Summary
Using phylogenetics and transcriptomics, we show that independent origins of both CAM and C4 photosynthesis in Caryophyllales co-opted the same genes for PEPC through similar adaptive changes.
Key words: C4 photosynthesis, CAM photosynthesis, co-option, evolution, phosphoenolpyruvate carboxylase (PEPC), phylogenetics.
Abstract
CAM and C4 photosynthesis are two key plant adaptations that have evolved independently multiple times, and are especially prevalent in particular groups of plants, including the Caryophyllales. We investigate the origin of photosynthetic PEPC, a key enzyme of both the CAM and C4 pathways. We combine phylogenetic analyses of genes encoding PEPC with analyses of RNA sequence data of Portulaca, the only plants known to perform both CAM and C4 photosynthesis. Three distinct gene lineages encoding PEPC exist in eudicots (namely ppc-1E1, ppc-1E2 and ppc-2), one of which (ppc-1E1) was recurrently recruited for use in both CAM and C4 photosynthesis within the Caryophyllales. This gene is present in multiple copies in the cacti and relatives, including Portulaca. The PEPC involved in the CAM and C4 cycles of Portulaca are encoded by closely related yet distinct genes. The CAM-specific gene is similar to genes from related CAM taxa, suggesting that CAM has evolved before C4 in these species. The similar origin of PEPC and other genes involved in the CAM and C4 cycles highlights the shared early steps of evolutionary trajectories towards CAM and C4, which probably diverged irreversibly only during the optimization of CAM and C4 phenotypes.
Introduction
During the evolutionary diversification of organisms, ecological selection pressures sometimes lead to the emergence of similar phenotypes in distantly related species. A good example of such convergent evolution is the recurrent emergence of CO2-concentrating mechanisms (CCMs) as an adaptation to environmental CO2 depletion (Raven et al., 2008). CCMs have arisen through the assembly of novel biochemical pathways, which increase the internal concentration of CO2 around Rubisco before its fixation by the C3 photosynthetic cycle (Christin and Osborne, 2013). In flowering plants, the most frequent and successful CCMs are C4 and CAM photosynthesis (Keeley and Rundel, 2003). The C4 and CAM CCMs differ in the overall mechanism of atmospheric CO2 concentration, but the biochemical cycles are similar (Osmond, 1978; Hatch, 1987). Plants of both types fix inorganic carbon by phosphoenolpyruvate carboxylase (PEPC). In C4 plants, the resulting acid is typically modified and transported to another cell, where CO2 is released to feed the C3 cycle that is active within these cells (Hatch, 1987; Sage et al., 2012). In CAM plants, a similar CCM occurs in which the initial carboxylation by PEPC and subsequent decarboxylation and refixation by Rubisco are temporally rather than physically separated. At night, PEPC fixes inorganic carbon into organic compounds, which are stored as malate in the vacuole until daytime, when the cycle is completed and CO2 is released to supply the C3 cycle (Osmond, 1978; Borland and Taybi, 2004).
The CAM and C4 pathways are complex traits, involving dozens of enzymes that fulfil different functions compared with the isoforms in the C3 ancestors (Bräutigam et al., 2011; Gowik et al., 2011). For the constituent enzymes investigated so far, the new function requires alterations in the expression pattern and/or its catalytic properties (Finnegan et al., 1999; Tausta et al., 2002; Svensson et al., 2003; Gowik and Westhoff, 2011; Ludwig, 2011). For instance, the PEPC enzyme is ubiquitous in plants, where multiple isoforms are responsible for various non-photosynthetic functions (Lepiniec et al., 1994; Aubry et al., 2011; Gowik and Westhoff, 2011). The C4-specific forms evolved from non-photosynthetic genes through adaptation of the catalytic properties to the new metabolic context (Dong et al., 1998; Gowik and Westhoff, 2011). In particular, the affinity of PEPC for its substrate PEP was decreased, and its sensitivity to feedback inhibition by malate was reduced (Bauwe and Chollet, 1986; Bläsing et al., 2000; Svensson et al., 2003; Gowik et al., 2006; Lara et al., 2006). In C4 grasses and sedges, this was achieved through numerous adaptive amino acid changes (Christin et al., 2007; Besnard et al., 2009).
Both the CAM and C4 pathways also require a specialized leaf anatomy (Hattersley, 1984; Nelson et al., 2005). Despite this apparent complexity, the C4 trait evolved a minimum of 62 times independently in flowering plants (Sage et al., 2011). While a precise tally is not yet available, it is likely that the total number of CAM origins will be even greater (Edwards and Ogburn, 2012). These numerous origins of novel photosynthetic types are, however, not evenly distributed across the phylogeny of flowering plants. While certain major lineages completely lack CCMs, others present a large number of independent origins of CAM, C4 or both (Crayn et al., 2004; Sage et al., 2011; Edwards and Ogburn, 2012). The high occurrence of C4 origins in some groups of angiosperms has been explained by different factors, with an emphasis on ecological and anatomical predispositions, as well as possible genomic predispositions (Sage, 2001; Monson, 2003; Christin et al. , 2013a, 2013b; Griffiths et al., 2013). The existence of anatomical and/or ecological predispositions for CAM evolution might similarly explain the repeated incidence of this CCM in some groups (Edwards and Ogburn, 2012), although this has never been rigorously tested.
The frequency of occurrence of CCMs is particularly high in the eudicot clade Caryophyllales, which encompasses many C4 origins (Kadereit et al., 2003; Christin et al., 2011; Sage et al., 2011; Kadereit et al., 2012) and also several CAM lineages, including constitutive CAM (e.g. cacti) and facultative CAM types that can switch to CAM depending on environmental conditions (Guralnick and Ting, 1987; Guralnick et al., 2008; Nyffeler et al., 2008). Of special interest are species of Portulaca, which are the only plants known to be capable of performing both C4 and CAM cycles (Koch and Kennedy, 1980, 1982; Guralnick et al., 2002). The majority of Portulaca species are C4 plants, with the associated anatomical specialization, but several species exhibit CAM-like physiology when grown in water-limited conditions (Kraybill and Martin, 1996; Guralnick et al., 2002). Exposure to drought triggers physiological and biochemical changes in these Portulaca species, with different expression levels and catalytic properties of several C4/CAM enzymes, and slight alterations in their leaf anatomy (Mazen, 1996, 2000; Lara et al., 2003, 2004). In molecular phylogenies, Portulaca is nested within Portulacineae (Nyffeler and Eggli, 2010; Ocampo and Columbus, 2010; Arakaki et al., 2011), and is apparently the only C4 member of this group that is otherwise rich in species with varying degrees of CAM photosynthesis, the best known of which are cacti. Despite these intriguing patterns, neither sequence nor expression data for known C4/CAM genes have been analysed in Portulaca or its relatives.
The large number of CCM origins within Caryophyllales might suggest that this clade is especially prone to transitions between photosynthetic types. However, the history of photosynthetic transitions within the clade is still unclear. In particular, it is unknown if CAM and C4 origins represent completely independent evolutionary phenomena, or are distinct end-points to a partially shared evolutionary trajectory (Edwards and Ogburn, 2012). To increase our understanding of CCM origins in Caryophyllales, we studied the evolution of genes encoding PEPC, a key enzyme common to all CAM and C4 cycles. In addition, we investigated in detail the transcriptome of Portulaca individuals operating either the C4 or CAM cycles. Our comparative analyses shed new light on the shared history of genes involved in CAM and C4 photosynthesis.
Material and methods
Diversity of genes encoding PEPC in plants
To reconstruct the history of the multigene family encoding PEPC, complete cDNA sequences available in the GenBank database were first retrieved. These were used as a query of BLAST searches against completely sequenced nuclear genomes available on Phytozome (Goodstein et al., 2012). This initial dataset was increased by screening genomic DNA from representatives of diverse Caryophyllales lineages and photosynthetic types through PCR to isolate genes encoding PEPC (Supplementary Tables S1 and S2, available at JXB online). These DNAs were first screened with primers ppc-1204-For and ppc-2890-Rev previously used to isolate PEPC genes from Molluginaceae, a family from Caryophyllales (Christin et al., 2011). These primers amplify a fragment encompassing exons 8 to 10, which represents about half of the whole coding sequence and is known to include major determinants of the C4-specific properties of PEPC (Bläsing et al., 2000; Engelmann et al., 2002). PCR, cloning and sequencing were performed as previously described (Christin et al., 2011). Preliminary analyses indicated that numerous PEPC genes were present in some Caryophyllales genomes. To increase the likelihood of sampling specific copies, additional primers were designed with the purpose of increasing the PCR specificity for certain gene lineages (Supplementary Table S3, available at JXB online). These additional PCRs were conducted as for the primers ppc-1204-For and ppc-2890-Rev. The PCR products were purified and directly sequenced with one of the primers used for the PCR. Chromatograms were visually inspected and PCR products were cloned only if multiple genes were detected by the presence of overlapping peaks.
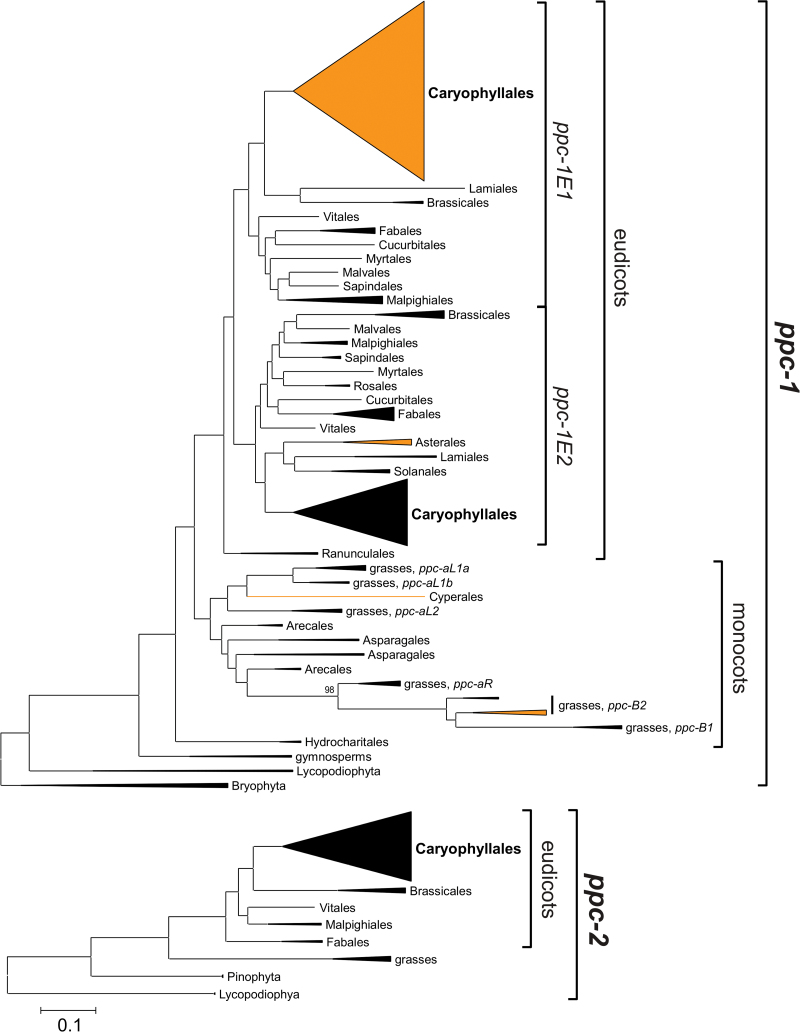
Exons were identified by homology to annotated sequences and following the GT-AG rule. All coding sequences, retrieved from public databases or isolated through PCR, were translated into protein sequences and aligned with ClustalW (Thompson et al., 1994). The alignment was visually inspected, manually refined, and replaced with the corresponding nucleotide sequences, which were later used for analyses. A preliminary phylogenetic tree identified two distantly related groups of PEPC encoding genes that diverged before the evolution of land plants (ppc-1 and ppc-2; Fig. 1), each represented by ferns, gymnosperms, and flowering plants. Despite clear homology, the two groups were highly divergent, leading to ambiguities in the alignment. Each group was consequently analysed separately.
Fig. 1.
Phylogenetic relationships among PEPC-encoding genes from land plants. The phylogenetic trees were obtained through Bayesian inference on each main gene lineage; ppc-1 and ppc-2. Taxonomic groups are compressed, with the size of triangles proportional to the number of sequences in the group. Gene lineages and main clades of flowering plants are delimited on the right. Clades containing some genes with a Ser780 are in orange. Details, including support values, are available in Supplementary Figs S1 and S2 (available at JXB online). The scale bar represents expected substitutions per site. (This figure is available in colour at JXB online.)
Transcriptome analysis of Portulaca oleracea
The species Portulaca oleracea was selected for transcriptome analyses to identify CAM- and C4-specific genes, with a special focus on PEPC. Seeds originating from Syria were provided by the USDA-ARS (GRIN accession: Grif 14515). Plants were grown in 3-inch pots of equal-parts gravel/calcinated clay perlite mix, and in a Conviron E7/2 plant growth chamber (Conviron Ltd., Winnipeg, Manitoba, Canada) with 14 hours of daylight. The growth chamber was illuminated with twelve 32-W fluorescent lamps and four 60-W incandescent lamps. Temperature was kept at 22°C from 3h before dark until after 3h of light, and was increased to 28°C for the middle portion of the light period. The position of pots within the growth chamber was randomized daily.
On the first day of the experiment (5 March 2012), all plants were bottom-watered and seedlings were split into two groups. One group was bottom-watered every 2–4 days, while the other group was bottom-watered less than once a week (Supplementary Table S4, available at JXB online). Nutrients were added to the water periodically at a concentration of 1:100 (w/v) of K:P:N=20:20:20. After 1 month under these conditions, leaves were collected, flash-frozen in liquid nitrogen, and stored at –80°C. Two individuals per group were sampled twice, after 4h of light and after 2h of dark. To control for stress effects that might have been induced by leaf cutting, the first individual from each group was sampled first during the day and then the following night, while the second individual was sampled first during the night and then the following day. An equivalent number of young and mature leaves were collected from the light and dark samples. Two additional individuals per group (watered frequently and watered occasionally) were sampled for acid titration (see below). One individual of each group was sampled first at the end of the dark period (1h before light) and then at the end of the light cycle (2h before dark). The sampling order was inverted for the second individual of the same group.
RNA was extracted from several leaf fragments using the FastRNA™ Pro Green Kit (MP Biomedicals, OH, USA). Several RNA extractions per sample (one individual in one condition) were pooled and prepared for sequencing using the Illumina TruSeq mRNA Sample Prep Kit (Illumina Inc., San Diego, CA, USA). Fragments of the cDNA libraries between 400 and 450bp long were selected, and the different samples were marked with specific bar-codes, multiplexed and sequenced using an Illumina HiSeq 2000 instrument, as paired-end 100bp reads.
Titratable acidity
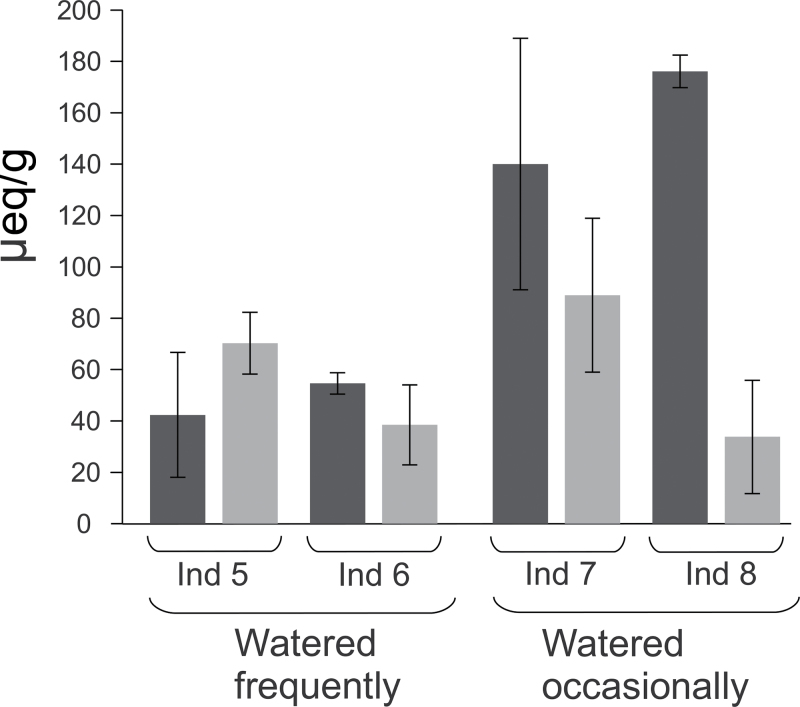
The titratable acidity was measured at the end of the dark phase and at the end of the light phase. An accumulation of acids during the night followed by consumption during the day is indicative of a CAM cycle (Silvera et al., 2005). Three leaves were analysed for each of the eight combinations of individuals/sampling time. Frozen leaves were weighed and immediately ground briefly in mortar and pestle and then boiled in 50ml 20% ethanol. After cooling, pH was slowly brought to 7 by adding 0.1 N NaOH in 5 or 10μl increments. The titratable acidity was calculated as the amount of H+ equivalents required to neutralize the leaf extracts (Silvera et al., 2005). Despite variability among individuals and leaves, the plants watered occasionally showed a clear increase of titratable acidity at the end of the dark period (Fig. 2). These results confirm that less frequent watering triggered a CAM physiology in Portulaca oleracea.
Fig. 2.
Effect of the water regime on titratable acidity. The titratable acidity (in microequivalents per gram of frozen weight, μeq/g) is indicated for samples taken at the end of the dark phase (dark grey) and at the end of the light phase (light grey). The error bars indicate standard deviations over three replicates.
Illumina data assembly and phylogenetic annotation
The reads corresponding to each of the eight different samples were assembled individually using the software Trinity (Grabherr et al., 2011) as implemented in the Agalma pipeline (Dunn et al., 2013). Reads from each Illumina run were mapped against the corresponding individual assembly using the software Bowtie 2 (Langmead and Salzberg, 2012) and its mixed model, which allows unpaired alignments when paired alignments fail. Only one of the best alignments chosen randomly was reported per read. The number of reads aligned to each contig was used to compute reads per million of alignable reads (rpm). Sequencing and mapping statistics are given in Supplementary Table S5 (available at JXB online).
The relative transcript abundance of each gene within each of the C4-related multigene families was estimated by assigning contigs to gene lineages based on phylogenetic analyses (Christin et al., 2013a). For genes encoding C4-related enzymes (Supplementary Table S6, available at JXB online), sequences available in Genbank for Caryophyllales and other plant lineages were extracted based on their annotation. A BLAST search was used to extract homologous loci for predicted cDNA from several completely sequenced genomes (Arabidopsis thaliana, Brachypodium distachyon, Carica papaya, Glycine max, Oryza sativa, Populus trichocarpa, Sellaginella moellendorffii, Sorghum bicolor, Selaginella moellendorffii and Vitis vinifera). For each C4-related enzyme, the dataset assembled from sequences retrieved from Genbank and complete genomes was used as a query of a BLAST search against each of the assembled transcriptomes. The longest matching region for each contig was extracted from the BLAST results when larger than 50bp. Each of these was successively aligned to the reference dataset using Muscle (Edgar, 2004). The resulting alignment was used to infer one phylogenetic tree per contig under maximum likelihood as implemented in PhyML (Guindon and Gascuel, 2003) using a GTR+G model. The resulting phylogenetic trees were inspected and each contig was assigned to a gene lineage if clearly nested within. The rpm values of all contigs assigned to a given gene lineage were summed to obtain an estimate of the transcript abundance of the gene lineage.
The assignment of contigs to gene lineages was repeated with a reference dataset composed of exons 8–10 of genes encoding PEPC. Caryophyllales sequences isolated by PCR or retrieved from Genbank were used if complete for the studied fragment and assigned to the gene lineage ppc-1E1 based on phylogenetic analyses (see Results). In order to increase the size of this dataset, the corresponding segment of matching contigs from Portulaca transcriptomes that represented at least half of the studied fragment were manually aligned with the dataset. Three contigs that covered the whole studied segment and which were clearly different from each other, as well as from Portulaca genes isolated by PCR, were added to the reference dataset. All matching Portulaca contigs were then successively placed in a phylogeny with this reference dataset and the total rpm values of each ppc-1E1 gene lineage was computed as described above.
Screening of ppc genes from other Portulaca transcriptomes
The 1KP project has sequenced transcriptomes from 1000 different species of plants, including several Portulaca species (http://www.onekp.com/; Johnson et al., 2012). RNA for Portulaca species was extracted from leaves sampled during the day. Of these, P. cryptopetala is a putative C3–C4 intermediate species (Voznesenskaya et al., 2010). Three additional clades of Portulaca are represented in the 1KP project (clades based on Ocampo and Columbus, 2012 and Ocampo et al., 2013); ‘Oleracea’ (P. molokiniensis, P. oleracea, P. suffruticosa), ‘Pilosa’ (P. grandiflora, P. amilis), and ‘Umbraticola’ (P. umbraticola). Sequences of the different Portulaca species corresponding to ppc-1E1 were retrieved through a BLAST search with ppc-1E1 isolated in the present study used as a query. Sequences were considered further only if they aligned with the reference dataset along more than 500bp. The selected sequences were aligned with the ppc-1 sequences isolated from genomic DNA or extracted from the P. oleracea transcriptome generated in this study (see above). The alignment was manually refined and only the longest of groups of identical sequences from the 1KP data was used for analyses.
Phylogenetic analyses and codon models
The two datasets composed of all ppc-1 and ppc-2 genes retrieved from available databases or generated in this study were used to infer phylogenetic trees with MrBayes 3.2.1 (Ronquist and Huelsenbeck, 2003). The general time reversible model of nucleotide substitution with a gamma-shape parameter and a proportion of invariants (GTR+G+I) was used. For ppc-2, two parallel analyses each composed of four chains were run for 20000000 generations, sampling a tree every 1000 generations after a burn-in period of 10000000 generations. Due to slow convergence of parallel runs, the number of parallel chains was increased to sixteen for analyses of ppc-1. For this dataset, two different analyses were run for 10000000, sampling a tree each 1000 generations after a burn-in period of 4000000 generations. Consensus trees were computed from all trees sampled after the burn-in period. Convergence of the analyses and adequacy of the burn-in period were determined using Tracer (Rambaut and Drummond, 2007).
In order to represent the rate of amino acid changes, the topologies inferred from the whole ppc-1 and ppc-2 datasets were used to infer branch lengths based on amino acid sequences while keeping the topology constant. These analyses were performed in PhyML, under a JTT+G substitution model. Statistical tests of adaptive evolution were also conducted on the group of ppc-1E1 sequences from Caryophyllales using codon models implemented in codeml of the PAML package (Yang, 2007). These models use the ratio of non-synonymous mutation rate per synonymous mutation rate (ω) as a proxy of selective pressures. An ω smaller than 1 indicates purifying selection, a value of 1 indicates relaxed selection, and a value greater than 1 indicates adaptive evolution. Different models allow ω to vary among sites of the protein or among both sites and branches of the phylogeny (Yang and Nielsen, 2002). The site model without adaptive evolution (M1a) was compared with the site model assuming adaptive evolution on some sites but on all branches (M2a), as well as to several branch-site models assuming adaptive evolution only on some sites and on some branches (referred to as foreground branches; model A). In these branch-site models, foreground branches have to be defined a priori. Different sets of foreground branches were successively selected, and the likelihoods were compared using Akaike information criteria (AIC). In this first model (a), foreground branches were defined as each branch leading to a group of putative C4 forms, to the group of sequences belonging to the CAM genera Mesembryanthemum and Drosanthemum, or to the group of sequences belonging to Portulaca and present at high transcript abundance in the night samples (putative CAM form; see Results). In the other models, entire gene lineages present in Portulacineae were successively added to the foreground branches: (b) branches a + ppc-1E1c, (c) branches b + ppc-1E1d + ppc-1E1e, (d) branches c + ppc-1E1b, and (e) branches d + ppc-1E1a.
Since the evolutionary history of C4 photosynthesis in the genus Portulaca is debated (Ocampo et al., 2013), the group of Portulaca sequences that contains putative C4 forms (ppc-1E1a and ppc-1E1a’) was analysed in more detail. Parallel adaptive genetic changes can bias the phylogenetic reconstruction when they occur in closely related taxa, and considering only third positions of codons can help recover the true phylogenetic relationships (Christin et al., 2007, 2012). To avoid a potential bias due to adaptive evolution, a phylogenetic tree was inferred as described above but considering only third positions of codons from Portulaca ppc-1E1a and ppc-1E1a’ sequences. The inferred topology was used to test alternative scenarios of adaptive evolution using the codon models described above. In addition, to the site models M1a and M2a, different branch-site models (A) were compared. Foreground branches were set following three evolutionary scenarios: adaptive evolution at the base of Portulaca (on the branch leading to ppc-1E1a’; single C4 optimization), at the base of each C4 clade of Portulaca ppc-1E1a’ (multiple C4 optimizations); and finally at the base of each C4 and C3–C4 clade of Portulaca ppc-1E1a’ (multiple C4 and C3–C4 optimizations).
Results
PEPC multigene family
The phylogenetic tree inferred using all genes encoding PEPC retrieved from GenBank, the 1KP project, or generated in this study showed that a gene duplication occurred before the emergence of land plants and produced two groups of distantly related genes present in most plant genomes (ppc-1 and ppc-2; Fig. 1; Gowik and Westhoff, 2011). One of these groups (ppc-1) contains all the C4-specific PEPC genes documented so far (Rao et al., 2002; Svensson et al., 2003; Gowik et al., 2006; Christin et al., 2007, 2011; Besnard et al., 2009; Gowik and Westhoff, 2011). The phylogenetic relationships in each group are compatible with the species relationships predicted from other markers (Supplementary Figs S1 and S2, available at JXB online; Soltis et al., 2011). The gene ppc-2 is present as a single copy in all the species considered (Fig. S2). No gene duplication of ppc-1 is detectable before the split of eudicots and monocots, but several gene duplications led to the six gene lineages present in grass genomes as shown previously (namely ppc-aL1a, ppc-aL1b, ppc-aL2, ppc-aR, ppc-B1, and ppc-B2; Fig. S1; Christin and Besnard 2009). In addition, a gene duplication occurred soon after the early diversification of eudicots, leading to two gene lineages present in most eudicots and named ppc-1E1 and ppc-1E2 (Fig. 1 and Supplementary Fig. S1, available at JXB online; these correspond to ppc-1 and ppc-2 in Christin et al., 2011).
Diversification of ppc-1E1 in Caryophyllales
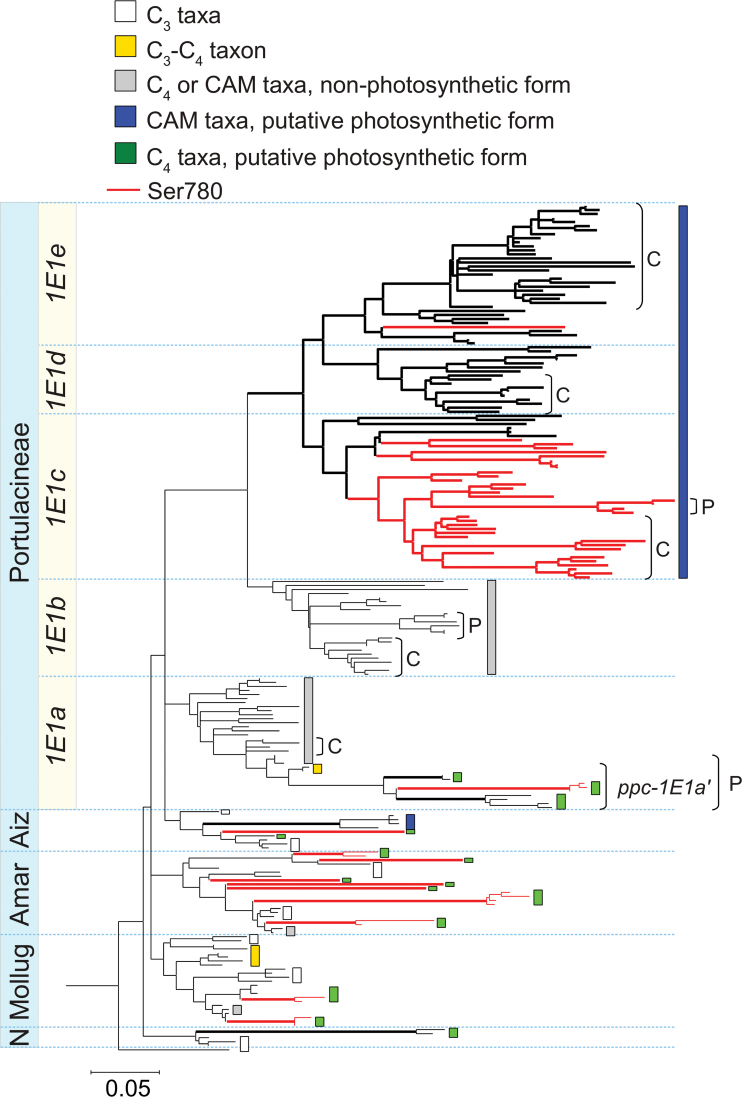
Genes belonging to the three ppc gene lineages present in eudicots were isolated from core Caryophyllales through PCR screening of genomic DNA (Supplementary Tables S1 and S2, available at JXB online). The phylogenetic trees of Caryophyllales ppc-2 and ppc-1E2 were consistent with published phylogenies based on chloroplast and nuclear markers (Brockington et al., 2009; Arakaki et al., 2011; Kadereit et al., 2012) and no ancient gene duplication was detectable (Supplementary Figs S1 and S2, available at JXB online). On the other hand, ppc-1E1 is present in a high number of copies in members of the Portulacineae (namely ppc1E1a to ppc-1E1e in Fig. 3 and Supplementary Fig. S1, available at JXB online), indicating that this gene lineage was repeatedly duplicated during the early diversification of this clade. The species relationships deduced from each of these gene lineages are consistent with those deduced from chloroplast markers (Arakaki et al., 2011). The ppc-1E1a gene lineage includes sequences isolated previously from cDNA of two members of Portulacineae, Pereskia aculatea and Selenicereus vitii (Gehrig et al., 2001).
Fig. 3.
Evolution of ppc-1E1 in Caryophyllales. The topology was inferred on nucleotide sequences, but branch lengths were estimated based on amino acid sequences. The branch lengths inferred on nucleotide sequences, together with all species names and support values, are available in Fig. S1. Groups of genes encoding a Ser780 are highlighted by red branches. Branches where some sites underwent an excess of non-synonymous mutations according to the best model are thicker. Putative C4 forms are delimited in green and putative CAM forms in blue. These were identified based either on transcript abundances in specific conditions, on the literature, or on an excess of amino acid changes in C4/CAM species. Genes of C4 or CAM taxa that represent putative non-photosynthetic duplicates are delimited in grey, those of C3 taxa in white, and those of C3–C4 taxa in yellow. Families outside Portulacineae and gene lineages for Portulacineae are indicated on the left: N, Nyctaginaceae; Mollug, Molluginaceae; Amar, Amaranthaceae; Aiz, Aizoaceae. Subclades of interest are indicated on the right: P, Portulaca; C, cacti. The full phylogenetic tree is available in Fig. S3. The scale bar represents expected substitutions per site. (This figure is available in colour at JXB online.)
The ppc-1E1 lineage contains genes encoding putative C4-specific forms in Alternanthera (Gowik et al., 2006), Bienertia and Suaeda (Lara et al., 2006), as well as genes encoding putative CAM-specific forms in Mesembryanthemum (Rickers et al., 1989). Most C4-specific PEPC genes studied previously encode a serine residue at the position homologous to position 780 of the maize PEPC protein (numbering based on Zea mays sequence CAA33317; hereafter referred to as Ser780), although this Ser780 is not necessary for the C4 function (Rao et al., 2008). In Flaveria, Ser780 has been shown to be a major determinant of the C4 properties of PEPC (Bläsing et al., 2000; Engelmann et al., 2002; Svensson et al., 2003). The homologous position is occupied by a conserved alanine in all characterized PEPC genes not involved in C4 photosynthesis (Svensson et al., 2003; Christin et al., 2007, 2011; Besnard et al., 2009). In the present dataset, a Ser780 is encoded by ppc-1E1 genes from the C4 taxa in Amaranthaceae, Aizoaceae, and Molluginaceae, while it is not encoded by homologous genes of C3 taxa of the same families and from other gene lineages (Fig. 3 and Supplementary Figs S1 and S2, available at JXB online). This supports earlier suggestions that ppc-1E1 encodes the C4-specific PEPC in numerous C4 Caryophyllales (Gowik et al., 2006; Christin et al., 2011; Gowik and Westhoff, 2011). However, the Ser780 is not encoded by ppc-1E1 genes from some C4 taxa in Nyctaginaceae and Aizoaceae.
Most members of Portulacineae have the ability to perform some degree of CAM (Guralnick et al., 2008; Nyffeler et al., 2008). The ppc-1E1c genes of Portulacineae encode a Ser780 in most species (Fig. 3). Sequences isolated by PCR from cDNA extracted from the stems of Nopalea and Echinocereus at night belong to this gene lineage and encode a Ser780, which indicates that proteins encoded by ppc-1E1c might be involved in the CAM pathway of cacti. This is further supported by the isolation of ppc-1E1c sequences from cDNA of another cactus (Hylocereus undatus; NCBI accession JF966382). However, the Ser780 residue is not encoded by ppc-1E1c genes in the sampled Montiaceae and in several of the sampled Didiereaceae (Supplementary Fig. S1, available at JXB online). In addition, the ppc-1E1e from Ceraria (Didiereaceae; Supplementary Fig. S1, available at JXB online) also encodes a Ser780. All other Portulacineae ppc-1E1 gene copies and all ppc-2 and ppc-1E2 (with the exception of some sequences from Flaveria known to be involved in C4 photosynthesis; Svensson et al., 2003) encode the Ala780 typical of non-C4 genes.
Transcriptome from Portulaca oleracea
Differences in estimates of transcript abundance of C4-related genes between individuals grown under the same watering regime were small (Supplementary Table S6, available at JXB online). In the well-watered samples, some of the gene lineages encoding the enzymes of the C4 pathway (β-CA, PEPC, NAD-MDH, NADP-MDH, ALA-AT, ASP-AT, PPDK, NAD-ME and NADP-ME) were present at high transcript abundance during the day (Supplementary Table S6, available at JXB online). With the exception of NADP-ME, these correspond to the enzymes postulated to play a role in the C4 pathway of P. oleracea (Lara et al., 2004). After 2h in the dark, the transcript abundance of these enzymes remained substantial, although in most cases it was strongly reduced compared with the day sample (Supplementary Table S6, available at JXB online). The only exception is the gene for ALA-AT, which was present at higher transcript abundance in the dark than in the light. In addition, one of the individuals showed a slight increase in the transcript abundance of one of the genes for NADP-MDH at night while its abundance was extremely low in the other individual (Supplementary Table S6, available at JXB online).
In the samples watered less frequently, genes encoding the same enzymes were present at high transcript abundance during the day, although the levels were generally lower than in the well-watered sample (Supplementary Table S6, available at JXB online). At night, genes encoding all C4-related enzymes were present at lower abundance, with the exception of genes for PEPC and NADP-MDH, for which the abundance of one gene lineage increased to reach levels equivalent to, or even higher than, those observed for the well-watered samples during the day (Table 1 and Supplementary Table S6, available at JXB online). This is consistent with the nocturnal part of the purported CAM cycle of P. oleracea, which is based on these two enzymes (Lara et al., 2004). However, the nocturnal part was assumed to also involve carbonic anhydrase, but the transcript abundance of this enzyme is strongly reduced at night although it remains high (Supplementary Table S6, available at JXB online). The transcript abundances suggest that a C4 cycle is present in both well-watered and drought conditions, but it is complemented by a CAM cycle in drought conditions, as indicated previously (Mazen, 2000).
Table 1.
Transcript abundances in rpm of PEPC-encoding genes in Portulaca oleracea grown in different conditions
| Time | Day | Night | Day | Night | ||||
|---|---|---|---|---|---|---|---|---|
| Condition | Watered frequently | Watered occasionally | ||||||
| Individual | 1 | 2 | 1 | 2 | 3 | 4 | 3 | 4 |
| ppc-2 | 13 | 14 | 6 | 1 | 5 | 30 | 7 | 17 |
| ppc-1E2 | 44 | 41 | 40 | 29 | 14 | 10 | 5 | 14 |
| ppc-1E1a | 131 | 139 | 157 | 192 | 167 | 196 | 185 | 173 |
| ppc-1E1b | 0 | 1 | 0 | 7 | 6 | 18 | 0 | 0 |
| ppc-1E1c | 3 | 0 | 820 | 1180 | 277 | 338 | 7602 | 6823 |
| pp c-1E1a’ | 9916 | 4697 | 6710 | 6052 | 7339 | 1868 | 1421 | 869 |
Detailed analysis of P. oleracea PEPC genes
The incorporation of contigs from the P. oleracea samples into the densely sampled Caryophyllales dataset allowed us to estimate the transcript abundance of each ppc-1E1 gene lineage (Table 1). In addition to ppc-1E2 and ppc-2, four distinct ppc-1E1 genes were isolated from the transcriptomes of P. oleracea, only one of which was also isolated from genomic DNA (ppc-1E1b). One of these four genes was clearly nested within ppc-1E1c and two in ppc-1E1a (Supplementary Fig. S1, available at JXB online). The phylogenetic relationships suggest a recent duplication of ppc-1E1a in Portulaca and one of the duplicates was named ppc-1E1a’.
The genes ppc-1, ppc-2, ppc-1E1a, and ppc-1E1b were present at similarly low abundances in all samples (Table 1). By contrast, ppc-1E1a’ was present at very high transcript abundances during the day in the well-watered samples (Table 1). This pattern is consistent with a function in the C4 pathway, which is moreover supported by the Ser780 encoded by the gene. The abundance of ppc-1E1a’ during the day decreased in one of the individuals that were watered less frequently and its abundance at night decreased in both individuals (Table 1). The gene ppc-1E1c was present at extremely low transcript abundances in the well-watered samples during the day. However, its abundance increased at night, and the nocturnal abundance was considerably higher in infrequently watered than in well-watered plants (Table 1). High nocturnal transcript abundance triggered by reduced water availability supports an involvement of the encoded enzyme in the CAM pathway of P. oleracea. This gene also encodes a Ser780.
Distribution of ppc-1E1 genes in other Portulaca species and evidence of adaptive evolution
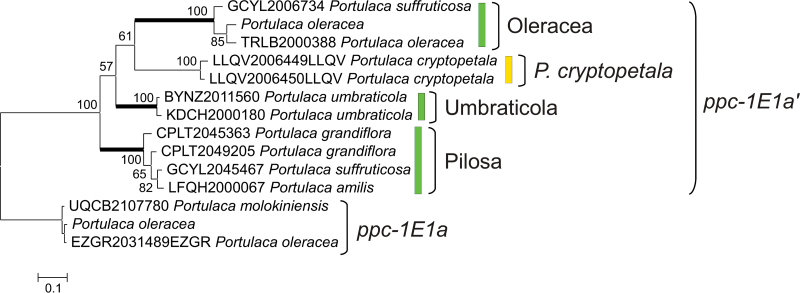
Using the 1KP transcriptome data, sequences corresponding to the genes ppc-1E1a and ppc-1E1c were retrieved from the Oleracea clade, while sequences corresponding to ppc-1E1b were retrieved from both Oleracea and Pilosa clades (Supplementary Fig. S1, available at JXB online). The RNA for this project was isolated during the day and putative CAM-specific genes would likely be missed. The putative C4 gene ppc-1E1a’ was retrieved from the four Portulaca clades. One sequence attributed to P. suffruticosa was nested within ppc-1E1a’ of the Pilosa clade (Fig. 4 and Supplementary Fig. S1, available at JXB online), which might indicate a biologically relevant phenomenon (e.g. hybridization) or a methodological problem (e.g. cross-contamination). Since these sequences were retrieved from leaf RNA isolated during the day, the presence of transcripts corresponding to ppc-1E1a’ is compatible with the hypothesis that this gene is involved in C4 photosynthesis of these different species.
Fig. 4.
Evolution of C4-specific PEPC genes in Portulaca. This phylogeny of Portulaca ppc-1E1a/ppc-1E1a’ was inferred from third positions of codons. Bayesian support values are indicated near branches and clades are delimited on the right. Putative C4 forms are highlighted in green and putative C3–C4 forms in yellow. Thick branches represent inferred episodes of adaptive evolution. The scale bar represents expected substitutions per site. (This figure is available in colour at JXB online.)
The branch lengths estimated from amino acid sequences strongly vary among clades (Fig. 3 and Supplementary Fig. S3, available at JXB online). Since the variation is more pronounced than with nucleotide sequences (Supplementary Figs S1 and S2, available at JXB online), it indicates an excess of non-synonymous mutations. In Molluginaceae and Amaranthaceae, clear increases in the rate of amino acid substitutions occurred on branches leading to genes encoding putative C4-specific enzymes. In Aizoaceae, a similar acceleration is visible on the branch leading to the putative C4 gene of Trianthema, but also to the genes of the CAM plant Mesembryanthemum crystallinum. In addition, the branch leading to the C4 Nyctaginaceae Boerhavia underwent many amino acid changes (Fig. 3), which is suggestive of functional divergence, potentially as an adaptation to the C4 context. These ppc-1E1 genes likely encode proteins involved in CCMs, but do not encode the Ser780. This indicates that changes happened in different parts of the coding sequences. The monophyletic group composed of Portulacineae ppc-1E1c, ppc-1E1d, and ppc-1E1e is also characterized by increased rates of amino acid substitutions, with further increases in some branches, such as those leading to ppc-1E1c from Portulaca, but also the cacti (Fig. 3 and Supplementary Fig. S3, available at JXB online). Within Portulacineae ppc-1E1a’, long branches lead to the putative C4-specific genes of Portulaca. However, most of the amino acid substitutions occurred after the divergence of the four clades (sensu Ocampo and Columbus, 2012) and comparatively few happened on the branch leading to the C3–C4 P. cryptopetala (Fig. 3).
The action of adaptive evolution on some branches of the phylogeny is supported by codon models. While the model assuming positive selection on some sites but all branches of Caryophyllales ppc-1E1 was not better than the null model, assuming increased rates of non-synonymous changes on some sites but some branches only led to a very significant increase in likelihood (Table 2). While all the tested sets of foreground branches led to a significant increase of likelihood, the model assuming increased rates of non-synonymous mutations in the whole of Portulacineae ppc-1E1c, ppc-1E1d, and ppc-1E1e gene lineages in addition to C4 and CAM clades outside of Portulacineae produced the best AIC (Table 2). In this model, 16.8% of sites were estimated to undergo more non-synonymous mutations in the selected foreground branches, although the optimized dN/dS ratio was not different from 1.
Table 2.
Codon models for ppc-1E1 of Caryophyllales
| Model | Foreground branches | Number of parameters | Log- likelihood | AIC score |
|---|---|---|---|---|
| M1aa | – | 399 | –49702 | 10202 |
| M2ab | – | 401 | –49702 | 10206 |
| Ac | (a) each C4 and CAM groupsd | 401 | –49588 | 99978 |
| Ac | (b) a + ppc-1E1c | 401 | –49523 | 99848 |
| Ac | (c) b + ppc-1E1d + ppc-1E1e | 401 | –49383 | 99568 |
| Ac | (d) c + ppc-1E1b | 401 | –49416 | 99634 |
| Ac | (e) d + ppc-1E1a | 401 | –49556 | 99914 |
a Site model without adaptive evolution.
b Site model with adaptive evolution.
c Branch-site model with adaptive evolution.
d Except for Portulacineae other than Portulaca.
In the phylogenetic tree inferred from all nucleotides, P. cryptopetala ppc-1E1a’ is sister to all other Portulaca (Supplementary Fig. S1, available at JXB online), but this species is sister to the Oleracea clade in the tree inferred on third positions of codons (Fig. 4), as expected based on other markers (Ocampo and Columbus, 2012; Ocampo et al., 2013). The model assuming adaptive evolution in the entire Portulaca ppc-1E1a/ppc-1E1a’ clade was not different from the model without adaptive selection. Similarly, assuming adaptive evolution at the base of ppc-1E1a’ did not improve the likelihood. However, assuming adaptive evolution at the base of each C4 clade of ppc-1E1a’ significantly improved the model (χ2=38.2, df=2, P <0.00001), which indicates that 10.7% of the sites have evolved under adaptive evolution on these branches, with a dN/dS ratio of 1.35. The model assuming adaptive evolution on branches leading to both C4 and C3–C4 clades was also better than the null model, but not as good as the model without adaptive evolution on the C3–C4 branch (difference of AIC >18).
Discussion
Increased rates of amino acid changes in both C4 and CAM origins
The evolution of genes encoding PEPC in the Caryophyllales is characterized by increased rates of amino acid substitutions and the recurrence of several amino acid changes previously detected in C4 monocots (e.g. E572Q, H665N, and A780S; Christin et al., 2007; Besnard et al., 2009). These increased rates of amino acid change are not limited to C4 taxa, but are also observed in CAM lineages, including Aizoaceae and Portulacineae species (Fig. 3), and the excess of non-synonymous mutations on these branches was confirmed with codon models (Table 2). C4- and CAM-specific PEPC differ in the timing of their activity, but the catalytic challenges they face in the two cycles are similar, because in both cases the concentrations of both substrates and products are greatly increased. The evolution of both C4- and CAM-specific PEPC consequently required adaptive mutations, some of which are shared among multiple origins, while several are probably specific to one or a few clades and might depend on the other amino acid mutations undergone by the coding sequence of the gene before its co-option.
In Portulacineae, the ppc-1E1 gene lineage is present in five copies, which appeared through several rounds of gene duplications (Fig. 3 and Supplementary Fig. S1, available at JXB online). The three most recent copies, namely ppc-1E1c, ppc-1E1d, and ppc-1E1e, are all characterized by increased rates of amino acid substitutions (Fig. 3 and Table 2). The Portulacineae encompass species with different degrees of CAM metabolism (Guralnick and Jackson, 2001; Nyffeler et al., 2008). The high number of ppc-1E1 copies could have promoted neofunctionalization of the genes by relaxing selective constraints, facilitating the diversification of photosynthetic types in this group. A gradual upregulation of the CCM over time would have triggered successive periods of adaptive genetic changes in response to modifications of the catalytic environment, explaining the high rates of amino acid substitutions sustained in the entire clade (Fig. 3). For instance, the accumulation of mutations on the branches leading to ppc-1E1c of CAM-constitutive cacti (Cactoideae and Opuntioideae; Fig. 3) could be linked to the evolution of a more efficient CAM pathway in these taxa. This contrasts with the evolution of C4-specific PEPC where adaptive changes are concentrated at the base of each C4 group (Fig. 3; Christin et al., 2007; Besnard et al., 2009), and might indicate that the optimization of PEPC for the CAM function is spread over a longer time period.
C4 origins in Portulaca within a CAM-like context
The putative gene encoding the CAM-specific PEPC in Portulaca belongs to the ppc-1E1c gene lineage (Fig. 3). This gene lineage is characterized by increased rates of amino acid substitutions in other CAM taxa, such as cacti. This suggests that members of this ppc-1E1c gene lineage may have been already involved in some type of CAM metabolism before the divergence of Portulaca and cacti. On the other hand, the putative C4-specific genes of Portulaca belong to the ppc-1E1a gene lineage, which is duplicated in these taxa. One of the duplicates was likely co-opted for C4 photosynthesis after the gene duplication. Other members of the ppc-1E1a gene lineage, including the second duplicate of Portulaca, underwent mutations that generated amino acid substitutions at the same rate as genes from C3 species (Fig. 3). Codon models confirmed that adaptive non-synonymous mutations did not occur on these genes, but were restricted to some members of the ppc-1E1a′ duplicate, which is specific to Portulaca. The evolution of C4-specific genes in Portulaca likely co-opted a non-CCM gene through numerous changes in the coding sequences. Therefore, the distribution of high rates of non-synonymous substitutions indicates that the evolution of C4-specific PEPC occurred after the divergence of Portulaca from other Portulacineae while the CAM-specific properties of the gene used by Portulaca were inherited from the common ancestor of Portulaca and the cacti.
Because Portulaca species are nested within a predominantly CAM lineage (Guralnick and Jackson, 2001; Nyfeller et al., 2008), it has been previously hypothesized that the C4 pathway of these taxa evolved from an ancestral CAM type (Sage, 2002). This hypothesis is corroborated by the evolutionary history of PEPC genes, with the CAM-specific gene of Portulaca being similar to CAM forms of other species, such as cacti (Fig. 3), while the C4-specific PEPC has been recruited from non-photosynthetic forms. For the other enzymes of the CAM and C4 pathways, Portulaca uses the same genes in both conditions (Supplementary Table S6, available at JXB online). This indicates that, for many enzymes of the CCM, the evolution of one CCM from the other does not require the co-option of new genes. However, since the timing of activity differs between the CCMs, modifications in the regulation of the genes are probably still required. For many of these genes, this may be possible because they are involved with both the C3 pathway and the decarboxylation phase of the C4 pathway that operates during the day in both the CAM and C4 pathways. In the case of PEPC, however, co-option of the CAM-specific gene into the C4 cycle might have been hampered by the distinct regulatory cascades controlling the transcription and translation of the CAM form at night and the C4 form during the day (Jiao and Chollet, 1991; Nimmo, 2003), leading to the recruitment of a distinct gene (namely ppc-1E1a’).
C4 evolution within Portulaca
Portulaca species do not form a homogeneous C4 group, but include a C3–C4 species and several C4 clades that differ in their C4-associated anatomical types and decarboxylating enzymes used for the C4 cycle (Voznesenskaya et al., 2010; Ocampo et al., 2013). In phylogenetic trees of Portulaca species, the C3–C4 taxon is nested within otherwise C4 lineages (Ocampo and Columbus, 2012), which might be interpreted as evidence for a C4 to C3–C4 reversion (Ocampo et al., 2013). However, evolutionary transitions between photosynthetic types are difficult to reconstruct based on species relationships, and C4-related phenotypic and genetic variation can help differentiate alternative scenarios (Christin et al., 2010; Hancock and Edwards, 2014). In the case of Portulaca, the PEPC gene of the C3–C4 P. cryptopetala is nested within those of different C4 clades (Fig. 4). If PEPC had been optimized once for a C4 function at the base of Portulaca, this would have occurred through adaptive evolution on the branch sustaining the whole clade. Such a scenario is ruled out, however, by modelling of codon transitions, which strongly favour a model with adaptive evolution restricted to the branches at the base of each of the three C4 clades (Fig. 4). This shows that the putative C4-specific PEPC of the three C4 clades included in this study underwent adaptive amino acid changes after their divergence, and after their separation from the lineage of P. cryptopetala. For instance, the Ser780 is restricted to genes from the Oleracea clade, while orthologous genes from members of the Pilosa clade underwent other amino acid substitutions that are shared with C4 monocots (e.g. A531P, S761A; Christin et al., 2007). These results show that the optimization of PEPC for a function in C4 photosynthesis occurred independently in each C4 clade, and refutes a C4 to C3–C4 reversal in P. cryptopetala.
In addition to independent optimizations of PEPC genes for C4 photosynthesis, variation exists in the C4-associated anatomy and biochemistry among C4 clades of Portulaca (Voznesenskaya et al., 2010; Ocampo et al., 2013). Based on these observations and our results, the most likely scenario is the addition of a C3–C4 suite of traits over an ancestral CAM-like type in the common ancestor of Portulaca. This C3–C4 type might have been co-opted several times independently for the evolution of a more efficient C4 trait, as suggested for Molluginaceae (Christin et al., 2011). A gradual increase of PEPC activity during the day might then have occurred concomitantly with the development of a more C4-like anatomy, characterized by a high bundle sheath to mesophyll ratio. One way to achieve this state is through high vein densities. Some members of Portulaca belong to a handful of lineages in the Portulacineae to have evolved high vein densities via the rearrangement of leaf vasculature into a three-dimensional configuration (Voznesenskaya et al., 2010; Ocampo et al., 2013; Ogburn and Edwards, 2013). While most of these vein rearrangements were associated with large increases in succulence, in Portulaca it may have allowed the acquisition of an optimized C4 CCM.
Conclusions
Caryophyllales is a hotspot of photosynthetic transitions, with at least 23 C4 and multiple CAM origins. Of three PEPC gene lineages present in eudicots (ppc-1E1, ppc-1E2, and ppc-2; Fig. 1), only ppc-1E1 was recurrently recruited into the C4 pathway, suggesting that this gene lineage was more suitable for a C4 function (Christin et al., 2013a). The evidence provided here also supports recruitment of the same gene lineage into CAM metabolism, which suggests that the same capacitated genes present in the common C3 ancestor were co-opted by the numerous CCM origins of Caryophyllales. The evolvability of one CCM compared to the other might depend on the ecology and leaf anatomy of the C3 ancestor of each lineage (Sage, 2002; Edwards and Ogburn, 2012; Edwards and Donoghue, 2013). However, in some cases, evolutionary bridges between the two photosynthetic types exist, as illustrated by Portulaca. This shows that while ancestral conditions could influence the evolutionary trajectories of the descendants, the determinism is not perfect and one photosynthetic type can be co-opted to evolve the other.
Supplementary material
Supplementary data are available at JXB online.
Figure S1. Phylogenetic relationships among ppc-1 genes. This phylogenetic tree was obtained through Bayesian inference on nucleotide sequences. Names of taxonomic groups and gene lineages are indicated on the right. Branches in lineages presenting a Ser780 are highlighted in red. Bayesian support values are indicated near branches. Asterisks indicate putative pseudogenes with one or several stop codons in the coding sequence. Black circles indicate sequences that were isolated from cacti cDNA. (A) Complete phylogenetic tree; (B) ppc-1E2 of Caryophyllales; (C, D) ppc-1E1 of Caryophyllales.
Figure S2. Phylogenetic relationships among ppc-2 genes. This phylogenetic tree was obtained through Bayesian inference on nucleotide sequences. Names of taxonomic groups are indicated on the right. Bayesian support values are indicated near branches.
Figure S3. Amino acid changes on genes encoding PEPC. The topology was inferred on nucleotide sequences, but branch lengths were estimated based on amino acid sequences. The branch lengths inferred on nucleotide sequences, together with all species names and support values, are available in Figs S1 and S2. The names of the main groups are indicated on the right. Groups of genes containing a Ser780 are highlighted by red branches. The asterisk highlights a pseudogene with multiple stop codons.
Table S1. Sample of Caryophyllales (excluding Portulacineae) used for analyses of PEPC-encoding genes.
Table S2. List of Portulacineae genes encoding PEPC analysed.
Table S3. Additional primers used for PCR amplification of Caryophyllales genes encoding PEPC.
Table S4. Water treatment of Portulaca oleracea plants.
Table S5. Sequencing and mapping statistics.
Table S6. Expression levels in rpm of C4-related genes in day and night samples of Portulaca oleracea plants grown in different conditions.
Acknowledgements
This work was funded by Marie Curie International Outgoing Fellowship 252569 to PAC and a National Science Foundation grant (DEB-1026611) and Brown University Salomon Faculty Research Award to EJE. This research was conducted using computational resources at the Center for Computation and Visualization, Brown University. The 1000 Plants (1KP) initiative, led by GKSW, is funded by the Alberta Ministry of Enterprise and Advanced Education, Alberta Innovates Technology Futures (AITF) Innovates Centre of Research Excellence (iCORE), Musea Ventures, and BGI-Shenzhen. The authors thank Professor Gerry Edwards and Professor Neal Stewart for providing seeds of several Portulaca species used to generate transcriptome data.
Glossary
Abbreviations:
- AIC
Akaike information criteria
- CCM
CO2-concentrating mechanism
- PEPC
phosphoenolpyruvate carboxylase
- rpm
reads per million of alignable reads.
References
- Arakaki M, Christin PA, Nyffeler R, Lendel A, Eggli U, Ogburn RM, Spriggs E, Moore MJ, Edwards EJ. 2011. Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proceedings of the National Academy of Sciences USA 108, 8379–8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. Journal of Experimental Botany 62, 3049–3059 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Chollet R. 1986. Kinetic properties of phosphoenolpyruvate carboxylase from C3, C4, and C3–C4 intermediate species of Flaveria (Asteraceae). Plant Physiology 82, 695–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Muasya AM, Russier F, Roalson EH, Salamin N, Christin PA. 2009. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Molecular Biology and Evolution 26, 1909–1919 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Westhoff P, Svensson P. 2000. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant of C4-specific characteristics. Journal of Biological Chemistry 275, 27917–27923 [DOI] [PubMed] [Google Scholar]
- Borland AM, Taybi T. 2004. Synchronization of metabolic processes in plants with Crassulacean acid metabolism. Journal of Experimental Botany 55, 1255–1265 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington SF, Alexandre R, Ramdial J, Moore MJ, Crawley S, Dhingra A, Kilu K, Soltis DE, Soltis PS. 2009. Phylogeny of the Caryophyllales sensu lato: revisiting hypotheses on pollination biology and perianth differentiation in the core Caryophyllales. International Journal of Plant Sciences 170, 627–643 [Google Scholar]
- Christin PA, Besnard G. 2009. Two independent C4 origins in Aristidoideae (Poaceae) revealed by the recruitment of distinct phosphoenolpyruvate carboxylase genes. American Journal of Botany 96, 2234–2239 [DOI] [PubMed] [Google Scholar]
- Christin PA, Besnard G, Edwards EJ, Salamin N. 2012. Effect of genetic convergence on phylogenetic inference. Molecular Phylogenetics and Evolution 62, 921–927 [DOI] [PubMed] [Google Scholar]
- Christin PA, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP. 2013a. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biology and Evolution 5, 2174–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Freckleton RP, Osborne CP. 2010. Can phylogenetics identify C4 origins and reversals? Trends in Ecology and Evolution 25, 403–409 [DOI] [PubMed] [Google Scholar]
- Christin PA, Osborne CP. 2013. The recurrent assembly of C4 photosynthesis, an evolutionary tale. Photosynthesis Research 117, 163–175 [DOI] [PubMed] [Google Scholar]
- Christin PA, Osborne CP, Chatelet DS, Columbus JT, Besnard G, Hodkinson TR, Garrison LM, Vorontsova MS, Edwards EJ. 2013b. Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proceedings of the National Academy of Sciences USA 110, 1381–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Sage TL, Edwards EJ, Ogburn RM, Khoshravesh R, Sage RF. 2011. Complex evolutionary transitions and the significance of C3–C4 intermediate forms of photosynthesis in Molluginaceae. Evolution 65, 643–660 [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Savolainen V, Duvall MR, Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Current Biology 17, 1241–1247 [DOI] [PubMed] [Google Scholar]
- Crayn DM, Winter K, Smith JAC. 2004. Multiple origins of crassulacean acid metabolism and the epiphytic habit in the Neotropical family Bromeliaceae. Proceedings of the National Academy of Sciences USA 101, 3703–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong LY, Masuda T, Kawamura T, Hata S, Izui K. 1998. Cloning, expression, and characterization of a root-form phosphoenolpyurvate carboxylase from Zea mays: comparison with the C4-form enzyme. Plant and Cell Physiology 39, 865–873 [DOI] [PubMed] [Google Scholar]
- Dunn CW, Howison M, Zapata F. 2013. Agalma: an automated phylogenomics workflow. BMC Bioinformatics 14, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Donoghue MJ. 2013. Is it easy to move and easy to evolve? Evolutionary accessibility and adaptation. Journal of Experimental Botany 64, 4047–4052 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Ogburn RM. 2012. Angiosperm responses to a low-CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories. International Journal of Plant Sciences 173, 724–733 [Google Scholar]
- Engelmann S, Bläsing OE, Westhoff P, Svensson P. 2002. Serine 774 and amino acids 296 to 437 comprise the major C4 determinants of the C4 phosphoenolpyruvate carboxylase of Flaveria trinervia . FEBS Letters 524, 11–14 [DOI] [PubMed] [Google Scholar]
- Finnegan PM, Suzuki S, Ludwig M, Burnell JN. 1999. Phosphoenolpyruvate carboxykinase in the C4 monocot Urochloa panicoides is encoded by four differentially expressed genes. Plant Physiology 120, 1033–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig H, Heute V, Kluge M. 2001. New partial sequences of phosphoenolpyruvate carboxylase as molecular phylogenetic markers. Molecular Phylogenetics and Evolution 20, 262–274 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Bräutigam A, Weber KL, Weber APM, Westhof P. 2011. Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23, 2087–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Engelmann S, Bläsing OE, Raghavendra AS, Westhoff P. 2006. Evolution of C4 phosphoenolpyruvate carboxylase in the genus Alternanthera: gene families and the enzymatic characteristics of the C4 isozyme and its orthologues in C3 and C3/C4 Alternantheras. Planta 223, 359–368 [DOI] [PubMed] [Google Scholar]
- Gowik U, Westhoff P. 2011. C4-phosphoenolpyruvate carboxylase. In: Rhagavendra AS, Sage RF, eds. C4 photosynthesis and related CO2 concentrating mechanisms. Advances in Photosynthesis and Respiration, Vol 32 Dordrecht: Springer, 257–275 [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotechnology 29, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths H, Weller G, Toy LMF, Dennis RJ. 2013. You’re so vein: bundle sheath physiology, phylogeny and evolution in C3 and C4 plants. Plant Cell and Environment 36, 249–261 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704 [DOI] [PubMed] [Google Scholar]
- Guralnick LJ, Cline A, Smith M, Sage RF. 2008. Evolutionary physiology: the extent of C4 and CAM photosynthesis in the genera Anacampseros and Grahamia of the Portulacaceae. Journal of Experimental Botany 59, 1735–1742 [DOI] [PubMed] [Google Scholar]
- Guralnick LJ, Edwards G, Ku MSB, Hockema B, Franceschi VR. 2002. Photosynthesis and anatomical characteristics in the C4-crassulacean acid metabolism-cycling plant Portulaca grandiflora . Functional Plant Biology 29, 763–773 [DOI] [PubMed] [Google Scholar]
- Guralnick LJ, Jackson M. 2001. The occurrence and phylogenetics of CAM in the Portulacaceae. International Journal of Plant Sciences 162, 257–262 [Google Scholar]
- Guralnick LJ, Ting IP. 1987. Physiological changes in Portulacaria afra (L.) Jacq. during a summer drought and rewatering. Plant Physiology 85, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock L, Edwards EJ. 2014. Phylogeny and the inference of evolutionary trajectories. Journal of Experimental Botany 65, 3491–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106 [Google Scholar]
- Hattersley PW. 1984. Characterization of C4 type leaf anatomy in grasses (Poaceae), mesophyll-bundle sheath area ratios. Annals of Botany 53, 163–179 [Google Scholar]
- Jiao JA, Chollet R. 1991. Posttranslational regulation of phosphoenolpyruvate carboxylase in C4 and Crassulacean Acid Metabolism plants. Plant Physiology 95, 981–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MTJ, Carpenter EJ, Tian Z, et al. 2012. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLOS ONE 7, e50226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit G, Ackerly D, Pirie MD. 2012. A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proceedings of the Royal Society of London Series B 279, 3304–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit G, Borsch T, Weising K, Freitag H. 2003. Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. International Journal of Plant Sciences 164, 959–986 [Google Scholar]
- Keeley JE, Rundel PW. 2003. Evolution of CAM and C4 carbon-concentrating mechanisms. International Journal of Plant Sciences 164, S55–S77 [Google Scholar]
- Koch K, Kennedy RA. 1980. Characteristics of crassulacean acid metabolism in the succulent C4 dicot, Portulaca oleracea L. Plant Physiology 65, 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K, Kennedy RA. 1982. Crassulacean acid metabolism in the succulent C4 dicot, Portulaca oleracea L. under natural environmental conditions. Plant Physiology 69, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraybill AA, Martin CE. 1996. Crassulacean acid metabolism in three species of the C4 genus Portulaca. International Journal of Plant Sciences 157, 103–109 [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara MV, Chuong SDX, Akhani H, Andreo CS, Edwards GE. 2006. Species having C4 single-cell-type photosynthesis in the Chenopodiaceae family evolved a photosynthetic phosphoenolpyruvate carboxylase like that of Kranz-type C4 species. Plant Physiology 142, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara MV, Disante KB, Podesta FE, Andreo CS, Drincovich MF. 2003. Induction of a Crassulacean acid like metabolism in the C4 succulent plant, Portulaca oleracea L.: physiological and morphological changes are accompanied by specific modifications in phosphoenolpyruvate carboxylase. Photosynthesis Research 77, 241–254 [DOI] [PubMed] [Google Scholar]
- Lara MV, Drincovich MF, Andrea CS. 2004. Induction of a crassulacean acid-like metabolism in the C4 succulent plant, Portulaca oleracea L.: study of enzymes involved in carbon fixation and carbohydrate metabolism. Plant Cell and Environment 45, 618–626 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Vidal J, Chollet R, Gadal P, Cretin C. 1994. Phosphoenolpyruvate carboxylase: structure, function, regulation and evolution. Plant Science 99, 111–124 [Google Scholar]
- Ludwig M. 2011. The molecular evolution of β-carbonic anhydrase in Flaveria . Journal of Experimental Botany 62, 3071–3081 [DOI] [PubMed] [Google Scholar]
- Mazen AMA. 1996. Changes in levels of phosphoenolpyurvate carboxylase with induction of Crassulacean acid metabolism (CAM)-like behavior in the C4 plant Portulaca oleracea . Physiologia Plantarum 98, 111–116 [Google Scholar]
- Mazen AMA. 2000. Changes in properties of phosphoenolpyruvate carboxylase with induction of Crassulacean Acid Metabolism (CAM) in the C4 plant Portulaca olearacea . Photosynthetica 38, 385–391 [Google Scholar]
- Monson RK. 2003. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. International Journal of Plant Sciences 164, S43–S54 [Google Scholar]
- Nelson EA, Sage TL, Sage RF. 2005. Functional leaf anatomy of plants with crassulacean acid metabolism. Functional Plant Biology 32, 409–419 [DOI] [PubMed] [Google Scholar]
- Nimmo HG. 2003. How to tell the time: the regulation of phosphoenolpyruvate carboxylase in Crassulacean acid metabolism (CAM) plants. Biochemical Society Transactions 31, 728–730 [DOI] [PubMed] [Google Scholar]
- Nyffeler R, Eggli U. 2010. Disintegrating Portulacaceae: a new familial classification of the suborder Portulacineae (Caryophyllales) based on molecular and morphological data. Taxon 59, 227–240 [Google Scholar]
- Nyffeler R, Eggli U, Ogburn M, Edwards EJ. 2008. Variations on a theme: repeated evolution of succulent life forms in the Portulacineae (Caryophyllales). Haseltonia 14, 26–36 [Google Scholar]
- Ocampo G, Columbus JT. 2010. Molecular phylogenetics of suborder Cactineae (Caryophyllales), including insights into photosynthetic diversification and historical biogeography. American Journal of Botany 97, 1827–1847 [DOI] [PubMed] [Google Scholar]
- Ocampo G, Columbus JT. 2012. Molecular phylogenetics, historical biogeography, and chromosome number evolution of Portulaca (Portulacaceae). Molecular Phylogenetics and Evolution 63, 97–112 [DOI] [PubMed] [Google Scholar]
- Ocampo G, Koteyeva NK, Voznesenskaya EV, Edwards GE, Sage TL, Sage RF, Columbus JT. 2013. Evolution of leaf anatomy and photosynthetic pathways in Portulacaceae. American Journal of Botany 100, 2388–2402 [DOI] [PubMed] [Google Scholar]
- Ogburn RM, Edwards EJ. 2013. Repeated origin of three-dimensional leaf venation releases constraints on the evolution of succulence in plants. Current Biology 23, 722–726 [DOI] [PubMed] [Google Scholar]
- Osmond CB. 1978. Crassulacean acid metabolism. A curiosity in context. Annual Reviews of Plant Physiology 29, 379–414 [Google Scholar]
- Rambaut A, Drummond AJ. 2007. Tracer v1.4, available at http://beadt.bio.ed.ac.uk/Tracer
- Rao SK, Magnin NC, Reiskind JB, Bowes G. 2002. Photosynthetic and other phosphoenolpyruvate carboxylase isoforms in the single-cell, facultative C4 system of Hydrilla verticillata . Plant Physiology 130, 876–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SK, Reiskind JB, Bowes G. 2008. Kinetic analyses of recombinant isoforms of phosphoenolpyruvate carboxylase from Hydrilla verticillata leaves and the impact of substituting a C4-signature serine. Plant Science 174, 475–483 [Google Scholar]
- Raven JA, Cockell CS, De La Rocha CS. 2008. The evolution of inorganic carbon concentrating mechanisms in photosynthesis. Philosophical Transactions of the Royal Society of London Series B 363, 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickers J, Cushman JC, Michalowski CB, Schmitt JM, Bohnert HJ. 1989. Expression of the CAM-form of phospho(enol)pyruvate carboxylase and nucleotide sequence of a full length cDNA of Mesembryanthemum crystallinum . Molecular and General Genetics 215, 447–454 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sage RF. 2001. Environmental and evolutionary preconditions for the origin and diversification of the C4 photosynthetic syndrome. Plant Biology 3, 202–213 [Google Scholar]
- Sage RF. 2002. Are crassulacean acid metabolism and C4 photosynthesis incompatible? Functional Plant Biology 29, 775–785 [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin PA, Edwards EJ. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany 62, 3155–3169 [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis. Annual Reviews of Plant Biology 63, 19–47 [DOI] [PubMed] [Google Scholar]
- Silvera K, Santiago LS, Winter K. 2005. Distribution of crassulacean acid metabolism in orchids of Panama: evidence of selection for weak and strong modes. Functional Plant Biology 32, 397–407 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Smith SA, Cellinese N, et al. 2011. Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany 98, 704–730 [DOI] [PubMed] [Google Scholar]
- Svensson P, Bläsing OE, Westhoff P. 2003. Evolution of C4 phosphoenolpyruvate carboxylase. Archives of Biochemistry and Biophysics 414, 180–188 [DOI] [PubMed] [Google Scholar]
- Tausta SL, Miller Coyle H, Rothermel B, Stifel V, Nelson T. 2002. Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Molecular Biology 50, 635–652 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DJ, Gibson TJ. 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and matrix choice. Nucleic Acids Research 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyava NK, Edwards GE, Ocampo G. 2010. Revealing diversity in structural and biochemical forms of C4 photosynthesis in genus Portulaca L. (Portulacaceae). Journal of Experimental Botany 61, 3647–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24, 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R. 2002. Codon-substitutions models for detecting molecular adaptation at individual sites along specific lineages. Molecular Biology and Evolution 19, 908–917 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.