Summary
The measured differential expression of genes between bundle sheath and mesophyll cells at successive developmental stages of the maize leaf is used to identify C4-photosynthesis-related candidates.
Key words: Bundle sheath, C4 photosynthesis, Kranz, leaf development, mesophyll, transcriptome.
Abstract
The comparison of the cell-specific transcriptomes of bundle sheath (BS) and mesophyll (M) cells from successive developmental stages of maize (Zea mays) leaves reveals that the number of genes preferentially transcribed in one cell type or the other varies considerably from the sink–source transition to mature photosynthetic stages. The number of differentially expressed (DE) genes is maximal at a stage well before full maturity, including those that encode key functions for C4 photosynthesis. The developmental dynamics of BS/M differential expression can be used to identify candidates for other C4-related functions and to simplify the identification of specific pathways members from otherwise complex gene families. A significant portion of the candidates for C4-related transcription factors identified with this developmental DE strategy overlap with those identified in studies using alternative strategies, thus providing independent support for their potential importance.
Introduction
C4 photosynthesis has evolved independently in many plant groups, both monocot and dicot (Sage et al., 2011; Aliscioni et al., 2012; Denton et al., 2013). The majority of existing C4 species rely on the metabolic cooperation of leaf bundle sheath (BS) and mesophyll (M) cells with a Kranz-type anatomy surrounding the venation (Langdale, 2011). In this two-cell metabolism, M cells perform primary carbon assimilation and adjacent BS cells perform a linked primary carbon reduction. M cells fix atmospheric CO2 into C4 acids, which are passed through plasmodesmata to adjacent BS cells and decarboxylated. The BS is specialized with a permeability boundary to retain the released CO2 and to exclude O2, thereby enhancing the Rubisco-initiated carbon reduction steps (Nelson, 2010). BS cells seem to be developmentally related to the endodermal cells that surround the vasculature of the root and stem (Slewinski, 2013).
Many studies have evaluated the regulation of individual genes whose products are BS- or M-specific, and several studies have compared the transcriptomes of mature BS and M cells (Hibberd and Covshoff, 2010; Langdale, 2011). However, mature tissues provide limited insight into the developmental mechanisms that produce mature BS and M cells. Whole-leaf transcriptome and proteome studies of sequential developmental stages from the base to tip of the young maize leaf (Nelson, 2011) revealed a developmentally dynamic process in the appearance and disappearance of transcripts, transcription factors and other proteins, with a significant portion reduced to low level or absent in mature tissue (Majeran et al., 2010; Li et al., 2010; Pick et al., 2011). This suggests that some important factors may appear and act at early stages only.
In this study we compare the transcriptomes of maize BS and M cells at three successive developmental stages: sink–source transition (SST), maturing, and mature. In addition to resolving the timing of processes during the differentiation of the two cell types, the dynamics of differential BS/M expression of genes can be used as a filter for identifying new candidates for C4-related genes. It also provides a means to select pathway candidates among gene family members. We compare candidate C4-related genes identified by our BS/M laser microdissection (LM)-based strategy with the candidates identified by alternative strategies. Consensus candidates identified by multiple methods warrant further study.
Materials and methods
Cell and RNA isolation
Maize Mo17 and B73 leaf sections were collected from synchronously grown plants and preserved in 100% ice-cold acetone as described in Li et al., 2010. After fixation in paraffin, BS and M cells were captured using a PixCell IIe or Veritas LM system (Life Technologies) and the RNA recovered with a Picopure RNA isolation kit (Life Technologies) according the to the manufacturer’s instructions. A sample of 50–100ng of total RNA was amplified through one round using the RiboAmp HS plus RNA amplification kit (Life Technologies). RNA quality was monitored using a Bioanalyzer 2100 (Agilent).
Library preparation and Illumina sequence analysis
Library construction, Illumina sequencing, and sequence analysis were done as previously described (Li et al., 2010). The background level was estimated by calculating the alignment density in genomic regions that are at least 10kb away from any exon. Based on the estimated background level and the Poisson model, we calculated the P-value for testing whether this gene is expressed or not (whether the observed counts arose from background noise or they were true signal) by calculating the probability of observing the observed counts or more counts if all reads mapped to this gene were background noise. We controlled the false discovery rate (FDR) at 0.1% using Benjamini and Hochberg’s procedure (Benjamini and Hochberg, 1995). Raw datasets are available at NCBI Gene Expression Omnibus (www.ncbi.nim.nih.gov/geo), accession GSE 54272.
DESeq (Anders and Huber, 2010), an R package, was used with upper quartile normalization method (Bullard et al., 2010) to test for differential expression between BS and M cells in each stage. The Benjamini and Hochberg method was applied to the list of resulting P-values to control FDR (Benjamini and Hochberg, 1995). Genes that were differentially expressed between BS and M cells (FDR<0.01) were then separated into BS or M expression using log2-fold change of BS/M. The functional annotation of identified genes was based on the Mapman pathways (http://mapman.gabipd.org/web/guest/mapmanstore—Zm_B73_5b_FGS_cds_2012download) with manual corrections and further refinement using Putative Orthologous Groups (http://cas-pogs.uoregon.edu/#/) (Walker et al., 2007; Tomcal et al., 2013) and Phytozome (http://www.phytozome.net/). Protein data were compared using the Plant Proteome Database- PPDB (http://ppdb.tc.cornell.edu/) (Sun et al., 2009). Homeologue identification was performed using data provided by http://skraelingmountain.com/datasets.php (Schnable et al., 2012).
For comparison of identified transcription factors from other reports, all gene names were checked with B73 RefGen_v2 to ensure that the maize identifications were consistent.
Results
The expression of many genes differentially expressed between BS and M cells peaks and declines before maturity during C4 leaf development.
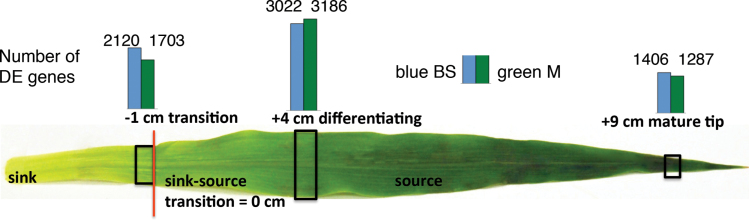
We inventoried the transcripts present in BS and mesophyll M cells at three sites along the developmental gradient of a nine-day-old 3rd maize leaf, using LM and RNA-seq, as described in Li et al. (2010). The three sites corresponded to SST, maturing, and mature developmental stages, located at –1cm, +4cm and +9cm (leaf tip), relative to the completion of the sink–source transition, which we previously showed is located at the point touched by the ligule of the 2nd leaf (Li et al., 2010; Majeran et al., 2010). The SST is exposed to a lower level of light than the two succeeding stages.
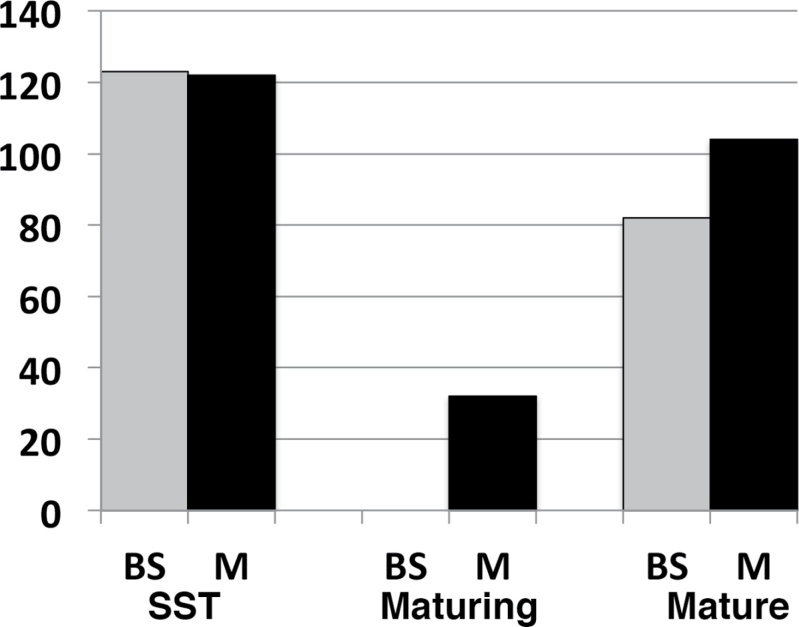
Expressed genes above background were determined as described in materials and methods; false discovery rate (FDR) was controlled at 0.1%, 27 819 genes were expressed, based on the filtered gene set of maize assembly v2. For each stage, the BS- and M-differentially expressed (DE) genes were identified by comparing the transcript levels between BS and M cells at each stage, using the R package, DESeq, as described in materials and methods. This analysis identified 7994 genes that were either BS- or M-enriched in expression (Supplementary dataset S1). In this report, DE is defined as those genes whose transcripts were significantly higher in one cell type versus the other as determined by DESeq. BS- or M-enriched (or –biased) genes are defined as those whose transcripts were more abundant in that cell type versus the other; “-specific” genes are defined as those that were exclusively detected in one type. The majority of DE genes (~85%) ranged from 2-fold enriched to completely specific for BS or M. The number of genes with cell-enriched transcripts was greatest during the maturing stage, and was markedly less in the mature stage (Fig. 1). Transcripts of 1703, 3186, and 1287 genes were M-cell-enriched in the SST, maturing, and mature stages, respectively, a total of 4154 unique genes that were enriched in M cells in at least one stage. Transcripts of 2120, 3022, and 1406 were BS-enriched, a total of 4043 unique genes that were enriched in BS cells in at least one stage.
Fig. 1.
Developmental dynamics of BS/M differential expression (DE). Three sections from the third leaf of a 9-day-old maize plant representing locations of sink–source transition (SST), maturing, and mature developmental stages, the source material for BS and M LM capture. Details of sample site location, anatomy and ultrastructure are in Li et al. (2010) and Majeran et al. (2010). Blue bars above the leaf represent the number of DE genes found to be BS-biased and green bars represent the number of M-biased DE genes.
Most DE genes for BS and M cells are not absolutely cell-specific, reinforcing observations in many organisms that cell-specific traits are produced through a combination of transcriptional and translational mechanisms (e.g. Ramskold et al., 2009). The C4 carbon shuttle genes, all transcribed at a high level, exhibit a range of BS-M transcriptional ratios in the three developmental stages. Comparisons of BS and M transcriptomes isolated from a single stage by the same LM method reveal a broad range of ratios of differential expression for different genes, suggesting that the BS-M distributions are not a cross-contamination artefact. It is also consistent with the distributions observed in proteomic measurements performed on mechanically separated cells from the same sources (Friso et al., 2010; Majeran et al., 2010). As shown previously, there is good correlation between the BS/M transcript ratio and the protein BS/M ratio of photosynthesis-related genes (Li et al., 2010).
C4-associated metabolic genes are BS/M-cell-specific from initial expression near leaf sink–source transition, peak in level at midblade, decline at mature tip
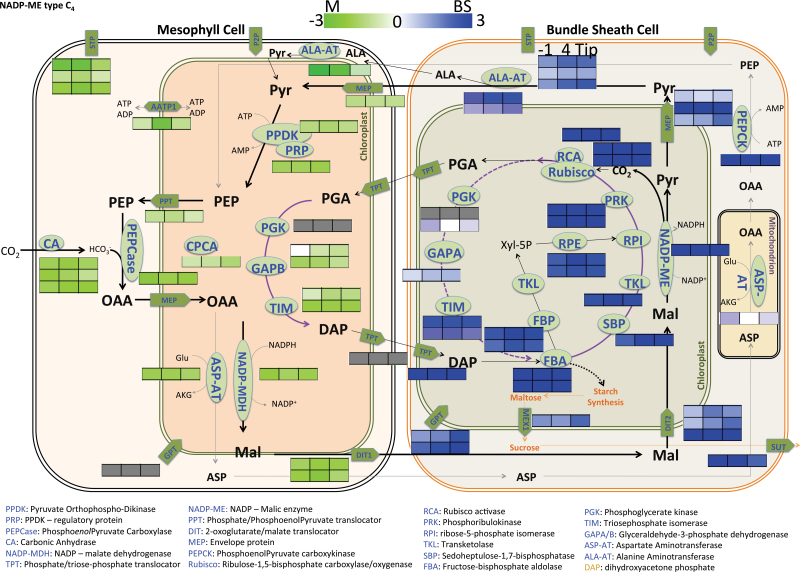
Genes encoding proteins known to be either BS- or M-specific in mature leaves exhibit a transcriptional bias at the first stage sampled, the SST stage, where Kranz anatomy is evident, but cells have not yet transitioned to a photosynthetically active state (Majeran et al., 2010). For instance, transcripts for the core C4 pathway enzymes are all at least two-fold more highly expressed in one cell type versus the other in the SST stage, which is surrounded and shaded by an older leaf (e.g. log2 BS/M for MDH=–2, PepC=–2.2, NADP-ME=3.1, TKL= 2.6) (Fig. 2 and Li et al., 2010). This pattern is also true for cell-type-biased genes in photorespiration, starch and sucrose metabolism, sulfur assimilation, and other pathways that become active following the SST (Supplementary Fig. S1). In the subsequent well-illuminated stages (maturing and mature), the level of expression increases, but the cell type bias is maintained.
Fig. 2.
C4 cycle genes required for cell cooperation are cell-biased at SST. The C4 cycle between BS and M cells relies on numerous enzymes expressed in either cell type. Heat maps of three boxes next to enzyme names represent the log2-transformed BS/M expression ratio at the three developmental stages sampled (SST, maturing, mature). Each row indicates the results from a gene with the same proposed activity. Green signifies an M-biased ratio and blue represents a BS-biased ratio. Modified from Li et al., 2010 and detailed expression data is found in Supplementary dataset 2.
Many DE transcripts encoding products that function in mature leaves peak in abundance at maturing stage before declining significantly at mature stage. We do not present here a K-means clustering of DE genes by their expression dynamics over the three stages, as the peak-and-decline developmental dynamic was previously documented for individual genes at the whole-leaf transcriptome level (Li et al., 2010; Pick et al., 2011). The abundance of most of the corresponding proteins does not follow this pattern. Instead, C4 photosynthesis-associated proteins and other metabolic pathway proteins generally remain abundant at the tip of the leaf, without the decline observed for their transcripts (Friso et al., 2010; Majeran et al., 2010).
Members of the same gene family are differentially expressed in BS and M cells
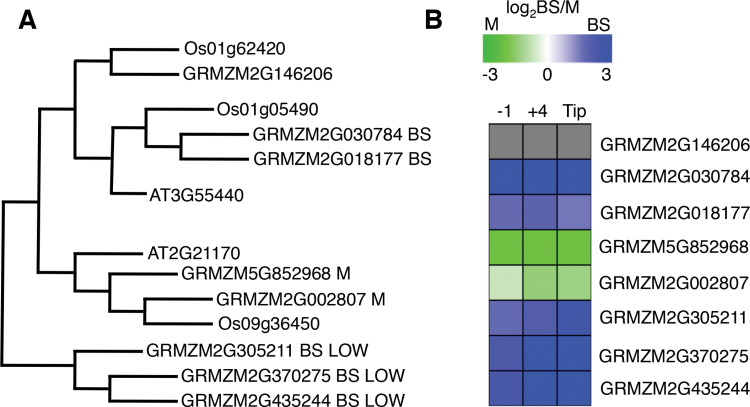
Cell expression ratios and cell-specific co-expression patterns can be used as filters to infer the BS/M-relevant member(s) of otherwise redundant gene families annotated with a general function. For example, some gene families that encode activities found in both BS and M cells seem to have subfunctionalized, with distinct members driving expression in each cell type (e.g. triosephosphate isomerase, TIM), whereas others use the same members in both cells (e.g. PGK) (Figs 2 and 3). In the case of TIM, a gene family encodes activities for steps in glycolysis and also in the production of d-glyceraldehyde 3-phosphate (DAP), a Calvin cycle intermediate, in both BS and M cells. Two closely related TIM genes (GRMZM2G002807 and GRMZM5G852968) are highly expressed in M cells and are maize homeologues (Schnable et al., 2011). The other TIM genes have a more complex relationship. Of the five BS DE genes in the same POG (putative orthologue group; Walker et al., 2007; Tomcal et al., 2013), two are highly expressed (GRMZM2G030784, GRMZM2G018177) and three are expressed at a much lower level (~10× less) but still with a BS-enhanced ratio (GRMZM2G370275, GRMZM2G435244, GRMZM2G305211). The two more highly expressed BS-enriched TIMs are also homeologues, but the three less expressed genes are not. Transcript amounts from a sixth gene, encoding the cytosolic form of TIM (GRMZM2G146206) are high at the leaf base (all-cell sample) but low in BS and M cells of subsequent stages. The cytosolic protein and its transcripts are both BS- biased. Generally, the similarity of expression patterns among gene family members can be predicted by their sequence similarity dendrogram (Fig. 3A, Supplementary dataset S2). The divergence of expression among such closely related gene family members suggests that comparison of their cis-regulatory regions may reveal motifs for cell-specific expression.
Fig. 3.
TIM family members assigned to BS or M. (A) TIM family putative orthologue group, POG, represented by phenetic tree generated from protein MUSCLE alignment and using UPGMA (unweighted pair group method and arithmetic mean) (Edgar, 2004; Tomcal et al., 2013). Members with similar cell-biased expression cluster together. (B) DE heat map of TIM genes. Grey boxes for the cytosolic form of TIM represent no DE data. Green signifies an M-biased ratio and blue represents a BS-biased ratio based on the log2-transformed BS/M ratio for each section. Detailed expression data is found in Supplementary dataset 2.
Additional examples of gene families that contain both BS and M-enriched family members were found in metabolic pathways both related and unrelated to C4 photosynthesis. For example, five closely related pyruvate kinase genes exhibit distinct expression patterns in the leaf, including two M-biased, two BS-biased, and one BS-M unbiased. Of the five cytosolic glutamine synthetases (GS1), transcripts for two are M-enriched and three are BS-enriched (Cren and Hirel, 1999). The presence of GS activity in both BS and M cells has been documented (Becker et al., 2000) and the transcript patterns correspond well with protein location and relative abundance (Martin et al., 2006; Sun et al., 2009). In contrast, a single DE glutamate dehydrogenase gene (gdh1) is BS-expressed and the corresponding protein is found only in the BS (Friso et al., 2010; Majeran et al., 2010; Supplementary dataset S2).
Use of developmental and cell-type co-expression to infer gene function and interactions
The developmental dynamics of cell-type specificity can permit the assignment of putative functions of members of large families before direct functional tests. As an example of the inference of function, our data supports the current understanding of an additional decarboxylation cycle involving phosphoenolpyruvate carboxykinase (PEPCK), alanine aminotransferase (AlaAT) and aspartate aminotransferase (AspAT) (Leegood and Walker, 2003; Pick et al., 2011). PEPCK (GRMZM2G001696) was found to be highly expressed in BS in all three sections with the highest expression at the tip, which is unusual for a C4 gene (Fig. 2, Supplementary dataset S2). The DE genes include one AspAT isoform expressed in M (GRMZM5G836910) but not the one (GRMZM2G094712) detected by another study in mature BS cells (Chang et al., 2012). We find GRMZM2G094712 is BS-enriched only at SST, the stage at which it is expressed at the highest level, and similar to the AspAT (GRMZM2G033799) that was DE in this experiment. Neither of the BS AspATs have high transcript accumulation profiles in this experiment, but both proteins have been identified in BS cells (Friso et al., 2010; Majeran et al., 2010), suggesting regulation is post-transcriptional. Among the large AlaAT gene family, at least five genes meet our DE criteria. One M-biased member (GRMZM2G088064) exhibits the dynamic pattern of C4 genes, starting at SST, peaking at maturing stage, and declining at mature. The corresponding protein is found in field-grown leaves, but was not identified in the proteome of M cells (Friso et al., 2010; Majeran et al., 2010). Of the remaining AlaAT activities, transcripts for all are BS-enriched; three were found in the proteome of BS cells (Friso et al., 2010; Majeran et al., 2010). Two AlaAT genes (GRMZM2G053999, GRMZM5G840582) exhibit the levels and developmental patterns of C4-related genes.
As another example of the inference of gene function based on cell-specific expression pattern, aquaporins constitute large families of transporters with many leaf-expressed members. Aquaporins were initially associated with water homeostasis, but some are involved in transport of solutes such as CO2 (Uehlein et al., 2003; Hanba et al., 2004). Water transport is crucial for photosynthesis and phloem loading. In the maize leaf, BS cells are surrounded by a suberin boundary similar to the Casparian strip that surrounds root endodermal cells and makes the movement of water aquaporin-dependent (Amodeo et al., 1999; Johansson et al., 2000). The aquaporin family in maize has over 30 members in four major subfamilies that play important roles in the leaf (Chaumont et al., 2001; Johanson et al., 2001; Heinen et al., 2009).
We observe 16 BS-biased and 4 M-biased aquaporin family genes, of which a subset is expressed at high levels beginning at SST, consistent with the building of infrastructure for subsequent C4 processes (Supplementary dataset S2). Members of the PIP (plasma membrane intrinsic protein) subfamily were the most abundant in either cell type, although all four major subfamilies were represented in the BS DE list. The BS-enriched DE genes included seven PIP members. The M-enriched DE genes included four PIPs with differing dynamic patterns, only one of which was M-enriched at all developmental stages. Six BS and two M-biased PIPs are closely related by sequence but were not found to be maize homeologues. The two M-enriched DE PIPs (GRMZM2G154628, GRMZM2G081192) are in the same sub-cluster and both show a pattern of highest expression at the SST stage. The BS-M biases we observe for transcripts for the 21 aquaporins agree with those found in mature stage in another study (Chang et al., 2012), except for two for which we find a lower BS/M ratio at mature stage, and three for which we observe a differing specificity at an earlier stage. Transcripts for PIPs are abundant at the leaf base (Li et al., 2010) where PIPs are likely to be incorporated into cell walls during cell division; the SST cell-type-enhanced accumulation we observe may be a continuation of this process.
Cell-specific co-expression information that includes early stages can help to associate cognate pairs of interacting proteins among multiple candidates in gene families, even if expression levels are low. For instance, Aux/IAA repressors interact with auxin response factor (ARF) transcription factors in response to auxin (Hagen and Guilfoyle, 2002). Both partners in this interaction come from gene families, making possible many theoretical pairings. A cell-specific co-expression strategy was previously employed in Arabidopsis to identify candidate pairings (Gandotra et al., 2013). Our cell-type co-expression data considerably limits the candidate partners in BS and M cells. For example, in M cells, one ARF and six Aux/IAAs are preferentially expressed at the SST stage (Supplementary dataset S2). The ARF and three Aux/IAAs continue to show M expression through the mature stage at the tip of the leaf. In BS cells, three ARFs and two Aux/IAAs are preferentially expressed at the SST stage and only one ARF and one Aux/IAA remain BS-expressed out to the mature tip.
The most highly expressed DE transcripts are abundant at all stages
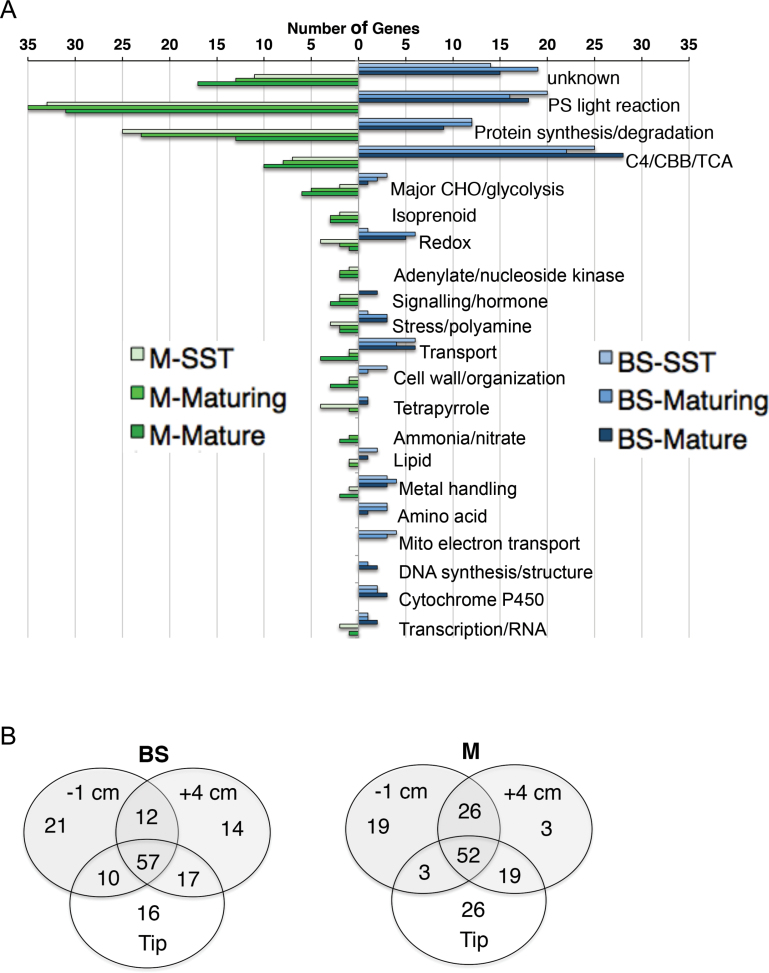
As transcripts for C4 enzymes are all expressed at a high level throughout the green blade, it is likely that genes encoding supportive functions such as transporters might also be distinguished by their high transcript accumulation profiles. For example, in maturing stage, we detect C4 NADP-malic enzyme (ME) with a normalized expression value of 1178 and C3 NADP-ME with value of 3. Among BS- or M-enhanced transcripts, we compared the functional categories of the 100 most highly expressed genes at each stage (Fig. 4A). Note that long transcripts are not over-represented in this analysis because the LM-RNA-seq strategy includes an RNA amplification step that biases reads to the 3’ ends of transcripts. As for the major C4 enzymes, cell-type-enriched gene expression is apparent in the SST stage. The majority of highly expressed genes remain in the top-100 class throughout the three stages (Fig. 4A and Supplementary dataset S3). As expected, most are photosynthesis-associated, but protein synthesis and degradation and redox are also represented. Several transporters are highly expressed at all stages, including two aquaporins and two mitochondrial membrane transporters (DiT2 and AAC) all of which are BS-enriched. The C4-related DiT1 transporter is M-enhanced and highly expressed in all stages (Taniguchi et al., 2004).
Fig. 4.
Most highly expressed DE genes present throughout blade. (A) The BS or M DE lists were filtered for the top 100 most highly expressed DE genes in each of the three developmental stages. For each section, genes were placed in Mapman categories and plotted by number of members. The categories with the most members were as expected and remained high throughout in both cell types. (B) Venn diagram of genes in the top-100 DE set for each developmental stage. Many genes were found in top-100 of all three stages. The top-100 set of maturing stage shared the most with those of the other two stages. In this set, only 14% of BS genes and 3% of M genes were maturing stage-unique. In contrast, 19% of M genes in SST and 26% of M genes in the mature stage top-100 sets were stage-unique.
Several of the highest cell-enriched transcripts are associated with activities proposed to support C4 photosynthesis. Transcripts for the subunits of the NAD(P)H dehydrogenase (NDH) complex are notably high in BS cells at all three stages, consistent with the high protein levels previously observed in BS chloroplasts (Darie et al., 2006; Majeran et al., 2008). The corresponding activity is proposed to be important for preventing CO2 leakage from the BS when ME decarboxylation is faster than Rubisco carboxylation (Takabayashi et al., 2005). M cells accumulate high levels of transcripts for isoprenoid and tetrapyrrole-pathway activities that produce chlorophyll and other structural components for photosynthesis and respiration. Many of the highly expressed and cell-biased transcripts encoding currently unknown activities or structures may be additional components that support C4 capacity.
A subset of BS-M differentially expressed (DE) genes were only among the top-100 most highly expressed for a single stage. In M cells, transcripts from 19 DE genes accumulate to very high levels only in the SST stage (Fig. 4B). Three are highly abundant at maturing stage only and 26 at mature stage only. In BS cells, 21, 14, and 16 are at very high levels in SST, maturing, and mature stages only. Although found to be differentially expressed, most of these very highly expressed genes are not absolutely cell-specific. Proteomic data from the mature stage (Friso et al., 2010; Majeran et al., 2010) provides evidence for 69% of the corresponding predicted proteins in BS cells and 47% in M cells, with distribution ratios consistent with the transcript ratios.
Those very highly expressed only at SST stage include BS-enriched genes involved in lipid synthesis, a plastid development protein, and chloroplast chaperonins, and M-enriched genes for four ribosomal proteins, two tetrapyrrole synthesis proteins, and other chloroplast-related proteins, all presumably associated with building chloroplast capacity. In contrast, the M-enriched transcripts that were in the top-100 set only at mature stage are chloroplast-related, but seem to be dedicated to regulation or protection. Among these are transcripts for fibrillin, a plastoglobule protein thought to protect against stress (Singh and McNellis, 2011); peroxiredoxin, an antioxidant enzyme; and pyruvate phosphate dikinase (PPDK) regulatory protein, which controls PPDK activity by phosphorylation (Burnell and Chastain, 2006; Chastain et al., 2008; Friso et al., 2010). BS-enriched transcripts at this stage include genes associated with energy capture (H(+)-ATPase, GPT2 transporter), photorespiration (glycine cleavage H protein) and sulfate transport.
Stage-enhanced DE transcripts: SST stage includes putative BS secondary wall cellulose synthase; mature stage-enhanced DE genes are enriched in stress-related classes
The SST and mature stages exhibited notable examples of DE transcripts that are very low or nearly absent at the other two stages; the maturing stage exhibited few such examples (Fig. 5). Gene lists were generated such that the normalized expression value of less than 0.2 in the other two sections was considered non-expressed. For most of these stage-enhanced genes, the level of expression is much less than that for C4 pathway genes (Supplementary dataset S4). The following are highlights for the two stages:
Fig. 5.
DE genes expressed in only one stage. DE genes that were expressed in only one of the three stages are plotted for each cell type. The number is highest for the SST stage and smallest for maturing stage for both cell types.
Sink–source transition stage
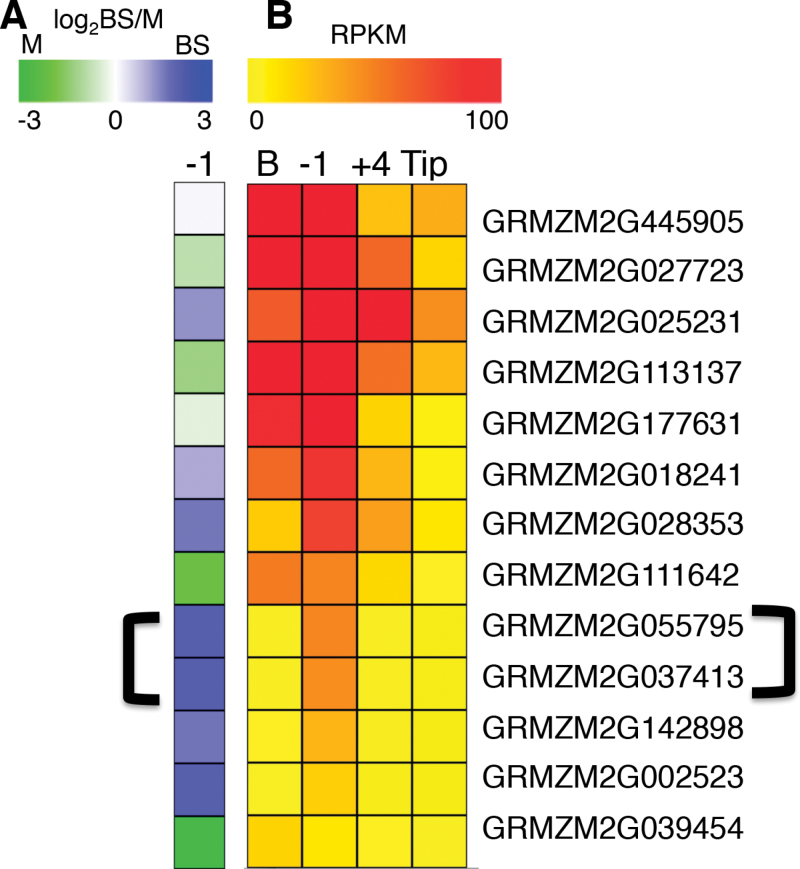
The SST stage-enhanced transcripts include BS-specific sucrose synthase, consistent with the prevailing sink metabolism, and cellulose synthase (CES), perhaps associated with strengthening of BS physical properties. According to corresponding proteomic data from the same source material (Majeran et al., 2010), levels of this sucrose synthase protein decline from (earlier stage) leaf base to SST and subsequent stages. The SST stage uniquely exhibits transcripts from numerous secondary wall synthesis-associated genes. Among seven cell wall-related BS-enhanced genes are two maize CES genes closely related to each other and to the CESA8/IRX1 Arabidopsis gene, which causes enhanced drought and osmotic stress tolerance when mutant (Turner and Somerville, 1997; Taylor et al., 2003). Secondary cell walls are laid down following cell expansion to enhance mechanical strength (Endler and Persson, 2011). All but one (GRMZM2G039454) of the many maize CES homologues in the CES POG are most highly expressed in the SST stage (Fig. 6). The most highly expressed (GRMZM2G445905) is not DE, but equal in both cell types. Also consistent with this SST-localized and BS-specific developmental pattern are transcripts for the maize homologue of the secondary wall master regulator NST1 (GRMZM2G171395), three lignin biosynthetic genes and a laccase, suggesting that a burst of BS-specific wall strengthening occurs immediately before blade emergence (Mitsuda et al., 2007; Berthet et al., 2011; Wang and Dixon, 2012). Also sharing this SST-localized, BS-specific pattern are three RALFs (rapid alkalinization factors) generally associated with growth inhibition (Bedinger et al., 2010), and here perhaps terminating cell expansion for BS cells before the source transition.
Fig. 6.
Cellulose synthase genes expressed in BS at SST stage. (A) A heat map representing the log2 BS/M ratio for a family of related CesA homologues where blue represents BS and green M. Two BS expressed members indicated by a bracket were found only expressed in the SST stage. (B) Heat map from low (yellow) to high (red) represents the whole section RNA-seq data (Li et al., 2010) for the same genes. Note that the two bracketed BS genes are also only present in the SST stage.
Several other sink-related genes are expressed in the SST stage and decline in subsequent stages. These include invertase 2 (IVR2, GRMZM2G089836, M-enriched), which is active in sink tissues (Kim et al., 2000), and ZmABI39 (GRMZM2G172621, BS/M equal) a member of the ABI3-VP1 family that is in the same orthologue group as the Arabidopsis gene HSI2, a sink tissue repressor of seed maturation genes (Tsukagoshi et al., 2005). Several amino acid transporters serve sink tissue in Arabidopsis (Su et al., 2004; Hammes et al., 2006). In maize, one amino acid transporter candidate (GRMZM2G404888, BS-biased at SST) is expressed most highly in the leaf base and SST stages (Li et al., 2010; Supplementary dataset S2).
A sink-tissue-associated calcium-dependent kinase family (CPDK, CPK) is important in stress response (Ludwig et al., 2004). In maize, 40 CPKs are identified (Kong et al., 2013), four of which (ZmCPK5, 23, 29, 30) show expression levels highest in the leaf base and decrease in subsequent stages (Li et al., 2010; Sekhon et al., 2011). We observe that ZmCPK29 and ZmCPK30 (homeologues) are M-enhanced at SST, ZmCPK23 is BS-enhanced, and ZmCPK5 is uniformly expressed (Supplementary dataset S2).
Mature stage
The mature stage seems to undergo a decline in photosynthesis-related transcripts and a rise in stress-related transcripts, as has been noted in two previous transcriptome studies (Li et al., 2010; Pick et al., 2011). We observe 104 genes that are mature- and M-enhanced and 82 genes that are mature- and BS-enhanced. The M-enhanced list is rich in maize genes in POGs associated with stress responses in Arabidopsis. For example, the maize gene GRMZM2G066870 corresponds to Arabidopsis AT3G05880 (RARE-COLD-INDUCIBLE 2A) a gene induced by low temperatures, dehydration, salt stress, and ABA (abscisic acid) (Capel et al., 1997; Nylander et al., 2001). Of the five transcription factors included in the M-specific group, four have Arabidopsis homologues that are induced by stress conditions including H2O2 (At1g10585), jasmonic acid (JA; AtWRKY50-At5g26170; Gao et al., 2011), salt and other abiotic and biotic stresses (AtWRKY33-At2g38470; Birkenbihl et al., 2012; Maekawa et al., 2012), carbon/nitrogen status, and various abiotic stresses (bHLH92- At5g43650; Jiang et al., 2009). Also in the M-specific list are a raffinose synthase gene potentially related to stress (Nishizawa et al., 2008) and several ARD genes likely to act in a stress-related methionine salvage pathway, as described below in the Discussion. The BS-specific list includes a smaller number of genes potentially related to stress.
Differential expression of C4-related genes
DE patterns in BS and M cells beginning at an early stage may be a common feature of genes with significant roles in C4 metabolism and Kranz anatomy, as shown for the well-characterized C4 pathway enzymes. The following are examples in support of this hypothesis. The TFs Golden2 (G2, GRMZM2G087804) and Golden Like1 (GLK1, GRMZM2G026833) influence BS/M chloroplast specialization, and are cell-enriched (G2 in BS, GLK1 in M; Hall et al., 1998; Rossini et al., 2001). Both ZmG2 and ZmGLK1 are expressed in the leaf base, but peak at the mature stage (Li et al., 2010). Two transcriptome studies (Li et al., 2010; Chang et al., 2012) confirm the DE patterns for mature stage BS and M cells. We find that these patterns are present at all three developmental stages, as expected for regulators of cell specification (log2 BS/M G2: 0.93, 2.36, 1.76; GLK1: –1.8, –1.4, –1.7). Another example, the mitochondrial transporter A BOUT DE SOUFFLE (BOU) influences photorespiration in Arabidopsis (Eisenhut et al., 2013), and the maize homologue is likely to be C4-related. We find that ZmBOU (GRMZM2G024823) is expressed robustly in a highly cell-biased fashion at all three stages, from SST to mature (log2 BS/M, 1.6, 3.2, 2.8).
The RBCL RNA S1-Binding domain protein (RLSB, GRMZM2G087628), binds the mRNA of the large subunit of Rubisco (rbcL) and influences rbcL expression (Bowman et al., 2013). Although RLSB protein is detected only in BS chloroplasts, a transcriptome study detected the presence of transcript in M cells (Chang et al., 2012). We find this pattern is developmentally dynamic: the SST stage exhibits a BS-bias (log2 BS/M 0.3), maturing stage an M-bias (log2 BS/M –1.3), and mature stage a slight M-bias (log2 BS/M –0.2). Possibly consistent with this, a transgenic knockdown of the RLSB transcript in maize had a large effect early on, but fades as the leaf matures (Bowman et al., 2013).
As a final example of a C4-related DE pattern, the maize Scarecrow TF homologue ZmSCR was shown recently to be critical for Kranz patterning of BS and M cells (Slewinski et al., 2012). We observe that ZmSCR (GRMZM2G131516) is significantly BS-biased at all three developmental stages sampled (log2 BS/M 0.4, 0.7, 0.9); it is particularly highly expressed in the leaf base (Li et al., 2010).
The consistently BS/M-biased patterns of expression of these and other C4-related genes suggests that further interrogation of our data set based on DE patterns can be used to infer other likely candidates with roles in Kranz anatomy and C4 metabolism
BS/M specificity as a filter for TF candidates with C4 roles
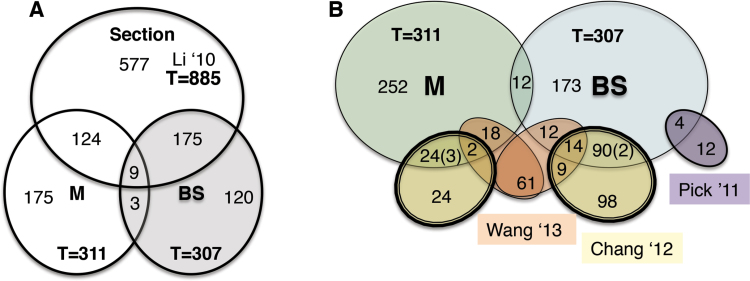
Previously, 938 genes annotated as transcription factors (TFs) were identified as expressed at some stage during leaf development (Li et al., 2010). To compare this list with the BS/M DE-identified TFs, the previously generated list was first converted to B73 RefGen_v2 (5b.60) from v1 (4a.53), resulting in 885 possible leaf blade TFs that could be directly compared. The cell-specific transcriptomes in this study are from the same material and stages, with the exception of the leaf base, which is not amenable to LM. We find that 20% of the TF transcripts are BS-biased and 14% are M-biased (Fig 7A and Supplementary dataset S5). Transcripts for eight of nine TFs initiate expression in SST stage with M-enhanced expression and switch to BS-enriched in later stages. A significant number of the TFs detected in cell-specific transcriptomes had not been detected in the whole leaf transcriptomes, suggesting that the cell-specific method is more sensitive to low levels of expression.
Fig. 7.
Comparing strategies for transcription factor identification. (A) Comparison of BS and M DE-identified TFs with those previously identified from the whole sectional data, after elimination of any genes not found in B73 RefGen_v2 (Li et al., 2010). More BS transcription factors overlap than M. The overlap between the BS and M represents expression patterns that switched DE cell-bias over the developmental gradient. (B) Comparison of BS and M DE-identified transcription factors with other published C4 transcription factor lists. Yellow ovals represent data from Chang et al., 2012; orange ovals represent data from Wang et al., 2013; and purple oval represents data from Pick et al., 2011. In the overlap between the Chang et al., 2012 data and the LM data (this study) the numbers in parenthesis represent expression patterns that changed cell-bias over the developmental gradient and thus were not restricted to the cell type identified by cell separation.
Several other studies used alternative approaches to identify candidate C4-related TFs (Pick et al., 2011; Chang et al., 2012; Wang et al., 2013), enabling comparison for consensus (Fig. 7B and Supplementary dataset S5). Note that these studies all used comparable versions of maize genome annotation. One study (Pick et al., 2011) obtained stage-specific but non-cell-specific transcriptomes from the maize leaf developmental gradient and identified candidate TFs with the same developmental dynamic as one of three key C4 pathway enzymes. That study identified TFs based on co-expression with the NADP-malic enzyme; we find four are BS-enhanced, although none of the other alternative methods identified them as C4-related candidates (Fig. 7B).
Another cell-specific transcriptome study (Chang et al., 2012), employing mechanical isolation of BS and M cells from more mature leaves of maize cultivar White Crystal, identified cell-specific TFs using a rigorous criterion for specificity (Ri>.99). 40% were also detected in our DE lists, which accept a greater window of differential expression in BS and M cells. Our analysis identified comparable numbers of BS and M-specific candidate TFs, whereas the study of Chang et al. (2012) identified many fewer M-specific candidates. For five (2%) of the genes, we detected significant transcript in the opposite cell type with a changing developmental dynamic (i.e. our study-GRMZM2G336533: log2 BS/M SST=0.8, maturing=0.9, mature=–1.1; Chang et al. 2012 study; M cell-specific).
A third study (Wang et al., 2013) employed the comparison of transcriptomes of leaf primordia and husk leaves to identify genes active at early times establishing the C4 Kranz features, which are diminished or lacking in husk leaves. Although comparing different stages and materials, the results of this study overlapped with our results to a remarkable extent. Our DE data identified 46 TFs of the 116 C4-related candidates found by this method (40%; 26 BS-specific, 20 M-specific). Of the remaining 70, we measure non-cell-specific expression of 58 (83%) in the leaf blade and leaf-base expression of an additional 8 (Li et al., 2010). We find that most of the 46 cell-specific TFs are expressed at a high level at the SST stage, suggesting that they may be dedicated to late C4-related development, such as refinement of BS-M cell differentiation and barriers.
The subset of TFs that were identified by all three strategies was detected with the same cell-specificity by both mechanical cell isolation and microdissection. 14 BS-specific and two M-specific TFs were identified in three studies and 178 were identified in at least two studies. The consensus of these studies provides support for a considerable number of candidate TFs that might be profitably investigated further for specific roles in C4 development and function.
Discussion
Cell-specific transcriptomes from successive developmental stages provide novel tools
We show that cell-specific transcriptomes from BS and M cells at successive developmental stages can augment the insights into C4-related processes that can be gained from BS/M studies of a single mature stage or from multiple stages of whole leaves. First, our studies resolve the early BS- and M-specificity of transcripts for C4-related processes, before the sink–source transition and before significant illumination. The early expression of C4-related genes was observed previously in whole leaves (Li et al., 2010; Pick et al., 2011); our data show that cell-specificity is in place from these earliest stages. Second, these transcriptomes enable the assignment of roles to C4-related gene family members primarily expressed at an early developmental stage, the matching of candidate interaction partners that are active early, and the identification of C4-related transcription factors, all of which may be absent or difficult to detect in later mature stages. Third, the availability of multiple stages from the same leaves enables the profiling of dynamic cell-specificity for individual genes, some of which are present or cell-specific only in a single stage. Finally, the transcriptome stages reveal a succession of cell-specific activities and roles not previously viewed by other studies, as described below.
Evidence of stress-related transcription in Kranz cell in mature (tip) stage
We observed several stress-related genes among the highest expressed genes in mature stage BS and M cells. Mature M cells accumulate transcripts from four acireductone dioxygenases (ARD), which have been associated with methionine salvage in Arabidopsis. In the ARD-dependent Yang Cycle (Yang and Hoffman, 1984), S-adenosyl methionine (SAM) is converted to 5-methyl-thioadenosine (MTA). There are four ARD genes in Arabidopsis, three of which (AtARD1, 2, 3) are expressed in phloem cells along with other identified genes in the cycle (Pommerrenig et al., 2011). AtARD4 is expressed in the phloem and BS cells.
In maize, our transcriptome data suggest that the entire methionine cycle may function in cooperating BS and M cells; data is lacking for phloem. We observe M-enhanced expression in mature stage of four ZmARD genes, all homologous to AtARD4. For the single maize gene (GRMZM2G165998) homologous to AtARD1, 2 and 3, we observe strong BS-enhanced expression at all stages, consistent with detection of the protein in BS cells (Sun et al., 2009). For methionine salvage steps from MTA to SAM, the homologous maize genes are either BS-enhanced or equal in both cell types. However, the two relevant activities for MTA steps in maize—an S-adenosylmethionine decarboxylase (GRMZM2G154397) and an arginine decarboxylase (GRMZM2G374302)—seem to be M-enhanced. Both steps generate CO2 and lead to putrescine, which is associated with stress tolerance and regulation of photosynthesis (Kusano et al., 2008; Ioannidis et al., 2012). Polyamines also protect against oxidative damage (Rhee et al., 2007).
Transcripts for several additional stress-related activities are notably cell-specific and abundant at the mature tip. Among these are those for trehalose metabolism (BS-specific and M-specific members), hexokinase signalling (BS-specific), AKIN10 homologues (M-specific at SST, BS-specific at maturing/mature; Fragoso et al., 2009), and a variety of genes related to reactive oxygen species.
The presence of stress-related transcripts is unlikely to be an artefact of experimental conditions that cause leaf tips to dehydrate or senesce prematurely, as they are observed in many independent studies, with plants grown under varying conditions (Li et al., 2010; Pick et al., 2011; Chang et al., 2012). Instead, the encoded activities are likely to belong to a battery of developmental mechanisms that address the potentially damaging by-products of peak photosynthetic and assimilatory activities.
Use of homeologous genes in maize for C4-related processes
The filtered lists contain many pairs of homologous genes. These genes may be homeologues (syn: ohnologues, syntenic paralogues) resulting from an ancient whole genome duplication in the maize lineage (Schnable et al., 2009). The maize whole genome duplication has been dated to 5–12 million years ago (Swigonova et al., 2004) and is shared by the sister genus Tripsacum, but not by the core Andropogoneae (Bomblies and Doebley, 2005), placing the whole genome duplication shortly after the origin of C4 photosynthesis (Aliscioni et al., 2012). Previous work in the crucifers has demonstrated that retention of whole genome duplicates may act as a way for plants to increase flux through whole metabolic pathways (Bekaert et al., 2011), a trait likely under high levels of selection following the transition to a new photosynthetic method. A second model predicts that gene pairs descended from an ancestral gene expressed in both the M and BS might subfunctionalize (Force et al., 1999) with one gene copy specializing in M expression/function and the other in BS expression/function.
In the case of the TIM family, the model of increasing metabolic capacity is more consistent with our observations than the subfunctionalization model. The TIM family contains two pairs of homeologous genes (four total genes). Each gene pair shares a conserved pattern of expression, with one pair of genes expressed preferentially in the M and the other expressed preferentially in the BS. Another example is the two sets of transcription factors identified in three studies (this study; Chang et al., 2012; Wang et al., 2013). Only one pair (Dof-like) are homeologues and they have very similar expression in this dataset. About a quarter of the closely examined genes (100) that were found in various filtered DE lists were homeologues and were expressed in a similar pattern. This observation suggests that homeologue retention may be an active mechanism to increase protein levels of some activities for C4 cycle efficiency and should be investigated more thoroughly.
Comparison of transcriptome data from alternative isolation methods for BS and M cells
Studies of BS and M cells from maize and other C4 species have employed a variety of methods to distinguish the contributions of the two cell types, sometimes with varying results, as described in the Results. The LM method we employed has the advantage that M and BS cells can be isolated from the same preparation, using the same technique. As has been suggested, some level of cross-contamination of adjacent cell types is probably unavoidable, influencing the absolute value of BS-M differences, but not the detection of significant differences. In this study, we evaluated the entire class of differentially expressed genes, from ratios of expression near 1 to absolutely cell-specific, with >85% with a ratio greater than 2. For individual genes, we find mostly good correlation with the BS-M distribution ratios found in proteomic studies of the same source material (Friso et al., 2010; Majeran et al., 2010), with the caveat that the abundance of a significant number of proteins differs from their relative transcript abundance. Our transcriptome data for BS cells is generally consistent with mature stage BS transcriptome data produced in a recent study employing mechanical cell separation (Chang et al., 2012). However, our data for M cells was less consistent with the M cell transcriptome data from that study, which employed a protoplasting method to isolate M cells. Despite the somewhat different values produced by the alternative methods, genes and patterns identified independently by multiple methods are likely to be the strongest candidates for involvement in C4-related and other cell-specific pathways.
Supplementary data
Supplementary data are available on JXB online.
Dataset S1. All BS and M DE genes.
Dataset S2. DE genes for C4 and other genes mentioned in text.
Dataset S3. The top 100 most highly expressed DE genes in each section.
Dataset S4. DE genes expressed in only one section.
Dataset S5. Transcription factor identification comparison.
Figure S1. Heat maps of log2BS/M for metabolic pathways related to C4.
Acknowledgements
The authors would like to thank James Schnable for discussions about maize homeologues and Zenghong Ding for analyses not included in this paper. This work was supported by National Science Foundation (US) Plant Genome Research Program awards 0701736 and 1127017 to TN, TPB, PL, and QS.
Glossary
Abbreviations:
- BS
bundle sheath
- DE
differentially expressed
- LM
laser microdissection
- M
mesophyll
- PPDP
plant proteome database (http://ppdb.tc.cornell.edu/)
- SST
sink–source transition.
References
- Aliscioni S, Bell HL, Besnard G, et al. 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. The New Phytologist 193, 304–312 [DOI] [PubMed] [Google Scholar]
- Amodeo G, Dorr R, Vallejo A, Sutka M, Parisi M. 1999. Radial and axial water transport in the sugar beet storage root. Journal of Experimental Botany 50, 509–516 [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TW, Carrayol E, Hirel B. 2000. Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 211, 800–806 [DOI] [PubMed] [Google Scholar]
- Bedinger PA, Pearce G, Covey PA. 2010. RALFs: Peptide regulators of plant growth. Plant Signaling & Behavior 5, 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC. 2011. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. The Plant Cell 23, 1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate — a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological 57, 289–300 [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, et al. 2011. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. The Plant Cell 23, 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE. 2012. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiology 159, 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Doebley JF. 2005. Molecular evolution of FLORICAULA/LEAFY orthologs in the Andropogoneae (Poaceae). Molecular Biology and Evolution 22, 1082–1094 [DOI] [PubMed] [Google Scholar]
- Bowman SM, Patel M, Yerramsetty P, Mure CM, Zielinski AM, Bruenn JA, Berry JO. 2013. A novel RNA binding protein affects rbcL gene expression and is specific to bundle sheath chloroplasts in C4 plants. BMC Plant Biology 13, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN, Chastain CJ. 2006. Cloning and expression of maize-leaf pyruvate, Pi dikinase regulatory protein gene. Biochemical and Biophysical Research Communications 345, 675–680 [DOI] [PubMed] [Google Scholar]
- Capel J, Jarillo JA, Salinas J, Martínez-Zapater JM. 1997. Two homologous low-temperature-inducible genes from Arabidopsis encode highly hydrophobic proteins. Plant Physiology 115, 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-M, Liu W-Y, Shih AC-C, et al. 2012. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiology 160, 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain CJ, Xu W, Parsley K, Sarath G, Hibberd JM, Chollet R. 2008. The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis possess a novel, unprecedented Ser/Thr protein kinase primary structure. The Plant Journal 53, 854–863 [DOI] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R. 2001. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiology 125, 1206–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cren M, Hirel B. 1999. Glutamine synthetase in higher plants regulation of gene and protein expression from the organ to the cell. Plant and Cell Physiology 40, 1187–1193 [Google Scholar]
- Darie CC, De Pascalis L, Mutschler B, Haehnel W. 2006. Studies of the Ndh complex and photosystem II from mesophyll and bundle sheath chloroplasts of the C4-type plant Zea mays . Journal of Plant Physiology 163, 800–808 [DOI] [PubMed] [Google Scholar]
- Denton AK, Simon R, Weber AP. 2013. C4 photosynthesis: from evolutionary analyses to strategies for synthetic reconstruction of the trait. Current Opinion in Plant Biology 16, 315–321 [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Planchais S, Cabassa C, et al. 2013. Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. The Plant Journal 73, 836–849 [DOI] [PubMed] [Google Scholar]
- Endler A, Persson S. 2011. Cellulose synthases and synthesis in Arabidopsis . Molecular Plant 4, 199–211 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso S, Espindola L, Paez-Valencia J, Gamboa A, Camacho Y, Martinez-Marajas E, Coello P. 2009. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiology 149, 1906–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. 2010. Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiology 152, 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra N, Coughlan SJ, Nelson T. 2013. The Arabidopsis leaf provascular cell transcriptome is enriched in genes with roles in vein patterning. The Plant Journal 74, 48–58 [DOI] [PubMed] [Google Scholar]
- Gao Q-M, Venugopal S, Navarre D, Kachroo A. 2011. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiology 155, 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. 2002. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Molecular Biology 49, 373–385 [PubMed] [Google Scholar]
- Hall LN, Roth R, Brutnell TP, Langdale JA. 1998. Cellular differentiation in the maize leaf is disrupted by bundle sheath defective mutations. Symposia of the Society for Experimental Biology 51, 27–31 [PubMed] [Google Scholar]
- Hammes UZ, Nielsen E, Honaas LA, Taylor CG, Schachtman DP. 2006. AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis . The Plant Journal 48, 414–426 [DOI] [PubMed] [Google Scholar]
- Hanba YT, Shibasaka M, Hayashi Y, Hayakawa T, Kasamo K, Terashima I, Katsuhara M. 2004. Overexpression of the barley aquaporin HvPIP2;1 increases internal CO(2) conductance and CO(2) assimilation in the leaves of transgenic rice plants. Plant and Cell Physiology 45, 521–529 [DOI] [PubMed] [Google Scholar]
- Heinen RB, Ye Q, Chaumont F. 2009. Role of aquaporins in leaf physiology. Journal of Experimental Botany 60, 2971–2985 [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology 61, 181–207 [DOI] [PubMed] [Google Scholar]
- Ioannidis NE, Cruz JA, Kotzabasis K, Kramer DM. 2012. Evidence that putrescine modulates the higher plant photosynthetic proton circuit. PloS One 7, e29864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Deyholos MK. 2009. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Molecular Genetics and Genomics 282, 503–516 [DOI] [PubMed] [Google Scholar]
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P. 2001. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiology 126, 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P. 2000. The role of aquaporins in cellular and whole plant water balance. Biochimica et Biophysica Acta 1465, 324–342 [DOI] [PubMed] [Google Scholar]
- Kim J-Y, Mahé A, Brangeon J, Prioul J-L. 2000. A maize vacuolar invertase, IVR2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiology 124, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Lv W, Jiang S, Zhang D, Cai G, Pan J, Li D. 2013. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano T, Berberich T, Tateda C, Takahashi Y. 2008. Polyamines: essential factors for growth and survival. Planta 228, 367–381 [DOI] [PubMed] [Google Scholar]
- Langdale JA. 2011. C4 Cycles: past, present, and future research on C4 photosynthesis. The Plant Cell 23, 3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC, Walker RP. 2003. Regulation and roles of phosphoenolpyruvate carboxykinase in plants. Archives of Biochemistry and Biophysics 414, 204–210 [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42, 1060–1067 [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JDG. 2004. CDPK-mediated signalling pathways: specificity and cross-talk. Journal of Experimental Botany 55, 181–188 [DOI] [PubMed] [Google Scholar]
- Maekawa S, Sato T, Asada Y, Yasuda S, Yoshida M, Chiba Y, Yamaguchi J. 2012. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Molecular Biology 79, 217–227 [DOI] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, et al. 2010. Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. The Plant Cell 22, 3509–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Zybailov B, Ytterberg AJ, Dunsmore J, Sun Q, van Wijk KJ. 2008. Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Molecular & Cellular Proteomics 7, 1609–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Lee J, Kichey T, et al. 2006. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. The Plant Cell 18, 3252–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. 2007. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis . The Plant Cell 19, 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T. 2010. Development of leaves in C4 plants: Anatomical features that support C4 metabolism. In: Raghavendra AS, Sage RF, eds. C4 Photosynthesis and Related CO2 Concentrating Mechanisms. Netherlands: Springer, 147–159 [Google Scholar]
- Nelson T. 2011. The grass leaf developmental gradient as a platform for a systems understanding of the anatomical specialization of C4 leaves. Journal of Experimental Botany 62, 3039–3048 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. 2008. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiology 147, 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander M, Heino P, Helenius E, Palva ET, Ronne H, Welin BV. 2001. The low-temperature- and salt-induced RCI2A gene of Arabidopsis complements the sodium sensitivity caused by a deletion of the homologous yeast gene SNA1. Plant Molecular Biology 45, 341–352 [DOI] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. The Plant Cell 23, 4208–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B, Feussner K, Zierer W, Rabinovych V, Klebl F, Feussner I, Sauer N. 2011. Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis . The Plant Cell 23, 1904–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R. 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. Plos Computational Biology 5, e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HJ, Kim E-J, Lee JK. 2007. Physiological polyamines: simple primordial stress molecules. Journal of Cellular and Molecular Medicine 11, 685–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini L, Cribb L, Martin DJ, Langdale JA. 2001. The maize Golden2 gene defines a novel class of transcriptional regulators in plants. The Plant Cell 13, 1231–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF, Christin P-A, Edwards EJ. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany 62, 3155–3169 [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M, Lyons E. 2012. Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biology and Evolution 4, 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M. 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proceedings of the National Academy of Sciences, USA 108, 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sekhon RW, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. 2011. Genome-wide atlas of transcription during maize development. The Plant Journal 66, 553–563 [DOI] [PubMed] [Google Scholar]
- Singh DK, McNellis TW. 2011. Fibrillin protein function: the tip of the iceberg? Trends in Plant Science 16, 432–441 [DOI] [PubMed] [Google Scholar]
- Slewinski TL. 2013. Using evolution as a guide to engineer Kranz-type C4 photosynthesis. Frontiers in Plant Science 4, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski TL, Anderson AA, Zhang C, Turgeon R. 2012. Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant and Cell Physiology 53, 2030–2037 [DOI] [PubMed] [Google Scholar]
- Su Y–H, Frommer WB, Ludewig U. 2004. Molecular and functional characterization of a family of amino acid transporters from Arabidopsis . Plant Physiology 136, 3104–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PDB, van Wijk KJ. 2009. PPDB, the plant proteomics database at Cornell. Nucleic Acids Research 37, D969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z, Lai JS, Ma JX, Ramakrishna W, Llaca V, Bennetzen JL, Messing J. 2004. Close split of sorghum and maize genome progenitors. Genome Research 14, 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi A, Kishine M, Asada K, Endo T, Sato F. 2005. Differential use of two cyclic electron flows around photosystem I for driving CO2-concentration mechanism in C4 photosynthesis. Proceedings of the National Academy of Sciences, USA 102, 16898–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Nagasaki J, Kawasaki M, Miyake H, Sugiyama T, Taniguchi M. 2004. Differentiation of dicarboxylate transporters in mesophyll and bundle sheath chloroplasts of maize. Plant and Cell Physiology 45, 187–200 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. 2003. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences, USA 100, 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomcal M, Stiffler N, Barkan A. 2013. POGs2: a web portal for facilitate cross-species inferences about protein architecture and function in plants. PLoS One 8, e82569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K. 2005. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiology 138, 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. 1997. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. The Plant Cell 9, 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R. 2003. The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425, 734–737 [DOI] [PubMed] [Google Scholar]
- Walker NS, Stiffler N, Barkan A. 2007. POGs/PlantRBP: a resource for comparative genomics in plants. Nucleic Acids Research 35, D852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-Z, Dixon RA. 2012. On-off switches for secondary cell wall biosynthesis. Molecular Plant 5, 297–303 [DOI] [PubMed] [Google Scholar]
- Wang P, Kelly S, Fouracre JP, Langdale JA. 2013. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. The Plant Journal 75, 656–670 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher-plants. Annual Review of Plant Physiology and Plant Molecular Biology 35, 155–189 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.