Abstract
Currently there are several methods to extract bacterial DNA based on
different principles. However, the amount and the quality of the DNA obtained by each
one of those methods is highly variable and microorganism dependent, as illustrated
by coagulase-negative staphylococci (CoNS) which have a thick cell wall that is
difficult to lyse. This study was designed to compare the quality and the amount of
CoNS DNA, extracted by four different techniques: two in-house protocols and two
commercial kits. DNA amount and quality determination was performed through
spectrophotometry. The extracted DNA was also analyzed using agarose gel
electrophoresis and by PCR. 267 isolates of CoNS were used in this study. The column
method and thermal lyses showed better results with regard to DNA quality (mean ratio
of A260/280 = 1.95) and average concentration of DNA (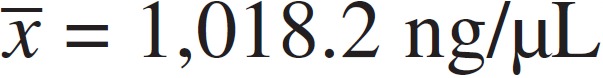 ), respectively. All four methods tested provided appropriate DNA
for PCR amplification, but with different yields. DNA quality is important since it
allows the application of a large number of molecular biology techniques, and also
it's storage for a longer period of time. In this sense the extraction method based
on an extraction column presented the best results for CoNS.
), respectively. All four methods tested provided appropriate DNA
for PCR amplification, but with different yields. DNA quality is important since it
allows the application of a large number of molecular biology techniques, and also
it's storage for a longer period of time. In this sense the extraction method based
on an extraction column presented the best results for CoNS.
Keywords: DNA extraction, Coagulase-negative Staphylococci (CoNS), Spectrophotometry
Abstract
Atualmente, para extrair o DNA bacteriano, existem diversos métodos
baseados em diferentes princípios. Entretanto, a quantidade e qualidade do DNA obtido
por cada um destes métodos é variável e depende do tipo de micro-organismo em
questão; os estafilococos coagulase-negativos (CoNS), por exemplo, possuem parede
celular espessa difícil de lisar. O objetivo deste estudo foi comparar a quantidade e
a qualidade do DNA extraído de isolados clínicos de CoNS utilizando quatro
metodologias diferentes: dois protocolos caseiros e dois kits comerciais. A
determinação da quantidade e da qualidade do DNA foi realizada por
espectrofotometria. O DNA extraído também foi analisado em eletroforese em gel de
agarose e por PCR. A concentração média de DNA foi mais alta no método de lise
térmica (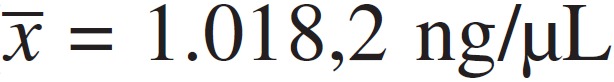 ). Entretanto, com relação à qualidade do DNA, o kit comercial que
utiliza um método de extração baseado em uma coluna de separação apresentou melhor
resultado (média da relação A260/280 = 1,95). As quatro técnicas testadas forneceram
DNA passível de amplificação por PCR, porém com diferentes rendimentos. A qualidade
do DNA extraído de bactérias é importante, pois possibilita a realização de maior
número de técnicas de biologia molecular e também armazenamento do material por maior
período de tempo. Neste sentido, a técnica de extração por coluna de separação
apresentou melhor desempenho frente aos CoNS.
). Entretanto, com relação à qualidade do DNA, o kit comercial que
utiliza um método de extração baseado em uma coluna de separação apresentou melhor
resultado (média da relação A260/280 = 1,95). As quatro técnicas testadas forneceram
DNA passível de amplificação por PCR, porém com diferentes rendimentos. A qualidade
do DNA extraído de bactérias é importante, pois possibilita a realização de maior
número de técnicas de biologia molecular e também armazenamento do material por maior
período de tempo. Neste sentido, a técnica de extração por coluna de separação
apresentou melhor desempenho frente aos CoNS.
INTRODUCTION
In recent years, numerous methods developed by the molecular biology have been applied on the scientific research in clinical microbiology. On those methods, the genetic material most widely used is DNA. Therefore, for an effective DNA extraction there are several methods based on different principles4. However, the amount and the quality of the DNA obtained for each one of those methods are variable, becoming an important factor in the type of molecular method that will be used, and on the subsequent storage of this material18. Moreover, to access bacterial DNA it is necessary to disrupt the cell wall, which presents distinct characteristics among different types of microorganisms13.
Coagulase-negative staphylococci (CoNS) are gram-positive cocci often considered as culture contaminants, since many species are members of the human skin microbiota and mucous membranes. However, CoNS are increasingly being recognized as agents of clinically important infections. In some recent studies they have been considered the main responsible for infections related to medical devices and surgical sites9,12. Furthermore, those microorganisms are also common as agents of the bovine mastitis, as well as infections in humans related to contaminated animal products7,16.
CoNS have a rigid cell wall that can be difficult to lyse, and special enzymes and methods have been developed to address this problem2,18. The gram-positive cell wall is composed of a complex reticulate of peptideoglycan, teichoic acid, polysaccharides, and other proteins, whereas the gram-negative cell wall is thinner with a simpler reticular pattern13. Methods used for genomic DNA isolation from gram-negative bacteria are not always successful with gram-positive2,6,11. This is important because a suitable method of DNA isolation and recovery is one of the most important prerequisites for molecular tests18,19.
The aim of this study was to select a method that provides the best DNA from culture strains of CoNS clinical isolates. The main parameters considered were those related to storage for future testing in genomic libraries and application in molecular biology tests for scientific research or clinical diagnosis. To achieve this, the quality and the quantity of the DNA extracted from CoNS cultures through four different techniques were analyzed. The quality and amount of DNA was determined by spectrophotometry. The visualization of pattern bands in agarose gel electrophoresis was performed to evaluate the DNA integrity and a PCR to test the possibility of amplification of the different types of materials.
MATERIALS AND METHODS
267 CoNS from infections in patients admitted at the “Santa Casa de Misericórdia de Porto Alegre Medical Center” were selected. Those microorganisms were obtained from 2002 to 2004 mainly from bacteremia, but also from other types of infections such as surgical wounds, and infections related to indwelling devices. Species identification was previously performed by SECCHI et al. and published in 200815 (Table 1). Each isolate was stored on skimmed milk (Difco Skim Milk, Becton Dickinson) and was cultivated in tryptone soy agar (TSA) (Tryptone Soy Agar, Oxoid) for 24 hours prior to the extraction procedures. One or two isolated colonies were then grown in two mL brain heart infusion broth (BHI, Merck) for 16 hours; one mL of this suspension was centrifuged at 7000 x g/5 min and the bacterial pellet obtained used as a template in all methods.
Table 1. Species and mean yield of DNA quantity (ng/µL) of each method.
| Specie | No. of isolates (%) | Extraction method | |||
| Boiling | Phenol-chloroform | Extraction column | Salting out | ||
| S. epidermidis | 107 (40.1) | 1173.3 | 67.3 | 100.1 | 30.1 |
| S. haemolyticus | 91 (34.2) | 1181.8 | 202.1 | 35.1 | 16.4 |
| S. hominis | 27 (10.1) | 989.4 | 61.6 | 34.5 | 14.0 |
| S. warneri | 15 (5.6) | 1002.2 | NT | 12.9 | 86.9 |
| S. capitis | 7 (2.6) | 1016.5 | 89.2 | 15 | 21.1 |
| S. saprophyticus | 6 (2.2) | 1052* | 50.6 | 54.3 | NT |
| S. cohnii | 4 (1.5) | 825* | 28* | 41.7 | 18.9* |
| S. caprae | 3 (1.1) | NT | 40* | 28.8 | NT |
| S. xylosus | 3 (1.1) | NT | 23 | 31.6 | NT |
| S. sciuri | 2 (0.7) | NT | NT | 86.5 | NT |
| S. lugdunensis | 1 (0.4) | NT | NT | 32* | NT |
| S. auricularis | 1 (0.4) | NT | NT | 41* | NT |
| Total | 267 (100.0) | 1018.2 | 87.8 | 65.5 | 28.2 |
Just one sample tested; NT = not tested.
Four methods were performed, the first one, popularly known as boiling, was based on ALEXOPOULOU et al. 1 with no relevant modifications. This procedure was performed on 50 randomly selected samples. The second, phenol-chloroform, it was based on SILVA & SILVA16 with just one relevant modification: 20 µL of lysozyme (100 mg mL–1) (Sigma) were added to the enzymatic lysis suspension. The phenol-chloroform used was Equilibrated Phenol, pH 8.0, purchased from USB Corporation. This procedure was applied to 70 randomly selected samples. The third method was based on extraction columns and was performed according to the kit manual (QIAamp DNA mini kit, Qiagen). Lysozyme and lysostaphin were used together on the enzymatic lyses step. One modification was introduced: after the deproteinization step with proteinase K, 4 µL of RNase A (100 mg mL–1) (BioAmerica) were added to it and then incubated at 70 °C for 10 min. All the samples (n = 267) were submitted to this extraction method. Finally, we also tested a salting out method following the kit manual (Wizard® Genomic DNA Purification Kit, Promega) recommendations for gram-positive bacteria without modifications. This procedure was performed on 50 randomly selected samples. The usual bacterial amount used in all methods had a cell growth incubation period of 16 hours in one mL of brain heart infusion broth (BHI) (Merck).
After extraction procedures, we verify the presence and integrity of the DNA on the extracted material in agarose gel electrophoresis. Five microliters of DNA and five µL of loading buffer (buffer type IV14) were mixed in a 0.2 mL tube. This mixture was placed into the gel wells and subjected to a voltage of six volts/cm for 20 min in a horizontal electrophoresis system (PowerPac Basic, Bio-Rad). After that, DNA visualization and photo documentation were performed by the system MF-Chemi BIS (BioAmerica, Inc.). The extracted material of each method was also analyzed by spectrophotometry. Two µL of DNA extracted from each sample were placed directly on the spectrophotometer (NanoDrop, 2000, Thermo Cientific) and measures in 230, 260 and 280 nm were performed. The system software provides the DNA concentration in ng/µL and automatically calculates the absorption ratio 260/280 (A260/280) and 260/230 (A260/230). Lastly, we performed amplification by PCR using the pair of primers described by JENSEN et al. 8: G1 (GAA GTC GTA ACA AGG) and L1 (CAA GGC ATC CAC CGT) (Integrated DNA Technologies, Coralville, USA). This pair of primers was selected from conserved sequences from the adjacent 16S and 23S genes8. The amplification reaction was performed on a LifePro Thermal Cycler (Hangzhou Bioer Technology Co. Ltda, Hangzhou, China). The reagent concentrations and the reaction conditions were the same ones used by COUTO et al. 3. Based on the results of DNA quantification found by spectrophotometry, approximately 50 ng of DNA were used on each PCR reaction. The amplification was checked in agarose gel electrophoresis under the same conditions described above. Each sample was tested once. The major costs of reagents per sample are shown in Table 2.
Table 2. Main cost of reagents per sample of each method in U. S. dollars (US$).
| Method | Kit | Lysostaphin | Lisozyme | Proteinase K | Equilibrated phenol | Total |
|---|---|---|---|---|---|---|
| Boiling | NR | NR | NR | 0.51 | NR | 0.51 |
| Phenol-chloroform | NR | 13.80 | 0.08 | 0.50 | 0.15 | 14.53 |
| Column | 6.15 | 24.85 | 0.14 | * | NR | 31.14 |
| Salting out | 2.79 | 20.71 | 0.03 | NR | NR | 23.53 |
NR: not required;
: provided in the kit.
To obtain values of mean and standard deviation, calculations of descriptive measures were used. The quantitative variables were tested for normality by the Kolmogorov-Smirnov test. Mann-Whitney U test was used to compare results among species. Values of the quantity and quality of the DNA from the four extraction methods were compared by the Kruskal-Wallis test; multiple comparisons between methods were analyzed using Dunn's test. Kruskal-Wallis and Dunn's test were performed with the support of the program GraphPad Prism (GraphPad, version 5) and other tests, with the program Statistical Package for the Social Sciences (SPSS Statistics, version 17). The significance level used was 5%.
RESULTS AND DISCUSSION
The increasing use of molecular methods on clinical microbiology further emphasizes the necessity of efficient DNA purification, free of proteins and high molecular weight cellular debris5,10. Several studies have shown the importance of the parameter “quality” in obtaining better results on techniques like RAPD (Random Amplification of Polymorphic DNA) and RFLP (Restriction Fragment Length Polymorphism)1,4,11,17. Furthermore, differences between gram-positive and gram-negative bacteria should also be considered when choosing the DNA extraction method10, a fact that prevents the use of many low-cost protocols indicated for any type of microorganisms11. This study aimed to identify the most suitable method for DNA extraction for CoNS evaluating mainly the quality of the material obtained from four different methodologies.
The agarose gel electrophoresis showed better results for the samples
extracted by the method of extraction column. Although some samples evidenced DNA
fragmentation, on almost all samples extracted by this method it was possible to
visualize the DNA. Samples extracted by boiling, phenol-chloroform and salting out
showed the same quality range, but inconstant presence. On the other hand, comparing the
four methods by spectrophotometry, samples extracted by boiling demonstrated more DNA
than the others (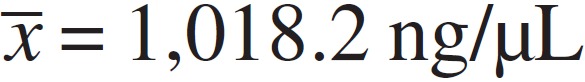 ) (Table 1). The average amount
of DNA extracted by the phenol-chloroform method was a little bit higher than by
extraction column, 87.8 and 65.5 ng/µL, respectively, but this was not statistically
significant (p > 0.05). Likewise, according to Kruskall-Wallis test,
the average A280/230 and A260/230 differences was statistically significant between all
four methods (p < 0.0001); except for average A260/280 between
extraction column and salting out and the average A260/230 between phenol-chloroform and
salting out (p > 0.05) (Table
3). Finally, all four methods provided effective DNA for PCR amplification
with the pair of primers used. The percentage of positive amplification results in one
single reaction performed was 94% for the extraction column method, 91% for the salting
out, 83% for the boiling, and 71% for the phenol-chloroform.
) (Table 1). The average amount
of DNA extracted by the phenol-chloroform method was a little bit higher than by
extraction column, 87.8 and 65.5 ng/µL, respectively, but this was not statistically
significant (p > 0.05). Likewise, according to Kruskall-Wallis test,
the average A280/230 and A260/230 differences was statistically significant between all
four methods (p < 0.0001); except for average A260/280 between
extraction column and salting out and the average A260/230 between phenol-chloroform and
salting out (p > 0.05) (Table
3). Finally, all four methods provided effective DNA for PCR amplification
with the pair of primers used. The percentage of positive amplification results in one
single reaction performed was 94% for the extraction column method, 91% for the salting
out, 83% for the boiling, and 71% for the phenol-chloroform.
Table 3. Mean results of each extraction method regarding parameters evaluated.
| Parameter | Boiling | Phenol-chloroform | Extraction column | Salting out | p |
|---|---|---|---|---|---|
| Agarose gel electrophoresis | Variable* | Variable* | Positive | Variable* | |
| Amount of DNA in ng/µL | 1018.2 (168.8) | 87.8 (70.2) | 65.5 (64.5) | 28.2 (32.5) | < 0.0001 |
| A260/280 | 0.42 (0.03) | 1.42 (0.15) | 1.95 (0.26) | 1.82 (0.44) | < 0.0001 |
| A260/230 | 2.99 (0.72) | 0.66 (0.31) | 1.34 (0.66) | 0.81 (0.45) | < 0.0001 |
| PCR | Positive | Positive | Positive | Positive | |
| Yeld | 200 µL | 100 µL | 200 µL | 100 µL |
Some samples showed no visible bands; ( ) = Standard deviation.
The bands visualized by the four extraction methods in agarose gel electrophoresis showed the same quality parameters. The great majority of the bands were well defined and not fragmented. However, some samples, mainly the ones extracted by boiling, phenol-chloroform, and salting out showed no visible bands on the gel. Surprisingly, in the spectrophotometry those samples showed to hold an amount of DNA detectable using the method of agarose gel electrophoresis. Regarding agarose gel electrophoresis, similar results were found by NOGUEIRA et al. 11 and as reported by them the absence of bands on the gel was not determinant for obtaining a successful PCR. On the boiling method, the high protein contamination can lead to an overestimation of the real concentration of DNA14. Besides that, DNA fragmentation by the high temperatures used on this method can be responsible for the low visualization of bands on agarose gels.
Analysis by spectrophotometry can reveal important information about the DNA extracted from one sample. Besides determining the DNA amount (ng/µL), the A260/280 can be used to assess the DNA purity concerning the protein contamination. Pure DNA preparations have a A260/280 of 1.8. Similarly, significant absorbance at 230 nm decreases the values of A260/230 and indicates contamination with polyphenols and organic compounds14. So, although the boiling method presented the greatest average amount of DNA extracted, this may not be true since the very low value of A260/280 (0.42) evidences a high protein contamination (Table 3). In the same way, looking for A260/230 we also can see a low value (0.66) for the phenol-chloroform extraction method. In this case, this high presence of organic solvents can be responsible for the low PCR amplification (71%). The extraction column method showed the best results regarding quality and purity of the DNA extracted, which partially explains the better results obtained on the PCR amplification.
Calculations of the standard deviation values demonstrated that the quality of DNA obtained in each one of the four methods did not change much (normal distribution). In contrast, the quantitative values on phenol-chloroform, extraction column, and salting out had higher variation resulting on a significant standard deviation (Table 2). The reason for this is the different amount of DNA obtained in each species. Comparing each quantitative result with the respective specie, some interesting observations can be made. The mean amount of DNA extracted by the extraction column method from Staphylococcus epidermidis was 100.1 ng/µL, greater than the overall average of other species (p < 0.001) (Table 1). In the specific case of this method, this finding is important because Staphylococcus epidermidis is the most common CoNS clinical isolate12. Nevertheless, the boiling method didn't show significant differences among species. On the other hand, Staphylococcus haemolyticus and Staphylococcus warneri had the largest average number by phenol-chloroform and salting out, respectively and Staphylococcus capitis, the lower by extraction column. However, our results cannot explain those findings and further studies are needed to investigate the reason of those variations.
Using the pair of primers described by JENSEN et al. 8 and the DNA obtained by the methods used in this study we were able to perform the proposed PCR. However, using similar methods of boiling and phenol-chloroform, NOGUEIRA et al. 11 found no positive results in a RAPD-PCR for the gram-positive samples, only for the gram-negative. Nevertheless, ALEXOPOULOU et al. 1 published a study in 2006 in which they successfully used the same boiling method used by us and performed a CoNS identification by RFLP-PCR.
Currently there are numerous in-house protocols and commercial kits based on different principles for DNA extraction from bacteria. Nevertheless, the number of comparative studies between these methods is still scarce5,10. Taking the difficulty to lyse the CoNS cell wall into account, we evaluated four methods based on different principles and found significant differences between the quality and the amount of the DNA extracted. Quality of the DNA extracted is important when choosing the extraction method to be used for those species, especially if we consider posterior sequencing, long term storage or the construction of genomic libraries for future tests. Considering these parameters extraction column method has proven to be the best alternative when extracting DNA from CoNS. However, when working with CoNS the choice of the best method should be made thinking about both the purpose of the investigation and the future perspectives, taking into consideration the methods yield and the resources available. In-house protocols are cheaper and can be used mainly for quick determinations, but the quality of the material makes commercial kits worthwhile in deeper investigations. We hope that the results presented in this study could help other researches or professionals to make this important decision.
Acknowledgments
The authors thanks to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Rio Grande do Sul (FAPERGS) for financial support.
Footnotes
Author Contributions KCR and TGSP participated in the discussions during the study, elaboration of statistical analyses, figure and tables, and helped to draft the manuscript. CFO designed the study, carried out the molecular biology procedures and drafted the manuscript. AR participated in the design of the study, elaboration of statistical analyses, figure and tables and helped to carry out the spectrophotometry analyses. PAd’A participated in the design and coordination of the study. All authors read and approved the final manuscript.
REFERENCES
- 1.Alexopoulou K, Foka A, Petinaki E, Jelastopulu E, Dimitracopoulos G, Spiliopoulou I. Comparison of two commercial methods with PCR restriction fragment length polymorphism of the tuf gene in the identification of coagulase-negative staphylococci. Lett Appl Microbiol. 2006;43:450–454. doi: 10.1111/j.1472-765X.2006.01964.x. [DOI] [PubMed] [Google Scholar]
- 2.Cheng HR, Jiang N. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 2006;28:55–59. doi: 10.1007/s10529-005-4688-z. [DOI] [PubMed] [Google Scholar]
- 3.Couto I, Pereira S, Miragaia M, Sanches IS, Lencastre H de. Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR. J Clin Microbiol. 2001;39:3099–3103. doi: 10.1128/JCM.39.9.3099-3103.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes BA. Introducing a molecular test into the clinical microbiology laboratory – development, evaluation, and validation. Arch Pathol Lab Med. 2003;127:1106–1111. doi: 10.5858/2003-127-1106-IAMTIT. [DOI] [PubMed] [Google Scholar]
- 5.Giraffa G, Rossetti L, Neviani E. An evaluation of chelex-based DNA purification protocols for the typing of lactic acid bacteria. J Microbiol Methods. 2000;42:175–184. doi: 10.1016/s0167-7012(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 6.Goldenberger D, Perschil I, Ritzler M, Altwegg M. A simple “universal” DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. Genome Res. 1995;4:368–370. doi: 10.1101/gr.4.6.368. [DOI] [PubMed] [Google Scholar]
- 7.Huber H, Ziegler D, Pflüger V, Vogel G, Zweifel C, Stephan R. Prevalence and characteristics of methicillin resistant coagulase-negative staphylococci from livestock, chicken carcasses, bulk tank milk, minced meat, and contact persons. BMC Vet Res. 2011;7:6. doi: 10.1186/1746-6148-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen MA, Webster JA, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloos WE, Bannerman TL. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McOrist AL, Jackson M, Bird AR. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J Microbiol Methods. 2002;50:131–139. doi: 10.1016/s0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 11.Nogueira CAM, Momesso CAS, Machado RLD, Almeida MTG de, Rossit ARB. Desempenho de kits comerciais e protocolos laboratoriais para a extração de DNA genômico bacteriano. Rev Panam Infectol. 2004;6:35–38. [Google Scholar]
- 12.Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009;23:73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Salton MR. Studies of the bacterial cell wall. IV. The composition of the cell walls of some Gram-positive and Gram-negative bacteria. Biochim Biophys Acta. 1953;10:512–523. doi: 10.1016/0006-3002(53)90296-0. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Russel DW. Quantitation of nucleic acids. In: Sambrook J, Russell DW, editors. Molecular cloning: a laboratory manual. 3rd ed. Vol. 2001. New York: Cold Spring Harbor Laboratory Press; pp. A8.19–A8.24. [Google Scholar]
- 15.Secchi C, Antunes AL, Perez LR, Cantarelli VV, d'Azevedo PA. Identification and detection of methicillin resistance in non-epidermidis coagulase-negative staphylococci. Braz J Infect Dis. 2008;12:316–320. doi: 10.1590/s1413-86702008000400012. [DOI] [PubMed] [Google Scholar]
- 16.Silva ER, Silva N. Coagulase gene typing of Staphylococcus aureus isolated from cows with mastitis in southeastern Brazil. Can J Vet Res. 2005;69:260–264. [PMC free article] [PubMed] [Google Scholar]
- 17.Tyler KD, Wang G, Tyler SD, Johnson WM. Factors affecting reliability and reproducibility of amplification based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilfinger W. Extended bactozol enzyme solution™ lysis procedure for the isolation of genomic DNA from recalcitrant gram-positive bacteria. Molecular Research Center Technical Bulletin No. 8. [[Cited 2012 October]]. Available from: URL: http://mrcgene.com/tb8-grampositive.htm.
- 19.Yang G, Erdman DE, Kodani M, Kools J, Bowen MD, Fields BS. Comparison of commercial systems for extraction of nucleic acids from DNA/RNA respiratory pathogens. J Virol Methods. 2011;171:195–199. doi: 10.1016/j.jviromet.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Wilfinger W. Extended bactozol enzyme solution™ lysis procedure for the isolation of genomic DNA from recalcitrant gram-positive bacteria. Molecular Research Center Technical Bulletin No. 8. [[Cited 2012 October]]. Available from: URL: http://mrcgene.com/tb8-grampositive.htm.


