Abstract
Hepatic stimulator substance (HSS), also known as augmenter of liver regeneration (ALR), acts as a hepatotrophic growth factor to promote liver regeneration after liver damage or partial hepatectomy. However, the expression and function of HSS during liver development in mammals remain largely unknown. In this work, the hepatoblasts were isolated from mice at embryonic day 13.5 (E13.5), and HSS expression and its role during hepatoblast maturation were investigated. The results showed that HSS expression was enhanced in the hepatoblasts compared with mouse primary hepatocytes. HSS expression (23 kDa) was significantly decreased if the hepatoblast maturation was induced by a combination of oncostatin M (OSM), dexamethasone (DEX), and hepatocyte growth factor (HGF). We also found that knockdown of HSS expression (mainly 23-kDa isoform) by siRNA promoted hepatoblast maturation and also activated the signal transducer and activator of transcription 3 (STAT3) phosphorylation levels. However, if STAT3 activity was blocked by a small-molecule inhibitor Stattic, then hepatocyte maturation could be abolished, suggesting that STAT3 was most likely a potential molecule responsible for HSS signaling. In summary, our results demonstrated for the first time that HSS might be an active factor participating in the regulation of liver development and hepatocyte maturation.
Introduction
Liver development in mice begins at embryonic day 8.5 (E8.5) from an area of the primitive gut endoderm that is specified by signals from the cardiac mesoderm and the surrounding mesenchyme [1]. These signals ultimately result in the proliferation of hepatoblasts followed by their migration into the surrounding mesenchyme. At approximately E13.5, the hepatoblasts begin to give rise to mature hepatocytes in the liver parenchyma, whereas they differentiate into cholangiocytes in the periportal area. During the late fetal and neonatal stages, the liver initiates to express several genes associated with liver maturation, such as glucose-6-phosphatase (G6Pase) and tyrosine amino transferase (TAT), and begins to establish the architecture of the liver lobules.
Throughout liver embryology, a few of liver-specific transcription factors have been identified and their functions in controlling differentiation during development have been elucidated. Among them, hepatocyte nuclear factor 4α (HNF4α) expression is found to increase in hepatoblasts at the ninth day of gestation (E9.0) and HNF4α deficiency in fetal hepatoblasts might lead to a shutdown of expression of many hepatic enzymes, yielding to hepatic abnormal morphology [2]. Meanwhile, CCAAT/enhancer binding protein (C/EBP) factors first appear at E9.5 and gene knockout of C/EBPα causes neonatal death in mice due to hypoglycemia as result of the impaired hepatocyte maturation and defective glycogen storage [3]. Recently, C/EBPα and C/EBPβ also have been reported as the markers of early liver development [4]. In our previous studies, we found that the HNF4α and C/EBPα, which play important roles in liver development, could downregulate hepatic stimulator substance (HSS) expression [5,6].
HSS was first identified by LaBrequce in the liver of weanling rats or regenerating livers of rats in 1975 [7]. Partial purification of HSS predicted that it has molecular weight of ca. 15 kDa with iso-homodimer form [8]. HSS stimulates specifically liver cells or hepatoma cells to proliferation. This action of HSS is characterized with the tissue-specific, but nonspecies specific manner [9]. Later, it was found that HSS could promote primary hepatocyte growth only when it was combined used with epidermal growth factor (EGF) or transforming growth factor alpha (TGF-α) [10], indicating an amplification capacity of HSS for EGF or TGF-α action. Owing to this feature, HSS was then nominated as augmenter of liver regeneration (ALR). ALR cDNA was first cloned by Starzl's lab in 1995 [11] and its molecular biology was extensively studied as well [12]. It is demonstrated that ALR gene has high homology with a known gene in yeast as erv1 (essential for respiratory vertebrate) that makes it possible to rename ALR gene as Gfer (Growth factor erv1-like). As previous reports, ALR protein presents two splicing forms with large molecule of 23 kDa and small molecules of 15 kDa. ALR with small molecules was considered as a cytokine. Upon binding to its receptor [13], it produces a cellular proliferative signaling via mitogen-activated protein kinase (MAPK) pathway and initiates the growth response by activating EGF receptor (EGFR) [14]. While ALR with large molecule size is mainly located in the mitochondrial intermembrane space, contributing to the biogenesis of cytosolic Fe/S proteins and to cellular iron homeostasis as well [15]. Based upon the crystallization of recombinant ALR protein, it is confirmed that ALR with large molecule size is a mammalian FAD-dependent sulfhydryl oxidase (SOX2) with cytochrome c reductase activity [16], indicating that ALR might be related with mitochondrial biogenesis and metabolism. Recently, Gandhi et al. also reported that ALR is critically important for the survival of hepatocytes by its association with mitochondria and regulation of ATP synthesis [17,18].
Although accumulative information about HSS/ALR on liver protection, proliferation, and regeneration has been obtained, its function on the liver development remains largely unknown. Few studies have explored the role of ALR during liver development. Most recently, Li et al. showed that ALR promotes liver outgrowth during zebrafish hepatogenesis [19]. However, the zebrafish embryonic liver does not provide a comprehensive understanding of liver development in vertebrates, and the results obtained from zebrafish might not be applicable to mammalian liver development [20]. Moreover, Dayoub et al. reported that transcription factors Nrf2 (nuclear factor erythroid 2-related factor 2) and Foxa2 (forkhead box protein A2) could regulate ALR expression, especially the later is also a very important transcription factor in liver development [21,22]. All these information suggest that HSS/ALR might play an important role during regulation of liver development; therefore, this question is required to further be clarified in liver model of mammals.
The hepatoblast has been proposed as an ideal cell system to study liver development and differentiation due to its high proliferation rate and its potential to differentiate into hepatocytes and cholangiocytes [23]. In this study, we are aiming to demonstrate whether HSS/ALR is involved in regulation of liver development. Our results confirm that this factor is actually participated in early development and maturation of liver through signal transducer and activator of transcription 3 (STAT3) pathway.
Materials and Methods
Animals
C57BL/6 mice were purchased from the Academy of Military Medical Sciences (Beijing, China) and maintained under controlled conditions with a 12-h light-dark cycle. All the issues related to the animal maintenance and surgical operations were performed in accordance with the guidelines of the Chinese Council on Animal Care and with the approval of the Ethics Committee of Capital Medical University (Beijing, China). The age of the embryos was determined by the number of days after the appearance of the vaginal plug; noon on the day that the vaginal plug appeared was considered 0.5 days of gestation. The embryos were isolated from the uteruses of pregnant mice on day 13.5 of gestation.
Isolation and culture of hepatoblasts from fetal livers
Isolation of fetal hepatoblasts from embryonic mouse livers and the cell cultures were performed as previously described [24]. All animals were treated humanely, and the experimental protocols used were approved by the Human and Animal Ethics Committee, Capital Medical University. Briefly, after deep anesthesia, the pregnant mice were sacrificed by cervical dislocation. Gravid uteri were dissected into 1×phosphate-buffered saline (PBS; 130 mM NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4·H2O, pH 7.2). Using a stereo microscope (SZX12 system; Olympus Optical Co., Ltd.), the livers were carefully dissected and isolated from the fetal uterus. Then, the embryonic liver tissues were minced and dissociated with liver digestion medium (0.25% trypsin/ethylene diamine tetraacetic acid [EDTA] solution) followed by hemolysis with hypotonic buffer (16 mM Tris and 200 mM NH4Cl). The cells were maintained in mouse embryo fibroblast-coated tissue culture dishes with hepatocyte basal culture medium (Dulbecco's modified Eagle's medium [DMEM]; Gibco Life Technology) supplemented with 10% fetal bovine serum (FBS; HyClone), 2 mM L-glutamine (Cyagen Biosciences, Inc.), 1×nonessential amino acid solution (Cyagen Biosciences, Inc.), 100 U/mL streptomycin and 100 U/mL penicillin (Beijing Solarbio Science & Technology Co., Ltd.), and 28.6 μM β-mercaptoethanol (Sigma-Aldrich). The cultures were maintained at 37°C in a humidified 5% CO2 atmosphere in air. The culture media were replaced every 2 days. After ∼7–10 days in culture, colonies could be observed with an endodermal morphology, that is, polyhedral cells with phase-bright boarders. The colonies were selectively detached from culture by scraping with a cell scraper and digested with 0.05% trypsin/EDTA. After several passages, the cultured cells were removed onto six-well plates precoated with 0.1% gelatin (Cyagen Biosciences, Inc.).
Immunofluorescence
The hepatoblasts derived from E13.5 mice were seeded into six-well plates coated with 0.1% gelatin. When the plates reached 90% confluence, the cells were fixed with 4% paraformaldehyde in 0.1 M sodium phosphate buffer at pH 7.4 at room temperature for 30 min. After being permeabilized with 0.5% Triton X-100 in PBS and blocked with normal serum, the cells were incubated with an anti-ALR antibody (diluted 1:200; Santa Cruz Biotechnology), an anti-albumin antibody (ALB, diluted 1:100; Santa Cruz Biotechnology), an anti-cytokeratin 18 antibody (CK-18, diluted 1:100; Abcam), an anti-gamma-glutamyl transpeptidase antibody (GGT, diluted 1:100; Abcam), or an anti-cytokeratin 19 antibody (CK-19, diluted 1:100; Abcam) at 4°C overnight. IgG controls were used to control for background staining intensity. After three washes in PBS, the primary antibodies were detected with the corresponding Alex Fluor 594-conjugated anti-Rabbit IgG (diluted 1:500; Invitrogen) or Alex Fluor 488-conjugated anti-Goat IgG (diluted 1:500; Invitrogen) at 37°C for 30 min. The nuclei were stained in a 10 μg/mL solution of DAPI. The sections were examined under a Leica fluorescence microscope (DM5000 B; Leica Microsystems).
Flow cytometry
To characterize the phenotype of the cells that we isolated, we used flow cytometry to detect the expression of the surface markers of the cells. The hepatoblast suspension was prepared by trypsin/EDTA, and then 1×105–106 cells were stained sequentially with an allophycocyanin (APC)-Cy™ 7-conjugated anti-CD45 mAb, a fluorescein isothiocyanate (FITC)–conjugated anti-CD34 mAb, a phycoerythrin (PE)–conjugated anti-CD44 mAb, and an APC-conjugated anti-CD117 mAb (all from BD Bioscience). For surface antigen DLK detection, the cells were incubated with a rabbit anti-delta-like protein antibody (DLK; Abcam). The background fluorescence level of the cell suspensions was estimated using the appropriate IgG control. After three washes with PBS, cell fluorescence was analyzed with an FACS Calibur flow cytometer (Becton Dickinson) using CellQuest Software (BD Bioscience).
Xenograft assays
To exclude tumorigenesis of the hepatoblasts, 5×106 cells were suspended in 200 μL PBS and injected subcutaneously into the right upper forelimb of 4-week-old male athymic BALB/c mice to evaluate whether xenograft tumors could be generated. HepG2 cells were prepared and handled similarly as a control. Tumor growth was examined daily for at least 5 weeks.
Isolation of mouse hepatocytes
Mouse primary hepatocytes were isolated from male C57BL/6 mice (8–10 weeks old) by a two-step collagenase perfusion [25]. The viability of freshly isolated hepatocytes was more than 90%, as confirmed by trypan blue exclusion. The isolated hepatocytes were suspended in William's E medium supplemented with 10% FBS, 100 μg/mL streptomycin, and 100 units/mL penicillin and then plated in collagen-coated six-well culture plates at a density of 3×105 cells/well. After the cell attachment, the medium was removed and replaced with a fresh medium containing 0.5% FBS. And the culture media were replaced every 2 days.
In vitro differentiation
For induction of hepatoblast maturation, the cells were plated at 3×105 cells per well in six-well plates. When 85% of confluence was reached, the cells were washed twice with PBS and subsequently cultured in basal medium combined with 10 ng/mL OSM (R&D Systems, Inc.), 20 ng/mL mouse HGF (R&D Systems, Inc.), and 100 nM DEX (Sigma-Aldrich). The differentiation media were changed every 2 days.
The cholangiocyte differentiation was established by plating the hepatoblasts on a layer of Matrigel (BD Biosciences). Pure Matrigel (200 μL) was spread on each well of the 12-well plates and allowed to settle at 37°C for at least 30 min. The cells were then plated at a density of 2×105 cells/cm2 and cultured in basal medium. The basal medium was changed every 2 days.
RNA extraction and quantitative real-time PCR
Total RNA extraction, first-strand cDNA synthesis, and real-time PCR (qRT-PCR) were performed as described previously [6]. qRT-PCR was performed to assess the gene expression of premature hepatocyte markers, such as AFP and DLK, and mature hepatocyte markers, such as ALB, TAT, TO (tryptophan-2, 3-dioxygenase), CPS (carbamoyl phosphate synthetase), and G6Pase. Meanwhile, the gene expression of cholangiocyte markers, such as CK-7, CK-18, and CK-19, was analyzed in parallel. The primers used for the identification of gene expression were designed based on the mouse mRNA sequences and are listed in Table 1. The amplicon expression in each sample was normalized to β-actin. After normalization, the expression of the genes was quantified using a 2−ΔΔCt calculation [26].
Table 1.
Primer Sequences Used for Real-Time Polymerase Chain Reaction
| Gene | Strand | Primer sequence (5′→3′) |
|---|---|---|
| ALR | Sense | CGAGTATCCTTCAGCCAGTG |
| Antisense | AGCCGTCCTTCCAGCCGTCA | |
| AFP | Sense | AGCTCAGCGAGGAGAAATGGT |
| Antisense | GTTCACAGGGCTTGCTTCATTC | |
| ALB | Sense | AACCCAACCACCTTTATG |
| Antisense | GACGGACAGATGAGACCA | |
| DLK | Sense | CGGGAAATTCTGCGAAATAG |
| Antisense | TGTGCAGGAGCATTCGTACT | |
| G6Pase | Sense | TCCACCTTGACACTACACCC |
| Antisense | TGCTGAGTTCTCCCTTGC | |
| TO | Sense | TGGAAAGGGACCTGGAAT |
| Antisense | ACGACGACTGTCATACCG | |
| TAT | Sense | TGCTGGATGTTCGCGTCAATA |
| Antisense | CGGCTTCACCTTCATGTTGTC | |
| CPS | Sense | AGCACCCGCATCTACGC |
| Antisense | AGCCCTTTCAGTGTCTTATCC | |
| CK-7 | Sense | CATTGAGATCGCCACCTACC |
| Antisense | ACAGGTCCCATTCCGTCTC | |
| CK-18 | Sense | GAGGCAGAGATTGCCACCTA |
| Antisense | AGGGCATCGTTGAGACTGA | |
| CK-19 | Sense | AGTCCCAGCTCAGCATGAA |
| Antisense | TAACGGGCCTCCGTCTCT | |
| β-Actin | Sense | ACCCACACTGTGCCCATCTA |
| Antisense | GCCACAGGATTCCATACCCA |
Western blot
Total cellular proteins were extracted using cell lysis buffer (20 mM Tris/HCl, 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, and protease inhibitor cocktail, pH 7.5). The protein concentrations of the lysates were determined according to the bicinchoninic acid (BCA) method using a protein assay kit (Pierce Biotechnology). Samples containing 100 μg of protein were separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (Millipore) for western blot analysis. Primary antibodies against α-fetoprotein (AFP, diluted 1:2000; Santa Cruz Biotechnology), ALB (diluted 1:2000), ALR (diluted 1:500), hepatic nuclear factor 4α (HNF4α, diluted 1:2000; Abcam), or glyceraldehyde 3-phosphate dehydrogenase (GAPDH, diluted 1:10,000; KangChen) were used. The membranes were washed and incubated with secondary antibodies conjugated to horseradish peroxidase. Immunoreactive proteins were detected by enhanced chemiluminescence (ECL; Applygen Technologies, Inc.). The relative density of the protein bands was quantitatively determined with ImageJ software (National Institutes of Health).
Glycogen assay
Regulation of the blood glucose level is another important function of mature hepatocytes. To examine whether hepatocytes that had been subjected to maturation induction by a combination of oncostain M, dexamethasone and hepatocyte growth factor (ODH) could produce and store glycogen, we analyzed the accumulation of intracellular glycogen in vitro by the periodic-acid-Schiff (PAS) staining method. The cultures were fixed in Carnoy's fixative at room temperature for 10 min and oxidized in 1% periodic acid for 15 min. After oxidation, the cultures were rinsed three times with distilled water and then treated with Schiff's reagent (Sigma-Aldrich) for 30 min. After rinsing with distilled water for 5 min, the PAS-positive cells and whole cells were examined under a light microscope (DMIL LED; Leica Microsystems).
ALB and urea detection
Serum albumin is the most abundant protein synthesized by hepatocytes, and its production starts in the early stage of liver development and reaches the maximum level in the adult liver [27]. Therefore, albumin secretion is a critical function of mature hepatocytes. The concentrations of ALB and urea secreted into the culture media were analyzed using the automated biochemical analyzer (BS-200; Mindray), according to the manufacturer's instructions.
siRNA treatment
The hepatoblasts were transfected with ALR-specific siRNAs or nontargeting control (scrambled) siRNAs (Dharmacon) according to standard protocols. Briefly, the cells were replated in six-well plates (3×105 cells per well) and grown in DMEM hepatocyte basal culture medium without antibiotics for 24 h to 70%–80% confluence. To prepare the transfection complex, DharmaFECT-4 transfection reagent (4 μL per well) was incubated with the ALR siRNAs or the scrambled siRNAs in antibiotic-free and serum-free medium for 30 min at room temperature. The cells were then incubated with the siRNA-DharmaFECT-4 complexes at 37°C for 24 h. After 24 h, the transfection mixture was replaced by DMEM hepatocyte basal culture medium and cultured for 6 days. The sequences of ALR siRNAs and scrambled siRNAs are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd).
Data analysis
The results of multiple observations are presented as the means±SDs of at least three independent experiments. The data were analyzed with the statistical software SPSS 11.5 (IBM), and differences between various groups were analyzed by one-way analysis of variance. *P<0.05 was considered statistically significant.
Results
Identification of hepatoblasts isolated from fetal liver buds
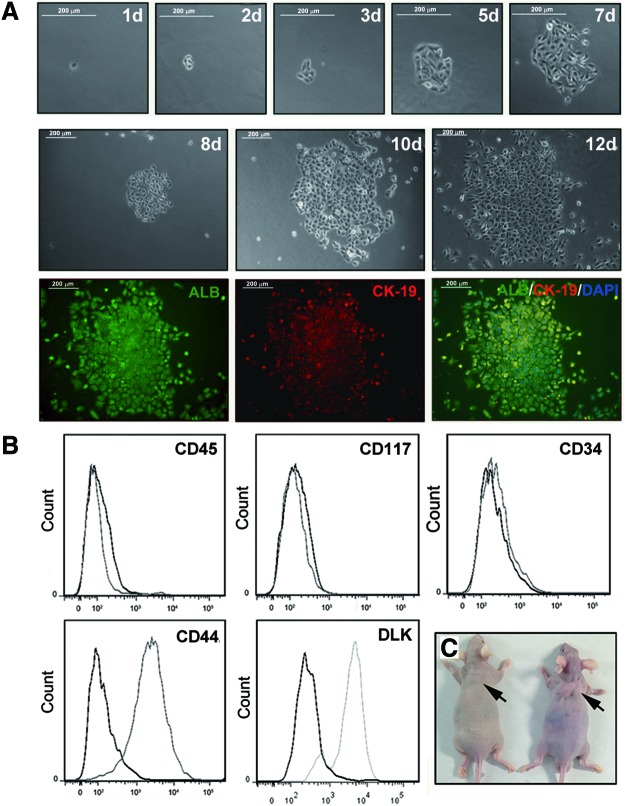
The hepatoblast is a good tool to study ALR expression during liver development. To examine ALR expression during hepatocyte maturation, we isolated hepatoblast cells from E13.5 fetal livers and successfully cultured the cells for a period of time about 2 or 3 weeks. To verify that the hepatoblast cells are capable of bipotential differentiation, they were seeded into six-well culture plates as that an individual cell was adequately isolated from each other. As shown in Fig. 1A, in the colony formed out of a single hepatoblast, the cells expressed both ALB and CK-19, indicating that the hepatoblasts we isolated are featured with bipotential progenitor cells. To further examine whether the hepatoblasts that we isolated exhibited the features of liver progenitor cells, several cell markers were evaluated by immunofluorescence staining. As shown in Fig. 1B, the cells did not express CD34, CD45, or CD117, indicating that they might not have originated from hematopoietic cells. However, 90% of the cells were CD44 positive and 87% of the cells were DLK positive, suggesting that these cells possessed the features of mesenchymal cells and hepatoblasts. Meanwhile, the hepatoblasts isolated from the fetal livers were characterized as bipotential progenitors capable of differentiating into mature hepatocytes and cholangiocytes in vitro (Supplementary Figs. S1 and S2). Moreover, the tumorigenic potential of these hepatic progenitor cells was examined using a xenograft assay. Figure 1C showed that the progenitors caused no tumors 5 weeks after injection. As a control, the HepG2 cells that were inoculated into the control mice apparently generated tumors. These results demonstrated that the hepatoblasts that were isolated fulfilled the criteria of hepatic progenitors.
FIG. 1.
Characterization of the hepatoblasts. (A) The hepatoblasts isolated from E13.5 fetal livers were cultured on six-well plates coated with type-I collagen at a density of 50 cells/cm2 in the presence of hepatocyte growth factor and epidermal growth factor. A single cell was selected and monitored for colony formation. Phase-contrast images of the culture taken from day 1 to 12 after plating are shown. After 12 days of culture, the colony was double-stained for ALB (green) and CK-19 (red). And nuclei (blue) were stained with DAPI. A large colony derived from a single progenitor cell consisted of ALB+ and CK-19+ cells. Scale bar=200 μm. (B) FACS analysis for surface markers, such as CD34, CD44, CD45, CD117, and DLK. Thin curves: specific antibody staining. Thick curves: isotype control antibody. The data are representative of three independent experiments. (C) In vivo tumorigenesis. A total of 5×106 hepatoblasts or HepG2 as control were injected into the right upper flank region of immunodeficient mice. The HepG2 cells induced tumors after 5 weeks in the immunodeficient mice, while the hepatoblasts caused no tumors. The arrows show the position of injection of HepG2 cells and hepatoblasts. Color images available online at www.liebertpub.com/scd
ALR expression decreased with hepatoblast maturation
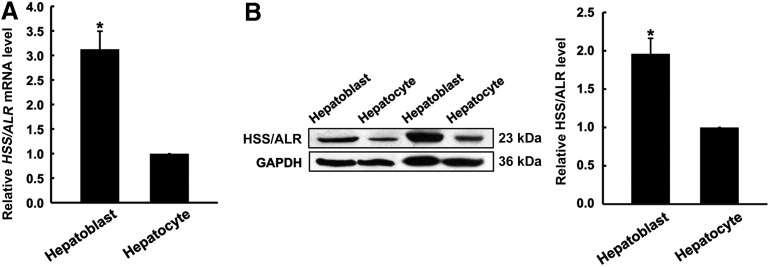
HSS/ALR has been reported as a hepatotrophic factor for liver growth [28] and as a survival factor in hepatocyte protection [17]. However, there are few reports of ALR expression during hepatocyte maturation. Therefore, ALR expression was examined in the hepatoblasts and the primary hepatocytes. As shown in Fig. 2A, ALR mRNA was highly expressed in the hepatoblasts, and this expression was markedly decreased by 62% in the mature hepatocytes. Western blot analysis confirmed the results of qRT-PCR (Fig. 2B). These results clearly indicated that the ALR expression was reduced in mature hepatocytes.
FIG. 2.
Augmenter of liver regeneration (ALR) expression in the hepatoblasts. (A) The ALR mRNA expression in the mouse fetal hepatoblasts and mouse adult primary hepatocytes was measured relative to β-actin by quantitative real-time PCR (qRT-PCR). The results are the means±SDs of five independent experiments from different livers: *P<0.05 compared with the hepatocytes. (B) The ALR protein levels in the hepatoblasts and hepatocytes were analyzed by western blot. Samples containing 100 μg of protein extract were loaded and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE). Western blotting was performed using antibodies against ALR and GAPDH. The relative density of the ALR band was normalized to GAPDH. The values are expressed as the means±SDs of four independent experiments. The values are expressed as a ratio of the band intensities of the hepatocytes.
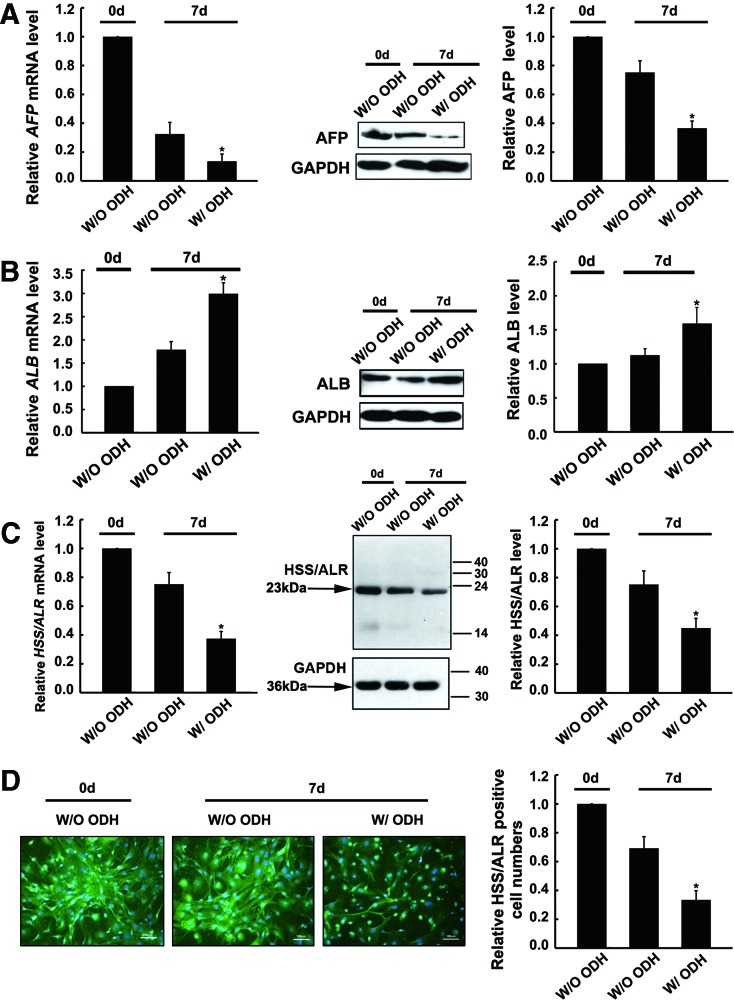
To verify whether the decrease in ALR expression was actually associated with the hepatocyte maturation process, the hepatoblasts were stimulated to mature via ODH and the ALR expression was simultaneously examined. The combined use of ODH was reported to successfully induce the conversion of hepatic progenitor cells into mature hepatocytes [29]. In the current study, the maturation incubation was performed with OSM (10 ng/mL) and DEX (100 nM) combined with HGF (20 ng/mL) for 7 days. As shown in Fig. 3A and B, after treatment with this combination of ODH, the hepatoblasts were able to differentiate into mature hepatocytes in vitro, as demonstrated by the changes in their cellular markers, for example, the ALB mRNA and protein levels, which are associated with mature hepatocytes, were significantly increased, while the AFP mRNA and protein levels, which are associated with progenitors, were dramatically decreased. Additionally, other features of mature hepatocytes could be observed (see Supplementary Fig. S1; morphology, glycogen storage, and biochemical index). As shown in Fig. 3C, the induction of hepatocyte maturation by ODH, the ALR mRNA and protein expression was obviously decreased by 61% and by 49%, respectively. In addition to the results of PCR and western blot, immunofluorescence showed that many of the cells were ALR positive day 0 of hepatoblast culture; that is, without induction; however, after ODH induction for 7 days, the number of ALR-positive cells significantly decreased by 65% (Fig. 3D). Western blot analysis also showed the 23-kDa isoform of ALR was mainly decreased in the hepatoblast maturation. These results demonstrated that ALR expression decreased during hepatoblast maturation.
FIG. 3.
Alteration of ALR expression during hepatoblast maturation. (A) AFP mRNA and protein levels after oncostatin M, dexamethasone and hepatocyte growth factor (ODH) induction. The AFP mRNA level was measured relative to β-actin, and the relative density of the AFP band was normalized to GAPDH. All of the results are the means±SDs (n=4). *P<0.05 compared with the cells at day 0 without ODH. (B) ALB mRNA and protein levels. All the values are expressed as the means±SDs of four independent experiments. *P<0.05 compared with the cells on day 0 without ODH. (C) The hepatoblasts were induced with ODH for 7 days. At the end of the incubation, ALR expression was detected by qRT-PCR and western blot. The values are expressed as the means±SDs of four independent experiments. GAPDH was used as the loading control in three independent experiments. *P<0.05 compared with the cells at day 0 without ODH. (D) The ALR expression was detected by immunofluorescence staining in the cells incubated with or without ODH. The cells, treated with or without ODH for 7 days, were stained with an anti-ALR antibody. ALR expression without ODH induction at day 0 was considered as the basal level. Nuclei (blue) were stained with DAPI. Scale bar=100 μm. Color images available online at www.liebertpub.com/scd
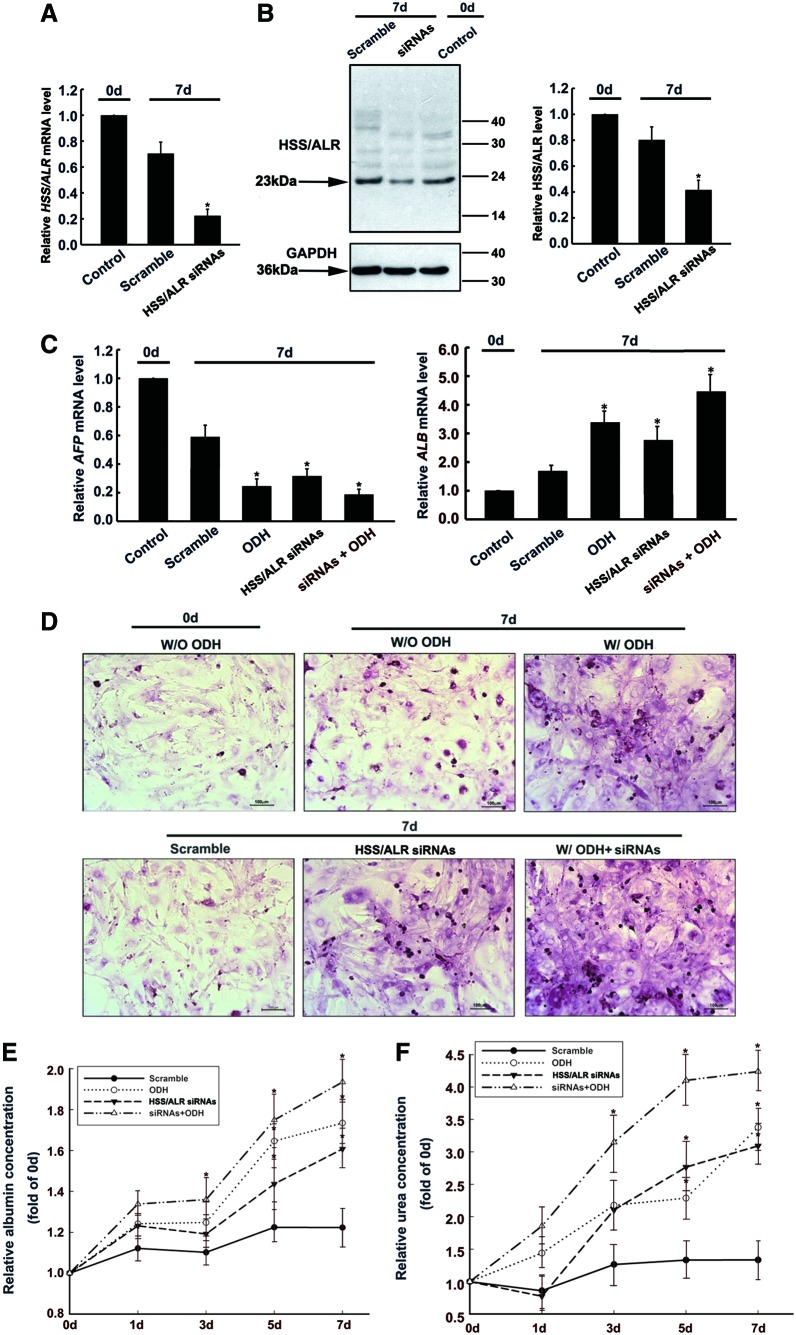
ALR siRNAs promoted hepatoblast maturation
As mentioned earlier, ALR expression decreased during hepatoblast maturation. Therefore, we were interested in whether the inhibition of ALR expression could accelerate hepatoblast maturation. We used interfering RNAs (siRNAs) to knockdown the expression of ALR. As shown in Fig. 4A and B, siRNA transfection inhibited ALR mRNA and protein expression in the hepatoblasts by 70% and 50%, respectively, compared with the scramble siRNA-transfected cells. And the 23-kDa isoform of ALR was mainly decreased after ALR siRNA transfection. Meanwhile, ALR siRNAs could promote hepatoblast maturation, indicating a 71% reduction in AFP mRNA expression and a 2.6-fold increase in ALB expression (Fig. 4C). Further, the hepatoblasts subjected to the ALR siRNAs displayed a considerable ability to synthesize glycogen and urea (Fig. 4D–F) compared with hepatoblasts without siRNAs; both of these characteristics are features of mature hepatocytes. It is interesting to note that if the hepatoblasts were transfected with ALR siRNAs, then the inhibition of ALR expression could also strengthen the inductive effect provided by ODH (Fig. 4C). These results strongly suggest that the downregulation of ALR expression might promote hepatoblast conversion into mature hepatocytes.
FIG. 4.
Hepatocyte maturation after ALR downregulation. The hepatoblasts were transfected with scrambled siRNAs or ALR siRNAs for 7 days. (A) The ALR mRNA level was measured after the transfection in the hepatoblasts by qRT-PCR. The values are expressed as the means±SDs of four independent experiments. *P<0.05 compared with the control cells at day 0 without ALR siRNAs. (B) The ALR protein level was measured by western blot after transfection. GAPDH served as a loading control. The intensities of each signal were analyzed by densitometry. The results are the means±SDs for four independent experiments. *P<0.05 compared with the control cells at day 0 without ALR siRNAs. (C) The AFP and ALB mRNA levels were measured by qRT-PCR after ALR siRNA transfection or ODH induction. The values are expressed as the means±SDs of four independent experiments. *P<0.05 compared with the control cells on day 0 without ALR siRNA or ODH induction. (D) Intracellular glycogen contents in the hepatoblasts subjected to ALR siRNAs analyzed by PAS staining. The untransfected hepatoblasts without ODH induction at day 0 represented the basal level of the glycogen content. Glycogen is shown in magenta. Scale bar=100 μm. (E) Albumin secretion was detected in the ALR siRNA and scrambled siRNA cells. The secretion of albumin by hepatoblasts treated with ODH was taken as a positive control. The values are expressed as the means±SDs of four independent experiments. *P<0.05 compared with the scrambled groups. (F) Urea synthesis was determined in the ALR siRNA- or ODH-induced hepatoblasts at different time points. The values are expressed as the means±SDs of four independent experiments. *P<0.05 compared with the scrambled groups. Color images available online at www.liebertpub.com/scd
ALR siRNAs promoted hepatoblast maturation identical to ODH induction
As mentioned earlier, ODH could effectively induce the maturation of hepatoblasts into hepatocytes. However, the knockdown of ALR expression by siRNAs was also able to promote hepatoblast maturation (Fig. 4A–C). Therefore, it would be interesting to investigate hepatoblast maturation stimulated by both of these approaches within one experiment. It is possible that either knockdown of ALR by siRNAs or ODH induction is stronger stimulus for hepatoblast maturation because the hepatoblasts lost their primitive markers and expressed the markers exclusively observed in mature hepatocytes (Fig. 4C). In addition to the cell markers, ALR knockdown stimulated the hepatoblasts to mature into functional hepatocytes capable of albumin secretion and urea metabolism, and identical to results obtained with ODH induction (Fig. 4E, F). Meanwhile, a combination of ALR siRNA transfection and ODH induction could further strengthen hepatoblast maturation when compared with ODH or ALR siRNA treatment alone (Fig. 4C–F). However, it should be noted that ALR siRNAs did not lead to a sharp increase in urea secretion at day 1, as observed with ODH induction, suggesting that ALR inhibition is not a rapid stimulus of the hepatocyte maturation process.
The signaling pathways involved in hepatoblast maturation resulting from ALR inhibition by siRNA
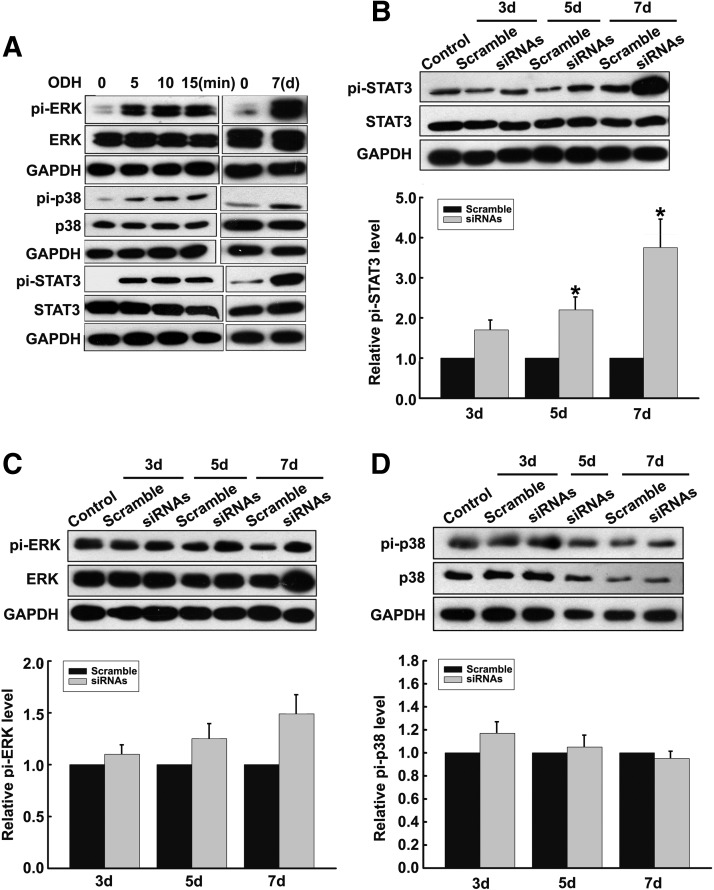
After confirming that ALR might participate in hepatocyte maturation, we were interested in identifying the signaling molecule(s) responsible for the maturation process. Initially, the phosphorylation levels of ERK, p38, and STAT3, which are the most significant components of liver maturation, were analyzed in the ODH-induced or ALR siRNA-transfected hepatoblast cells. As shown in Fig. 5A, ERK, p38, and STAT3 were rapidly phosphorylated within 5 min after ODH treatment, which is consistent with previous reports [30], and the phosphorylation of these molecules was maintained for 7 days. However, in the ALR siRNA cells, only the phosphorylation of STAT3 was significantly increased from day 5, reaching a 3.8-fold increase at day 7, compared with transfection with the scrambled control (Fig. 5B). The phosphorylation of the other two molecules (p38 and ERK) was not markedly altered in the ALR siRNA cells (Fig. 5C, D), suggesting that the signaling pathway stimulated by ALR knockdown during hepatocyte maturation might differ from that associated with ODH stimulation.
FIG. 5.
Signaling molecule phosphorylation in hepatoblasts with ALR downregulation. (A) Phosphorylation of ERK, p38, and STAT3 in hepatoblasts induced with ODH was detected by western blot at 0, 5, 10, 15 min, and day 7. (B–D) Phosphorylation of ERK, p38, and STAT3 in hepatoblasts transfected with ALR siRNAs was detected by western blot at 3, 5, and 7 days. The values from the untransfected cells at day 0 were considered as the control. The STAT3 phosphorylation markedly increased after transfection for 5 and 7 days, while the phosphorylation of ERK and p38 did not change significantly. The results are the means±SDs of four independent experiments. *P<0.05 compared with the scrambled groups at different time points.
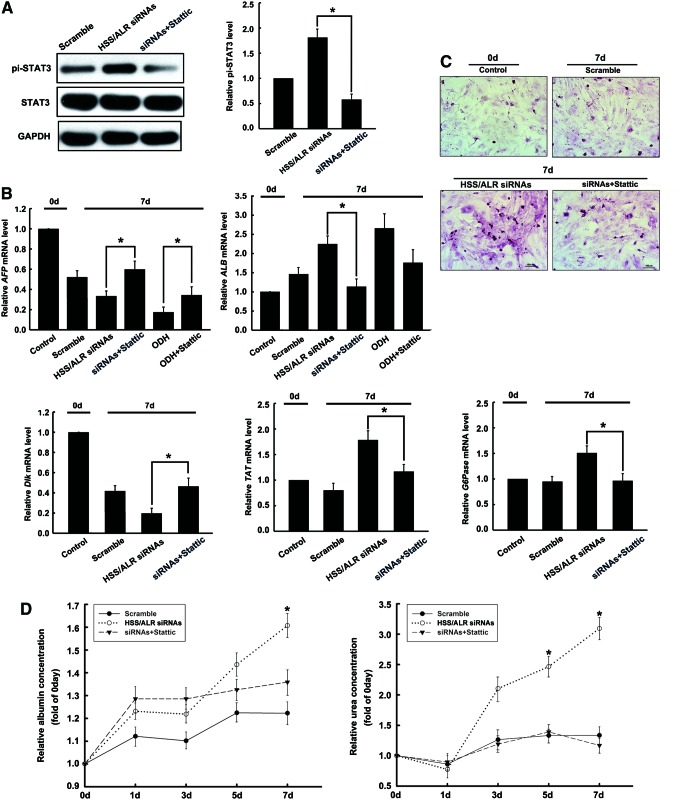
To further confirm the maturation-promoting role of STAT3 signaling in the ALR siRNA hepatoblasts, Stattic, a specific inhibitor of STAT3, was used to investigate whether the inactivation of STAT3 could weaken the conversion of hepatoblasts into mature hepatocytes. After transfection with ALR siRNAs for 24 h, Stattic was added to the cells at a concentration of 4 μM, and the hepatoblasts were incubated for 6 days. As shown in Fig. 6A, Stattic could inhibit ALR siRNA-induced STAT3 phosphorylation. As a consequence, hepatocyte maturation was hampered; for example, the levels of AFP and DLK mRNA, which were previously reduced as result of ALR siRNAs, were elevated, and the levels of ALB, TAT, and G6Pase mRNA, which were expressed by mature hepatocytes, decreased dramatically (Fig. 6C). Moreover, other characteristics presented by mature hepatocytes, such as glycogen storage, urea synthesis, and albumin secretion, were coincidently decreased (Fig. 6D, E).
FIG. 6.
The inhibition of maturation in ALR-knockdown hepatocytes transfected with siRNAs and treated with a STAT3 inhibitor. (A) Inhibition of STAT3 phosphorylation by Stattic. After transfection with ALR siRNAs for 24 h, the hepatoblasts were incubated with Stattic at a concentration of 4 μM for 6 days, and then STAT3 phosphorylation was detected by western blot. The results are the means±SDs from four independent experiments. *P<0.05 compared with the ALR siRNA hepatoblasts without Stattic. (B) Changes in the expression of hepatic marker genes caused by ALR siRNAs or ODH with Stattic treatment. After 7 days of culture, total RNA was extracted from hepatoblasts in the absence or presence of Stattic. The expression of immature hepatocyte markers (AFP and DLK) in the ALR siRNA hepatocytes was increased after Stattic treatment. In contrast, the mature hepatocyte markers (ALB, TAT, and G6Pase) were downregulated. The expression of AFP in ODH-induced hepatoblasts was increased after Stattic treatment, while ALB expression was not changed significantly. The results are the means±SDs (n=4). *P<0.05 compared with the ALR siRNA hepatoblasts without Stattic treatment. (C) The intracellular glycogen content in hepatoblasts was increased when ALR was downregulated. However, the increase in glycogen content in ALR siRNA hepatoblasts was reversed by Stattic. Glycogen is shown in magenta. Scale bar=100 μm. (D) Albumin secretion and urea production in the hepatoblasts. Similarly, the increase observed in the ALR-downregulated hepatoblasts was abolished if the hepatoblasts were treated with Stattic. The values are expressed as the means±SDs of four independent experiments. *P<0.05 represents a significant difference in the ALR-downregulated hepatoblasts resulting from treatment with Stattic. Color images available online at www.liebertpub.com/scd
Discussion
ALR promotes liver regeneration (LR) and maintains the viability of hepatocytes [17,18]. However, its expression and role in mammalian fetal liver development have not been thoroughly examined. Therefore, we first isolated hepatoblast cells from fetal livers at E13.5 in mice and established a culture system to examine ALR expression during hepatoblast maturation. The hepatoblasts that we isolated did not express the hematopoietic cell markers CD34, CD45, and CD117, but they expressed high levels of the mesenchymal cell marker CD44 and the hepatoblast marker DLK, suggesting that these cells displayed the characteristics of progenitors and had the potential to differentiate further. After induction with ODH, the hepatoblasts matured into hepatocytes expressing ALB, TAT, TO, CPS, and G6Pase; meanwhile, the expression of the progenitor markers AFP and DLK was decreased (Supplementary Fig. S1). The morphological and functional parameters indicated that these hepatoblast cells were able to mature into mature hepatocytes or cholangiocytes in vitro.
In this study, we demonstrated first that ALR was highly expressed in the mouse hepatoblasts, and the expression was decreased dramatically as the cells gave rise to mature hepatocytes (Figs. 2 and 3). ALR was shown to be highly expressed in fetal livers [31]. Meanwhile, the ALR/Gfer gene was also enriched in many other stem cells, such as mouse embryonic stem cells (ESCs) [32,33] and neuronal and hematopoietic stem cells (HSCs) [34]. In HSCs, the high expression of Gfer could restrict the abnormal HSC proliferation through its inhibition of Jab1-mediated turnover of p27kip1 [34]. In ESCs, Gfer plays an essential role in the maintenance of murine ESC pluripotency by preserving the structural and functional integrity of the mitochondria based on modulation of the key mitochondrial fission factor Drp1 (dynamic-related protein 1) [35]. And recently, Li et al. demonstrated that ALR is highly expressed in fetal livers and plays a developmental role in zebrafish [19]. All these findings provide the new time line and new insight that allow us to expand our viewing on this so-called liver-specific growth promoter. More important in this report is that we have identified the high expression of ALR and its 23-kDa isoform might be functionally regulated to participate in the mouse hepatic progenitor cell maturation. In addition, Li reported a role of ALR in fetal liver development based upon an experiment carried out in zebrafish, and information about ALR in regulation of maturating hepatic progenitor cells in mammals is still lacking. Therefore, our finding here in mammalian animal model has strengthened the value of Gfer or ALR in liver development. Moreover, our results demonstrated that 23-kDa isoform of ALR appears to be responsible for mature regulation of liver progenitors induced by ODH.
Interestingly, in the current study, we also observed that a decrease in ALR (mainly 23 kDa) expression could promote mouse hepatoblast maturation (Fig. 4). The 23-kDa isoform of ALR does affect ATP synthesis and cell survival similar to what have been confirmed in mature hepatocytes (data not shown) [17]. But ALR in hepatoblasts seems not to closely associate with apoptosis, which was similar with zebrafish liver cells without affecting apoptosis. However, Li et al. also demonstrated that the decrease in ALR expression had a negligible influence on hepatoblast determination or differentiation to hepatocytes during zebrafish liver development. We believe that the contradictory results are due to the use of a different animal model, different method, and different localization of ALR in the cells. In this study, siRNA strategy is applied to knockdown ALR expression in the mouse hepatoblasts, while the antisense morpholino oligonucleotides were used in zebrafish. More importantly, there would be differential results produced by zebrafish and mouse if both species are used to explore the mechanisms of liver development. Essentially, in mice, the liver is differentiated from endoderm and requires induction from the adjacent cardiac mesoderm. However, the development of zebrafish embryonic liver does not require regulatory signals from the cardiac mesoderm [20]. In addition, instead of mitochondrial residence for ALR in zebrafish liver cells, the 23-kDa ALR was found to localize in the cytosol of mouse hepatoblasts that we had isolated. So, the different localization of ALR in the cells may be accordance with different functions during liver development. For example, the Sonic hedgehog (Shh), also a key gene in the regulation of liver development, was found to be highly expressed in the DLK+ hepatoblasts from mouse fetal livers, and inhibition of Shh in vitro could potentiate the hepatic differentiation of hepatoblasts, which is similar to ALR [36]. However, the Shh specifically stimulated endocrine pancreatic development during the early stage of zebrafish development, with little effect on liver development [37,38].
The phosphorylation of ERK, P38, and STAT3 is believed to play important roles during liver development and hepatic progenitor cell maturation [29]; hence, the phosphorylation levels of these three molecules after ODH induction or ALR siRNA transfection were detected. As a result, we confirm that the phosphorylation of STAT3 was significantly increased during the ALR siRNA-induced hepatoblast maturation, and the enhanced STAT3 phosphorylation and hepatoblast maturation as well provided by ALR downregulation could be reversed by Stattic, an inhibitor of STAT3, suggesting that STAT3 could be a key molecule during the ALR siRNA-induced hepatoblast maturation. Meanwhile, as shown in Fig. 6B, Stattic appears to attenuate the hepatoblast maturation manifested by a reduction in AFP expression caused by ODH induction (Fig. 6B); however, the ALB expression remained insignificantly changed, suggesting that there might be other signaling molecules taking effect during the maturation process induced by ODH.
The STAT3 signaling pathway plays important roles in various biological responses, including cell growth, differentiation, and apoptosis [39]. Miyajima et al. demonstrated that the STAT3 signaling pathways could downregulate the expression of cyclin D in mouse fetal liver cells, which are undergoing maturation processes including a reduction of their self-renewal capacities [40]. Other evidence addresses the involvement of STAT3 in the regulation of liver development, indicating that STAT3 triggers the onset of the epithelial-mesenchymal transitions (EMTs) [41,42]. The EMT is known to occur when tissues are constructed during liver development [43]. Dayoub et al. demonstrated that the cytosolic ALR reduces hepatoma cell migration and augments epithelial growth and, therefore, may act as an EMT-reversing protein [44]. Very recently, we also found that a decrease in ALR expression could inhibit E-cadherin in hepatocytes [45]. And in our previous study, we found that the ALR (15 kDa)–provoked activation of ERK1/2 (extracellular signal-regulated kinases, ERK1/2) could negatively inhibit tyrosine phosphorylation in STAT3 [46]. In current study, we demonstrated that 23-kDa isoform of ALR is involved in regulation of liver cell maturation. It is unclear by what pathway does 23-kDa ALR regulate STAT3 phosphorylation, since, as measured by western blot, the activities of ERK and p38 remain unchanged after knockdown of ALR (23 kDa) (Fig. 5). Therefore, we hypothesize that there might be an intermediate signal molecule other than ERK1/2 responsible for activation of STAT3. It should be taken into note that, although STAT3 might mainly be responsible for HSS/ALR signaling pathway for regulation of liver maturation, it could not exclude other alternative pathways. The precious mechanism and signal pathways related in detail are required to further be addressed.
In summary, the present article reports for the first time that ALR downregulation is involved in the hepatoblast maturation process in vitro, which is highly related to the activity of the STAT3 molecule.
Supplementary Material
Acknowledgments
This work was supported by the National Key Basic Research Program of China (973 program, 2010CB534903) and National Natural Science Foundation of China (30900826 and 30470643).
Author Disclosure Statement
The authors indicate no potential conflicts of interest.
References
- 1.Jung J. (1999). Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science 284:1998–2003 [DOI] [PubMed] [Google Scholar]
- 2.Watt AJ, Garrison WD. and Duncan SA. (2003). HNF4: a central regulator of hepatocyte differentiation and function. Hepatology 37:1249–1253 [DOI] [PubMed] [Google Scholar]
- 3.Rastegar M, Rousseau GG. and Lemaigre FP. (2000). CCAAT/enhancer-binding protein-alpha is a component of the growth hormone-regulated network of liver transcription factors. Endocrinology 141:1686–1692 [DOI] [PubMed] [Google Scholar]
- 4.Westmacott A, Burke ZD, Oliver G, Slack JM. and Tosh D. (2006). C/EBPalpha and C/EBPbeta are markers of early liver development. Int J Dev Biol 50:653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo D, Dong LY, Wu Y, Yang L. and An W. (2008). Down-regulation of hepatic nuclear factor 4alpha on expression of human hepatic stimulator substance via its action on the proximal promoter in HepG2 cells. Biochem J 415:111–121 [DOI] [PubMed] [Google Scholar]
- 6.Dong LY, Sun G, Jiang L, Shao L, Hu Y, Jiang Y, Wang Y. and An W. (2010). Epidermal growth factor down-regulates the expression of human hepatic stimulator substance via CCAAT/enhancer-binding protein beta in HepG2 cells. Biochem J 431:277–287 [DOI] [PubMed] [Google Scholar]
- 7.LaBrecque DR. and Pesch LA. (1975). Preparation and partial characterization of hepatic regenerative stimulator substance (SS) from rat liver. J Physiol 248:273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaBrecque DR, Steele G, Fogerty S, Wilson M. and Barton J. (1987). Purification and physical-chemical characterization of hepatic stimulator substance. Hepatology 7:100–106 [DOI] [PubMed] [Google Scholar]
- 9.LaBrecque DR. (1991). Hepatic stimulator substance. Discovery, characteristics and mechanism of action. Dig Dis Sci 36:669–673 [DOI] [PubMed] [Google Scholar]
- 10.Fleig WE. and Hoss G. (1989). Partial purification of rat hepatic stimulator substance and characterization of its action on hepatoma cells and normal hepatocytes. Hepatology 9:240–248 [DOI] [PubMed] [Google Scholar]
- 11.Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter KA. and Starzl TE. (1995). Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc Natl Acad Sci U S A 92:3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giorda R, Hagiya M, Seki T, Shimonishi M, Sakai H, Michaelson J, Francavilla A, Starzl TE. and Trucco M. (1996). Analysis of the structure and expression of the augmenter of liver regeneration (ALR) gene. Mol Med 2:97–108 [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Yang X, Zhang Y, Wang Q, Chen H, Wei H, Xing G, Xie L, Hu Z, et al. (1999). Identification and characterization of receptor for mammalian hepatopoietin that is homologous to yeast ERV1. J Biol Chem 274:11469–11472 [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Li M, Xing G, Hu Z, Wang Q, Dong C, Wei H, Fan G, Chen J, et al. (2000). Stimulation of the mitogen-activated protein kinase cascade and tyrosine phosphorylation of the epidermal growth factor receptor by hepatopoietin. J Biol Chem 275:37443–37447 [DOI] [PubMed] [Google Scholar]
- 15.Lange H, Lisowsky T, Gerber J, Muhlenhoff U, Kispal G. and Lill R. (2001). An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep 2:715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CK, Dailey TA, Dailey HA, Wang BC. and Rose JP. (2003). The crystal structure of augmenter of liver regeneration: a mammalian FAD-dependent sulfhydryl oxidase. Protein Sci 12:1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thirunavukkarasu C, Wang LF, Harvey SA, Watkins SC, Chaillet JR, Prelich J, Starzl TE. and Gandhi CR. (2008). Augmenter of liver regeneration: an important intracellular survival factor for hepatocytes. J Hepatol 48:578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi CR. (2012). Augmenter of liver regeneration. Fibrogenesis Tissue Repair 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Farooq M, Sheng D, Chandramouli C, Lan T, Mahajan NK, Kini RM, Hong Y, Lisowsky T. and Ge R. (2012). Augmenter of liver regeneration (alr) promotes liver outgrowth during zebrafish hepatogenesis. PLoS One 7:e30835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao T. and Peng J. (2009). Liver development in zebrafish (Danio rerio). J Genet Genomics 36:325–334 [DOI] [PubMed] [Google Scholar]
- 21.Dayoub R, Groitl P, Dobner T, Bosserhoff AK, Schlitt HJ. and Weiss TS. (2010). Foxa2 (HNF-3beta) regulates expression of hepatotrophic factor ALR in liver cells. Biochem Biophys Res Commun 395:465–470 [DOI] [PubMed] [Google Scholar]
- 22.Dayoub R, Vogel A, Schuett J, Lupke M, Spieker SM, Kettern N, Hildt E, Melter M. and Weiss TS. (2013). Nrf2 activates augmenter of liver regeneration (ALR) via antioxidant response element and links oxidative stress to liver regeneration. Mol Med 19:237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dabeva MD, Petkov PM, Sandhu J, Oren R, Laconi E, Hurston E. and Shafritz DA. (2000). Proliferation and differentiation of fetal liver epithelial progenitor cells after transplantation into adult rat liver. Am J Pathol 156:2017–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogler LE. (1997). Selective bipotential differentiation of mouse embryonic hepatoblasts in vitro. Am J Pathol 150:591–602 [PMC free article] [PubMed] [Google Scholar]
- 25.de Longueville F, Atienzar FA, Marcq L, Dufrane S, Evrard S, Wouters L, Leroux F, Bertholet V, Gerin B, et al. (2003). Use of a low-density microarray for studying gene expression patterns induced by hepatotoxicants on primary cultures of rat hepatocytes. Toxicol Sci 75:378–392 [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ. and Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 27.Tilghman SM. and Belayew A. (1982). Transcriptional control of the murine albumin/alpha-fetoprotein locus during development. Proc Natl Acad Sci U S A 79:5254–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y, Fu YL, Yu M, Yue PB, Ge CH, Xu WX, Zhan YQ, Li CY, Li W, et al. (2009). Human augmenter of liver regeneration is important for hepatoma cell viability and resistance to radiation-induced oxidative stress. Free Radic Biol Med 47:1057–1066 [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Li W, Liu B, Wang P, Li W. and Zhang H. (2012). Efficient generation of functional hepatocyte-like cells from human fetal hepatic progenitor cells in vitro. J Cell Physiol 227:2051–2058 [DOI] [PubMed] [Google Scholar]
- 30.Kamiya A, Kinoshita T. and Miyajima A. (2001). Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett 492:90–94 [DOI] [PubMed] [Google Scholar]
- 31.He F, Wu C, Tu Q. and Xing G. (1993). Human hepatic stimulator substance: a product of gene expression of human fetal liver tissue. Hepatology 17:225–229 [PubMed] [Google Scholar]
- 32.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA. and Lemischka IR. (2002). A stem cell molecular signature. Science 298:601–604 [DOI] [PubMed] [Google Scholar]
- 33.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC. and Melton. DA. (2002). “Stemness”.: transcriptional profiling of embryonic and adult stem cells. Science 298:597–600 [DOI] [PubMed] [Google Scholar]
- 34.Teng EC, Todd LR, Ribar TJ, Lento W, Dimascio L, Means AR. and Sankar U. (2011). Gfer inhibits Jab1-mediated degradation of p27kip1 to restrict proliferation of hematopoietic stem cells. Mol Biol Cell 22:1312–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd LR, Damin MN, Gomathinayagam R, Horn SR, Means AR. and Sankar U. (2010). Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell 21:1225–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirose Y, Itoh T. and Miyajima A. (2009). Hedgehog signal activation coordinates proliferation and differentiation of fetal liver progenitor cells. Exp Cell Res 315:2648–2657 [DOI] [PubMed] [Google Scholar]
- 37.diIorio PJ, Moss JB, Sbrogna JL, Karlstrom RO. and Moss LG. (2002). Sonic hedgehog is required early in pancreatic islet development. Dev Biol 244:75–84 [DOI] [PubMed] [Google Scholar]
- 38.Roy S, Qiao T, Wolff C. and Ingham PW. (2001). Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr Biol 11:1358–1363 [DOI] [PubMed] [Google Scholar]
- 39.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K. and Hirano T. (1996). Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 5:449–460 [DOI] [PubMed] [Google Scholar]
- 40.Matsui T, Kinoshita T, Hirano T, Yokota T. and Miyajima A. (2002). STAT3 down-regulates the expression of cyclin D during liver development. J Biol Chem 277:36167–36173 [DOI] [PubMed] [Google Scholar]
- 41.Yamashita S, Miyagi C, Carmany-Rampey A, Shimizu T, Fujii R, Schier AF. and Hirano T. (2002). Stat3 controls cell movements during Zebrafish gastrulation. Dev Cell 2:363–375 [DOI] [PubMed] [Google Scholar]
- 42.Yang Y, Pan X, Lei W, Wang J, Shi J, Li F. and Song J. (2006). Regulation of transforming growth factor-beta 1-induced apoptosis and epithelial-to-mesenchymal transition by protein kinase A and signal transducers and activators of transcription 3. Cancer Res 66:8617–8624 [DOI] [PubMed] [Google Scholar]
- 43.Choi SS. and Diehl AM. (2009). Epithelial-to-mesenchymal transitions in the liver. Hepatology 50:2007–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dayoub R, Wagner H, Bataille F, Stoltzing O, Spruss T, Buechler C, Schlitt HJ. and Weiss TS. (2011). Liver regeneration associated protein (ALR) exhibits antimetastatic potential in hepatocellular carcinoma. Mol Med 17:221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Dong LY, Sun G. and An W. (2014). Downregulation of hepatic stimulator substance during the early phase of liver regeneration inhibits E-cadherin expression in mice. Int J Biochem Cell Biol 47:38–46 [DOI] [PubMed] [Google Scholar]
- 46.Tian ZJ. and An W. (2004). ERK1/2 contributes negative regulation to STAT3 activity in HSS-transfected HepG2 cells. Cell Res 14:141–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.