Abstract
The responses of long-term growth of plants under elevated CO2 have been studied extensively. Comparatively, the responses of plants to subambient CO2 concentrations have not been well studied. This study aims to investigate the responses of the model C3 plant, Arabidopsis thaliana, to low CO2 at the molecular level. Results showed that low CO2 dramatically decreased biomass productivity, together with delayed flowering and increased stomatal density. Furthermore, alteration of thylakoid stacking in both bundle sheath and mesophyll cells, upregulation of PEPC and PEPC-K together with altered expression of a number of regulators known involved in photosynthesis development were observed. These responses to low CO2 are discussed with regard to the fitness of C3 plants under low CO2. This work also briefly discusses the relevance of the data to C4 photosynthesis evolution.
Key words: Arabidopsis, C4 photosynthesis, evolution, low CO2, photorespiration, stress responses.
Introduction
The response of plants grown in lower CO2 concentrations has been much less studied than responses to elevated CO2 concentrations (Long et al., 2004, 2006; Ainsworth and Long, 2005; Gerhart and Ward, 2010). Among these limited studies, some have demonstrated that a large genetic variation in response to low CO2 exists among Arabidopsis accessions. For example, Sharma et al. (1979) screened 33 Arabidopsis accessions for survival time under limiting CO2 when grown side by side with C4 plants (Zea mays L.) in an air-tight chamber where CO2 concentration was reduced to below the compensation point of C3 plants and found a 1–2-week difference in the survival time in different accessions and also found substantial genetic segregation among F2 parents, with extreme differences in survival time near the CO2 compensation point.
Arabidopsis genotypes from different elevations show significant variation in the response of seed number when grown at low CO2 (20 Pa) (Ward and Strain, 1997). Ward et al. (2000) performed an artificial selection experiment using Arabidopsis for high seed number over five generations at low CO2 (20 Pa, or 200 ppm); the selected populations produced 25% more seeds and 35% more biomass on average than control populations which were randomly selected at the fifth generation when grown at low CO2. In addition, Ward and Kelly (2004) also observed a high level of genetic variation in survival, reproductive output, and total seed production among the Arabidopsis genotypes when grown at low CO2 (200 ppm). All these studies suggest that Arabidopsis has adaptive phenotypic plasticity in response to low CO2.
In a carbon starvation experiment, 5-week-old Arabidopsis rosettes treated with ambient (350 ppm) CO2 or compensation point (<50 ppm) CO2 were collected in the light for 4h to investigate responses to changing endogenous sugar concentrations in rosettes at the gene expression level using the GeneChip Arabidopsis ATH1 genome array (Bläsing et al., 2005). However, these studies have not addressed the mechanism of long-term responses of plants to low CO2.
This study conducted a survey of responses of C3 plants to long-term low CO2 treatments at the molecular level. Arabidopsis was chosen as the model system because its genome has been fully sequenced and is still the best annotated plant genome to date (The Arabidopsis Genome Initiative, 2000); the well-annotated Arabidopsis genome facilitates analysis of global gene expression using RNA-Seq technology. This study sequenced the transcriptome of 6-week old Arabidopsis seedlings grown under ambient CO2 (380 ppm) or low CO2 (100 ppm). The results are discussed with particular reference to the significance of the altered gene expression to the fitness of C3 plants under low CO2. The relevance of low CO2 to C4 evolution is also briefly discussed.
Materials and methods
Plant growth and harvest
Arabidopsis thaliana Columbia-0 (Col-0) seeds were imbibed in 0.1% (w/v) agar solution and incubated at 4 °C for 2 d to break dormancy. Imbibed seeds were germinated and grown in Pindstrup soil in a Percival incubator (NC-350HC-LC, Nihonika, Japan) in which CO2 gas can be accurately and stably controlled in the range of 100–3000 ppm. CO2 concentrations 100 and 380 ppm were applied in two separate chambers and maintained throughout this study. CO2 concentrations were monitored and maintained throughout the experiments. Plants were grown under a 8/16h light/dark cycle (photosynthetic photon flux density 150 μmol m–2 s–1) at 21 °C and 70% relative humidity. After 4 weeks, the photoperiod was changed to a 16/8h light/dark cycle for a further 2 weeks. On day 42, samples were taken during the middle of the light period and mature expanded rosette leaves from 10–15 individual plants were harvested, immediately frozen in liquid nitrogen, and stored at –80 °C until use. The samples were taken from 12 individual pots.
Morphological data collection
Scanning electron microscopy and transmission electron microscopy were used to observe the changes of ultrastructure by low CO2. The number of stomata was counted in four fields of view from the fully expanded leaves of no less than eight individual plants for each treatment (Supplmentary Fig. S1 available at JXB online).
RNA preparation and sequencing
Total RNA was prepared with TRIzol (Invitrogen Life Technologies, Shanghai, China), according to the manufacturer’s instructions. Following extraction, total RNA was purified using a RNeasy Mini Kit including on-column DNase digestion (Qiagen, Shanghai, China). Purified RNA was checked for integrity and quality using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The cDNA library was constructed for sequencing as described in Illumina TruSeqTM RNA sample preparation version 2 guide (catalog no. RS-930–1021). Sequencing was performed using a Illumina HiSeq 2000 (Illumina, San Diego, USA).
Mapping and quantification of sequence reads
Clean reads were mapped onto the latest A. thaliana Col-0 genome assembly (TAIR 10) or a minimal set of coding sequences of the TAIR 9 genome release (Gowik et al., 2011) using the bowtie version 0.12.7 (Langmead et al., 2009). The best hit of each read with a maximum of three nucleotide mismatches was used (-v 3 --best). The raw digital gene expression counts were normalized using the RPKM (reads/kb/million) method (Mortazavi et al., 2008; Nagalakshmi et al., 2008; Supplementary Tables S1 and S2 available at JXB online).
To identify differentially expressed genes, an expression profile matrix was built which integrated the digital gene expression count for each gene in each library, total gene count for each condition were used as background to check if a gene is significantly differentially expressed in low and CO2 normal conditions by applying the chi-squares test. A FDR-corrected P-value was calculated using the formula where i represents the ascending order of P-values, p(i) represents the ith P-value, C represents a chosen constant, and N represents the size of dataset (Benjamini and Hochberg, 1995). Significantly differentially expressed genes were picked following the criteria P<0.001, FDR<0.025, |log2Ratio|≥1.2.
Results
Effects of long-term low CO2 on biomass growth, stomata density, and chloroplast ultrastructure
CO2 is the major source of carbon for photosynthesis and plays a vital role in plant growth. High CO2 often increases the growth and reproduction of C3 annuals, whereas low CO2 decreases growth (Ward et al., 2000; Ward, 2005). Previous studies showed that minimum CO2 concentrations between 180 and 200 ppm during the Last Glacial Maximum were already stressful on modern C3 plants (Dippery et al., 1995; Ward, 2005); therefore, this work set low CO2 concentration as 100 ppm. Arabidopsis plants grown at 100 ppm for 6 weeks were much smaller than those grown under normal CO2 (380 ppm) (Fig. 1). In addition, low CO2 led to a slight delay in flowering time (data not shown). The results showed that low CO2 (100 ppm) had a dramatic impact on the growth of the C3 plant Arabidopsis.
Fig. 1.
Arabidopsis thaliana Col-0 grown under normal CO2 (380 ppm) and low CO2 (100 ppm) for 4 weeks (A; 8/16h light/dark cycle (photosynthetic photon flux density 150 μmol m–2 s–1, 21 °C, 70% relative humidity) and for 6 weeks (B; 4 weeks under conditions as for A plus 2 weeks under a 16/8h light/dark cycle).
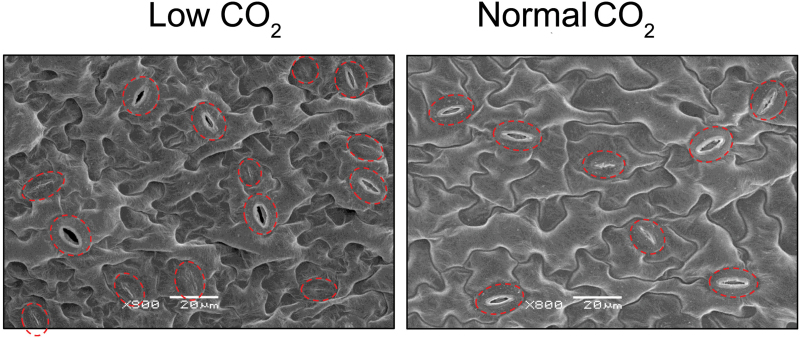
Stomata control the entry of CO2 into the leaves of plants for photosynthesis. There is a strong inverse correlation between atmospheric CO2 and stomatal density (the number of stomata per unit area) (Franks et al., 2012). This work examined the stomatal density of abaxial (lower) leaf blade epidermis of Arabidopsis plants grown at either low CO2 or normal CO2 for 6 weeks (Supplmentary Fig. S1). As expected, stomatal density was significantly higher (mean±SE 509±59mm–2) in plants grown at low CO2 compared to plants at normal CO2 (297±54mm–2) (Fig. 2).
Fig. 2.
Effect of low atmospheric CO2 on stomatal density. Representative scanning electron micrographs of abaxial (lower) leaf blade epidermis of Arabidopsis grown under low CO2 (100 ppm) or normal CO2 (380 ppm) for 6 weeks. Dashed lines indicate stomata. Bars, 20 μm.
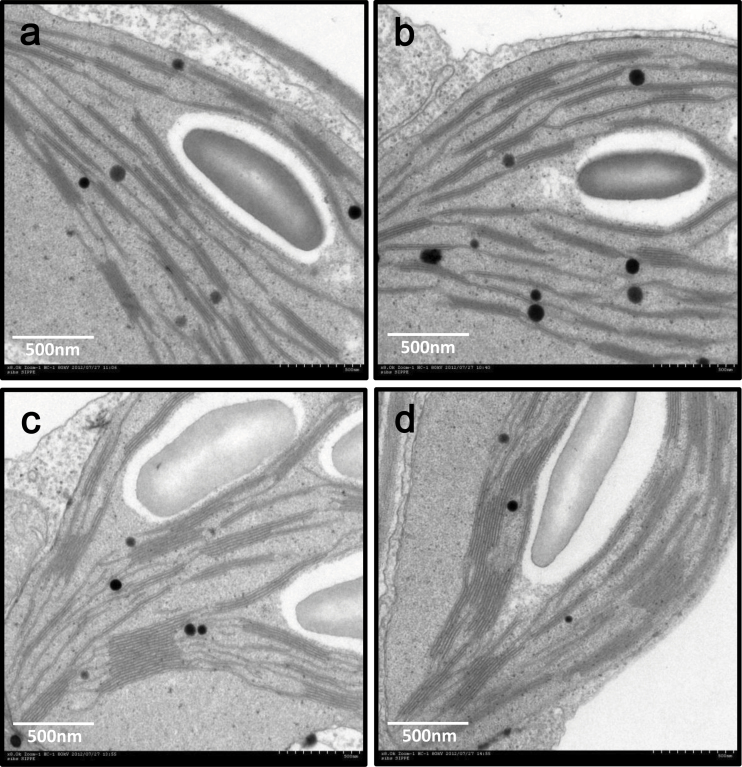
In plants, photosynthesis occurs exclusively in the chloroplast, and the photosystems (PSI and PSII) exist on the thylakoid membrane inside a chloroplast. PSII is limited to granal thylakoids, while PSI exists exclusively in the thylakoids exposed to the stroma (Albertsson, 1995; Dekker and Boekema, 2005; Sakamoto et al., 2008). The ultrastructure of mature leaves under low CO2 were examined using transmission electron microscopy, and the size and the arrangement of bundle sheath cells and mesophyll cells was not changed, while Arabidopsis grown under low CO2 showed decreased stacking in chloroplast grana in both mesophyll and bundle sheath cells under low CO2 compared to normal CO2 (Fig. 3).
Fig. 3.
Effect of low CO2 on chloroplast ultrastructure. Representative transmission electron micrographs of ultrastructure of Arabidopsis grown under low CO2 (A, B) or normal CO2 (C, D) for 6 weeks: (A and C) mesophyll cell; (B and D) bundle sheath cell. Bars, 500nm.
Some C4-cycle genes were upregulated under low CO2
The mRNA-seq analysis to compare transcriptomes between closely related C4 and C3 species within the genus Flaveria and Cleome using Arabidopsis as the reference genome defined a list of enzymes, transporters, and regulatory proteins required for the core C4 cycle (Bräutigam et al., 2011; Gowik et al., 2011). It has been reported that Arabidopsis shows the characteristics of C4 photosynthesis in midveins (Brown et al., 2010), but nothing is known about the plasticity of these characteristics.
In order to check whether C4-related characteristics can be regulated by low CO2 stress, the transcript abundances of putative C4-related genes were examined. The transcript encoding the enzyme phosphoenolpyruvate carboxylase (PEPC, At2g42600) showed 2.10-fold higher transcript abundance, followed by PEPC kinase (PEPC-K, At1g08650) with a 1.99-fold increase in abundance (Table 1 and Supplementary Table S3 available at JXB online). In addition, the transcript abundances for the genes encoding alanine aminotransferase (At1g17290), chloroplast NAD-dependent malate dehydrogenase (At3g47520), pyruvate orthophosphate dikinase regulatory protein (At4g21210), inorganic pyrophosphatase 2 (At2g18230), chloroplast dicarboxylate transporter 1 (At5g12860) and 2 (At5g64280) showed trends of upregulation but their fold changes were less than 2.
Table 1.
Transcription abundance of C4-cycle genes and C4-related transportersReads were mapped onto the latest Arabidopsis thaliana Col-0 genome assembly (gene mapping) or a minimal set of coding sequences of the TAIR 9 genome release (core set mapping) using bowtie. Low: low CO2, 100 ppm; Nor: normal CO2, 380 ppm; rpkm, reads per kilobase per million mapped reads. AlaAT, alanine aminotransferase; AspAT, aspartate amino transferase; cpNAD-MDH, chloroplast NAD-dependent malate dehydrogenase; Dit, chloroplast dicarboxylate transporter; PEPC, phosphoenolpyruvate carboxylase; PEPC-K, PEPC kinase; PEP-CK, PEP carboxykinase; PPA2, inorganic pyrophosphatase 2; PPDK-RP, pyruvate orthophosphate dikinase regulatory protein; PPT1, phosphoenolpyruvate/phosphate translocator 1; TPT, triose phosphate transporter; –, no expression detected.
| Gene ID | Protein | Gene mapping | Core set mapping | ||||
|---|---|---|---|---|---|---|---|
| Low (rpkm) | Nor (rpkm) | Fold change | Low (rpkm) | Nor (rpkm) | Fold change | ||
| At1g08650 | PEPC-K | 26.220 | 13.206 | 1.985 | 30.156 | 15.230 | 1.980 |
| At1g17290 | AlaAT | 56.839 | 48.904 | 1.162 | 65.884 | 57.025 | 1.155 |
| At1g62800 | AspAT | 0.707 | 2.388 | 0.296 | 0.916 | 2.701 | 0.339 |
| At2g18230 | PPA2 | 10.288 | 6.468 | 1.591 | 11.812 | 7.459 | 1.584 |
| At2g42600 | PEPC | 197.604 | 93.979 | 2.103 | 227.099 | 108.523 | 2.093 |
| At3g47520 | cpNAD-MDH | 74.044 | 64.394 | 1.150 | 85.015 | 74.261 | 1.145 |
| At4g21210 | PPDK-RP | 213.075 | 191.911 | 1.110 | 244.822 | 221.204 | 1.107 |
| At4g37870 | PEP-CK | 26.606 | 43.413 | 0.613 | 30.589 | 50.065 | 0.611 |
| At5g12860 | Dit1 | 356.959 | 275.153 | 1.297 | 409.991 | 317.354 | 1.292 |
| At5g33320 | PPT1 | – | – | – | 44.181 | 50.558 | 0.874 |
| At5g46110 | TPT | 508.799 | 533.917 | 0.953 | 584.330 | 615.779 | 0.949 |
| At5g64280 | Dit2 | 27.003 | 20.059 | 1.346 | – | – | – |
Photorespiratory genes showed trends of upregulation under low CO2
Low atmospheric CO2 concentration would increase photorespiration, so this work also examined the transcript abundances of photorespiration genes. Nearly all genes showed trends of upregulation in plants grown under low CO2 compared with those under normal CO2 (Table 2 and Supplementary Table S4 available at JXB online), except for the gene encoding glycine decarboxylase L-protein (mtLPD1; At1g48030); however, the fold changes were all less than 2. The differential responses of genes involved in the photosynthetic light reactions, Calvin Benson cycle, and ABA and IAA metabolisms were shown in Supplementary Tables S8–11 available at JXB online.
Table 2.
Transcription abundance of photorespiration genesThe genes in bold represent these that plays a major function in photorespiration and the knockout results in a low CO2-sensitive phenotype (Bauwe, 2011). Reads were mapped onto the latest Arabidopsis thaliana Col-0 genome assembly (gene mapping) or a minimal set of coding sequences of the TAIR 9 genome release (core set mapping) using bowtie. Low: low CO2, 100 ppm; Nor: normal CO2, 380 ppm; rpkm, reads per kilobase per million mapped reads.
| Gene ID | Enzyme | Gene | Gene mapping | Core set mapping | ||||
|---|---|---|---|---|---|---|---|---|
| Low (rpkm) | Nor (rpkm) | Fold change | Low (rpkm) | Nor (rpkm) | Fold change | |||
| At1g11860 | Glycine decarboxylase T-protein | GLDT1 | 909.564 | 789.090 | 1.153 | 1044.524 | 909.989 | 1.148 |
| At1g23310 | Glutamate:glyoxylate aminotransferase | GGT1 | 461.752 | 399.672 | 1.155 | 538.428 | 465.798 | 1.156 |
| At1g48030 | Glycine decarboxylase L-protein | mtLPD1 | 250.934 | 270.703 | 0.927 | – | – | – |
| At1g68010 | Hydroxypyruvate reductases | HPR1 | 302.977 | 286.059 | 1.059 | 347.873 | 329.958 | 1.054 |
| At1g70580 | Glutamate:glyoxylate aminotransferase | GGT2 | 39.407 | 23.536 | 1.674 | – | – | – |
| At1g80380 | l-Glycerate 3-kinase | GLYK | 130.215 | 118.051 | 1.103 | 149.539 | 136.138 | 1.098 |
| At2g13360 | Alanine:glyoxylate aminotransferase | AGT1 | 1069.682 | 953.983 | 1.121 | 1228.446 | 1100.193 | 1.117 |
| At2g26080 | Glycine decarboxylase P-protein | GLDP2 | 87.272 | 75.611 | 1.154 | 216.751 | 173.881 | 1.247 |
| At3g14415 | Glycolate oxidase | GOX2 | 497.994 | 473.499 | 1.052 | 571.960 | 546.046 | 1.047 |
| At3g14420 | Glycolate oxidase | GOX1 | 698.380 | 617.984 | 1.130 | 801.954 | 712.742 | 1.125 |
| At4g33010 | Glycine decarboxylase P-protein | GLDP1 | 800.116 | 599.088 | 1.336 | – | – | – |
| At4g37930 | Serine hydroxymethyltransferase | SHM1 | 1188.138 | 842.873 | 1.410 | 1364.293 | 972.136 | 1.403 |
Chloroplast biogenesis- and maintenance-related genes showed differential expression in low CO2
Given the differential expression of genes involved in chloroplast biogenesis and maintenance between the C3 and C4 Flaveria species (Gowik et al., 2011) and the altered chloroplast ultrastructure between low CO2 and normal CO2 (Fig. 3), this work examined the transcript abundances of genes involved in chloroplast biogenesis and maintenance under low CO2 and compared them with previously identified genes differentially expressed between C3 and C4 species (Gowik et al., 2011) (Table 3 and Supplementary Table S5 available at JXB online). All the chloroplast biogenesis- and maintenance-related genes upregulated by low CO2 shown in Table 3 were also upregulated in C4 Flaveria species, and five genes downregulated by low CO2 (At5g52540, At1g52290, At5g20720, At2g32180, and At3g19820) were also downregulated in C4 Flaveria species (Gowik et al., 2011); however, only At44446, At5g52540, At3g17040, and At1g52290 showed a ratio of expression abundance greater than 2.
Table 3.
Transcript abundance of genes related to chloroplast biogenesis and maintenanceReads were mapped onto the latest Arabidopsis thaliana Col-0 genome assembly (gene mapping) or a minimal set of coding sequences of the TAIR 9 genome release (core set mapping) using bowtie. Low: low CO2, 100 ppm; Nor: normal CO2, 380 ppm; rpkm, reads per kilobase per million mapped reads.
| Gene ID | Protein | Gene mapping | Core set mapping | ||||
|---|---|---|---|---|---|---|---|
| Low (rpkm) | Nor (rpkm) | Fold change | Low (rpkm) | Nor (rpkm) | Fold change | ||
| At1g02560 | CLPP5 (nuclear-encoded CLP protease 5), protease subunit | 177.486 | 153.738 | 1.154 | 203.821 | 177.293 | 1.150 |
| At1g06430 | FTSH8 (cell-division protease ftsH-8) | 46.648 | 33.676 | 1.385 | 53.561 | 38.836 | 1.379 |
| At1g09340 | CRB (chloroplast RNA binding) | 477.334 | 377.221 | 1.265 | 548.065 | 435.050 | 1.260 |
| At1g10350 | Putative DnaJ heat-shock protein | 5.668 | 8.818 | 0.643 | 6.507 | 10.169 | 0.640 |
| At1g32080 | Putative membrane protein | 277.058 | 238.279 | 1.163 | 318.113 | 274.786 | 1.158 |
| At1g44446 | Chlorophyllide a oxygenase | 16.090 | 45.126 | 0.357 | 18.475 | 52.075 | 0.355 |
| At1g52290 | Protein kinase-like protein | 6.746 | 13.767 | 0.490 | – | – | – |
| At1g55490 | CPN60B (chaperonin 60 beta); RuBisCO large subunit-binding protein subunit beta | 294.626 | 236.862 | 1.244 | 346.733 | 285.802 | 1.213 |
| At1g62750 | SCO1(SNOWY COTYLEDON1); elongation factor EF-G | 369.741 | 244.662 | 1.511 | 443.343 | 295.894 | 1.498 |
| At1g74730 | Unknown protein | 233.905 | 183.326 | 1.276 | 268.981 | 211.675 | 1.271 |
| At2g03390 | uvrB/uvrC motif-containing protein | 43.955 | 31.538 | 1.394 | 50.468 | 36.370 | 1.388 |
| At2g30950 | VAR2 (VARIEGATED 2); cell-division protease ftsH-2 | 621.194 | 440.354 | 1.411 | 713.485 | 507.890 | 1.405 |
| At2g32180 | PTAC18 (plastid transcriptionally active 18) | 19.442 | 27.606 | 0.704 | 22.323 | 31.836 | 0.701 |
| At2g35490 | Putative plastid-lipid- associated protein 3 | 108.463 | 88.003 | 1.232 | 124.626 | 101.487 | 1.228 |
| At2g46100 | Nuclear transport factor 2 (NTF2) family protein | 63.534 | 49.599 | 1.281 | 72.948 | 57.419 | 1.270 |
| At3g17040 | HCF107 (high chlorophyll fluorescent 107) | 9.314 | 21.102 | 0.441 | 10.694 | 24.335 | 0.439 |
| At3g19820 | DWF1 (DWARF 1) | 77.847 | 87.660 | 0.888 | 89.382 | 101.091 | 0.884 |
| At3g24430 | HCF101 (high chlorophyll fluorescence 101) | 65.051 | 49.942 | 1.303 | 74.690 | 57.594 | 1.297 |
| At4g24190 | SHD (SHEPHERD)/HEAT SHOCK PROTEIN 90–7 | 73.310 | 64.708 | 1.133 | 84.173 | 74.623 | 1.128 |
| At5g12470 | Unknown protein | 52.805 | 35.339 | 1.494 | 60.629 | 40.753 | 1.488 |
| At5g20720 | CPN20 (chaperonin 20) | 247.229 | 356.642 | 0.693 | 283.940 | 411.333 | 0.690 |
| At5g42270 | VAR1 (VARIEGATED 1); cell-division protease ftsH-5 | 413.651 | 316.477 | 1.307 | 475.001 | 364.966 | 1.301 |
| At5g52540 | Unknown protein | 16.931 | 45.877 | 0.369 | 19.307 | 52.863 | 0.365 |
Of the genes showing a fold change more than 2, three (At1g44446, At3g17040, and At5g52540) were enriched in C4 Flaveria species compared to C3 species. PSII concentrations are well correlated with chlorophyll b synthesis (Bailey et al., 2001), and chlorophyllide a oxygenase (At1g44446) is considered a critical enzyme responsible for chlorophyll b synthesis (Yamasato et al., 2005). HCF107 (At3g17040) is a sequence-specific RNA-binding protein and remodels local RNA structure in a manner that accounts for its ability to enhance translation (Sane et al., 2005; Hammani et al., 2012). The hcf107 mutation in Arabidopsis leads to a defective PSII (Felder et al., 2001). Although many chloroplast-targeted DnaJ proteins have not been characterized, it has been hypothesized that chloroplast-targeted DnaJ proteins participate in protein folding, unfolding, and assembly processes, and some DnaJ proteins are involved in the stabilization of thylakoid membrane complexes such as photosystem II (Chen et al., 2010). Therefore, these three downregulated genes were related to reduced PSII and this is in agreement with the ultrastructural analysis (Fig. 3).
Differentially expressed transcription factors
Ten differentially expressed transcription factors were identified (|log2Ratio|≥1.2) (Table 4). Of these, GOLDEN2-LIKE2 (GLK2, At5g44190), of the GLK family which is involved in chloroplast development (Langdale, 2011), was significantly downregulated under low CO2. The GLK2 counterpart GLK1 (At2g20570) was also downregulated in low CO2 but to a lesser extent.
Table 4.
Differentially expressed transcription factors using Deseq softwareAP2-EREBP, Apetala 2 ethylene-responsive-element-binding proteins; C2H2, C2H2 zinc finger domain; G2-like, golden2-like; SBP, SQUAMOSA promoter-binding proteins. P<0.001, FDR<0.025, |log2Ratio|≥1.2.
| TF family name | TF locus ID | Gene name | Gene description |
|---|---|---|---|
| Upregulated under low CO2 | |||
| AP2-EREBP | At1g74930 | ORA47 (Octadecanoid derivative- responsive AP2/ERF-domain transcription factor 47) | ORA47 is a regulator of jasmonate biosynthesis (Pauwels and Goossens, 2008) |
| C2C2-GATA | At4g26150 | CGA1 (CYTOKININ-RESPONSIVE GATA FACTOR1) | CGA1 was regulated by light, nitrogen, cytokinin, and gibberellic acid, and modulated nitrogen assimilation, chloroplast development, and starch production (Bi et al., 2005; Naito et al., 2007; Mara and Irish, 2008; Richter et al., 2010; Hudson et al., 2011); CGA1 play a key role in chloroplast development, growth, and divison in Arabidopsis (Chiang et al., 2012) |
| AP2-EREBP | At4g34410 | RRTF1 (redox-responsive transcription factor 1) | RTF1 is involved in redox homeostasis under high light stress (Khandelwal et al., 2008) |
| AP2-EREBP | At5g05410 | DREB2A (dehydration-responsive element-binding protein 2A) | DREB2A is involved in dehydration- responsive gene expression and overexpression of an active form of DREB2A results in significant stress tolerance to dehydration and significant growth retardation (Sakuma et al., 2006) |
| C2H2 | At5g59820 | ZAT12 | Zat12 plays a central role in reactive oxygen and abiotic stress signalling in Arabidopsis and overexpression of Zat12 in Arabidopsis results in the enhanced expression of oxidative- and light stress-response transcripts (Davletova et al., 2005) |
| Downregulated under low CO2 | |||
| C2C2-CO-like | At1g49130 | COL8 (CONSTANS-LIKE 8) | Zinc finger (B-box type) family protein |
| SBP | At2g33810 | SPL3 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3) | SPL3 is involved in regulation of flowering and vegetative phase change (Cardon et al., 1997; Wu and Poethig, 2006; Yamaguchi et al., 2009) |
| C2C2-CO-like | At4g27310 | BBX28 | Zinc finger (B-box type) family protein |
| G2-like | At5g44190 | GLK2 (Golden2-like 2) | GLK2 is required for normal chloroplast development (Fitter et al., 2002); GLK2 together with GLK1 optimize photosynthetic capacity by integrating responses to variable enironmental and endogenous cues (Waters et al., 2009) |
| MADS | At5g62165 | AGL42 (AGAMOUS-LIKE 42) | AGL42 is involved in the floral transition and RNAi-directed downregulation of AGL24 results in late flowering (Yu et al., 2002) |
Waters et al. (2009) identified 20 most upregulated genes by GLK1 and GLK2 induction using inducible gene expression combined with transcriptome analysis. The current work assessed the alteration of these 20 primary targets of GLK gene action and found nearly that all of them, except COR15a (At2g42540) were downregulated (Table 5 and Supplementary Table S6 available at JXB online). COR15a was significantly induced under low CO2 instead, possibly because COR15a is an indirect, secondary target of GLK2 (Waters et al., 2009).
Table 5.
Transcript abundance of GLK-regulated genesReads were mapped onto the latest Arabidopsis thaliana Col-0 genome assembly (gene mapping) or a minimal set of coding sequences of the TAIR 9 genome release (core set mapping) using bowtie. The most upregulated genes by GLK1 and GLK2 induction identified by Waters et al. (2009) were examined and nearly all of them were downregulated by low CO2, except COR15a (At2g42540). Low: low CO2, 100 ppm; Nor: normal CO2, 380 ppm; rpkm, reads per kilobase per million mapped reads. CAO, chlorophyllide a oxygenase; CHLH, magnesium chelatase; COR15a, COLD-REGULATED 15A; GCN5 related, ornithine N-delta-acetyltransferase; GLK1, Golden2-like 1; GLK2, Golden2-like 2; Lhcb, light harvesting complex subunit; MRU1, mto responding up 1; PORB, NADPH:protochlorophyllide oxidoreductase B.
| Gene ID | Protein | Gene mapping | Core set mapping | ||||
|---|---|---|---|---|---|---|---|
| Low (rpkm) | Nor (rpkm) | Fold change | Low (rpkm) | Nor (rpkm) | Fold change | ||
| At1g15820 | Lhcb6 | 2153.237 | 3099.576 | 0.695 | 2472.919 | 3575.038 | 0.692 |
| At1g44446 | CAO | 16.090 | 45.126 | 0.357 | 18.475 | 52.075 | 0.355 |
| At1g76100 | Plastocyanin | 132.858 | 261.423 | 0.508 | – | – | – |
| At2g05070 | Lhcb2.2 | 431.847 | 1425.637 | 0.303 | – | – | – |
| At2g20570 | GLK1 | 32.786 | 59.366 | 0.552 | 37.677 | 68.503 | 0.550 |
| At2g34430 | Lhcb1.4 | 599.406 | 1663.045 | 0.360 | – | – | – |
| At2g35260 | Expressed protein | 74.776 | 98.821 | 0.757 | 85.888 | 114.002 | 0.753 |
| At2g39030 | GCN5 related | 0.140 | 3.243 | 0.043 | – | – | – |
| At2g42220 | Rhodanese-like domain-containing protein | 220.272 | 249.887 | 0.881 | 253.769 | 288.712 | 0.879 |
| At2g42540 | COR15a | 193.204 | 31.607 | 6.113 | 268.502 | 47.764 | 5.621 |
| At3g08940 | Lhcb4.2 | 326.811 | 1274.137 | 0.256 | – | – | – |
| At3g27690 | Lhcb2.4 | 136.883 | 414.191 | 0.330 | 157.608 | 478.391 | 0.329 |
| At3g56940 | Mg-Proto IX ME cyclase | 428.161 | 770.890 | 0.555 | 491.646 | 889.102 | 0.553 |
| At4g27440 | PORB | 400.475 | 873.866 | 0.458 | – | – | – |
| At5g13630 | CHLH | 456.682 | 432.350 | 1.056 | 524.401 | 498.633 | 1.052 |
| At5g35490 | MRU1 | 7.921 | 28.980 | 0.273 | 9.095 | 33.420 | 0.272 |
| At5g44190 | GLK2 | 5.722 | 26.922 | 0.213 | 6.570 | 31.047 | 0.212 |
| At5g54270 | Lhcb3 | 2240.644 | 3734.380 | 0.600 | 2573.689 | 4307.756 | 0.597 |
Stress-induced mutagenesis pathway was changed under low CO2
It has been shown that DNA double-strand break-dependent stress-induced mutagenesis is important to evolution, through producing more mutations under stress in Escherichia coli (Cirz et al., 2005; Shee et al., 2011; Al Mamun et al., 2012). As a severe stress, can low CO2 induce more mutagenesis in natural populations? This work examined the transcriptional changes in genes encoding products related to human DNA repair proteins and found that genes involved in damage sensing (At5g40450, At2g26980, At4g04720), photoreactivation (At3g15620), homologous recombination (At3g48190), nucleotide excision repair (At2g36490, At3g02060, At5g04560, At1g52500, At3g28030, At5g45400), and DNA polymerases (At4g32700, At1g67500) were upregulated by low CO2 (Table 6 and Supplementary Table S7 available at JXB online). These results suggest that low CO2 might induce a similar mechanism of DNA double-strand break-dependent stress-induced mutagenesis to promote evolution.
Discussion
This study, as far as is known for the first time, investigated responses to low CO2 at the transcriptome level in model plant Arabidopsis. Here, the observed changes of transcriptomics under low CO2 are briefly discussed, with particular reference to their potential significance for fitness of C3 plants under low CO2 and potential linkage to C4 photosynthesis evolution.
Low CO2 reduced productivity
Arabidopsis plants grown under low CO2 had extremely small stature compared with plants grown under normal CO2 (Fig. 1). This result is in accordance with previous studies on the effect of low CO2 on plant growth (Ward, 2005). Arabidopsis grown under low CO2 has about a 7-day delay in flowering time. This has also been observed earlier (Ward and Strain, 1997) and could be interpreted as a mechanism to allow for greater accumulation of stored reserves that could be allocated to reproduction, resulting in increased fitness under low CO2 (Sage and Coleman, 2001; Ward, 2005).
Many studies have shown that atmospheric CO2 concentration negatively regulates stomatal density (Woodward, 1987; Beerling et al., 2001; Franks and Beerling, 2009; Doheny-Adams et al., 2012; Franks et al., 2012). Paleontological research has suggested that the long-term decreases in atmospheric throughout the entire evolutionary history of vascular plants led to the evolution of high densities of small stomata in order to attain the highest g cmax values required to counter CO2 ‘starvation’ (Franks and Beerling, 2009; Franks et al., 2012). Stomata also exhibit short-term adaptive responses to atmospheric CO2 over much shorter timescales. For example, A. thaliana Col-0 grown at high CO2 (720 ppm) had reduced stomata density compared with those grown at ambient CO2 (360 ppm) (Lake et al., 2001). In the current work, plants grown under low CO2 developed leaves with higher stomatal density (over 60% increase compared to normal CO2; Fig. 2), suggesting that the plants developed a greater g cmax to counteract the CO2 limitation of photosynthesis. These results suggest that low CO2 is a severe stress to C3 plants and may greatly reduce C3 plant productivity.
Responses of genes involved in C4 photosynthesis and photorespiration under low CO2
In C4 plants, CO2 is initially fixed by the enzyme PEPC into a C4 acid and then transported to the site of Rubisco (Hatch, 1987). The only photosynthetic gene expression patterns common to all independently evolved C4 lineages are upregulation of PEPC and downregulation of Rubisco in mesophyll cells (Sinha and Kellogg, 1996; Langdale, 2011). Arabidopsis has four genes encoding PEPC, and AtPPC2 (At2g42600) is the only isoform expressed in leaves. Unlike the other three PEPCs, the expression of AtPPC2 is stable and has not been reported to be regulated by any stress (Sánchez et al., 2006; Doubnerová and Ryšlavá, 2011); however, the current work found that AtPPC2 was upregulated by low CO2 (Table 1). The regulators of photosynthetic genes are also crucial to maintain C4 photosynthesis: e.g. plant PEPC activity is further regulated through reversible phosphorylation by PEPC-K (Nimmo, 2003). Transcripts encoding the C4-specific regulatory factors PEPC-K and pyruvate orthophosphate dikinase regulatory protein were upregulated as well (Table 1). However, changes in other C4-related genes were less, with fold changes of less than 2.
When grown in low CO2, plants would experience relatively high levels of flux through the photorespiratory pathway because of the competitive reactions of Rubisco oxygenation. In this study, a trend of upregulation of the photorespiratory genes was observed in plants grown under low CO2 (Table 2), although most of the genes showed a fold change of less than 2. The recent study of transcriptome analysis using C3, C3–C4 intermediate, and C4 species of Flaveria found that transcript abundances for most genes related to photorespiration in the C3–C4 intermediate species Flaveria ramosissima were even higher than in the C3 species Flaveria robusta (Gowik et al., 2011), which is indicative of the importance of the photorespiratory pathway during the evolution of C4 photosynthesis. The different subunits of glycine decarboxylase showed altered expression, although the fold changes of these subunits were about 0.9–1.3.
Overall, the data from this study suggest that expression of PEPC and PEPC-K is increased under low CO2, which most likely reflects their potential role for refixation of photorespired CO2 under low CO2 (Sage et al., 2012). For most of the other C4 genes, although trends of upregulation were observed, the fold changes were less than 2. Although by using expression level changes of all genes under two conditions as background, this work obtained P-values much less than 0.01 for many C4-related genes, it is likely that lack of biological replicates could had potentially led to an overestimation of the reliability of statistical tests and caused problems in identifying significantly changed genes, especially when their fold changes were less than 2. Based on these, this work cannot state that low CO2 induced upregulation of C4 genes, except for those genes which showed fold changes over 2 (e.g. PEPC).
Readjustment of balance between light absorption and CO2 fixation under low CO2
These data on chloroplast ultrastructure and transcript abundance of genes involved in chloroplast biogenesis and maintenance are consistent with the model for long-term photosynthetic regulation by GLK proteins (Waters and Langdale, 2009). When light is high and atmospheric CO2 is limiting, the rate of CO2 fixation is insufficient to use all of the output of the light-harvesting reactions, resulting in an overly reduced photosynthetic electron transport. This triggers a decrease of GLK transcription (GLK1 and GLK2; Table 5). Since GLK transcription factors directly regulate a large suite of genes involved in light-harvesting and thylakoid protein complexes, especially those of PSII (Waters et al., 2009), the light-harvesting components in the thylakoid membrane LHCB2.2 (At2g05070), LHCB4.2 (At3g08940), Lhcb3 (At5g54270), Lhcb2.4 (At3g27690), and Lhcb1.4 (At2g34430) were downregulated under low CO2. In addition, the downregulation of the chlorophyllide a oxygenase gene led to the decrease of chlorophyll b synthesis. These results were consistent with the fewer and less-stacked grana observed and a higher proportion of non-stacked stromal lamellae, as observed in glk1 glk2 mutants (Fig. 3). Therefore, these observed expression changes in GLK and the genes regulated by GLK can be interpreted as reflecting the altered balance between CO2 fixation and light absorption.
Evolutionary implications of plants of to low CO2
Growing evidence suggests that all of the basic elements of C4 photosynthesis already existed in C3 plants. For example, all of the enzymes involved in C4 photosynthesis exist in C3 plants and play different roles in C3 plant metabolism (Aubry et al., 2011). Some elements controlling the cell specific expression of C4-related enzymes have been found in C3 plants (Brown et al., 2011). Moreover, typical C3 plants (e.g. tobacco and Arabidopsis) show the characteristics of C4 photosynthesis in midveins (Hibberd and Quick, 2002; Brown et al., 2010).
Can some features related to C4 photosynthesis be enhanced under some conditions in a C3 plant? This work showed that under low atmospheric CO2, A. thaliana Col-0 adjusted a series of biological processes, especially the upregulation of PEPC and PEPC-K gene expression, and also the altered expression of some transcription factors related to photosynthesis development, and the downregulation of light-harvesting and thylakoid protein complexes. Although this study also observed that the majority of the other C4-cycle genes were upregulated under low CO2 in Arabidopsis, their fold changes were less than 2 and therefore no firm statements regarding their changes can be made.
Experiments with more biological replicates and Arabidopsis accessions are still needed to firmly conclude whether low CO2 can induce upregulation of other C4-related genes. Therefore, the results from this paper do not support a scenario where low CO2 acts as a signal to induce C4 biochemical features in C3 plants. It is most likely that the upregulation of PEPC and PEPC-K might be a mechanism that C3 plants used to refix photorespired and respired CO2 and also to recapture the released ammonium from photorespiration and hence increase the competitive advantages under low CO2 conditions.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Measurement of stomatal density.
Supplementary Table S1. Gene mapping results.
Supplementary Table S2. Core-set gene mapping results.
Supplementary Table S3. Transcript abundance of C4 cycle genes and C4-related transporters.
Supplementary Table S4. Transcript abundance of photorespiration genes.
Supplementary Table S5. Transcript abundance of genes related to chloroplast biogenesis and maintenance.
Supplementary Table S6. The 20 most-upregulated genes following GLK2 induction.
Supplementary Table S7. Transcript abundance of DNA-repair genes.
Supplementary Table S8. Transcript abundance of photosynthesis genes.
Supplementary Table S9. Transcript abundance of Calvin Benson cycle genes.
Supplementary Table S10. Transcript abundance of ABA-metabolism genes.
Supplementary Table S11. Transcript abundance of auxin-metabolism genes.
Acknowledgements
The authors gratefully acknowledge Prof. Julian Hibberd and Paul Quick for his comments on earlier draft of this paper. The funding for the authors’ research has been provided by the Bill and Melinda Gates Foundation (grant no. OPP1014417), the Ministry of Science and Technology of China (grant no. 2011DFA31070), the National Natural Science Foundation of China (grant no. 31200267), and the Young Talent Frontier Program of Shanghai Institutes for Biology Sciences/Chinese Academy of Sciences (grant no. 09Y1C11501).
References
- Ainsworth EA, Long SP. 2005. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy. New Phytologist 165, 351–371 [DOI] [PubMed] [Google Scholar]
- Albertsson P-Å. 1995. The structure and function of the chloroplast photosynthetic membrane — a model for the domain organization. Photosynthesis Research 46, 141–149 [DOI] [PubMed] [Google Scholar]
- Al Mamun AAM, Lombardo M-J, Shee C, et al. 2012. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338, 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. Journal of Experimental Botany 62, 3049–3059 [DOI] [PubMed] [Google Scholar]
- Bailey S, Walters RG, Jansson S, Horton P. 2001. Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta 213, 794–801 [DOI] [PubMed] [Google Scholar]
- Bauwe H. 2011. Photorespiration: the bridge to C4 photosynthesis. In: Raghavendra AS, Sage RF, eds, C4 photosynthesis and related CO2 concentrating mechanisms. Dordrecht, The Netherlands: Springer; pp 81–108 [Google Scholar]
- Beerling DJ, Osborne CP, Chaloner WG. 2001. Evolution of leaf-form in land plants linked to atmospheric CO2 decline in the Late Palaeozoic era. Nature 410, 352–354 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 57, 289–300 [Google Scholar]
- Bi Y-M, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S. 2005. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. The Plant Journal 44, 680–692 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Gunther M, et al. 2005. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis . The Plant Cell 17, 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439 [DOI] [PubMed] [Google Scholar]
- Brown NJ, Palmer BG, Stanley S, et al. 2010. C4 acid decarboxylases required for C4 photosynthesis are active in the mid-vein of the C3 species Arabidopsis thaliana, and are important in sugar and amino acid metabolism. The Plant Journal 61, 122–133 [DOI] [PubMed] [Google Scholar]
- Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P. 1997. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3, a novel gene involved in the floral transition. The Plant Journal 12, 367–377 [DOI] [PubMed] [Google Scholar]
- Chen K-M, Holmström M, Raksajit W, Suorsa M, Piippo M, Aro E-M. 2010. Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana . BMC Plant Biology 10, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y-H, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE. 2012. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis . Plant Physiology 160, 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biology 3, e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. 2005. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis . Plant Physiology 139, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ. 2005. Supramolecular organization of thylakoid membrane proteins in green plants. Biochimica et Biophysica Acta 1706, 12–39 [DOI] [PubMed] [Google Scholar]
- Dippery JK, Tissue DT, Thomas RB, Strain BR. 1995. Effects of low and elevated CO2 on C3 and C4 annuals. I. Growth and biomass allocation. Oecologia 101, 13–20 [DOI] [PubMed] [Google Scholar]
- Doheny-Adams T, Hunt L, Franks PJ, Beerling DJ, Gray JE. 2012. Genetic manipulation of stomatal density influences stomatal size, plant growth and tolerance to restricted water supply across a growth carbon dioxide gradient. Philosophical Transactions of the Royal Society B 367, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doubnerová V, Ryšlavá H. 2011. What can enzymes of C4 photosynthesis do for C3 plants under stress? Plant Science 180, 575–583 [DOI] [PubMed] [Google Scholar]
- Felder S, Meierhoff K, Sane AP, Meurer J, Driemel C, Plücken H, Klaff P, Stein B, Bechtold N, Westhoff P. 2001. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. The Plant Cell 13, 2127–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. 2002. GLK gene pairs regulate chloroplast development in diverse plant species. The Plant Journal 31, 713–727 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Beerling DJ. 2009. Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proceedings of the National Academy of Sciences, USA 106, 10343–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJ, Leitch IJ, Ruszala EM, Hetherington AM, Beerling DJ. 2012. Physiological framework for adaptation of stomata to CO2 from glacial to future concentrations. Philosophical Transactions of the Royal Society B 367, 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart LM, Ward JK. 2010. Plant responses to low [CO2] of the past. New Phytologist 188, 674–695 [DOI] [PubMed] [Google Scholar]
- Gowik U, Bräutigam A, Weber KL, Weber APM, Westhoff P. 2011. Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? The Plant Cell 23, 2087–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Cook WB, Barkan A. 2012. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proceedings of the National Academy of Sciences, USA 109, 5651–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106 [Google Scholar]
- Hibberd JM, Quick WP. 2002. Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415, 451–454 [DOI] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi Y-M, Rothstein SJ. 2011. GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis . PLoS ONE 6, e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal A, Elvitigala T, Ghosh B, Quatrano RS. 2008. Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiology 148, 2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Quick WP, Beerling DJ, Woodward FI. 2001. Plant development: signals from mature to new leaves. Nature 411, 154. [DOI] [PubMed] [Google Scholar]
- Langdale JA. 2011. C4 cycles: past, present, and future research on C4 photosynthesis. The Plant Cell 23, 3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Ainsworth EA, Leakey ADB, Nosberger J, Ort DR. 2006. Food for thought: lower-than-expected crop yield stimulation with rising CO2 concentrations. Science 312, 1918–1921 [DOI] [PubMed] [Google Scholar]
- Long SP, Ainsworth EA, Rogers A, Ort DR. 2004. Rising atmospheric carbon dioxide: plants FACE their future. Annual Review of Plant Biology 55, 591–628 [DOI] [PubMed] [Google Scholar]
- Mara CD, Irish VF. 2008. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis . Plant Physiology 147, 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T. 2007. Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana . Bioscience, Biotechnology and Biochemistry 71, 1557–1560 [DOI] [PubMed] [Google Scholar]
- Nimmo HG. 2003. Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Archives of Biochemistry and Biophysics 414, 189–196 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Goossens A. 2008. Fine-tuning of early events in the jasmonate response. Plant Signaling and Behavior 3, 846–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. 2010. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes and Development 24, 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF, Coleman JR. 2001. Effects of low atmospheric CO2 on plants: more than a thing of the past. Trends in Plant Science 6, 18–24 [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and evolution of C4 photosynthesis. Annual Review of Plant Biologist 63, 19–47 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Miyagishima S-Y, Jarvis P. 2008. Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6, e0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Functional analysis of an Arabidopsis transcription tactor, DREB2A, involved in drought-responsive gene expression. The Plant Cell 18, 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R, Flores A, Cejudo FJ. 2006. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 223, 901–909 [DOI] [PubMed] [Google Scholar]
- Sane AP, Stein B, Westhoff P. 2005. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis . The Plant Journal 42, 720–730 [DOI] [PubMed] [Google Scholar]
- Sharma RK, Griffing B, Scholl RL. 1979. Variations among races of Arabidopsis thaliana (L.) Heynh for survival in limited carbon dioxide. Theoretical Applied Genetics 54, 11–15 [DOI] [PubMed] [Google Scholar]
- Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. 2011. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli . Proceedings of the National Academy of Sciences, USA 108, 13659–13664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NR, Kellogg EA. 1996. Parallelism and diversity in multiple origins of C4 photosynthesis in the grass family. American Journal of Botany 83, 1458–1470 [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature 408, 796–815 [DOI] [PubMed] [Google Scholar]
- Ward JK. 2005. Evolution and growth of plants in a low CO2 world. In: Ehleringer JR, Cerling TE, Dearing MD, eds, A history of atmospheric CO2 and its effects on plants, animals, and ecosystems. New York: Springer; pp 232–257 [Google Scholar]
- Ward JK, Antonovics J, Thomas RB, Strain BR. 2000. Is atmospheric CO2 a selective agent on model C3 annuals? Oecologia 123, 330–341 [DOI] [PubMed] [Google Scholar]
- Ward JK, Kelly JK. 2004. Scaling up evolutionary responses to elevated CO2, lessons from Arabidopsis . Ecology Letters 7, 427–440 [Google Scholar]
- Ward JK, Strain BR. 1997. Effects of low and elevated CO2 partial pressure on growth and reproduction of Arabidopsis thaliana from different elevations. Plant, Cell and Environment 20, 254–260 [Google Scholar]
- Waters MT, Langdale JA. 2009. The making of a chloroplast. EMBO Journal 28, 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. 2009. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis . The Plant Cell 21, 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward FI. 1987. Stomatal numbers are sensitive to increases in CO2 from pre-industrial levels. Nature 327, 617–618 [Google Scholar]
- Wu G, Poethig RS. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3 . Development 133, 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu M-F, Yang L, Wu G, Poethig RS, Wagner D. 2009. The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1 . Developmental Cell 17, 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasato A, Nagata N, Tanaka R, Tanaka A. 2005. The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll b accumulation in Arabidopsis . The Plant Cell 17, 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP. 2002. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proceedings of the National Academy of Sciences, USA 99, 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.