Summary
Multiple isoforms of phosphoenolpyruvate carboxylase genes were sequenced from related orchid species with distinct photosynthesic types. Phylogenetic analysis indicated that CAM-associated isoforms originated from gene duplications and adaptive sequence divergence.
Keywords: Crassulacean acid metabolism, gene duplication, Orchidaceae, Oncidiinae, phosphoenolpyruvate carboxylase, photosynthesis.
Abstract
Phosphoenolpyruvate carboxylase (PEPC) catalyses the initial fixation of atmospheric CO2 into oxaloacetate and subsequently malate. Nocturnal accumulation of malic acid within the vacuole of photosynthetic cells is a typical feature of plants that perform crassulacean acid metabolism (CAM). PEPC is a ubiquitous plant enzyme encoded by a small gene family, and each member encodes an isoform with specialized function. CAM-specific PEPC isoforms probably evolved from ancestral non-photosynthetic isoforms by gene duplication events and subsequent acquisition of transcriptional control elements that mediate increased leaf-specific or photosynthetic-tissue-specific mRNA expression. To understand the patterns of functional diversification related to the expression of CAM, ppc gene families and photosynthetic patterns were characterized in 11 closely related orchid species from the subtribe Oncidiinae with a range of photosynthetic pathways from C3 photosynthesis (Oncidium cheirophorum, Oncidium maduroi, Rossioglossum krameri, and Oncidium sotoanum) to weak CAM (Oncidium panamense, Oncidium sphacelatum, Gomesa flexuosa and Rossioglossum insleayi) and strong CAM (Rossioglossum ampliatum, Trichocentrum nanum, and Trichocentrum carthagenense). Phylogenetic analysis revealed the existence of two main ppc lineages in flowering plants, two main ppc lineages within the eudicots, and three ppc lineages within the Orchidaceae. Our results indicate that ppc gene family expansion within the Orchidaceae is likely to be the result of gene duplication events followed by adaptive sequence divergence. CAM-associated PEPC isoforms in the Orchidaceae probably evolved from several independent origins.
Introduction
Crassulacean acid metabolism (CAM) is one of three modes of photosynthesis found in vascular plants for the assimilation of atmospheric CO2. CAM differs from C3 and C4 photosynthesis in that CAM plants take up CO2 with low water expenditure at night when evaporative demand is low (Winter and Smith, 1996b ; Cushman, 2001). CAM is phylogenetically widespread across 343 genera and 35 plant families comprising more than 6% of flowering plant species (Griffiths, 1989; Smith and Winter, 1996; Holtum et al., 2007; Silvera et al., 2010a ). The multiple independent origins of CAM and the functional convergence of CAM traits in the many lineages in which it occurs suggest that the starting point for CAM evolution might have required relatively few genetic changes. However, this notion may be simplistic, given the complexity of the CAM pathway and the fact that several biochemical and anatomical requirements, and regulatory changes associated with gene expression patterns, are all tightly coordinated in CAM plants (Cushman et al., 2008; Silvera et al., 2010a ). To gain insight into the evolutionary history of genes recruited for CAM function, the study of taxa containing many closely related species with contrasting photosynthetic pathways is experimentally helpful. In this context, we use tropical orchids as a study group, because CAM is widespread among epiphytes within this large family of vascular plants (Winter and Smith, 1996a ; Silvera et al., 2009, 2010a, b ), and because species within the Orchidaceae exhibit a gradient of photosynthetic pathways ranging from C3 photosynthesis to weak- and strong-CAM modes (Silvera et al., 2005, 2010a ). Weak-CAM species show low-level CAM activity and typically obtain 5% or less of their carbon through the CAM pathway under well-watered conditions. Weak CAM appears to be common among neotropical orchid species (Silvera et al. 2005).
In vascular plants, phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) belongs to a multigene family, and each member encodes an enzyme with specialized functions (Gehrig et al., 1995, 1998, 2001; Chollet et al., 1996; Izui et al., 2004; O’Leary et al., 2011). In species performing C4 photosynthesis and CAM, one or more ppc genes encode isoforms of the enzyme that catalyse the fixation of atmospheric CO2 into C4-dicarboxylic acids. In CAM plants, CAM-specific isoforms catalyse the nocturnal, irreversible β-carboxylation of phosphoenolpyruvate in the presence of HCO3 – and Mg2+ yielding oxaloacetate and inorganic phosphate. Oxaloacetate is then converted to malate, which is stored as malic acid in the vacuole. During the subsequent day, malic acid is decarboxylated, resulting in CO2 release and refixation by ribulose-1,5-bisphosphate carboxylase/oxygenase. This CO2-concentrating mechanism, or ‘CO2 pump’, suppresses photorespiration and improves water-use efficiency relative to that of C3 and C4 plant species (Cushman and Bohnert, 1997). Cytosolic and chloroplastic PEPC enzymes are found in photosynthetic organisms from higher plants and green algae to cyanobacteria and photosynthetic bacteria, and also in non-photosynthetic bacteria and protozoa (Chollet et al., 1996; Izui et al., 2004). In all plants, ‘housekeeping’ or non-photosynthetic isoforms of PEPC catalyse anapleurotic reactions to replenish biosynthetic precursors for the Krebs cycle. PEPC has many other physiological roles in plants that include maintaining cellular pH, supplying carbon to N2-fixing legume root nodules, absorbing and transporting cations in roots, control of stomatal movements, fruit maturation, and seed germination (Latzko and Kelly, 1983; Chollet et al., 1996; Echevarria and Vidal, 2003; Izui et al., 2004; O’Leary et al., 2011; Singh et al., 2012). While PEPC is mainly a cytosolic enzyme, a plastid-localized version of the enzyme, rice Osppc4, has been described and is involved in providing organic acids for ammonium assimilation in leaves (Masumoto et al., 2010).
The currently available molecular data support the view that none of the C4 or CAM enzymes are unique to C4 or CAM plants (Westhoff and Gowik, 2004; Gowik and Westhoff, 2011), suggesting that these ubiquitous and functionally diverse isoforms served as starting points for the evolution of the C4 and CAM genes (Cushman and Bohnert, 1999; Monson, 1999). Furthermore, evidence from PEPC and comparative analysis of the C4-cycle enzymes in C3, C3–C4 intermediates, and C4 species in the genus Flaveria suggests that key amino acid residue changes are responsible for their acquisition of distinct kinetic and regulatory properties (Blasing et al., 2000, 2002; Westhoff and Gowik, 2004). Similarly, phylogenetic analysis indicates that multiple origins of C4 photosynthesis in grasses and sedges are the likely result of recurring selection acting on a few amino acid positions of the PEPC enzyme within and across taxonomic scales (Christin et al., 2007, 2012b ; Besnard et al., 2009). Phylogenetic analyses also indicate that there was a single PEPC origin before the divergence of bacteria and plant lineages (Chollet et al., 1996; Izui et al., 2004; Westhoff and Gowik, 2004). CAM-specific PEPC isoforms are thought to have first evolved in response to water deficit from ancestral non-photosynthetic isoforms by gene duplication, followed by acquisition of transcriptional control sequences that mediate leaf- or photosynthetic-tissue-specific increases in mRNA expression (Gehrig et al., 2001, 2005; Taybi et al., 2004). For example, seven distinct PEPC isoforms were recovered in the CAM species Kalanchoe pinnata (Lam.) Pers.: four isoforms from leaves and three from roots (Gehrig et al., 2005). In Mesembryanthemum crystallinum L., a CAM-specific isoform was expressed during the induction of CAM, in addition to an uninduced housekeeping isoform (Cushman and Bohnert, 1999). Based on comparative studies of PEPC in many plant taxa including orchids performing CAM, Gehrig et al. (2001) predicted the clustering of PEPC isoforms according to their taxonomic position and specific function.
Previously, full-length ppc genes were characterized from bacteria, several vascular plant species, cyanobacteria, and protozoa (Izui et al., 2004). The purpose of our study was to reconstruct the evolutionary history of PEPC in Orchidaceae. We have characterized the diversity of ppc genes in a phylogenetic context, using partial-length sequences from a closely related group of orchid species in the Oncidiinae that express photosynthetic pathways ranging from C3 photosynthesis to weak CAM and strong CAM. The results highlight the evolutionary diversification of ppc gene families and indicate the possible role of gene duplication and recruitment of ppc genes for CAM.
Materials and methods
Oncidiinae species and characterization
Eleven closely related species within the Oncidiinae with a range of photosynthetic pathways from C3 photosynthesis [Oncidium cheirophorum Rchb. f., Oncidium maduroi Dressler, Oncidium sotoanum R. Jimenez & Hagsater, and Rossioglossum krameri (Rchb. f.) M.W. Chase & N.H. Williams] to weak CAM [Gomesa flexuosa (G.Lodd.) M.W. Chase & N.H. Williams, Oncidium panamense Schltr., Oncidium sphacelatum Lindl., and Rossioglossum insleayi (Barker ex Lindl.) Garay & G.C. Kenn.] and strong CAM [Rossioglossum ampliatum (Lindl.) M.W. Chase & N.H. Williams, Trichocentrum carthagenense (Jacq.) M.W. Chase & N.H. Williams, and Trichocentrum nanum (Lindl.) M.W. Chase & N.H. Williams] were chosen as a study group for genetic studies of CAM based on carbon isotopic composition and titratable acidity measurements (Table 1; Silvera et al., 2005). Oncidiinae represents one of the most highly diverse clades of orchids from the neotropics, with variation in chromosome number, vegetative features, photosynthetic mechanisms, and floral characteristics (Chase et al., 2005). A phylogenetic reconstruction for these 11 species was performed using a matrix that included sequences of nuclear ribosomal DNA internal transcribed spacer 1 (nrITS-1 and -2), plastid DNA regions ycf1 (~1200bp portion from the 5′ end, and ~1500bp portion from 3′ end), matK, and the trnH–psbA intergenic spacer, as previously described (Neubig et al., 2012) in each species, except for O. maduroi, which included only data for nrITS-1 and -2 and ycf1.
Table 1.
Values of δ13C, leaf thickness, and titratable acidity for 11 species from the OncidiinaeTitratable acidity (∆H+) is represented as the difference between the mean±standard deviation of three replicates at morning and evening (Silvera et al., 2005, and this study). Species are listed in order based on ∆H+ from C3 photosynthesis to weak CAM and strong CAM. FW, fresh weight; NS, not significant.
| Species name | Leaf δ13C (‰) | Leaf thickness (mm) | H+ (evening) (µmol H+g–1 FW) | H+ (morning) (µmol H+g–1 FW) | ∆H+ | Photosynthetic pathway |
|---|---|---|---|---|---|---|
| Oncidium sotoanum | –25.2 | 0.25 | 2.9±0.4 | 2.7±1.6 | –0.2 NS | C3 |
| Oncidium cheirophorum | –27.4 | 0.36 | 29.9±3.3 | 30.2±2.7 | 0.3 NS | C3 |
| Rossioglossum krameri | –31.7 | 0.35 | 14.4±2.5 | 14.8±2.4 | 0.4 NS | C3 |
| Oncidium maduroi | –24.7 | 0.24 | 17.3±1.9 | 19.5±0.8 | 2.2 NS | C3 |
| Rossioglossum insleayi | –22.5 | 1.10 | 16.3±0.5 | 34.9±12 | 18.60a | Weak CAM |
| Oncidium panamense | –26.2 | 0.54 | 11.5±0.7 | 33.2±0.3 | 21.7a | Weak CAM |
| Oncidium sphacelatum | –27.9 | 0.53 | 8.3±6.3 | 31.2±2.1 | 22.9a | Weak CAM |
| Gomesa flexuosa | –24.4 | 0.26 | 37.9±9.4 | 74.0±10.4 | 36.1a | Weak CAM |
| Trichocentrum nanum | –17.2 | 3.40 | 18.8±4.1 | 57.3±5.2 | 38.50a | Strong CAM |
| Trichocentrum carthagenense | –12.2 | 2.32 | 12.5±0.4 | 77.3±3.4 | 64.8a | Strong CAM |
| Rossioglossum ampliatum | – 15.3 | 1.59 | 5.5±1.3 | 153.5±3.6 | 148.0a | Strong CAM |
a Denotes significance between means of the morning and evening at P<0.05 as determined by Student’s t-test. NS, not significant.
Gas-exchange measurements
Photosynthetic gas exchange was measured on attached, mature leaves of plants from each of the 11 orchid species targeted for this study. Measurements of 24h CO2 exchange demonstrate the proportions of CO2 fixed in the light and dark, respectively, and thus allow conclusions about the degree to which plants engage in CAM relative to C3 photosynthesis. Leaves were sealed inside a Plexiglass® cuvette located within a controlled-environment chamber (Environmental Growth Chambers, Chagrin Falls, Ohio, USA). The hole in the cuvette through which the leaf was inserted was sealed with a non-porous synthetic rubber sealant (Terostat VII; Henkel-Teroson, Heidelberg, Germany). Prior to measurements, plants were well watered, fertilized once per week with a commercial 20:20:20 and/or 16:32:16 (N:P:K) fertilizer solution, and maintained in an open greenhouse. The diel temperature range within the greenhouse varied from a minimum of 20 °C to a maximum of 32.2 °C, and relative humidity varied from 80 to 100%. Daily light availability ranged from 5 to 70% of full sunlight and corresponded roughly to the natural light exposure of these species in the field. Plants were kept in their growth medium, which consisted of lightweight pumice aggregates and coarse synthetic sponge, to avoid dehydration before experiments. Due to the small size of T. nanum leaves, an entire plant of this species, including roots, was placed inside the cuvette. Net CO2 exchange was measured continuously using a flowthrough gas-exchange system (Walz, Effeltrich, Germany) operating at an airflow rate of 1.3 l min–1 and monitored with a LI-6252 infrared gas analyser (Li-Cor, Lincoln, NE, USA) operating in the absolute mode for up to 5 d. More than one individual per species was measured. The temperature inside the cuvette and chamber was 25 °C during the day and 22 °C during the night under an ambient CO2 concentration with a dew point of 18 °C and a light intensity of 300 µmol m–2 s–1 during the 12h light period.
Titratable acidity determinations
Titratable acidity (∆H+) measurements were conducted as described previously and are presented as the difference between the mean±standard deviation of three replicate samples at dawn and dusk (Silvera et al., 2005). Nocturnal acid accumulation reflects the magnitude of dark CO2 fixation but does not provide information on the uptake of atmospheric CO2 via C3 photosynthesis in the light.
Plant material
Leaf samples for ppc gene analysis were obtained from mature leaves of mature orchids for 10 of the 11 species targeted for this study (O. cheirophorum was excluded from the gene analysis due to insufficient leaf material). Plants were grown in closed greenhouses at the University of Nevada, Reno, and the University of California, Riverside. Daily temperatures within the greenhouse varied from 17 to 35 °C, relative humidity varied from 40 to 80%, and the mean photon flux density was 200 µmol m–2 s–1. Plants were watered daily and nutrients were supplied once per week with a combination of slow-release fertilizer (Osmocote® 19-6-12 formula; Scotts Company, Marysville, OH, USA) and commercial fertilizer solution (Schultz® 19-31-17 formula; Spectrum Brands, Madison, WI, USA). Healthy leaves from each of 10 species were collected at 2 p.m. and 2 a.m. to account for putative circadian differences in the relative expression abundance of PEPC mRNAs. Root samples were also collected at the same times from mature plants and stored separately. All samples were flash frozen in liquid nitrogen immediately after harvesting, and stored at –80 °C until isolation of total RNA.
RNA extraction
Total RNA was extracted using an RNeasy Midi kit (Qiagen, Valencia, CA, USA) with a modified polyethylene glycol RNA extraction method including high-molecular-weight polyethylene glycol (Gehrig et al., 2000), which has proven successful for RNA isolation from succulent and non-succulent orchid tissues. RNA integrity was examined by 1% agarose gel electrophoresis, and RNA quality and concentration were examined using a NanoDrop ND-1000 UV-V spectrophotometer (NanoDrop Technologies, Rockland, DE, USA).
Reverse transcription (RT)-PCR amplification and cloning
An 1100bp fragment was amplified by RT-PCR using two degenerate primers, as defined previously for PEPC (Taybi et al., 2004; Gehrig et al., 2005; Kore-eda et al., 2005) and modified slightly for orchid specificity. The degenerate primers were: 5′-TCNGAYTCNGGVAARGAYGC-3′ (forward) and reverse 5′-GCDGCRATRCCYTTCATKG-3′ (reverse). Using a One-Step RT-PCR kit (Qiagen), 500ng of total RNA was reverse transcribed and amplified following the manufacturer’s instructions. Final concentrations of the reaction components were 400 µM for each dNTP, 1× Qiagen One-Step RT-PCR Buffer containing 12.5mM MgCl2, 2 µl of Qiagen One-Step RT-PCR Enzyme Mix, and 1 µM PEPC forward and reverse primers. The following temperature cycling conditions were used: reverse transcription at 50 °C for 30min, initial PCR activation at 95 °C for 15min, amplification for 39 cycles at 94 °C for 1min, 40 °C for 1min, and 72 °C for 1min, and final extension at 72 °C for 10min. RT-PCR was used to amplify a fragment of 1100bp encompassing the C-terminal third of the PEPC coding region. By using this partial sequence, distinct isoforms could be distinguished without the need to isolate the full 3000bp sequence, because this fragment was variable enough to allow differentiation among isoforms. The sequence also has a highly conserved active site that facilitates correct alignment and identification (Gehrig et al., 2001; Izui et al., 2004) (Supplementary Fig. S2 at JXB online). In this regard, the use of partial rather than full-length PEPC cDNA sequences has proven useful for molecular phylogenetic and taxonomic comparisons across species, thus saving time and financial resources for researchers interested in using PEPC as a molecular marker (Gehrig et al., 2001). The RT-PCR products were purified by agarose gel electrophoresis, recovered using a QIAquick Gel Extraction kit (Qiagen), cloned into the TA-TOPO cloning vector pCR2.1 vector system (InvitrogenTM Life Technologies, Carlsbad, CA, USA), and transformed into XL1-Blue or TOP10 competent Escherichia coli cells following the manufacturer’s instructions. Bacterial cells containing plasmids from 100–150 randomly selected clones per orchid species were grown in Terrific Broth liquid medium for 16h at 37 °C with vigorous shaking. The bacterial cells were then harvested by centrifugation at 13 000g, and the plasmids were purified using a Qiagen Plasmid Mini kit (Qiagen) following the manufacturer’s instructions. Selected cDNA clones were then analysed by EcoRI restriction enzyme digestion and electrophoresis on 1% agarose gels stained with 0.5 µg ml–1 of ethidium bromide. Selected plasmids were sequenced at the Nevada Genomics Center, University of Nevada, Reno, with a ABI BigDye™ Terminator Cycle Sequencing Ready Reaction kit, v3.1, and an ABI 3730 Sequence Analyzer (Applied Biosystems, Life Technologies, Grand Island, NY, USA), using the M13 forward (5′-TGTAAAACGACGGCCAGT-3′) and reverse (5′-GAGCGGATAACAATTTCACACAG-3′) primer sets. Over 1200 cDNA clones were sampled and sequenced from 10 of the 11 closely related Oncidiinae species targeted for this study, and 1000 cDNAs were selected and identified as PEPC isoforms (100 clones per species).
PEPC sequence analysis
Raw sequences were edited manually by removing vector sequences using MacVector v.11.1 software (MacVector, Cary, NC, USA). Forward and reverse PEPC fragments were then assembled in MacVector. Over 1200 assembled sequences were verified by identifying conserved amino acid sequences using the Basic Local Alignment Search Tool (BLAST) within the non-redundant database at the National Center for Biotechnology Information, and were translated into the corresponding amino acid sequences. Multiple sequence alignments for both nucleotide and protein sequences were used to visually identify distinct PEPC isoforms within each species separately. Sequences that were identical within a species were considered the same isoform. PEPC isoforms present in each species were then named based on their relative abundance, so that ppc1-o1 (letter ‘o’ for Oncidiinae) would correspond to the most abundantly transcribed isoform recovered by clone sampling in each species, followed by ppc1-o2, ppc1-o3, ppc1-o4, ppc1-o5, and ppc1-o6, respectively. By naming isoforms in this fashion, we could easily identify the isoforms that are abundantly expressed in leaves and roots when perfoming phylogenetic analysis. All isoforms were then renamed based on their position in the PEPC phylogenetic analysis (ppc1-M1-o1, ppc1-M1-o2, ppc1-M2-o1, ppc-M2-o2, and so forth).
Rapid amplification of cDNA ends (RACE) amplification
The 3′ ends of the PEPC cDNA fragments for three species (O. maduroi, O. panamense, and R. ampliatum) were recovered using the 3′ RACE system (SMARTTM RACE cDNA Amplification, BD Bioscience Clontech, Mountain View, USA) following the manufacturer’s instructions and using gene-specific primers (GSPs) based on sequences obtained by the initial degenerate RT-PCR (Supplementary Table S1 at JXB online). RACE and larger amplified cDNA products were then sequenced using a BigDye™ Terminator Sequencing kit and an ABI 3730 Sequence Analyzer in the Nevada Genomics Center at the University of Nevada, Reno. Sequences were edited manually for base-call inaccuracies, and vector sequences were removed using MacVector v.11.1 software. Sequences were then assembled into the 1100bp isoform fragments for each species using the assembly project function in MacVector. To identify sequencing errors or chimaeras that were potentially formed during RACE, GSPs were also designed to confirm isoform identities (Supplementary Table S2 at JXB online). We only performed RACE amplification in three of the 10 species for which tissue was abundantly available.
Phylogenetic analyses of ppc
Multiple sequence alignments that included 39 PEPC isoforms from the study species and 273 ppc sequences available in GenBank and Phytozome (Goodstein et al., 2012) were obtained using the MUSCLE function (Edgar, 2004) in MEGA v.5.2.2 (Tamura et al., 2011) and refined manually. The ppc sequences downloaded from GenBank included those from several orchid genera with CAM species, such as Angraecum, Dendrobium, Epidendrum, Leptotes, Microcoelia, Phalaenopsis, Solenagis, Taeniophyllum, and Vanilla. The ppc gene sequences from 63 plant genera and a sequence from the alga Chara fragilis Desv. (Supplementary Table S3 at JXB online) were also included in the alignment. Because phylogenetic trees based on ppc gene sequences from broad phylogenetic sampling have been shown consistently to produce two distinct lineages that are highly homologous but that diverged before the evolution of land plants (ppc-1 and ppc-2), we focused our analysis only on sequences from ppc-1, which contain all of the CAM and C4-specific ppc genes (Gowik and Westhoff, 2011; Christin et al., 2014). By using only ppc-1 genes, we avoided ambiguities in the alignment files that could then be reflected in the phylogenetic analysis (Christin et al., 2014). Phylogenetic trees were inferred with MrBayes v.3.2.1 (Ronquist and Huelsenbeck, 2003) following a general time-reversible model of nucleotide substitution with a γ-shaped parameter and a proportion of invariants (GTR+G+I). Two analyses were performed in parallel; each was composed of 16 chains, run for 20 000 000 generations, sampling a tree every 1000th generation after a burn-in period of 7 000 000 generations. A consensus tree was computed after the burn-in period. The convergence and appropriateness of the burn-in period were checked with Tracer (Rambaut and Drummond, 2007).
Results
Photosynthetic patterns and relationship of 11 Oncidiinae species
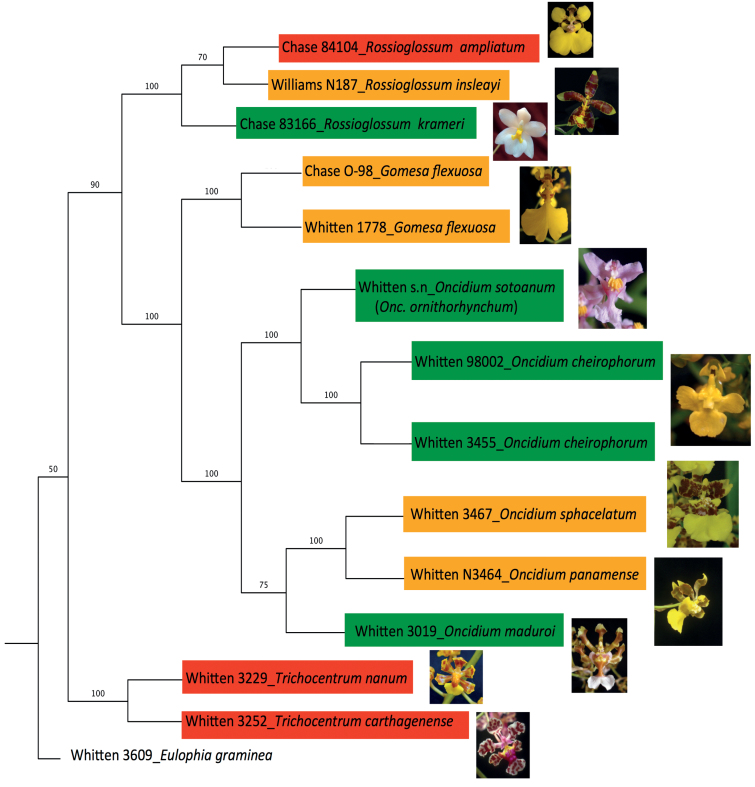
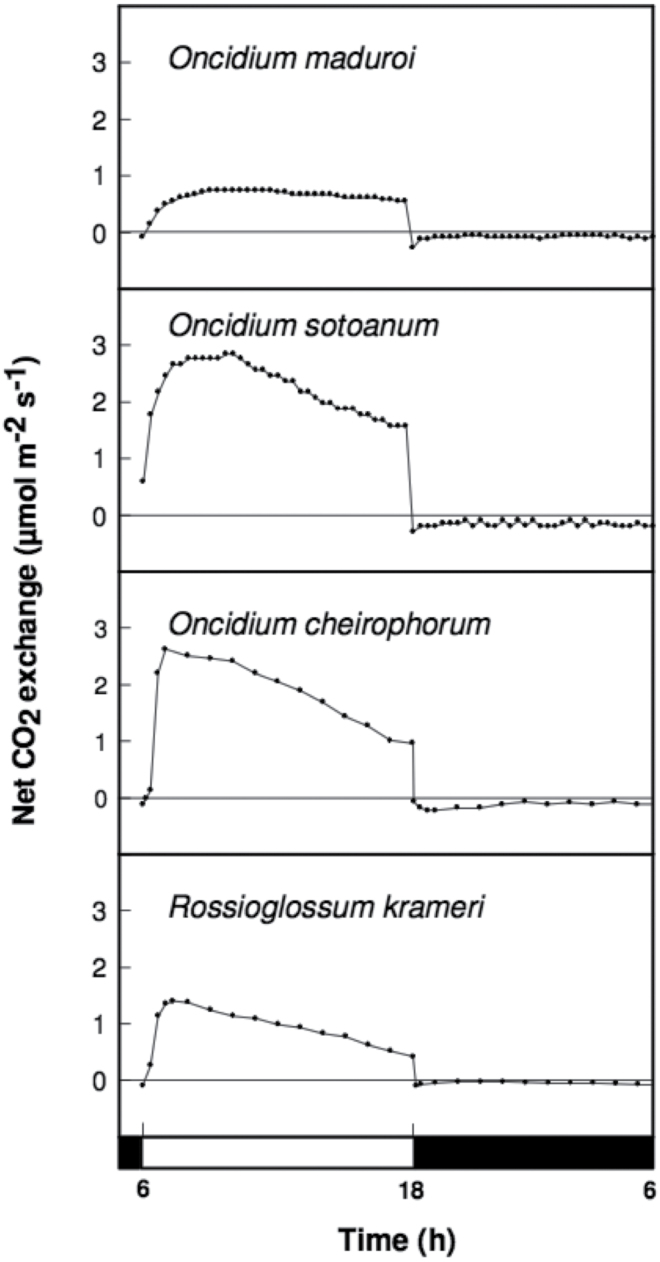
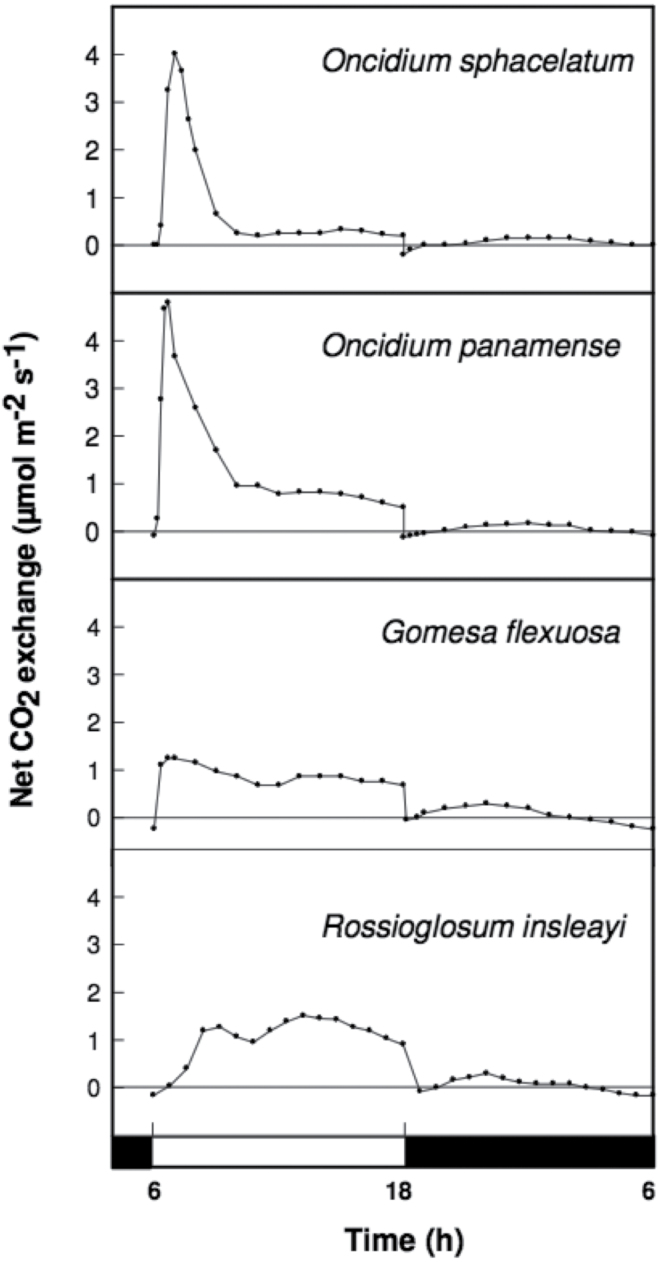
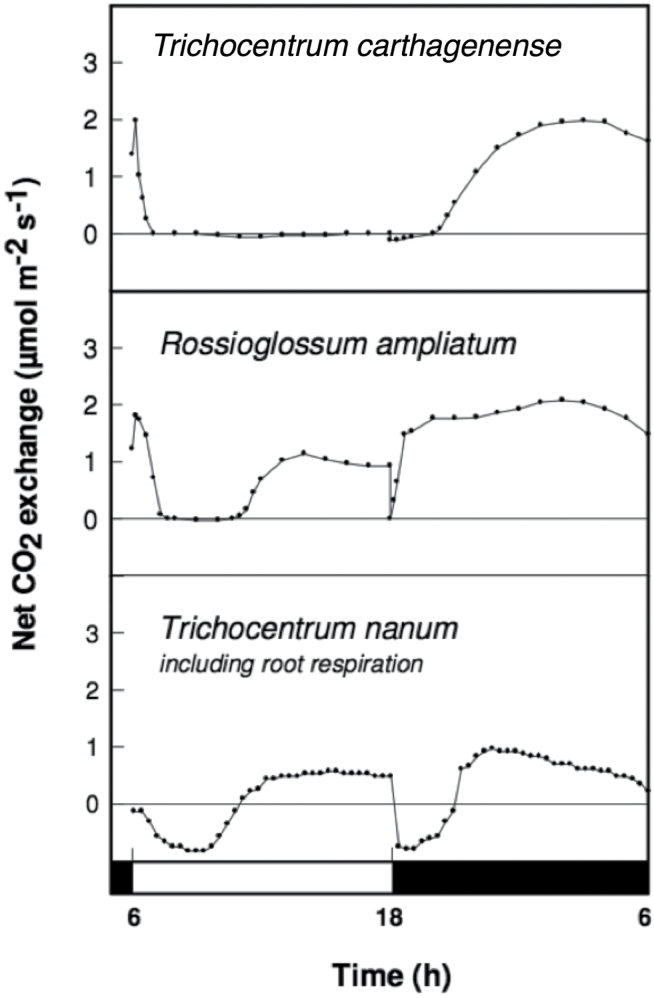
Among the 11 Oncidiinae species used in this study, the genus Rossioglossum contained C3, weak-CAM, and strong-CAM species. Oncidium was represented by both weak-CAM and C3 species, whereas Gomesa was represented by one weak-CAM species and Trichocentrum contained two strong-CAM species (Fig. 1). The phylogenetic relationship of the 11 Oncidiinae species used in this study (Fig. 1) followed the nomenclature proposed by Neubig et al. (2012) and is consistent with the phylogenetic relationships within Oncidiinae sensu Chase inferred from 590 species (Neubig et al., 2012). O. maduroi, O. sotoanum, O. cheirophorum, and R. krameri showed no nocturnal net CO2 uptake and therefore were considered C3 species (Fig. 2). G. flexuosa, O. panamense, O. sphacelatum, and R. insleayi clearly exhibited nocturnal net CO2 uptake, but its contribution to total carbon gain was small compared with CO2 uptake during the light period, so they were classified as weak-CAM species (Fig. 3). In these weak-CAM species, the majority of the CO2 was taken up during the day followed by a small amount of CO2 loss during the beginning of the night and limited net CO2 uptake throughout the night. R. ampliatum, T. carthagenense, and T. nanum were considered strong-CAM species based on their pronounced net CO2 uptake during the night (Fig. 4). T. nanum (a strong-CAM species; Fig. 4, bottom panel) showed daytime net CO2 uptake during the afternoon hours. However, the majority of its net CO2 uptake occurred during the middle of the night. Root respiration for T. nanum was included because the whole plant was measured within the cuvette due its small size. Compared with the other two strong-CAM species for which only leaf-gas exchange was determined, the presence of root respiration in the T. nanum experiment resulted in considerable CO2 loss during the early light and dark periods. Nonetheless, the strong-CAM character of the 24h CO2 gas-exchange pattern in this species was evident.
Fig. 1.
Phylogenetic relationship of 11 species from the Oncidiinae with photosynthetic pathways ranging from strong CAM (red shading) to weak CAM (yellow shading), and C3 photosynthesis (green shading). The phylogeny was reconstructed using sequences of nrITS-1 and -2, plastid DNA ycf1, matk, and the trnH–psbA intergeneric spacer. Eulophia graminea was used as the outgroup. Values on the branches represent bootstrap support (%). Accession numbers next to each species correspond to the voucher specimens deposited at the University of Florida Herbarium (FLAS). Representative images of corresponding floral morphology are shown to the right of each species designator. Photos by K. Silvera.
Fig. 2.
Continuous net CO2 exchange by C3 photosynthesis orchid species during a 12h light (open bar)/12h dark (filled bar) cycle period.
Fig. 3.
Continuous net CO2 exchange by weak-CAM orchid species during a 12h light (open bar)/12h dark (filled bar) cycle period.
Fig. 4.
Continuous net CO2 exchange by strong-CAM orchid species during a 12h light (open bar)/12h dark (filled bar) cycle period.
Identification of multiple PEPC isoforms within Oncidiinae species
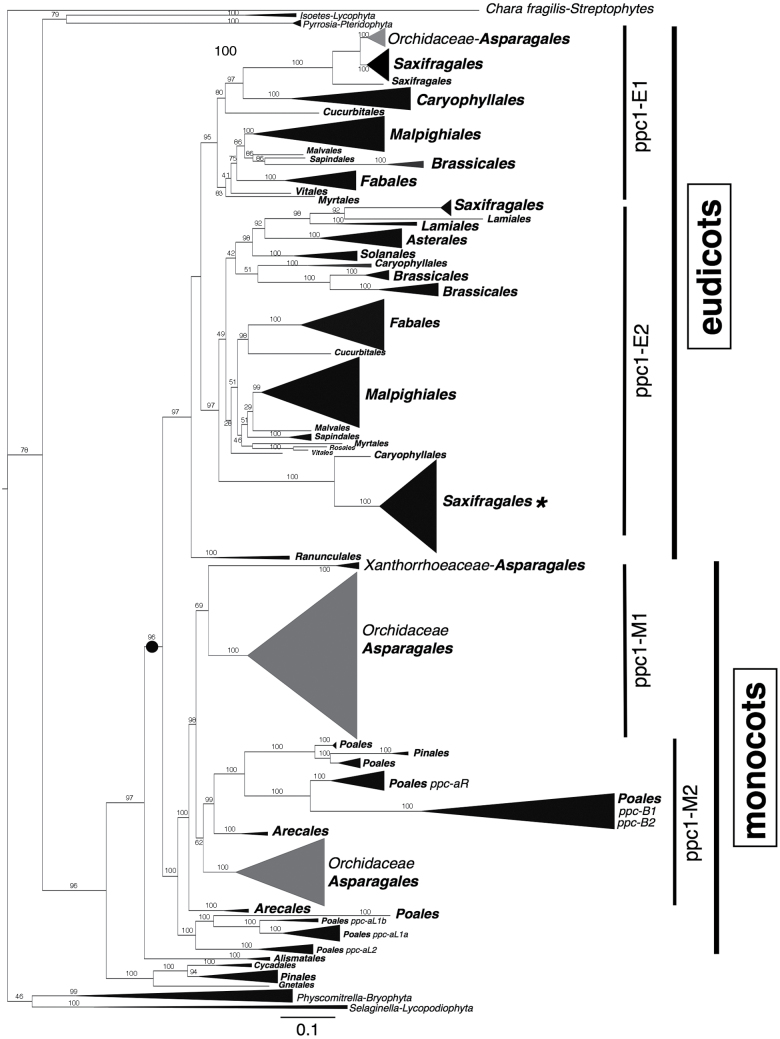
Transcripts of between two and six isoforms of PEPC were sufficiently abundant to be recovered from each of the 10 species studied, based on our sampling strategy. Species performing C3 photosynthesis expressed two to three isoforms, whereas species with weak CAM expressed three to four isoforms, and species with strong CAM expressed four to six isoforms (Table 2). Multiple nucleotide sequence alignment of the 1100bp PEPC fragments from 39 isoforms recovered in this study and 273 sequences downloaded from GenBank and Phytozome showed 162 informative sites, 192 sites without gaps, and 186 variable sites. The ppc-1 consensus tree generated from nucleotide sequence information from 312 species revealed two main lineages in flowering plants (eudicots and monocots; Fig. 5). The ppc sequences from the algal species C. fragilis, and those present in Bryophyta (Physcomitrella) and Lycopodiophyta (Selaginella), were sister to the ppc sequences from gymnosperms and angiosperms. The ppc lineage composed of Pyrrosia, a CAM fern (Fig. 5), and the aquatic CAM genus Isoetes was basal to the angiosperm and gymnosperm ppc lineages (Fig. 5).
Table 2.
PEPC isoform counts from 10 species from Oncidiinae based on relative abundance of clone samplingFour additional isoforms were recovered using GSP by RACE amplification.
| Species name | Isogene | Relative abundance (%) | Functional designation |
|---|---|---|---|
| Oncidium maduroi (C3) | ppc1-M1-o1 | 88 | C3 |
| ppc1-M2-o2 | 11 | C3 | |
| ppc1-M1-o3 | 1 | C3 | |
| Rossioglossum krameri (C3) | ppc1-M1-o1 | 92 | C3 |
| ppc1-M2-o2 | 8 | C3 | |
| Oncidium sotoanum (C3) | ppc1-M1-o1 | 86 | C3 |
| ppc1-M2-o2 | 10 | C3 | |
| ppc1-M1-o3 | 4 | C3 | |
| Gomesa flexuosa (weak CAM) | ppc1-M1-o1 | 66 | CAM |
| ppc1-M1-o2 | 31 | C3 | |
| ppc1-M1-o3 | 2 | C3 | |
| ppc1-M2-o4 | 1 | C3 | |
| Oncidium sphacelatum (weak CAM) | ppc1-M1-o1 | 96 | CAM |
| ppc1-M1-o2 | 2.5 | C3 | |
| ppc1-M1-o3 | 1 | C3 | |
| Oncidium panamense (weak CAM) | ppc1-M1-o1 | 73 | CAM |
| ppc1-M1-o2 | 19 | C3 | |
| ppc1-M2-o3 | 7.5 | C3 | |
| ppc1-M1-o4 | by RACE | C3 | |
| Rossioglossum insleayi (weak CAM) | ppc1-M1-o1 | 76 | CAM |
| ppc1-M1-o2 | 11 | C3 | |
| ppc1-M2-o3 | 10 | C3 | |
| ppc1-M1-o4 | 2 | C3 | |
| Rossioglossum ampliatum (CAM) | ppc1-M1-o1 | 70 | CAM |
| ppc1-M1-o2 | 20 | C3 /CAM | |
| ppc1-M2-o3 | 5 | C3 | |
| ppc1-M2-o4 | 5 | C3 | |
| ppc1-M1-o5 | by RACE | C3 | |
| ppc1-M2-o6 | by RACE | C3 | |
| Trichocentrum carthagenense (CAM) | ppc1-M1-o1 | 84 | CAM |
| ppc1-M1-o2 | 9 | C3 /CAM | |
| ppc1-M2-o3 | 4 | C3 | |
| ppc1-M1-o4 | 3 | C3 | |
| ppc1-M1-o5 | by RACE | C3 | |
| Trichocentrum nanum (CAM) | ppc1-M1-o1 | 59 | CAM |
| ppc1-M2-o2 | 20 | C3 /CAM | |
| ppc1-M1-o3 | 16 | C3 | |
| ppc1-M2-o4 | 4 | C3 | |
| ppc1-M1-o5 | 1 | C3 |
Fig. 5.
Phylogenetic relationship of 312 ppc-1 gene sequences with emphasis on the Orchidaceae (shaded in grey). The phylogenetic tree was obtained through Bayesian analysis. Taxonomic groups are compressed based on plant orders, with the size of the triangle proportional to the number of sequences present in each clade. Orders are represented in bold, and the main ppc lineages are depicted on the right. Two main ppc lineages are highlighted in the tree (eudicots and monocots). The filled circle represents the duplication event before the split between eudicots and monocots. The asterisk represents a sequence of Cycas revoluta nested within a Kalanchoe clade (Saxifragales). Detailed information is available in Supplementary Fig. S1 at JXB online. Bar, expected substitutions per site.
The eudicot lineage was monophyletic and composed of two well-supported clades (ppc1-E1 and ppc1-E2; Fig. 5 and Supplementary Fig. S1). The position of these clades was consistent with those described by Christin and Besnard (2009) and Christin et al. (2014). Gene duplication events that led to the six gene lineages of ppc in C4 grasses are shown within the monocot lineages (namely ppc-aL1a, ppc-aL1b, ppc-aL2, ppc-aR, ppc-B1, and ppc-B2, Figs 5 and 6) and follow the nomenclature proposed by Christin and Besnard (2009). The monocot lineage in the current study appeared to be paraphyletic because some monocot Orchidaceae ppc sequences were embedded within the eudicots ppc1-E1 lineage (Fig. 5, represented by the grey clade).
Fig. 6.
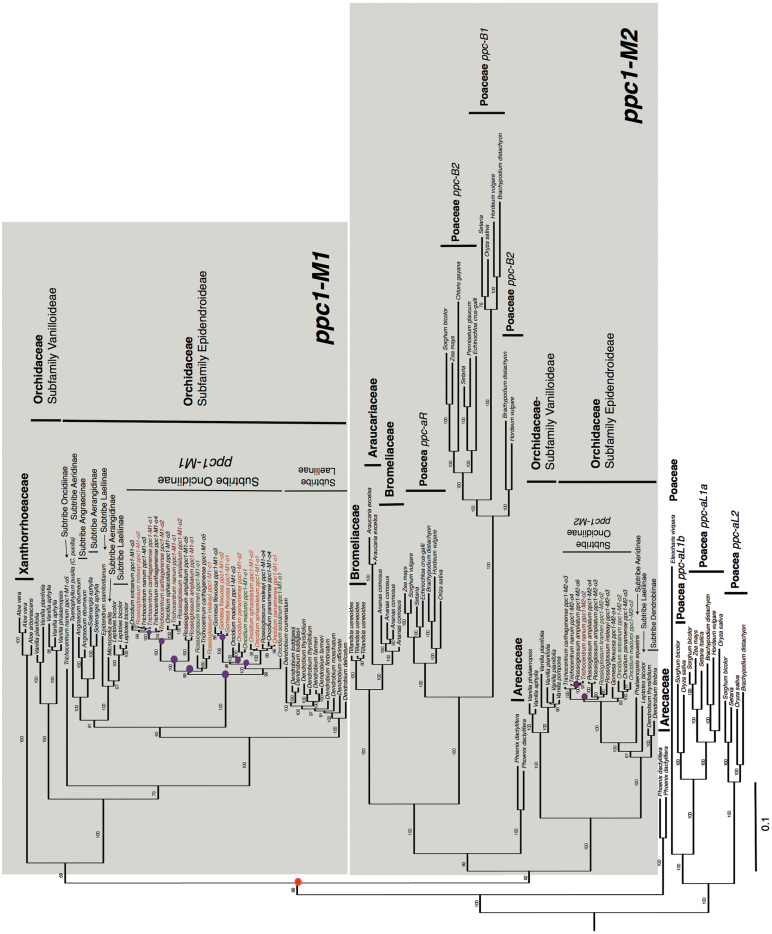
Phylogenetic relationship of monocot ppc1 sequences with emphasis on the Orchidaceae. The phylogenetic tree was obtained through Bayesian analysis. Plant families are represented in bold. Two lineages within monocots are highlighted in grey (ppc1-M1 and ppc1-M2). Orchid subfamilies and subtribes are depicted on the right. The two most abundantly transcribed ppc isoforms for C3, weak-CAM, and strong-CAM orchid species are highlighted in green, orange, and red, respectively. The red filled circle represents an early duplication event leading to lineages ppc1-M1 and ppc1-M2. Magenta filled circles represent duplication events within Orchidaceae lineages. Detailed information is available in Supplementary Fig. S1. Bar, expected substitutions per site.
Gene duplication events led to three ppc lineages in the Orchidaceae: two well-supported lineages within the monocot lineage (ppc1-M1 and ppc1-M2; Fig. 6), and a third lineage, which is embedded within the eudicot ppc1-E1 lineage (Fig. 5). The monocot lineage contained all of the 39 transcribed isoforms recovered in this study (ppc1-M1 and ppc1-M2; Fig. 6). The most abundantly transcribed isoform of all the Oncidiinae species recovered in this study grouped within the ppc1-M1 lineage (Fig. 6, ppc1-M1-o1 represented in red, orange, and green). The most abundantly transcribed isoforms for weak-CAM species (e.g. G. flexuosa ppc1-M1-o1 and ppc1-M1-o2, O. sphacelatum ppc1-M1-o1 and ppc1-M1-o2, O. panamense ppc1-M1-o1 and ppc1-M1-o2, and R. insleayi ppc1-M1-o1 and ppc1-M1-o2, represented in orange in Fig. 6) clustered together with the most abundantly transcribed isoforms for strong-CAM species (e.g. R. ampliatum ppc1-M1-o1 and ppc1-M1-o2, T. carthagenense ppc1-M1-o1 and ppc1-M1-o2, and T. nanum ppc1-M1-o1, represented in red in Fig. 6). The most abundantly transcribed isoform for C3 species (e.g. O. maduroi ppc1-M1-o1, R. krameri ppc1-M1-o1, and O. sotoanum ppc1-M1-o1, represented in green in Fig. 6) also clustered with the most abundantly transcribed isoforms for weak- and strong-CAM species. The phylogenetic similarity between the two most abundantly transcribed isoforms for weak- and strong-CAM species (ppc1-M1-o1 and ppc-M1-o2) and the most abundantly transcribed isoform for C3 species (ppc1-M1-o1) suggested that these orthologous isoforms have similar expression patterns and abundance and are probably involved in the same functions. The ppc sequences from Oncidiinae genera such as Gomesa, Oncidium, Rossioglossum, and Trichocentrum tended to separate into two distinct groups (ppc1-M1 and ppc1-M2), and this pattern was also consistent within orchid genera in which several isoforms were reported such as Vanilla, Leptotes, and Dendrobium (Fig. 6). Within the monocot lineage, Orchidaceae ppc sequences followed the species relationships in both M1 and M2 clusters. For example, ppc1-M1 was composed of ppc sequences from closely related Oncidiinae species (species within Gomesa, Oncidium, Rosioglossum, and Trichocentrum; Fig. 6), and ppc1-M1 was sister to ppc sequences from other species in the subfamily Epidendroideae (species within subtribes Laeliinae, Aeridinae, Angraecinae, Aerangidinae; Fig. 6). The ppc sequences from the subfamily Epidendroideae were sister to ppc sequences from Vanilla (subfamily Vanilloideae) and T. nanum ppc1-M1-o5 (Fig. 6). A gene duplication event probably occurred before the diversification of Oncidiinae ppc1-M1 and ppc1-M2 (Fig. 6, represented by a red filled circle). At least 10 gene duplication events were apparent within Oncidiinae ppc lineages and were related to ppc gene expansion in weak-CAM and strong-CAM species (Fig. 6, represented by magenta filled circles in ppc1-M1 and ppc1-M2). Eight gene duplication events occurred within Oncidiinae ppc1-M1, four of which were linked to gene family expansion in lineages leading to the weak-CAM species O. sphacelatum, O. panamense, and R. insleayi, and four were linked to gene family expansion in lineages leading to the strong-CAM species R. ampliatum, T. carthagenense, and T. nanum (Fig. 6). Within Orchidaceae ppc1-M2, two gene duplication events were linked to gene family expansion, one of which occurred in a lineage leading to R. ampliatum, and in another leading to T. nanum (Fig. 6, represented by filled circles in ppc1-M2).
Discussion
The Oncidiinae is one of the most diverse clades within the Orchidaceae, with a wide range of contrasting characteristics such as vegetative morphology, floral variation, chromosome number, and pollination systems (Neubig et al., 2012). Our results indicate that Oncidiinae also shows contrasting photosynthetic types, as demonstrated by gas exchange, titratable acidity, leaf thickness, and isotopic composition measurements (Table 1, Figs 2–4). Most species within the Oncidiinae are epiphytes in habitats with intermittent water availability, and many exhibit CAM photosynthesis (Silvera et al., 2009). The degree of CAM expression in orchid species correlates with leaf thickness or succulence (Winter et al., 1983), reduced intercellular air spaces, and large mesophyll cell size (Nelson et al., 2005), with a minimum necessary cell volume per unit leaf area for nocturnal acid storage. Therefore, thin-leaved, weak-CAM species are predicted to exhibit a limited degree of nocturnal net CO2 uptake (Fig. 3) when compared with thick-leaved, strong-CAM species (Fig. 4). Indeed, weak-CAM species in the Oncidiinae showed patterns of nocturnal CO2 uptake typical of CAM species but at a greatly reduced magnitude. Although their δ13C values were within the C3 range, these species exhibited statistically significant differences between evening and morning titratable acidity, which is indicative of weak CAM. Species with δ 13C values characteristic of C3 species can obtain up to one-third of their carbon via the CAM pathway (Winter and Holtum, 2002). In contrast, gas-exchange measurements for the thin-leaved C3 photosynthetic species O. maduroi, O. sotoanum, O. cheirophorum, and R. krameri showed net CO2 uptake exclusively during the daytime and a slight CO2 loss during the night (Fig. 2). Strong-CAM species with thick leaves such as R. ampliatum, T. carthagenense, and T. nanum exhibited most of their carbon gain at night (Fig. 4).
Differences in PEPC isoform numbers and relative mRNA abundance using 10 closely related Oncidiinae species were associated with the capacity to perform CAM, as measured by 24h net CO2 gas exchange. Based on our RT-PCR approach, as many as six isoforms were observed in weak- and strong-CAM species, and one to two putative CAM-specific PEPC isoforms were identified based on their relative abundance (Table 2) and position in the phylogenetic tree (Fig. 6, orthologous ppc1-M1-o1 and ppc-M1-o2 sequences represented in red and orange). The increase in number of PEPC isoforms associated with weak- and strong-CAM species could be the result of a single gene duplication event, gene duplication due to polyploidy, or different alleles of the same gene. Oncidiinae CAM species have a significantly higher DNA content compared with weak-CAM and C3 species (J.C. Cushman, unpublished data). A possible explanation for the expansion of the ppc gene family is that repeated genome duplication events leading to polyploidy in orchids have created an increased pool of duplicated genes and alleles suitable for CAM. Therefore, the presence of multiple genomes might confer an advantage for adaptive evolution (Hegarty and Hiscock, 2007). Genomic information is needed to verify the number of isoforms present in each species and the subsequent orthology or paralogy of the ppc sequences found in this study, and to identify missing gene lineages that could not be recovered with our PCR-based sampling approach. Even so, the present study suggests that the ppc gene family in Oncidiinae orchids has probably undergone gene family expansion during the evolutionary establishment of weak and strong CAM as evidenced by at least eight gene duplication events revealed by the phylogenetic tree analysis.
The identification of two distinct ppc clades in flowering plants indicates that these two lineages evolved independently of each other, and that this event occurred early in the diversification of ppc lineages (Christin and Besnard, 2009; Christin et al., 2012b ). Within the Oncidiinae, a gene duplication event occurred early in the diversification of ppc1-M1 and ppc1-M2 lineages. Several ppc1-M1 genes, presumably derived from gene duplication events (Fig. 6, represented by filled circles), evolved a role in CAM photosynthesis (Fig. 6, represented by red and orange sequences), whereas genes within ppc1-M2 maintained anapleurotic roles. This phylogenetic grouping of PEPC isoforms into distinct sister clades (ppc1-M1 and ppc1-M2) was also evident for other subtribes within the subfamily Epidendroideae (e.g. subtribe Laeliinae, subtribe Aeridinae, and subtribe Dendrobiinae; Fig. 6) and for the subfamily Vanilloideae PEPC isoforms.
Distinct Oncidiinae PEPC isoforms associated exclusively with root tissue were not identified in the current study. Gehrig et al. (2005) found three root isoforms of PEPC, each of which contained an insertion of 8 aa towards the C-terminal end of the enzyme in K. pinnata, a strong-CAM species. Putative root PEPC isoforms from the orchid species studied here showed isoforms that were identical to those found in leaves, and no insertions were evident. Because aerial roots in epiphytic orchids can engage in C3 photosynthesis (except for leafless orchids in which roots perform CAM; Winter et al., 1985), the ppc genes found in roots of epiphytic orchids are most likely the same as those found in leaves (Kwok-ki et al., 1983). Interestingly, T. nanum ppc1-M1-o5 (Fig. 6) was quite distinct from all other isoforms in the Oncidiinae ppc sequences, and was positioned close to Vanilla, a distantly related orchid group (Fig. 6). This very-low-abundance isoform might be difficult to recover, which might explain why it was not previously reported in closely related species. The many low-abundance isoforms (ppc1-o3 to ppc1-o6) found in this study might be the result of functional diversification of paralogous genes involved in non-photosynthetic PEPC functions (i.e. housekeeping or anapleurotic functions). The finding of a third ppc Orchidaceae lineage composed of three genera (Microcoelis, Leptotes, and Solenangis) outside the monocots and nested within the eudicots, and a Cycas revoluta ppc sequence nested within Kalanchoe sequences, is puzzling. Further investigation is needed to rule out the possibility that these sequences are the result of contamination or horizontal gene transfer. Additional quantitative and temporal expression analysis of mRNAs for all isoforms in this study is needed to confirm their putative functional contributions to CAM. Similarly, more genomic and transcriptomic data from orchid species are needed to confirm and identify ppc lineages in orchid genomes, and to recover isoforms that were missed due to potentially incomplete PCR-based sampling.
The PEPC isoforms most abundantly transcribed in C3 Oncidiinae species clustered closely with those of strong-CAM and weak-CAM species (Fig. 6, Oncidiinae species sequences highlighted in colors, ppc1-M1), suggesting that there is no specific convergence of amino acid changes or selective pressure towards amino acid changes linked to CAM function, as there is in C4 species (Christin et al., 2012b ). The most abundantly transcribed isoforms in cDNAs isolated from leaf and root photosynthetic tissues of C3, weak-CAM, and strong-CAM Oncidiinae species were orthologous, suggesting that these isoforms might be involved in similar functions and that they have a role in nocturnal CO2 uptake in species with weakly and strongly expressed CAM. All of the other Orchidaceae ppc sequences available from GenBank and used in this study belonged to strong-CAM species. There were no ppc sequences available from C3 orchid relatives, making it impossible to test whether ppc sequences from potentially closely related C3 species within the subfamilies Epidendroideae and Vanilloideae would have clustered in the same manner as those from strong-CAM species. This deficiency highlights the utility of conducting gene family surveys with closely related species with contrasting photosynthetic pathways to elucidate the molecular genetic underpinnings of photosynthetic pathway evolution.
Several scenarios could explain the diversification of PEPC isoforms. In one scenario, a ppc gene duplication event occurred early in the diversification of plants producing two clades: one clade with several duplicated sequences, one of which underwent recruitment for CAM through neofunctionalization, while the other clade contained sequences that retained the ancestral function. In general, neofunctionalizations require changes in gene expression (Bräutigam et al., 2011; Gowik and Westhoff, 2011) and/or amino acid substitutions to confer an entirely new function (Zhang, 2003). A second scenario involves a change in regulation. In this scenario, a gene duplication event from a ppc ancestral gene with dual functionality underwent regulatory changes that determined whether it would perform in CAM or C3 photosynthesis. This implies that neofunctionalization does not need to be linked to changes in amino acid positions, and that C3 paralogous genes can be recruited for CAM function through subfunctionalization, in which one of the duplicated genes becomes better at performing one of the functions of the progenitor genes (Hughes, 1999; Zhang, 2003). Perhaps the most likely scenario in the evolution of PEPC in orchids is the former, as suggested by the close clustering of ppc sequences from C3, weak-CAM, and strong-CAM species that exhibited greater transcript abundance (Fig. 6, orthologous ppc1-M1 genes, represented in red, orange, and green) relative to the other gene family members. This scenario suggests that increased transcript abundance occurs prior to the transition to the CAM phenotype. Also, the recurrent independent origin of CAM in distantly related plant clades (Keeley and Rundel, 2003), provides evidence that the evolution of CAM probably involves the use and modification of genes that are already present in the C3 ancestors of these species. Alternatively, cryptic genetic variants present in common ancestral populations could come to be expressed, and subsequently increase in frequency, in multiple descendent lineages under similar selection regimes (Barrett and Schluter, 2007; West-Eberhard et al., 2011). This hypothesis envisions the presence of unexpressed PEPC alleles suitable for CAM and/or C4 photosynthesis in the genome of ancestral distantly related C3 species for millions of years, which can then be incorporated into CAM or C4 species through increased expression of either formerly cryptic or universally adaptive genes. These alleles could then become fixed through natural selection in populations in which they are suited to a new photosynthetic mode (West-Eberhard et al., 2011; Christin et al., 2012a ). There is increasing evidence for the importance of cryptic and standing genetic variation in evolution (Gibson and Dworkin, 2004; Barrett and Schluter, 2007; McGuigan and Sgro, 2009) and no special mechanism is required for such variation to explain the convergent use of ancestral alleles in a new context.
Within the Orchidaceae, the presence of CAM is evolutionarily labile and prone to parallel evolution and reversal events especially within clades that contain large numbers of epiphytic species (subfamily Epidendroideae; Silvera et al., 2009, 2010a). The association of CAM with semi-arid or arid environments and microhabitats, or other stressful conditions, suggests a role for environmental influences in its recurrent origin and evolution (West-Eberhard, 2003). This suggestion is also supported by the observation that the extent of CAM expression often correlates with the degree of specialized adaptations to more xeric ecological niches (Kluge et al., 2001; Pierce et al., 2002; Zotz, 2004). The recurrent evolution of CAM reflects strong selection under conditions in which CAM might afford an advantage, as in the epiphytic habitat of orchids. Advantageous biochemical shifts and molecular genetic rearrangements could modulate changes in mRNA or protein expression patterns from C3 to CAM, and imply a direct role for environmental cues in allowing selection to act on variation in underlying genotypes. Also, structural precursors such as enlarged vacuoles and tight cell packing may need to be present for the evolutionary progression from C3 to CAM to occur (Sage, 2002). After CAM becomes established within a lineage, the expression of CAM can be plastic or environmentally sensitive, as illustrated by the existence of ‘facultative’, ‘inducible’, or ‘optional’ CAM species that engage in CAM in response to environmental stimuli such as water-deficit stress (Winter, 1985; Griffiths, 1988; Winter et al., 2008; Winter and Holtum, 2014). This hypothesis is also supported by the observation that the weak-CAM species contain increased numbers of gene duplication events compared with the C3 species, and that these species have added the novel capacity for net dark CO2 fixation to their continued capacity to express mostly C3 photosynthesis. This flexibility, combined with the ubiquity of enzymes required to perform CAM, might explain why multiple independent origins of CAM, as well as reversals, have been observed within the Orchidaceae (Silvera et al., 2009).
In summary, several lines of evidence presented here suggest that the evolution of CAM ppc genes in orchids involved gene duplication coupled with the recruitment of specific gene family member for photosynthetic pathway-specific functions. First, using our cloning approach, increased numbers of expressed PEPC isoforms were detected as sampling proceeded from C3 photosynthesis to weak-CAM and strong-CAM species within closely related species of the Oncidiinae. Secondly, phylogenetic analysis revealed that ppc genes with the greatest relative transcript abundance from C3, weak-CAM, and strong-CAM species grouped together. This observation suggests that the identified increases in PEPC mRNA expression typical of CAM-specific isogenes were acquired before the species diverged and might indicate parallel rather than convergent evolutionary tracks for these specific gene lineages.
The current Oncidiinae ppc dataset lays a strong foundation for future comparisons of gene lineages expressed in different tissues of C3, weak-CAM, and strong-CAM species. However, more comprehensive transcriptomic and genomic datasets based on deep-sequencing methods are required to identify potentially missing gene lineages and possible new PEPC isoforms. For example, gene family members with low-abundance transcripts identified in the current study might exhibit higher transcript abundance in other species, leading to a more detailed understanding of lineage-specific gene recruitment patterns among diverse CAM species. In addition to conducting detailed sampling of all possible tissues types, including leaf, pseudobulb, and root tissues, future works should include the sampling of these tissues over more detailed time courses in order to identify gene family members and lineages that have acquired pronounced diel or circadian mRNA expression patterns, which are likely to be useful diagnostic indicators for gene recruitment to CAM-specific function (Silvera et al., 2010a ). In conclusion, this study provides clear evidence for the roles of gene duplication and neofunctionalization within ppc gene lineages in the evolutionary progression of CAM within the Oncidiinae.
Supplementary data
Supplementary data is available at JXB online.
Supplementary Table S1. Gene-specific primer sets used for 3′ RACE amplification of the cDNA fragment coding for PEPC isoform for three species from Oncidiinae.
Supplementary Table S2. Gene-specific primer sets used for confirmation of the cDNA fragment recovered by RACE coding for PEPC isoform for three species from Oncidiinae.
Supplementary Table S3. The ppc sequence names used for phylogenetic analyses.
Supplementary Fig. S1. Phylogenetic tree of 312 ppc-1 gene sequences.
Supplementary Fig. S2. Protein sequence alignment for 28 PEPC isoforms and 11 PEPC isoforms recovered by RACE for 10 Oncidiinae species.
Acknowledgements
The authors thank Mark Whitten, Norris H. Williams, and Kurt M. Neubig for providing the Oncidiinae matrix to construct the phylogeny of species used in this study; Pascal-Antoine Christin for invaluable help with Bayesian analyses and for providing additional ppc plant sequences; Mary Jane West-Eberhard for comments and discussions to improve a previous version of the manuscript; Gaspar Silvera for supplying the orchid plant species; Cristina Milsner for assistance with PEPC cloning and sampling; the laboratories of Norm Ellstrand and Louis Santiago at the University of California, Riverside, for greenhouse assistance and support; and L. Santiago, B. Gulle Bilgi, M.A. Cushman, and two anonymous reviewers for helpful comments on the manuscript. This work was partially supported by funding from the Smithsonian Tropical Research Institute (to KS and KW), the US Environmental Protection Agency under the Greater Research Opportunities Graduate Program Agreement no. MA 91685201 (to KS), and the National Science Foundation NSF IOB-0543659 (to JCC). This publication was also made possible by the Panamanian Secretaria Nacional de Ciencia, Tecnología e Innovación (SENACYT), and by the National Institute of Health (NIH) Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources through its support of the Nevada Genomics, Proteomics, and Bioinformatics Centers. The views expressed in this publication are solely those of the authors and the EPA does not endorse any products or commercial services mentioned in this publication.
Glossary
Abbreviations:
- ∆H+
titratable acidity
- CAM
crassulacean acid metabolism
- GSP
gene-specific primer
- PEPC
phosphoenolpyruvate carboxylase
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription-PCR.
References
- Barrett RDH, Schluter D. 2007. Adaptation from standing genetic variation. Trends in Ecology & Evolution 23, 38–44 [DOI] [PubMed] [Google Scholar]
- Besnard G, Muasya AM, Russier F, Roalson EH, Salamin N, Christin PA. 2009. Phylogenomics of C4 photosynthesis in Sedges (Cyperaceae): multiple appearances and genetic convergence. Molecular Biology and Evolution 26, 1909–1919 [DOI] [PubMed] [Google Scholar]
- Blasing OE, Ernst K, Streubel M, Westhoff P, Svensson P. 2002. The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia—implications for the evolution of C4 photosynthesis. Planta 215, 448–456 [DOI] [PubMed] [Google Scholar]
- Blasing OE, Westhoff P, Svensson P. 2000. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. Journal of Biological Chemistry 275, 27917–27923 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M, Hanson L, Albert V, Whitten W, Williams N. 2005. Life history evolution and genome size in subtribe Oncidiinae (Orchidaceae). Annals of Botany 95, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R, Vidal J, Oleary MH. 1996. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 273–298 [DOI] [PubMed] [Google Scholar]
- Christin PA, Arakaki M, Osborne CP, Bräutigam A, Sage RF, Hibberd JM, Kelly S, Covshoff S, Wong GKS, Hancock L, Edward EJ. 2014. Shared origins of a key enzyme during the evolution of C4 and CAM metabolism. Journal of Experimental Botany 65, 3609–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Besnard G. 2009. Two independent C4 origins in Aristidoideae (Poaceae) revealed by the recruitment of distinct phosphoenolpyruvate carboxylase genes. American Journal of Botany 96, 2234–2239 [DOI] [PubMed] [Google Scholar]
- Christin PA, Edwards EJ, Besnard G, Boxall SF, Gregory R, Kellogg EA, Hartwell J, Osborne CP. 2012. a Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Current Biology 22, 445–449 [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Savolainen V, Duvall MR, Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Current Biology 17, 1241–1247 [DOI] [PubMed] [Google Scholar]
- Christin PA, Wallace MJ, Clayton H, Edwards EJ, Furbank RT, Hattersley PW, Sage RF, Macfarlane TD, Ludwig M. 2012. b Multiple photosynthetic transitions, polyploidy, and lateral gene transfer in the grass subtribe Neurachninae. Journal of Experimental Botany 63, 6297–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC. 2001. Crassulacean acid metabolism. A plastic photosynthetic adaptation to arid environments. Plant Physiology 127, 1439–1448 [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ. 1997. Molecular genetics of crassulacean acid metabolism. Plant Physiology 113, 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ. 1999. Crassulacean acid metabolism: molecular genetics. Annual Review of Plant Physiology and Plant Molecular Biology 50, 305–332 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Tillett RL, Wood JA, Branco JM, Schlauch KA. 2008. Large-scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and crassulacean acid metabolism (CAM). Journal of Experimental Botany 59, 1875–1894 [DOI] [PubMed] [Google Scholar]
- Echevarria C, Vidal J. 2003. The unique phosphoenolpyruvate carboxylase kinase. Plant Physiology and Biochemistry 41, 541–547 [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig H, Heute V, Kluge M. 2001. New partial sequences of phosphoenolpyruvate carboxylase as molecular phylogenetic markers. Molecular Phylogenetics and Evolution 20, 262–274 [DOI] [PubMed] [Google Scholar]
- Gehrig H, Taybi T, Kluge M, Brulfert J. 1995. Identification of multiple PEPC isogenes in leaves of the facultative crassulacean acid metabolism (CAM) plant Kalanchoe blossfeldiana Poelln cv Tom Thumb. FEBS Letters 377, 399–402 [DOI] [PubMed] [Google Scholar]
- Gehrig HH, Heute V, Kluge M. 1998. Toward a better knowledge of the molecular evolution of phosphoenolpyruvate carboxylase by comparison of partial cDNA sequences. Journal of Molecular Evolution 46, 107–114 [DOI] [PubMed] [Google Scholar]
- Gehrig HH, Winter K, Cushman J, Borland A, Taybi T. 2000. An improved RNA isolation method for succulent plant species rich in polyphenols and polysaccharides. Plant Molecular Biology Reporter 18, 369–376 [Google Scholar]
- Gehrig HH, Wood J, Cushman MA, Virgo A, Cushman JC, Winter K. 2005. Large gene family of phosphoenolpyruvate carboxylase in the crassulacean acid metabolism plant Kalanchoe pinnata (Crassulaceae). Functional Plant Biology 32, 467–472 [DOI] [PubMed] [Google Scholar]
- Gibson G, Dworkin I. 2004. Uncovering cryptic genetic variation. Nature Reviews Genetics 5, 681–690 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Westhoff P. 2011. The path from C3 to C4 photosynthesis. Plant Physiology 155, 56–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths H. 1988. Crassulacean acid metabolism: a re-appraisal of physiological plasticity in form and function. Advances in Botanical Research 15, 43–92 [Google Scholar]
- Griffiths H. 1989. Carbon dioxide concentrating mechanisms and the evolution of CAM in vascular epiphytes. In: Lüttge U, ed. Vascular plants as epiphytes. Berlin: Springer-Verlag, 42–86 [Google Scholar]
- Hegarty M, Hiscock S. 2007. Polyploidy: doubling up for evolutionary success. Current Biology 17, R927–R929 [DOI] [PubMed] [Google Scholar]
- Holtum JAM, Winter K, Weeks MA, Sexton TR. 2007. Crassulacean acid metabolism of the ZZ plant, Zamioculcas zamiifolia (Araceae). American Journal of Botany 94, 1670–1676 [DOI] [PubMed] [Google Scholar]
- Hughes AL. 1999. Adaptive evolution of genes and genomes. Oxford: Oxford University Press [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y. 2004. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annual Review of Plant Biology 55, 69–84 [DOI] [PubMed] [Google Scholar]
- Keeley JE, Rundel PW. 2003. Evolution of CAM and C4 carbon-concetrating mechanisms. International Journal of Plant Sciences 164, S55–S77 [Google Scholar]
- Kluge M, Razanoelisoa B, Brulfert J. 2001. Implications of genotypic diversity and plasticity in the ecophysiological success of CAM plants, examined by studies on the vegetation of Madagascar. Plant Biology 3, 214–222 [Google Scholar]
- Kore-eda S, Noake C, Ohishi M, Ohnishi J, Cushman JC. 2005. Transcriptional profiles of organellar metabolite transporters during induction of crassulacean acid metabolism in Mesembryanthemum crystallinum . Functional Plant Biology 32, 451–466 [DOI] [PubMed] [Google Scholar]
- Kwok-ki K, Hock-Hin Y, Choy-Sin H. 1983. The presence of photosynthetic machinery in aerial roots of leafy orchids. Plant and Cell Physiology 24, 1317–1321 [Google Scholar]
- Latzko E, Kelly G. 1983. The many-faced function of phosphoenolpyruvate carboxylase in C3 plants. Physiology Vegetation 21, 805–815 [Google Scholar]
- Masumoto C, Miyazawa SI, Ohkawa H, Fukuda T, Taniguchi Y, Murayama S, Kusano M, Saito K, Fukayama H, Miyao M. 2010. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proceedings of the National Academy of Sciences, USA 107, 5226–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan K, Sgro CM. 2009. Evolutionary consequences of cryptic genetic variation. Trends in Ecology & Evolution 24, 305–311 [DOI] [PubMed] [Google Scholar]
- Monson RK. 1999. Origins of C4 genes and evolutionary patterns in the C4 metabolic phenotype. In: Sage RF, Monson RK, eds. C4 Plant Biology. San Diego: Academic Press, 377–410 [Google Scholar]
- Nelson EA, Sage TL, Sage RF. 2005. Functional leaf anatomy of plants with crassulacean acid metabolism. Functional Plant Biology 32, 409–419 [DOI] [PubMed] [Google Scholar]
- Neubig KM, Whitten WM, Williams NH, Blanco MA, Endara L, Burleigh JG, Silvera K, Cushman JC. 2012. Generic recircumscription of Oncidiinae (Orchidaceae: Cymbidieae) based on maximum likelihood analysis of combined DNA datasets. Botanical Journal of the Linnean Society 168, 117–146 [Google Scholar]
- O’Leary B, Park J, Plaxton WC. 2011. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochemistry Journal 436, 15–34 [DOI] [PubMed] [Google Scholar]
- Pierce S, Winter K, Griffiths H. 2002. Carbon isotope ratio and the extent of daily CAM use by Bromeliaceae. New Phytologist 156, 75–83 [Google Scholar]
- Rambaut A, Drummond AJ. 2007. Tracer v.1.4. Available at: http://beast.bio.ed.ac.uk/
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sage RF. 2002. Are crassulacean acid metabolism and C4 photosynthesis incompatible? Functional Plant Biology 29, 775–785 [DOI] [PubMed] [Google Scholar]
- Silvera K, Neubig KM, Whitten WM, Williams NH, Winter K, Cushman JC. 2010. a Evolution along the crassulacean acid metabolism continuum. Functional Plant Biology 37, 995–1010 [Google Scholar]
- Silvera K, Santiago LS, Cushman JC, Winter K. 2009. Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiology 149, 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera K, Santiago LS, Cushman JC, Winter K. 2010. b The incidence of crassulacean acid metabolism in Orchidaceae derived from carbon isotope ratios: a checklist of the flora of Panama and Costa Rica. Botanical Journal of the Linnean Society 163, 194–222 [Google Scholar]
- Silvera K, Santiago LS, Winter K. 2005. Distribution of crassulacean acid metabolism in orchids of Panama: evidence of selection for weak and strong modes. Functional Plant Biology 32, 397–407 [DOI] [PubMed] [Google Scholar]
- Singh J, Reddy GM, Agarwal A, Chandrasekhar K, Sopory SK, Reddy MK, Kaul T. 2012. Molecular and structural analysis of C4-specific PEPC isoforms from Pennisetum glaucum plays a role in stress adaptation. Gene 500, 224–231 [DOI] [PubMed] [Google Scholar]
- Smith JAC, Winter K. 1996. Taxonomic distribution of crassulacean acid metabolism. In: Winter K, Smith JAC, eds. Crassulacean acid metabolism: biochemistry, ecophysiology and evolution. Berlin/Heidelberg: Springer-Verlag, 427–436 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Nimmo HG, Borland AM. 2004. Expression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase genes. Implications for genotypic capacity and phenotypic plasticity in the expression of crassulacean acid metabolism. Plant Physiology 135, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ, Smith JAC, Winter K. 2011. Photosynthesis, reorganized. Science 332, 311–312 [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. 2003. Developmental plasticity and evolution. Oxford: Oxford University Press [Google Scholar]
- Westhoff P, Gowik U. 2004. Evolution of C4 phosphoenolpyruvate carboxylase. Genes and proteins: a case study with the genus Flaveria . Annals of Botany 93, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K. 1985. Crassulacean acid metabolism. In: Barber J, Baker NR, eds. Photosynthetic mechanisms and the environment. Amsterdam: Elsevier, 329–387 [Google Scholar]
- Winter K, Garcia M, Holtum JAM. 2008. On the nature of facultative and constitutive CAM: environmental and developmental control of CAM expression during early growth of Clusia, Kalanchoe, and Opuntia . Journal of Experimental Botany 59, 1829–1840 [DOI] [PubMed] [Google Scholar]
- Winter K, Holtum JAM. 2002. How closely do the δC13 values of crassulacean acid metabolism plants reflect the proportion of CO2 fixed during day and night? Plant Physiology 129, 1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Holtum JAM. 2014. Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis. Journal of Experimental Botany 65, 3425–3441 [DOI] [PubMed] [Google Scholar]
- Winter K, Medina E, Garcia V, Mayoral ML, Muniz R. 1985. Crassulacean acid metabolism in roots of leafless orchid, Campylocentrum tyrridion Garay & Dunsterv. Journal of Plant Physiology 118, 73–78 [DOI] [PubMed] [Google Scholar]
- Winter K, Smith JAC. 1996. a Crassulacean acid metabolism: current status and perspectives. In: Winter K, Smith JAC, eds. Crassulacean Acid Metabolism: Biochemistry, Ecophysiology and Evolution. Berlin/Heidelberg: Springer-Verlag, 389–426 [Google Scholar]
- Winter K, Smith JAC. 1996. b An introduction to crassulacean acid metabolism: biochemical principles and ecological diversity. In: Winter K, Smith JAC, eds. Crassulacean acid metabolism. Berlin: Springer-Verlag, 1–13 [Google Scholar]
- Winter K, Wallace BJ, Stocker GC, Roksandic Z. 1983. Crassulacean acid metabolism in Australian vascular epiphytes and some related species. Oecologia 57, 129–141 [DOI] [PubMed] [Google Scholar]
- Zhang JZ. 2003. Evolution by gene duplication: an update. Trends in Ecology & Evolution 18, 292–298 [Google Scholar]
- Zotz G. 2004. How prevalent is crassulacean acid metabolism among vascular epiphytes? Oecologia 138, 184–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.