for 3-4 h. Neonatal mouse cartilage tissue was dispersed by pipetting, and single cells were inoculated onto type I collagen-coated multiwell dishes maintained in 10% FBS in αMEM. Rib cartilage fragments from 3 and 5 weeks old mice were partially digested with the initial collagenase treatment. We found that more than 4 hours collagenase treatment not only damaged cell viability, but also decreased yield of chondrocytes. To avoid those problems, we maintained partially digested cartilage fragments for 7-10 days in 10% FBS and 1 ng/ml recombinant basic FGF (R&D) and then digested again with 0.1% collagenase in for 2-3 h. Ascorbic acid (10 μg/ml) was added to the medium after cells reached confluency. Cartilage matrix content was determined by Alcian blue staining (pH 1.0).
Transfections
Single reporter assay: 4 × 104 cells per well were seeded onto a type I collagen coated 96 wells plate the day before transfection. Cells were maintained in 100 μl 10% FBS in MEM. Medium was changed and cells were immediately transfected with 0.2 μg/well of retinoic acid response element (RARE) luciferase reporter vector (Panomics, CA) and 0.2 μl of Lipofectamine LTX (Invitrogen, CA) according to the manufacturer’s protocol. Cultures were treated with indicated doses of all-trans retinoic acid (RA) (Sigma), RARγ agonist (VTP 204647) or RAR inverse agonist (INV) (AGN/VTP 194310) for 24 h after transfection; parallel control cultures received vehicle alone. Forty-eight hours later, cells were subjected to luciferase assay.
Dual reporter assay: Dual reporter assay system was employed to study the function of individual RAR and Zac1. Cell seeding and maintenance condition were same as those of single reporter assay. For gain of function experiments, 0.1 μg each of expression vector and RARE-luc and 1 ng phRG-SV40 (Promega) were co-transfected with 0.2 μl of Lipofectamine LTX. For loss of function experiments, 0.2 pmol of siRNA (non-specific siRNA mixture D-001810-10; SMARTpool siRNA mixture targeting mouse Zac1(Plagl1), E-046206; RARα, E-047625 and RARγ, E-040974, Dharmacon, CO), 30 ng RARE-luc and 1 ng phRG-SV40 (Promega) were transfected to each well with 0.2 μl Lipofectamine 2000 reagent (cat. No. 25087w pps, Invitrogen). Some cultures were treated with various retinoid derivatives from 24 to 48h after transfection. Forty-eight hours after transfection, cells were harvested and subjected to dual luciferase assay (Dual Luciferase Reporter Assay System, E1910, Promega). Transfection efficiency was normalized by Renilla luciferase activity generated by phRG-SV40.
Protein functional interaction assays
Interactions between RARγ and Zac1 were tested with Promega Checkmate mammalian two-hybrid system. RARγ and Zac1 subcloned to pACT and pBIND vectors respectively were transfected to AD293 cells with Lipofectamine LTX reagent. Total 0.2 μg of pACT, pBIND and pG5-luc (GAL4-luc reporter vector) were transfected at a ratio of 4:4:2.
Proteoglycan analysis
Newborn epiphyseal chondrocytes seeded at 2 × 104 cells/6 mm well were maintained in 10%FBS/αMEM. When confluent, cultures were preincubated for 24 hrs in αMEM/0.3 %FBS, treated with various retinoid concentrations for 4 h and labeled with 5 μCi/ml 35S-sulfate for 20 h. Proteoglycan synthesis was determined by measuring incorporation of 35S-sulfate into materials precipitated with cetylpyridinium chloride after protease digestion (Takebayashi et al., 1995).
To test RARγ or Zac1 over-expression on proteoglycan synthesis, chondrocytes were transfected with RARγ (pCMV-HA-RARγ), Zac1 (pCMV-Zac1) or insertless expression vector using Lipofectamine LTX reverse transfection protocol in which 0.1 μg of DNA and 0.4 μl Lipofectamine LTX in 20 μl Opti-MEM medium were dispensed into each well. Freshly isolated chondrocytes (5 × 104 cells) suspended in 10% FBS/αMEM medium were seeded onto the precoated wells. Six hours later, medium was changed to retinoid-depleted medium (0.5 % charcoal-treated serum, 1x ITS mixture and 0.5 % BSA in DMEM) and cultures were treated with retinoids or left untreated. Cells were lysed 24-48 hours after transfection for the reporter gene assay or labeled with 5 μCi/ml of 35S-sulfate for 20 hrs to measure proteoglycan synthesis. The parallel cultures that received identical treatment were used to determine DNA contents (Johnson-Wint and Wellis, 1982).
Results
RAR gene expression patterns
At the outset of this project, we determined the gene expression patterns of RARα, RARβ and RARγ in mouse limb growth plates, information that, surprisingly, was still lacking in the literature. In E15.5 embryos, RARα and RARβ were mildly expressed (Fig. 1A-C), but RARγ was very strongly expressed in resting, proliferative and pre-hypertrophic zones (Fig. 1D, arrow) and was down-regulated in hypertrophic zone (Fig. 1D, arrowhead). Similar overall patterns were seen in tibial growth plates in 1 week- and 3 week-old mice (Fig. 1E-H and 1I-L, respectively), and there was also appreciable RARα expression in perichondrium (Fig. 1F) and RARγ expression in primary spongiosa (Fig. 1L, arrowheads). Clearly, the RARs are differentially expressed in mammalian growth plates and RARγ is the most abundantly expressed family member.
Fig. 1.
RAR expression patterns in long bone growth plates.
(A-L) In situ hybridization analysis of RARα, RARβ or RARγ expression using sections of wild type E15.5 (A-D), 1 week-old (E-H) and 3 week-old (I-L) proximal tibial growth plates and 3 week-old RARβ/RARγ-deficient tibial growth plates (M-P). In (D), arrow points to upper growth plate zones, while arrowhead points to hypertrophic zone. In (L,P), double arrowheads point to primary spongiosa. OC, secondary ossification center. Size bars: A-D, 200 μm; E-H, 300 μm; and I-P, 75 μm.
Conditional compound RAR-deficient mice
To determine RAR roles in growth plate, we produced double RAR-deficient mice in cartilage by mating floxed RAR mice with Col2a1-Cre mice (Ovchinnikov et al., 2000) (all with mixed 129/SvPas-C57Bl/6 background). Mutant mice were born at Mendelian ratios. To determine efficiency and specificity of RAR deletion, DNA samples from rib and limb growth plate cartilaginous fragments and liver from newborn wild type (WT) and double RAR-deficient mice were processed for genomic PCR, using primers to distinguish wild type (W), floxed (F) and null (N) alleles (Fig. S1). RAR genes had been deleted in mutant cartilage, but not liver (Fig. S1). Efficiency of RARγ ablation in double RARα/RARγ– or RARβ/RARγ -deficient cartilage was nearly complete (Fig. S1), whereas efficiency of RARα and RARβ ablation averaged over 80% (Fig. S1). Southern hybridization also showed that RAR genes were deleted in cartilage, but not heart, liver, brain, spleen and bone marrow DNA; deletion efficiency was over 95% for RARγ and about 80% for RARα and RARβ (Fig. S2). In situ hybridization on tibial sections from 3 week-old RARβ/RARγ -deficient mice (Fig. 1M-P) confirmed that RARγ gene expression was markedly reduced in growth plate (Fig. 1P), but signal was still appreciable over primary spongiosa (Fig. 1P, arrowhead).
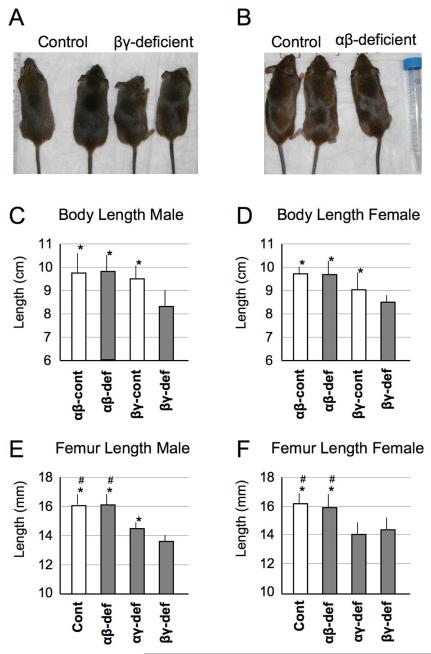
Given that mutant mice were born at Mendelian frequency and appeared normal at birth, we closely monitored them over age. Starting at about 3 weeks, we noted that RARα/RARγ– and RARβ/RARγ-deficient mice were consistently smaller than control littermates (heretofore, control littermates represent double floxed RAR mice lacking Col2a1-Cre) (Fig. 2A). By 2 months, overall growth retardation was pronounced in both mutant females and males and amounted to about 25% over controls (Fig. 2C,D). This trend was confirmed in limb skeletal elements such as femurs (Fig. 2E,F). Notably, mice deficient in RARα/RARβ genes were virtually similar to control littermates in both body size (Fig. 2B) and skeletal element length (Fig. 2C-F). In total, we examined over 115 mice from various litters (see Fig. 2 legend for details) and the above phenotypic changes in RARα/RARγ– and RARβ/RARγ-deficient mice were invariably seen. Clearly, deficiency of RARα/RARγ or RARβ/RARγ genes in cartilage causes significant skeletal growth retardation, whereas RARα/RARβ deficiency is tolerated.
Fig. 2.
Physical appearance and skeletal growth in control and RAR-deficient mice.
(A) The two RARβ/RARγ-deficient 5 week-old mice on the right (indicated as βγ-deficient) are visibly smaller than their control littermates on the left. (B) No obvious difference in body size is appreciable in 5 week-old control versus RARα/RARβ-deficient littermates. (C-F) Average body and femur lengths in male and female control and dual RAR-deficient mice at 8 weeks of age. Body length was measured from nose tip to sacral area. Numbers of mutant mice analyzed in this experiment are: RARα/RARβ-deficient (αβ-def, n=13) and control littermate (αβ-cont, n=19), RARβ/RARγ-deficient (βγ-def, n=24) and its control (βγ-cont, n=32), and RARα/RARγ-deficient (αγ-def, n=13) and its control littermate (n=14). Control in E and F are total of all three control littermates (n=65) Obtained data were subjected to the Mann-Whitney test. * p<0.05 compared to βγ-def; # p<0.05 compared to αγ-def
Growth plate organization and function
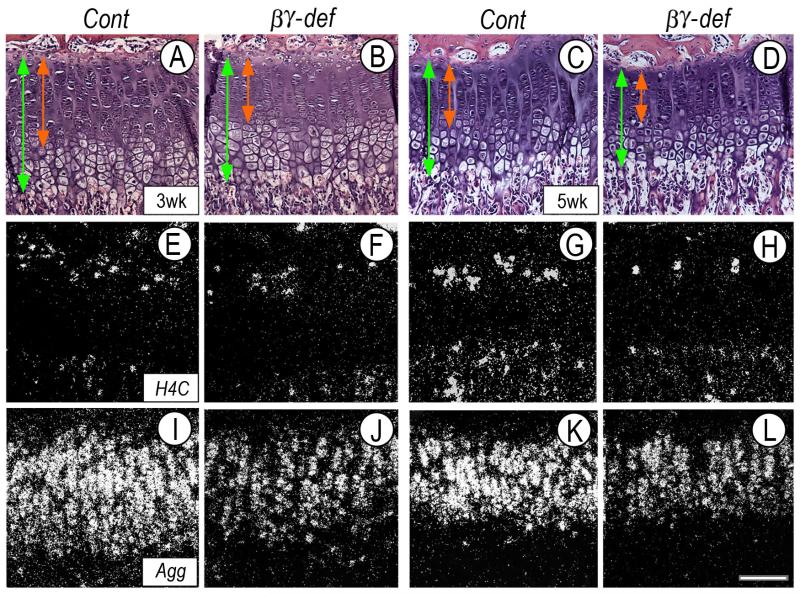
What could account for the growth retardation? Normally, growth plates sustain skeletal growth by: (a) proliferation of chondrocytes; (b) matrix synthesis and accumulation; and (c) chondrocyte hypertrophy (Hunziker, 1994; Wilsman et al., 1996). To determine whether and which of these parameters was affected, longitudinal serial sections of femoral and tibial growth plates from 3- and 5-week-old control and RARβ/RARγ-deficient littermates were processed for histological, histochemical and gene expression analyses. Interestingly, the height of mutant growth plates (Fig. 3B,D, vertical green line) was consistently shorter than that in control littermates at both ages (Fig. 3A,C, vertical green line). Shortening appeared to affect primarily the upper zones (Fig. 3B,D, vertical red line), while the hypertrophic zone seemed largely unaffected.
Fig. 3.
Analyses of growth plate organization, chondrocyte proliferation and aggrecan expression.
(A-L) Serial sections from proximal tibial growth plates of 3 week-old (A-B, E-F and I-J) or 5 week-old (C-D, G-H and K-L) control and RARβ/RARγ-deficient littermates were processed for histological analysis (A-D) and in situ hybridization for proliferation marker H4C (E-H) or aggrecan (Agg) (I-L). Note the reduction of mutant growth plate height (vertical green line) mostly affecting upper zones (red line), and more obvious in 5 week-old mice. Size bar, 75 μm.
To assess proliferation, companion sections were processed for close histological assessment and for gene expression of histone 4C (H4C), a general marker of mitotic cells. The average number of chondrocytes present in the proliferative zone in control 3- and 5-week-old tibial growth plates was 180 ± 13 and 179 ± 5, respectively. Interestingly, this number had decreased to 151 ± 15 and 150 ± 11 in RARβ/RARγ-deficient 3- and 5-week-old tibial growth plates (p<0.01). A similar trend was observed in companion sections processed for H4C expression; fewer positive chondrocytes were present in mutant proliferative zone (Fig. 3F,H, arrowheads) than control (Fig. 3E,G, arrows).
To monitor matrix synthesis and accumulation, we focused on aggrecan given its essential role in growth plate function. Aggrecan transcripts were extremely abundant in control growth plates, particularly in proliferative and pre-hypertrophic zones (Fig. 3I,K). In contrast, aggrecan expression was clearly reduced in RARβ/RARγ-deficient growth plates (Fig. 3J,L). These hybridizations were conducted with control and mutant growth plate sections mounted and processed on the same glass slides and thus, hybridization signal is directly comparable.
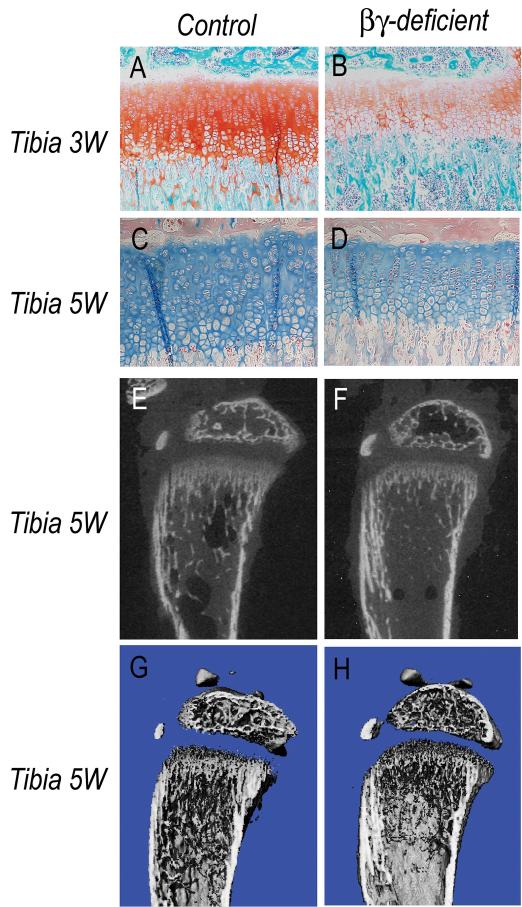
To visualize proteoglycans, growth plate sections from 3-week-old control and mutant littermates mounted on the same glass slides were stained with Safranin-O or Alcian Blue. In controls, staining was strong (Fig. 4A,C), but was clearly lower in RARβ/RARγ-deficient growth plates (Fig. 4B,D). Decreases in proteoglycan content were also observed at other sites in the same mutant animals including vertebral endplates and annulus fibrosus (Fig. S3).
Fig. 4.
Histochemical and μCT analyses of long bones.
(A-D) Longitudinal sections of proximal tibial long bone growth plates from 3 and 5 week-old control and RARβ/RARγ-deficient littermates were processed for histochemical proteoglycan staining with safranin O or alcian blue. Note the significant reduction in staining in mutant growth plates. (E-H) Long bones from 5 week-old control and RARβ/RARγ-deficient littermates were scanned by μCT and viewed as 2D pictures (E-F) or 3D cut-plane pictures (G-H). Note the significant decrease in trabecular bone in mutant samples.
Given that growth plate function is also required for endochondral ossification (Chan and Jacenko, 1998; Jacenko et al., 1993), we predicted that the above growth plate defects, and the decrease in aggrecan content in particular (Poole et al., 1982), should impair trabecular bone formation. Thus, we subjected tibias and femurs from control and RAR-deficient littermates to microcomputed tomography (μCT) to reveal mineralized tissue. Indeed, there was a clear trabecular bone reduction in RARβ/RARγ-deficient mice (Figures 4E-4H) and RARα/RARγ-deficient mice (Table S1), while bone structure and quality were essentially normal in RARα/RARβ-deficient mice (Table S1).
In summary, RARs are needed to sustain normal growth plate and chondrocyte function and bone formation and in particular, promote aggrecan expression and content.
RARs and proteoglycan expression
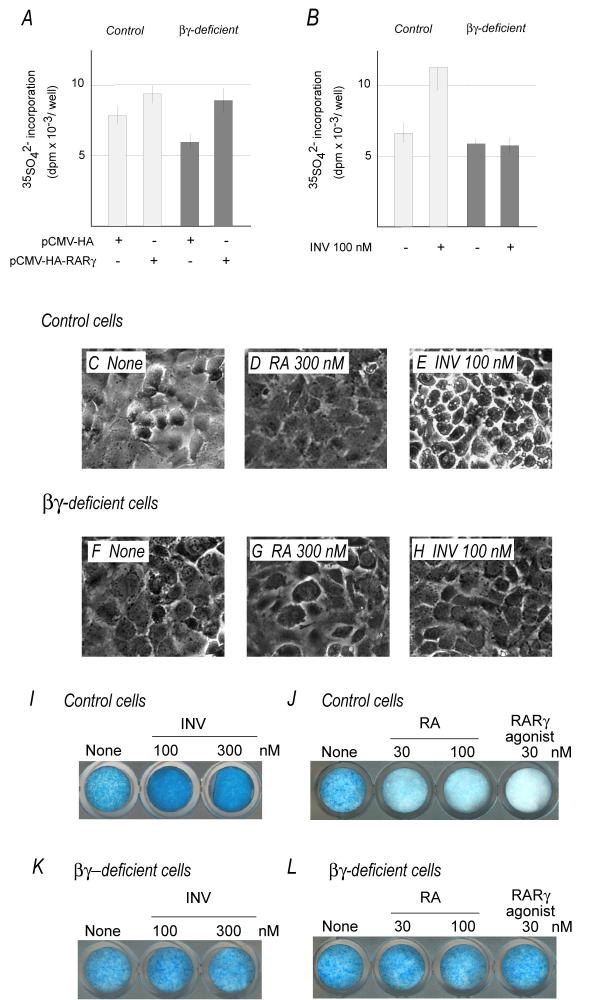
If RARs do benefit aggrecan gene expression and production, over-expression of RARs, and RARγ in particular, should increase such parameters. In addition, since the proliferative and pre-hypertrophic zones of growth plate (where RARγ and aggrecan are strongly co-expressed) are largely devoid of endogenous retinoids (von Schroeder and Heersche, 1998), RARγ should influence aggrecan expression as ligand-less factor. To test these interesting possibilities, epiphyseal chondrocytes from newborn wild type mice were transfected with a CMV-driven expression plasmid encoding full-length mouse RARγ (HA tagged) or control empty vector. Cells were maintained in medium containing 0.5% charcoal-stripped retinoid-free serum (Koyama et al., 1999) and were pulse-labeled with 35S-sulfate to measure proteoglycan production. Clearly, proteoglycan synthesis increased over 30% in RARγ transgene-expressing cultures compared to controls (Fig. 5A). Chondrocytes isolated from 3-4-week old ribs produced identical data (not shown). To confirm above data, chondrocytes from RARβ/RARγ-deficient mice were subjected to same experimentation. Over-expression of RARγ transgene did increase proteoglycan synthesis in these mutant cultures as well (Fig. 5A). Interestingly, baseline proteoglycan synthesis in untreated RARβ/RARγ-deficient cultures was consistently lower than wild-type cultures, likely reflecting lower endogenous RAR gene levels in mutant cells (Fig. 5A).
Fig. 5.
Proteoglycan synthesis and content following RARγ over-expression and inverse agonist treatment.
(A-B) Proteoglycan synthesis in control and RARβ/RARγ-deficient chondrocyte cultures over-expressing RARγ (pCMV-HA-RARγ) or following treatment with inverse agonist. (C-H) Phase microscopic appearance of control and RARβ/RARγ-deficient chondrocytes in untreated control cultures (None) and in companion cultures treated with 300 nM retinoic acid (RA) or 100 nM inverse agonist (INV). (I-L) Alcian blue staining of control and RARβ/RARγ-deficient cultures treated for 6 days with inverse agonist, RA or a RARγ-selective agonist. Consistency of results was established in three independent experiments.
When RARs are ligand-less, they function as transcriptional repressors (Koide et al., 2001; Kurosawa et al., 1995; Mannervik et al., 1999). Based on the data above, one would predict that experimental increase of RAR repressor function should lead to further increases in proteoglycan synthesis and levels. To test this intriguing idea, we resorted to the use of synthetic retinoids that are “inverse agonists”. Unlike standard agonists (that stimulate RAR transcriptional activity), inverse agonists stimulate RAR repressor function by reinforcing co-repressor binding (Germain et al., 2002; Gronemeyer et al., 2004; Klein et al., 1996; Thacher et al., 1999). Semi-confluent cultures in charcoal-treated medium were treated 100 nM inverse agonist (INV) for 2 days; companion cultures were treated with vehicle (EtOH). Incorporation of 35S-sulfate did nearly double in treated versus untreated cultures (Fig. 5B, white histograms). Companion cultures were treated with 100 or 300 nM of inverse agonist for 6 days (fresh drug added every two days) to see whether this prolonged treatment would result in higher proteoglycan content. Quite clearly, prolonged treatment markedly increased the amount of alcian blue-positive proteoglycan matrix compared to controls (Fig. 5I). Such increase was appreciable also by the phase microscopy appearance of treated chondrocytes that were surrounded by a bright and highly refractive contour (generated by pericellular proteoglycan accumulation) (Fig. 5E) compared to the more muted appearance of untreated chondrocytes (Fig. 5C).
If these increases in proteoglycan synthesis and accumulation reflect inverse agonist-dependent stimulation of repressor/co-repressor function, one would predict that (a) the opposite should be seen in cultures treated with standard retinoid agonists that induce ligand-dependent RAR transcriptional activity; and (b) no effects should be seen in RAR-deficient cultures. In line with these predictions, treatment of control chondrocyte cultures with all-trans-retinoic acid (RA) or a RARγ selective agonist: (i) drastically reduced proteoglycan accumulation (Fig. 5J); (ii) rendered the cells quite dull in appearance (Fig. 5D); and (iii) markedly decreased proteoglycan synthesis (not shown). In contrast, no appreciable changes in these parameters were seen in RARβ/RARγ-deficient cultures that failed to respond to RA or RARγ selective agonist (Fig. 5G,L) and inverse agonist (Fig. 5B,K).
Taken together, the above data strengthen the idea that RARs, and RARγ in particular, are beneficial for proteoglycan expression in chondrocytes when operating in a retinoid-free condition and thus possibly as ligand-less repressors.
Searching for RAR co-repressors
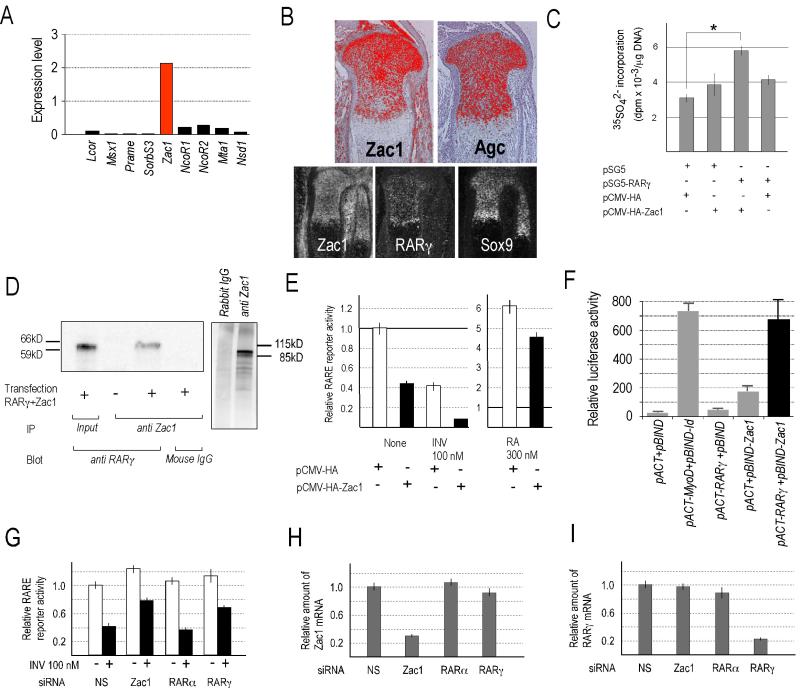
Which factors could possibly serve as RAR co-repressors in cartilage? In other systems the most widely studied co-repressors are NCoR-1 or NCoR-2/SMRT (Hermanson et al., 2002; Jepsen et al., 2007), but their possible function in growth plate and skeletal growth is largely unknown. In addition, studies showed that ligand-less RARα does recruit NCoR-1 and NCoR-2 efficiently, but RARβ and RARγ do not (Wong and Privalsky, 1998). Given that RARγ is the most abundantly expressed RAR in growth plate, we searched databases to identify potential RAR co-repressor candidates using the following criteria: (1) candidate should associate with nuclear hormone receptors and have repressor function; and (2) candidate should be expressed in skeletal tissue. This in silico search led us to Zac1 (zinc finger protein which regulates apoptosis and cell cycle arrest) (Spengler et al., 1997). Zac1 can act as transcriptional co-repressor or co-activator of nuclear hormone receptors (Huang and Stallcup, 2000) and interestingly, mice lacking Zac1 exhibit growth retardation and altered bone formation (Varrault et al., 2006). We found that Zac1 is very strongly expressed in mouse growth plate cartilage; NCoR1 and NCoR-2 are expressed also, but at more than 5-fold lower levels (Fig. 6A). In situ hybridization on mouse growth plates confirmed that Zac1 is strongly expressed and, strikingly, its expression pattern overlaps with that of RARγ, Sox9 and aggrecan (Fig. 6B). Indeed, proteoglycan synthesis in retinoid-free wild type chondrocyte cultures was stimulated by RARγ over-expression and was nearly doubled by over-expression of both RARγ and Zac1 (Fig. 6C). To see that Zac1 actually interacts with RARγ, we over-expressed RARγ and Zac1 in AD293 cell line and processed whole cell extracts for Zac1 immunoprecipitation followed by western blot for RARγ; clearly, Zac1 associates with RARγ (Fig. 6D). To test whether Zac1 influences RAR function at the RARE level, we co-transfected wild type chondrocyte cultures with an RARE reporter construct and a full-length mouse Zac1 expression vector; parallel cultures received only the RARE reporter construct. Clearly, basal RARE reporter activity was decreased by Zac1 over-expression and was further decreased by inverse agonist AGN co-treatment compared to control cells (Fig. 6E). Basal RARE reporter activity, however, was increased when endogenous Zac1 or RARγ expression was inhibited by siRNA (Fig. 6G). Inhibition of RARα expression had no effect, attesting importance of RARγ (Fig. 6G); specificity of siRNAs is demonstrated in Fig. 6H-6I.
Fig. 6.
Analyses of Zac1 expression patterns, interactions and function.
(A): Real-time PCR analysis of transcriptional co-factors in newborn tibial and femoral epiphyseal cartilage. Expression levels are shown relative to GAPDH. (B): Expression patterns of Zac1, aggrecan (Agc), RARγ and Sox9 in longitudinal sections of E15.5 mouse embryo tibias viewed at high (upper panels) and lower magnification (lower panels). (C): Proteoglycan synthesis in primary chondrocyte cultures following over-expression of Zac1 and/or RARγ. Cultures maintained in retinoid-free medium were pulse-labeled with 35-sulfate from 24 to 36 hrs after transfection. Graphs show average incorporation in triplicate cultures +/− SD. * p<0.05 by Student’s t-test comparing control empty vector transfected cultures vs Zac1 and RARγ transfected cultures. (D): Immunoprecipitation and immunoblot analysis of physical interaction between RARγ and Zac1 (left panel). Specificity of Zac1 antibodies was confirmed by immunoprecipitation of whole cell lysates compared to preimmune rabbit IgGs (right panel). (E): RARE reporter activity in primary cultured chondrocytes following Zac1 over-expression with or without treatment with inverse agonist AGN (left panel). Similar analyses with or without treatment with retinoic acid (RA) (right panel). (F): Mammalian two hybrid assays showing that RARγ and Zac1 interact to elicit very strong reporter activity (solid black column). pACT-MyoD and pBIND-Id were used as positive controls, and insert-less pACT and pBIND were negative controls. (G) RARE reporter activity in primary cultured chondrocytes following transfection of smart pool siRNA specific mixtures for Zac1, RARα or RARγ without (-) or with (+) co-treatment with 100 nM inverse agonist; non-specific (NS) was used in controls. (H-I) RT-PCR analysis of endogenous RNA levels for indicated genes following transfection of siRNAs for Zac1 (H) or RARγ (I); note that only the targeted RNA was decreased in each case. Transfection efficiency of all reporter assays was normalized by Renilla luciferase activity generated by co-transfected phRG-SV40 vector. Two independent siRNA transfection experiments and three independent experiments with all the other transfections were carried out and produced similar results.
Regulation of proteoglycan expression
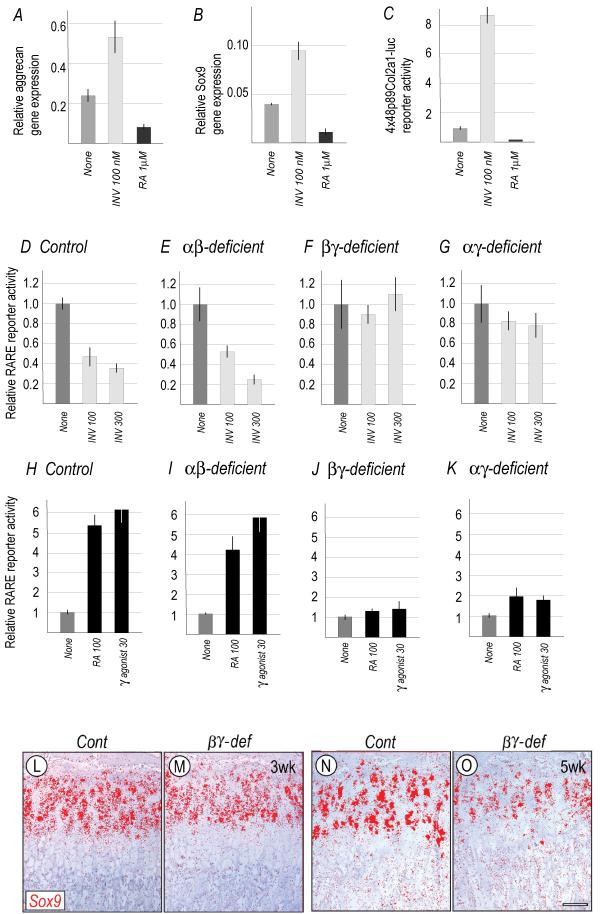
How could the apparent repressor function of RARs favor aggrecan gene expression? A reasonable possibility is that ligand-less RARs stimulate expression of Sox 9 which is a critical regulator of chondrogenesis (Bi et al., 1999) and is required for aggrecan expression (Bridgewater et al., 2003; Lefebvre and Smits, 2005). To assess this possibility, primary semi-confluent chondrocyte cultures were treated with 100 nM of inverse agonist (INV) for 2 days; for comparison, companion cultures were treated with a standard agonist (RA). Whole RNAs were processed for RT-PCR. Inverse agonist treatment did cause a concurrent stimulation of aggrecan and Sox9 expression (Fig. 7A,B), while RA inhibited both (Fig. 7A,B). The same trends were observed in cultures transfected with a Sox reporter construct that contains 4 × 48 bp Sox-binding Col2a1 enhancer (Zhou et al., 1998) (Fig. 7C). To verify that these pharmaceutical agents modulated RAR function at the genomic level and not via other pathways (Aggarwal et al., 2006; Alsayed et al., 2001), control and RAR-deficient chondrocyte cultures in standard medium were transfected with an RARE reporter plasmid and treated with inverse agonist, RA or RARγ-selective agonist. In control cultures, the inverse agonist markedly decreased baseline reporter activity (Fig. 7D) as seen above (see Fig. 6C), while RA or RARγ-selective agonist markedly stimulated it (Fig. 7H). The same results were obtained with RARα/RARβ-deficient cultures (Fig. 7E,I). However, no appreciable responses were seen in cultures deficient in RARβ/RARγ (Fig. 7F,J) or RARα/RARγ (Fig. 7G,K).
Fig. 7.
Analyses of aggrecan and Sox9 expression levels and patterns.
(A-C) Aggrecan and Sox9 expression levels (A-B) and 4x48bp Sox reporter activity (C) in primary chondrocyte cultures treated with 100 nM inverse agonist (INV) or 1 μM RA compared to untreated cultures (None). (D-K) RARE reporter activities in control, RARα/RARβ-, RARβ/RARγ- and RARα/RARγ-deficient chondrocyte cultures before (None) or after treatment with inverse agonist, RA or RARγ-selective agonist. All reporter assays were repeated 3-5 times and we obtained reproducible results. (L-O) In situ hybridization analysis of Sox9 expression in 3 week-old (L-M) and 5 week-old (N-O) proximal tibial growth plates from control (L and N) and RARβ/RARγ-deficient (M and O) littermates.
If the RARs regulate aggrecan expression via Sox9 as the above data imply, one would expect to see a decrease in Sox9 expression accompanying the decrease in aggrecan expression in RARβ/RARγ-deficient growth plates. In line with this prediction, there was a consistent and significant decrease in Sox9 transcripts in 3 and 5 week-old mutant versus control growth plates (Fig. 7L-O).
Discussion
The study provides fundamentally new insights into RAR roles in skeletal growth. We find that the RARs are differentially expressed in different zones of prenatal and postnatal mouse long bone growth plates, with RARγ being the most abundantly expressed member and its expression prominently characterizing the upper zones. Concurrent deficiency in RARα/RARγ or RARβ/RARγ in cartilage causes concurrent defects in growth plates, most notably a reduction in their overall height and a decrease in chondrocyte proliferation and aggrecan expression and content. These growth plate defects are associated with disturbances in primary spongiosa and trabecular bone that are substandard compared to those seen in control littermates. Since chondrocyte proliferation and matrix deposition are amongst the most critical factors in dictating growth rates (Hunziker, 1994; Wilsman et al., 1996), it is very likely that deficiencies in those two parameters directly led to skeletal growth retardation in mutant mice. It is clear then that the RARs have significant, but previously unappreciated roles in growth plate chondrocyte function and exert local influence on skeletal growth. The data correlate quite well and greatly extend previous findings obtained with general dual RARα/RARγ– or RARβ/RARγ-null mouse mice (Mendelsohn et al., 1994). The additional skeletal deformities seen in those mice and their neonatal lethality likely reflect broader systemic deficiencies due to general dual RAR deletion. On the other hand, the relatively mild defects in single RAR-null mutants reiterate the idea that other family members can compensate for lack of one RAR gene.
Postnatal growth retardation
Our data show that the skeletal growth retardation affecting RARα/RARγ- and RARβ/RARγ-deficient mice became apparent around 3 weeks of postnatal age, continued in adulthood and was not compen sated over further age. The apparent lack of growth retardation at younger age correlates well with our observation that the mutant mice were born at expected Mendelian frequencies and may thus have been largely refractory to RAR deficiency during prenatal developmental stages. It has long been appreciated that prenatal skeletal growth is largely controlled by local autocrine and paracrine factors including Indian hedgehog, whereas systemic factors become important postnatally (Fowden, 1995; Kronenberg, 2003; Nilsson et al., 2005). For example, growth hormone has no apparent role in mouse embryonic skeletal growth despite the fact that its receptor is expressed, but GH-deficiency causes postnatal growth retardation starting around 2 weeks of age. Related studies showed that concurrent deficiency in GH and insulin-like growth factor 1 (or liver IGF-1 and acid labile subunit) cause severe mouse skeletal growth retardation starting around 3 weeks postnatally (Lupu et al., 2001; Yakar et al., 2002). A similar trend was observed in transgenic mice expressing of the Achondroplasia mutant form of human FGF-R3; the mice were apparently normal during prenatal and neonatal stages, but became growth retarded starting around 4 weeks of age, with concurrent decrease in growth plate chondrocyte proliferation (Naski et al., 1998). Clearly, skeletal growth is a complex process that obeys a multitude of common and distinct pathways and mechanisms during prenatal and postnatal life. Our data show that RAR deficiency is well tolerated during prenatal and neonatal growth, but cannot be compensated and becomes deleterious to growth starting around 3 weeks of age by a combination of decreases in aggrecan production and deposition and chondrocyte proliferation. Whether the RAR deficiency affected the chondrocytes’ ability to respond to, and be influenced by, systemic factors remains to be established.
Retinoids and repressor function
Cartilage is unique among tissues for its avascular nature and growth plate cartilage is no exception, particularly in its upper zones. In fact, the growth plate is largely hypoxic and is quite sensitive to ablation of genes needed to cope with such an unusual metabolic status (Schipani et al., 2001). Given the absence of blood vessels, it is likely that the upper growth plate zones cannot receive liver-derived, blood-bourn and protein-bound retinoid precursors needed for intracellular production of biologically-active retinoids (Blaner and Olson, 1994) and would thus be largely devoid of such retinoids. Indeed, von Schroeder and Heersche showed with a widely-used RARE reporter mouse line (Rossant et al., 1991) that the upper zones of mouse limb growth plates are virtually devoid of reporter activity and hence active retinoids, while reporter activity is detectable in the lower hypertrophic zone and other retinoid-rich organs (von Schroeder and Heersche, 1998). We obtained similar evidence using a bioassay (Koyama et al., 1999) and have now confirmed it by direct measurements using LC/MS/MS procedures (Kane et al., 2005) (manuscript in preparation). Together, these studies suggest that RARs expressed in upper growth plate zones may be devoid of active ligands and could primarily, if not exclusively, exert ligand-less repressor function. Growth plate defects seen in our RARα/RARγ- or RARβ/RARγ-deficient mice would thus result from absence of such repressor function.
Nuclear receptor-mediated transcriptional repression is as important as transcriptional activation in regulating developmental processes (Kurosawa et al., 1995; Mannervik et al., 1999). For example, NCoR- or HDAC1-null mice die during gestation (Jepsen et al., 2000; Lagger et al., 2002), and ablation of HDAC4 (normally expressed by pre-hypertrophic chondrocytes) deranges growth plate chondrocyte maturation and causes premature hypertrophy (Vega et al., 2004). It is conceivable then that ligand-less RARs expressed in upper growth plate zones, and RARγ in particular, could cooperate with HDACs such as HDAC4 to repress target genes, maintain chondrocyte function in those zones including aggrecan expression, and allow growth plate and skeletal growth to proceed at normal rates. Interestingly, our data indicate that Sox9 gene expression is lower in RARβ/RARγ-deficient growth plates and is stimulated by inverse agonist treatment in cultured chondrocytes (along with a stimulation in proteoglycan synthesis). A recent relevant report is showing that ligand-bound RARs or TNFα suppress cartilage-matrix gene expression by limiting availability of p300, an essential co-factor for Sox9 activity (Rockel et al., 2008). The findings suggest that p300 available for activation of Sox9 increases when RARs are ligand-less. Thus, a normal in vivo role of RAR repressor function in growth plate could be to favor Sox 9 expression and function together with p300 and promotes the chondrocyte phenotype. A similar RAR repressor function, largely mediated by RARα, operates during limb mesenchymal chondrogenic cell differentiation and is required for initial up-regulation of Sox 9 gene expression (Weston et al., 2002). These authors suggested that RARα-dependent repressor function would directly block expression of “gene X”, a negative regulator of Sox9 gene expression. Though the nature of gene X in chondrogenesis remains unknown, progress has been made in other systems. For instance, Jepsen et al. reported recently that NCoR-2/SMRT and ligand-less RARs cooperate to directly repress expression of histone demethylase JMJD3 and in so doing, allow normal spatio-temporal progression of differentiation of neural stem cells into neurons (Jepsen et al., 2007).
Zac1 as a potential co-repressor
While the nature of putative direct targets of RAR repression in growth plate cartilage also awaits further work, our current data have identified Zac1 as a possible RAR co-factor. Our data not only show that Zac1 expression pattern is remarkably similar to that of RARγ in growth plate, but also that: Zac1 functionally associates with RARγ; Zac1 over-expression further boosts proteoglycan production in retinoid-free chondrocyte cultures; and Zac1 and RARγ co-operate to reduce transcription rates as revealed by RARE reporter assays. As noted above, Zac1 is an essential factors for many developmental processes (Spengler et al., 1997), and Zac1-null mice die at birth and exhibit significant defects including growth retardation and altered bone formation (Varrault et al., 2006). These authors performed a detailed in silico network gene analysis, carried out direct promoter and gene expression tests, and concluded that Zac1 is upstream of a number of systemic genes, including Igf2/H19, Cdkn1c and Dlk1, that would together control embryonic growth. Since the growth plates of the Zac1-null mice were not studied, the authors did not clarify what consequences the general ablation of Zac1 had for local chondrocyte and/or osteoblast function. Based on our data, we may surmise that absence of Zac1 could have diminished RAR repressor function in the growth plate, should have affected the same phenotypic traits deranged by conditional RAR ablation such as aggrecan expression, and may even have had additional systemically-driven consequences due to substandard expression of upstream genes.
Our two hybrid tests suggest that Zac1 and RARγ exhibit functional interaction, and this may possibly reflect physical interaction. Just as the RARs (Chambon, 1996), Zac1 is distinguished into several functional domains: a N-terminal zinc finger region; a central proline–rich region; a small and highly-conserved motif rich in hydrophobic amino acids (termed ΦXXΦΦ motif or NR box); and a C-terminal P/E-rich domain (Spengler et al., 1997). Using pull-down assays, Huang et al. showed that Zac1 interacts with the AF2 domain of androgen, estrogen and thyroid hormone receptors (Huang and Stallcup, 2000), and it is possible that Zac1 could interact with the same domain in RARs. Interestingly, our in silico analysis has not yet identified a consensus sequence in Zac1 for interaction with HDACs (unpublished data). Thus, possible Zac1/RAR complexes may recruit HDACs indirectly by association with other HDAC-binding factors or may use alternative strategies, including SUMOylation as shown for NCoR1 (Tiefenbach et al., 2006).
Derangement of growth plate function
The concurrent decreases in aggrecan expression, chondrocyte proliferation and trabecular bone deposition point to multiple important contributions of RARs in proper growth plate functioning. As our in vivo and in vitro data suggest, it is plausible that decreased RAR function may have led to decreased Sox 9 gene expression which would in turn cause decreased aggrecan expression and substandard matrix deposition. A very interesting and far-reaching implication of these findings and conclusions is that experimental stimulation of RAR repressor function in cartilage by genetic means or pharmacological agents (such as the one used in our study) could actually have therapeutic value and could be used to stabilize or improve the phenotype and matrix production of chondrocytes including articular chondrocytes.
What could account for the other growth plate changes in RAR-deficient mice? Chondrocyte proliferation is obviously essential for skeletal development and growth and counterbalances the loss of hypertrophic chondrocytes occurring at the chondro-osseous border. Chondrocyte proliferation is controlled by systemic factors (Nilsson et al., 2005) and local mechanisms such as those regulated by Indian hedgehog (Ihh) (Shimo et al., 2004; St-Jacques et al., 1999). Interestingly, treatment of growth plate chondrocyte cultures with retinoic acid (that induces RAR transcriptional activity) was found to strongly suppress Ihh expression (Wu et al., 2002). Thus, it is possible that under normal circumstances ligand-less RAR repressor function in upper growth plate zones may help to maintain physiologic Ihh expression levels.
Similarly, it remains to be clarified what caused the presence of substandard trabecular bone in the RARα/RARγ- or RARβ/RARγ-deficient mice. As pointed out above, it has long been appreciated that normal growth plate function and normal matrix composition are required for trabecular bone deposition. A first clue into such previously unsuspected paradigm came from the work of Poole and coworkers who originally showed that aggrecan persists in the calcified cartilage core of primary spongiosa trabeculae and is later removed by proteolytic action once the secondary spongiosa is laid down (Poole et al., 1982). A first genetic clue was provided by Jacenko and coworkers who showed that mice with altered growth plate collagen X function develop a skeleto-hematopoietic disease phenotype that includes a compressed growth plate, reduced primary spongiosa, and marrow hypoplasia (Gress and Jacenko, 2000). In an intriguing twist, it was realized recently that general deletion of RARγ, but not RARα, in mice actually causes bone marrow defects characterized by hematopoietic stem cell alterations and even a marked reduction in trabecular bone during aging (Purton et al., 2006; Walkley et al., 2007). It is pretty clear then that there are essential and intimate cross-talk interactions between growth plates and bone marrow required for physiologic progression of skeletal growth and development of hematopoietic and osteoprogenitor cells that can be deranged by malfunctioning of either growth plate or bone marrow.
Supplementary Material
Acknowledgements
We express our gratitude to: Allergan and Vitae Pharmaceuticals for providing the synthetic retinoids used in the study; Dr. Y. Yamada for providing Col2a1-Cre mice following approval by Dr. R. Behringer; and Dr. S. Murakami for providing a Sox9 reporter construct.
References
- Aggarwal S, Kim S-W, Cheon K, Tabassam FH, Yoon J-H, Koo JS. Nonclassical action of retinoic acid on the activation of the cAMP response element-binding protein in normal human bronchial epithelial cells. Mol. Biol. Cell. 2006;17 doi: 10.1091/mbc.E05-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsayed Y, Uddin S, Mahmud N, Lekmine F, Kalvakolanu DV, Minucci S, bokoch G, Platanias LC. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J. Biol. Chem. 2001;4012;276:4019. doi: 10.1074/jbc.M007431200. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is r equired for cartilage formation. Nature Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Blaner WS, Olson JA. Retinol and retinoic acid metabolism. In: Sporn MB, Roberts AB, Goodman DS, editors. The retinoids: Biology, Chemistry, and Medicine. Raven Press; New York, NY: 1994. pp. 229–255. [Google Scholar]
- Bour G, Plassat JL, Bauer F, Lalevee S, Rochette-Egly C. Vinexin beta interacts with the non-phosphorylaed AF-1 domain of retinoic receptor gamma (RARgamma) and represses RARgamma-mediated transcription. J. Biol. Chem. 2005;280:17027–17037. doi: 10.1074/jbc.M501344200. [DOI] [PubMed] [Google Scholar]
- Bridgewater LC, Walker MD, Miller GC, Ellison TA, Holsinger LD, Potter JL, Jackson TL, Chen RK, Winkel VL, Zhang Z, McKinney S, de Crombrugghe B. Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic Acids Res. 2003;31:1541–1553. doi: 10.1093/nar/gkg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- Chan D, Jacenko O. Phenotypic and biochemical consequences of collagen X mutations in mice and humans. Matrix Biol. 1998;17:169–184. doi: 10.1016/s0945-053x(98)90056-7. [DOI] [PubMed] [Google Scholar]
- Chapellier B, Mark M, Bastien J, Dierich A, LeMeur M, Chambon P, Ghyselinck NB. A conditional floxed (loxP-flanked) allele for the retinoic acid receptor beta (RARbeta) gene. Genesis. 2002a;32:91–94. doi: 10.1002/gene.10073. [DOI] [PubMed] [Google Scholar]
- Chapellier B, Mark M, Garnier JM, Dierich A, Chambon P, Ghyselinck NB. A conditional floxed (loxP-flanked) allele for the retinoic acid receptor gamma (RARgamma) gene. Genesis. 2002b;32:95–98. doi: 10.1002/gene.10072. [DOI] [PubMed] [Google Scholar]
- Chapellier B, Mark M, Garnier JM, LeMeur M, Chambon P, Ghyselinck NB. A conditional floxed (loxP-flanked) allele for the retinoic acid receptor alpha (RARalpha) gene. Genesis. 2002c;32:87–90. doi: 10.1002/gene.10071. [DOI] [PubMed] [Google Scholar]
- Chen JD, Umesono K, Evans RM. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc. Natl. Acad. Sci. USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epping MT, Wang LC, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. Lidand-dependent nuclear receptor corepressor LCoR functions by hystone deacetylase-dependent and -independent mechanisms. Mol. Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Fowden AL. Endocrine regulation of fetal growth. Reprod. Fertil. Dev. 1995;7:351–363. doi: 10.1071/rd9950351. [DOI] [PubMed] [Google Scholar]
- Germain P, Iyer J, Zechel C, Gronemeyer H. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature. 2002;415:187–192. doi: 10.1038/415187a. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Gress CJ, Jacenko O. Growth plate compressions and altered hematopoiesis in collagen X null mice. J. Cell Biol. 2000;149:983–993. doi: 10.1083/jcb.149.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson J-A, Laudet V. Principles of modulation of the nuclear receptor superfamily. Nature Rev. Drug Disc. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- Hauksdottir H, Farboud B, Privalsky ML. Retinoic acid receptors beta and gamma do not repress, but instead activate target gene expression in both the absence and presence of hormone ligands. Mol. Endocrinology. 2002;17:373–385. doi: 10.1210/me.2002-0340. [DOI] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 2002;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristic sof both corepressors and coactivators. EMBO J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-M, Stallcup MR. Mouse Zac1, a transcriptional coactivator and represor of nuclear receptors. Mol. Cell. Biol. 2000;20:1855–1867. doi: 10.1128/mcb.20.5.1855-1867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker EB. Mechanism of longitudinal bone growth and its regulation by growth plate chondrocytes. Microsc. Res. Tech. 1994;28:505–519. doi: 10.1002/jemt.1070280606. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Enomoto-Iwamoto M, Kurisu K. Actions of hedgehog proteins on skeletal cells. Crit. Rev. Oral Biol. Med. 1999;10:477–486. doi: 10.1177/10454411990100040401. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Higuchi Y, Koyama E, Enomoto-Iwamoto M, Yeh H, Abrams WR, Rosenbloom J, Pacifici M. Transcription factor ERG variants and functional diversification of chondrocytes during long bone development. J. Cell Biol. 2000;150:27–39. doi: 10.1083/jcb.150.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacenko O, Lu Valle P, Olsen BR. Spondylometaphyseal dysplasia in mice carrying a dominant negative mutation in a matrix protein specific for cartilage-to-bone transition. Nature. 1993;365:56–61. doi: 10.1038/365056a0. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurosawa R, Kumar V, Liu F, Seto E, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim H-J, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Johnson-Wint B, Wellis S. A rapid in situ deoxyribonucleic acid assay for determining cell number in culture and tissue. Anal. Biochem. 1982;122:338–344. doi: 10.1016/0003-2697(82)90292-5. [DOI] [PubMed] [Google Scholar]
- Kane MA, Chen N, Sparks S, Napoli JL. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem. J. 2005;388:363–369. doi: 10.1042/BJ20041867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ES, Pino ME, Johnson AT, Davies PJA, Napgal S, Scott SM, Krasinski G, Chandraratna RAS. Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J. Biol. Chem. 1996;271:22692–22696. doi: 10.1074/jbc.271.37.22692. [DOI] [PubMed] [Google Scholar]
- Koide T, Downes M, Chandraratna RAS, Blumberg B, Umesono K. Active repression of RAR signaling is required for head formation. Genes Dev. 2001;15:2111–2121. doi: 10.1101/gad.908801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama E, Golden EB, Kirsch T, Adams SL, Chandraratna RAS, Michaille J-J, Pacifici M. Retinoid signaling is required for chondrocyte maturation and endochondral bone formation during limb skeletogenesis. Dev. Biol. 1999;208:375–391. doi: 10.1006/dbio.1999.9207. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Kurosawa R, Soderstrom M, Horlein AJ, Halachmi S, Brown MA, Rosenfeld MG, Glass CK. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Research, Pt.C. 2005;75:200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P, Gansmuller A, Chambon P. Function of the retinoic acid receptors (RARs) during development. I. Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- Lupu F, Terwilliger JD, Lee K, Segre GV, Efstratiadis A. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 2001;229:141–162. doi: 10.1006/dbio.2000.9975. [DOI] [PubMed] [Google Scholar]
- Mannervik M, Nibu Y, Zhang H, Levine M. Transcriptional coregulators in development. Science. 1999;284:606–609. doi: 10.1126/science.284.5414.606. [DOI] [PubMed] [Google Scholar]
- Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, Vadlamudi RK, Kumar R. Transcription repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat. Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development. (II) Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Ruberte E, Chambon P. Retinoid receptors in vertebrate limb development. Dev. Biol. 1992;152:50–61. doi: 10.1016/0012-1606(92)90155-a. [DOI] [PubMed] [Google Scholar]
- Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- Nilsson O, Marino R, De Luca F, Phillip M, Baron J. Endocrine regulation of the growth plate. Horm. Res. 2005;64:157–165. doi: 10.1159/000088791. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- Poole AR, Pidoux I, Rosenberg L. Role of proteoglycans in endochondral ossification: immunofluorescent localization of link protein and proteoglycan monomer in bovine fetal epiphyseal growth plate. J. Cell Biol. 1982;92:249–260. doi: 10.1083/jcb.92.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton LE, Dworkin S, Olsen GH, Walkley CR, Fabb SA, Collins SJ, Chambon P. RAR(gamma) is critical for maintaining a balance between hematopietic stem cell self-renewal and differentiation. J. Exp. Med. 2006;203:1283–1293. doi: 10.1084/jem.20052105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel JS, Kudirka JC, Guzi AJ, Bernier SM. Regulation of Sox9 activity by crosstalk with nuclear factor-kB and retinoic acid receptors. Arthr. Res. Ther. 2008;10:R3. doi: 10.1186/ar2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Zirngibi R, Cado D, Shago M, Giguere V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight MC, Johnson RS. Hypoxia in cartilage: HIF-1α is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15:2865–2876. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz NB, Domowicz M. Chondrodysplasias due to proteoglycan defects. Glycobiology. 2002;12:57R–68R. doi: 10.1093/glycob/12.4.57r. [DOI] [PubMed] [Google Scholar]
- Shapses SA, Sandell LJ, Ratcliffe A. Differential rates of aggrecan synthesis and breakdown in different zones of the bovine growth plate. Matrix Biol. 1994;14:77–86. doi: 10.1016/0945-053x(94)90031-0. [DOI] [PubMed] [Google Scholar]
- Shimo T, Gentili C, Iwamoto M, Wu C, Koyama E, Pacifici M. Indian hedgehog and syndecan-3 coregulate chondrocyte proliferation and function during chick limb skeletogenesis. Dev. Dyn. 2004;229:607–617. doi: 10.1002/dvdy.20009. [DOI] [PubMed] [Google Scholar]
- Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, Journot L. Regulation of apoptosis and cell cycle arrest byZac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814–2725. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2076–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucov HM, Evans RM. Retinoic acid and retinoic acid receptors in development. Mol. Neurobiol. 1995;10:169–184. doi: 10.1007/BF02740674. [DOI] [PubMed] [Google Scholar]
- Takebayashi T, Iwamoto M, Jikko A, Matsumura T, Enomoto-Iwamoto M, Myoukai F, Koyama E, Yamaai T, Matsumoto K, Nakamura T, Kurisu K, Noji S. Hepatocyte growth factor/scatter factor modulates cell motility, proliferation and proteoglycan synthesis of chondrocytes. J. Cell Biol. 1995;129:1411–1419. doi: 10.1083/jcb.129.5.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamura Y, Otani T, Kanatani N, Koyama E, Kitagaki J, Komori T, Yamada Y, Costantini F, Wasisaka S, Pacifici M, Iwamoto M, Enomoto-Iwamoto M. Developmental regulation of Wnt/β-catenin signals is required for growth plate assembly, cartilage integrity, and endochondral ossification. J. Biol. Chem. 2005;280:19185–19195. doi: 10.1074/jbc.M414275200. [DOI] [PubMed] [Google Scholar]
- Thacher SM, Nagpal S, Klein ES, Arefieg T, Krasinski G, DiSepio D, Agarwal C, Johnson AT, Eckert RL, Chandraratna RAS. Cell type and gene-specific activity of the retinoid inverse agonist AGN 193109: divergent effects from agonist at retinoic acid receptor γ in human keratinocytes. Cell Growth Diff. 1999;10:255–262. [PubMed] [Google Scholar]
- Tiefenbach J, Novac N, Ducasse M, Eck M, Melchior F, Heinzel T. SUMOylation of the corepressor N-CoR modulates its capacity to repress transcription. Mol. Cell. Biol. 2006;17:1643–1651. doi: 10.1091/mbc.E05-07-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ul Haque MF, King LM, Krakow D, Cantor RM, Rusiniak ME, Swank RT, Superti-Furga A, Haque S, Abbas H, Ahmad W, Ahmad M, Cohn DH. Mutations in orthologous genes in human spondyloepimetaphyseal dysplasia and the brachymorphic mouse. Nature Genet. 1998;20:157–162. doi: 10.1038/2458. [DOI] [PubMed] [Google Scholar]
- van der Eerden BC, Karperien M, Wit JM. Systemic and local regulation of the growth plate. Endocr. Rev. 2003;24:782–801. doi: 10.1210/er.2002-0033. [DOI] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, Pavlidis P, Journot L. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev. Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzi C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- von Schroeder HP, Heersche JNM. Retinoic acid responsiveness of cells and tissues in developing fetal limbs evaluated in a RAREhsplacZ transgenic mouse model. J. Orthop. Res. 1998;16:355–364. doi: 10.1002/jor.1100160312. [DOI] [PubMed] [Google Scholar]
- Walkley CR, Olsen GH, Dworkin S, Fabb SA, Swann J, McArthur GA, Westmoreland SV, Chambon P, Scadden DT, Purton LE. A Microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor γ deficiency. Cell. 2007;129:1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Middleton F, Horton JA, Reichel L, Farnum CE, Damron TA. Microarray analysis of proliferative and hypertrophic growth plate zones identifies differential markers and signal pathways. Bone. 2004;35:1273–1293. doi: 10.1016/j.bone.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Kimata K, Line S, Strong D, Gao L, Kozak CA, Yamada Y. Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nature Genet. 1994;7:154–158. doi: 10.1038/ng0694-154. [DOI] [PubMed] [Google Scholar]
- Weston AD, Blumberg B, Underhill TM. Active repression by unligated retinoid receptors in development: less is sometimes more. J. Cell Biol. 2003;161:223–228. doi: 10.1083/jcb.200211117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston AD, Chandraratna RAS, Torchia J, Underhill TM. Requirement for RAR-mediated gene repression in skeletal progenitor differentiation. J. Cell Biol. 2002;158:39–51. doi: 10.1083/jcb.200112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsman NJ, Farnum CE, Green EM, Lieferman EM, Clayton MK. Cell cycle analysis of proliferative zone chondrocytes in growth plates elongating at different rates. J. Orthop. Res. 1996;14:562–572. doi: 10.1002/jor.1100140410. [DOI] [PubMed] [Google Scholar]
- Wong C-W, Privalsky ML. Transcriptional silencing is defined by isoform- and heterodimer-specific interactions between nuclear hormone receptors and corepressors. Mol. Cell. Biol. 1998;18:5724–5733. doi: 10.1128/mcb.18.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LN, Lu M, Genge BR, Guo GY, Nie D, Wuthier RE. Doscovery of sonic hedgehog expression in postnatal growth plate chondrocytes: differential regulation of sonic and indian hedgehog by retinoic acid. J. Cell. Biochem. 2002;87:173–187. doi: 10.1002/jcb.10285. [DOI] [PubMed] [Google Scholar]
- Yakar R, Rosen CJ, Beamer WG, Ackert-Bicknell CL, Wu Y, Liu JL, Ooi GT, Setser J, Frystyk J, Boisclair YR, LeRoith D. Circulating levels of IGF-1 directly regulate bone growth and density. J. Clin. Invest. 2002;110:771–781. doi: 10.1172/JCI15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Lefebvre V, Zhang Z, Eberspaecher H, de Crombrugghe B. Three HMG-like sequences within a 48-bp enhancer of the Col2a1 gene are required for cartilage-specific expression in vivo. J. Biol. Chem. 1998;273:14989–14997. doi: 10.1074/jbc.273.24.14989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.