Abstract
The Human Microbiome Project (HMP) is a global initiative undertaken to identify and characterize the collection of human-associated microorganisms at multiple anatomic sites (skin, mouth, nose, colon, vagina), and to determine how intra-individual and inter-individual alterations in the microbiome influence human health, immunity, and different disease states. In this review article, we summarize the key findings and applications of the HMP that may impact pharmacology and personalized therapeutics. We propose a microbiome cloud model, reflecting the temporal and spatial uncertainty of defining an individual's microbiome composition, with examples of how intra-individual variations (such as age and mode of delivery) shape the microbiome structure. Additionally, we discuss how this microbiome cloud concept explains the difficulty to define a core human microbiome and to classify individuals according to their biome types. Detailed examples are presented on microbiome changes related to colorectal cancer, antibiotic administration, and pharmacomicrobiomics, or drug–microbiome interactions, highlighting how an improved understanding of the human microbiome, and alterations thereof, may lead to the development of novel therapeutic agents, the modification of antibiotic policies and implementation, and improved health outcomes. Finally, the prospects of a collaborative computational microbiome research initiative in Africa are discussed.
Introduction: Emergence of Microbiomics
With every scientific breakthrough, whether it is a landmark discovery (such as the discovery of microbes as etiological agents of infectious diseases or DNA as the genetic material), a conceptual paradigm shift (such as Darwin's theory of evolution or Woese's three-domain tree of life), or a technical breakthrough (such as the polymerase chain reaction or high-throughput DNA sequencing), a set of satellite scientific branches and technological applications emerge, some of which persist and flourish, while others fade and ultimately perish.
With one such breakthrough, the Human Genome Project (HGP, URL: http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml), emerged what is now known as genome sciences and multi-omic technologies. The first draft of the human genome sequence (Lander et al., 2001; Venter et al., 2001) generated a suite of new subspecialty areas and analysis tools, together seeking to solve the mystery of how a relatively small number of genes account for all observed phenotypic variations among humans. As ensuing more complete and better-annotated versions of the human genome were being published, the emergent “-omic” bubble kept growing. The nascent scientific branches and technologies aimed at complementing human gene sequence data with multiple layers of functional information as well as data about genetic variations between individuals, communities, and races, in an attempt to explain the wide spectrum of diversity among human phenotypes.
Specifically, the efforts towards interpreting the human genome crystallized into four major endeavors:
1. Cataloguing human genome variations to understand the impact of these variations on health, disease, and response to therapy, exemplified by HapMap (International HapMap Consortium, 2003; Deloukas and Bentley, 2004), and by the Human Variome Project (HVP, URL: http://www.humanvariomeproject.org/) (Ring et al., 2006) and the collaborative environment around it (Ozdemir et al., 2011);
2. Understanding diversity in human phenotypes by determining the functional units of the human genome, their regulatory elements, and the variation in their regulation. These efforts are exemplified by the ENCyclopedia Of DNA Elements (ENCODE, URL: http://www.genome.gov/ENCODE) (ENCODE Project Consortium, 2011; Maher, 2012; Pennisi, 2012);
3. Modeling the human phenome from genomic data by reconstructing human metabolism [e.g., RECON (Thiele et al., 2013)] or by building entire systems-level models;
4. Understanding epigenetic variations and heritable variation in phenotypic traits, and the role of the environment in heritable epigenetic modifications (Richards et al., 2010).
Nevertheless, neither genetic, epigenetic, nor regulatory variations are sufficient to explain the complexity and diversity of human phenotypes (Davies, 2001; Zhang and Dolan, 2008). The hundred trillion microbes and viruses residing in every human body, which outnumber human cells and contribute at least 100 times more genes than those encoded on the human genome (Ley et al., 2006), offer an immense accessory pool for inter-individual genetic variation that has been underestimated and largely unexplored (Savage, 1977; Medini et al., 2008; Minot et al., 2011; Wylie et al., 2012). Thus, a natural extension of the HGP was the establishment of the Human Microbiome Project (HMP, URL: http://hmpdacc.org) (Turnbaugh et al., 2007; Peterson et al., 2009), which has benefited from tremendous advances in sequencing technology, metagenomics, and bioinformatics analysis tools. In our opinion, metagenomic-based efforts to identify and compare microbes all over the world, including human microbiome research, usher in a new era of microbiology, and amalgamate environmental and medical microbiology like never before.
In this article, we review the major findings of the HMP and associated studies; we focus on the impact of the HMP data on the human variome; and we explore their potential implications in the future of drug therapy and personalized medicine. We discuss three focus topics in further detail: microbiome and colorectal cancer, the impact of antibiotics on the microbiome, and drug–microbiome interactions. The PharmacoMicrobiomics web portal is described and presented as an initiative for collaborative research in Africa.
The Human Microbiome Project, Levels of Microbiome Variations, and Pharmacotherapy
Early anticipations
As soon as it was launched by the National Institutes of Health in 2007, and even before any sequence data were published, the HMP was anticipated to have a great impact on personalized medicine (Kinross et al., 2008; Wilson and Nicholson, 2009). Most of the attention, however, was given to the effect of the human microbiome on propensity to disease (Guarner and Malagelada, 2003; Kinross et al., 2012) [e.g., effects on obesity (Turnbaugh et al., 2006; Turnbaugh and Gordon, 2009) or inflammatory bowel disease (IBD) (Matto et al., 2005; Subramanian et al., 2006)] rather than its effects on therapeutic intervention. Even when the question of therapeutic intervention was brought up, prebiotics and probiotics, rather than pharmacological agents, were the center of attention (O'Mahony et al., 2001; Tuohy et al., 2009).
Meanwhile, a wealth of literature has long been available about the biotransformation of xenobiotics, notably by gut bacteria (reviewed in Sousa et al., 2008; Rizkallah et al., 2010; Johnson et al., 2012; Haiser and Turnbaugh, 2013). This valuable information is predominantly about drug metabolism by unknown human-associated microbes; however, only a few cases of inter-individual microbiome variations have been documented [e.g., digoxin (Mathan et al., 1989) and acetaminophen (Clayton et al., 2009)].
Current realizations
The human microbiome represents a variety of microbial species of different numbers and metabolic capabilities (Fig. 1). Apart from central metabolic pathways that might be shared by heterotrophic microbes and humans, hundreds of additional metabolic reactions greatly expand the predicted human metabolic reactome, thereby driving the human organism to rather function as an entire ecosystem, a supraorganism (Lederberg, 2000; Sleator, 2010; van Duynhoven et al., 2011) (Fig. 1).
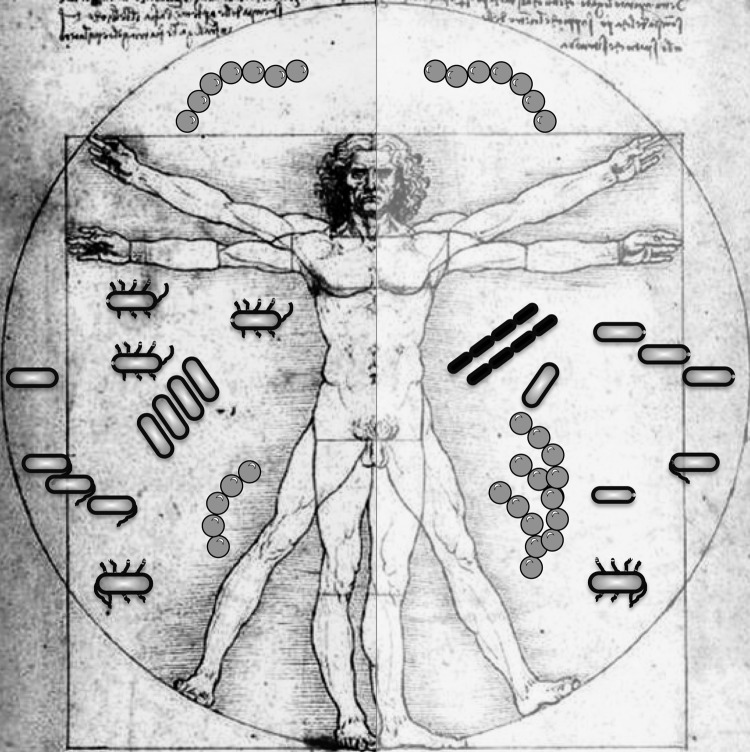
FIG. 1.
An illustration of the human supraorganism represented as a human body surrounded by a “microbiome cloud” at two health states, using Leonardo da Vinci's classic “Vitruvian Man.” From a genomic perspective, a human can be viewed as a relatively stable gene pool surrounded by a fluid microbial cloud or aura made up of a highly diverse set of microbes. In spite of evidence of memory that preserves the composition of an individual's microbiome, one's microbiome still varies continually with time, space, health state, and other intra-individual factors. Here, two different microbiome patterns are shown on the right and left of the same individual, representing variation in types and abundance of microbial species.
The HMP started in 2007 with some pilot studies to standardize the methodology and sequence reference bacterial genomes (Peterson et al., 2009; Proctor, 2011). The international effort grew fast, pioneered by the European Metagenomics of the Human Intestinal Tract (MetaHIT) consortium, who, in 2010, published a catalogue of the gut microbiome (Qin et al., 2010). Soon after, in 2012, the HMP published a full description of the composition and diversity of the microbiomes of five human body sites (gut, skin, nares, oral cavity, and vagina) (Human Microbiome Project Consortium, 2012) in addition to a growing collection of articles with more specific microbiome-related projects (URL: http://bit.ly/1d86hy2, eg., Gevers et al., 2012).
So far, most of the published data describing the so-called healthy human microbiome come from volunteers in Western countries. It will take more time to have a global representation of healthy microbiomes from humans living in different geographical areas and with different socioeconomic standards; yet some pilot attempts have been made (Yatsunenko et al., 2012; Lin et al., 2013)
The main features of the published human microbiome sequence are (Human Microbiome Project Consortium, 2012):
1. A tremendous diversity both on the genus and species level, with an added variability among closely related strains contributed by phages (Reyes et al., 2010) and other genomic islands;
2. Variable patterns of diversity within samples, with the most diverse microbial assortments being oral and distal gut samples, and the least diverse being vaginal microbiomes (Human Microbiome Project Consortium, 2012);
3. Within-subject variation was lower than between-subject variation over time in both organismal composition and metabolic function. Unlike diversity within samples, inter-individual diversity was highest in skin samples (possibly being the most exposed to rapid environmental changes);
4. Whereas microbial types varied widely to the extent of the absence of a universal taxon ubiquitous in all body habitats of all individuals, in most cases (except vaginal microbiome), metabolic pathways were stable so long as the subjects were healthy (Human Microbiome Project Consortium, 2012);
5. The absence of detrimental microbes in healthy microbiomes supports the hypothesis that microbiota undergo perturbations in configurations in disease states.
Microbiome variations
The microbiome not only expands the human-associated gene pool by orders of magnitudes, but is also more plastic or fluid than the human genome (Pflughoeft and Versalovic, 2012) (i.e., human-associated microbiota can be partly or fully exchanged) and more readily evolvable than most human cells (i.e., highly adaptive because of continuous exposure to human immune defenses, protozoal predation, phage attacks, dietary constituents, xenobiotics, antibiotics, and various toxins of bacterial, protozoal, and human origin) (Ley et al., 2006). Thus, the human supraorganism may be viewed as comprised of two constituents: (i) a relatively stable, inheritable human gene pool (usually stable over a human's lifetime with the exception of a few stable mutations, genomic imprinting, and cancers), surrounded by (ii) a cloud of a changeable, evolvable gene pool, supplied by a resident microbiota acquired after birth, whose composition varies with time, space, health, and hormonal state (Fig. 1). Here, we suggest a microbiome cloud model, reflecting the uncertainty of accurately and comprehensively defining the microbiome at a given time and space.
To systems pharmacologists, the concept of a stable gene pool associated with a fluid, evolvable, dispensable, exchangeable accessory gene repertoire is fascinating, as it strongly extends the established paradigm of pharmacogenomics. According to the extended paradigm, pharmacomicrobiomics, in contrast to pharmacogenomics, investigates multiple levels of variations that may affect medications in many more complex ways than the human genome variations do (Rizkallah et al., 2010). Microbiome variations do not merely occur between individuals, but, as suggested above, can be spatial, temporal, seasonal, developmental, hormonal, dietary, or drug-dependent within the same individual (Aziz, 2012; Ursell et al., 2012). Below we discuss examples of intra-individual variations that shape the microbiome structure, and we demonstrate how the concept of a microbiome cloud complicates cataloguing human microbiome variations, their classification into clusters or biome types, and the definition of a universal/core human microbiome.
Intra-individual variations shaping the microbiome structure
Microbial colonization of the newborn infant starts at the minute the birth process commences and is primarily affected with the mode of delivery (Dominguez-Bello et al., 2010). The complexity of the intestinal microbial communities evolves by age; by the end of first week of life, the gut microbiome becomes more complex than at birth, and by the end of first year of life, an adult-like complexity is observed (Palmer et al., 2007). Vaginally delivered infants possess, in all nonsterile anatomic sites, bacterial communities resembling their mothers' vaginal microbiotas. These communities are enriched in Lactobacillus, Prevotella, or Sneathia spp. In contrast, skin surface bacterial communities colonize all body habitats of Cesarean-delivered infants dominated by Staphylococcus, Corynebacterium, and Propionibacterium spp. (Dominguez-Bello et al., 2010).
Interestingly, the pioneer microbial communities acquired by birth influence the bacterial succession trajectory toward an adult complex and stable configuration. For example, the intestinal colonization by Lactobacillus, Bifidobacterium, and Bacteroides in infants born by C-section is delayed (Adlerberth et al., 2006). Moreover, vaginally acquired microbiota may act as a first line of defense protecting infants against the colonization of certain pathogens; as was the case where 64% to 82% of reported methicillin-resistant Staphylococcus aureus (MRSA) skin infections in newborns occurred in C-section delivered infants (Centers for Disease Control and Prevention (CDC), 2006). Furthermore, Cesarean-delivered babies were reported to be more susceptible to allergies and asthma (Bager et al., 2008), and the incidence of such allergies may be reduced by the administration of probiotics from birth to 6 months of age in C-section delivered babies—but not in vaginally delivered ones (Kuitunen et al., 2009).
The age effects are not limited to newborns. In a world with an unprecedented increase in global population over the age of 60, it is equally important to study the impact of the composition of the intestinal microbiota of older people on immunosenescence and frailty. The fecal microbiota of 178 elderly subjects (>65 years), grouped based on their residence location in the community (day-hospital or long-term residential care) was compared to that of 13 young healthy adults (Claesson et al., 2012). When classified by microbiota composition, defined as operational taxonomic unit (OTU) abundance, the majority of the long-stay subjects formed a distinct cluster separated from most community-dwelling and young healthy subjects. Microbiotas of the long-stay individuals were dominated by Bacteroidetes, whereas community-dwelling ones were enriched in Firmicutes (Claesson et al., 2011).
In an attempt to distinguish patterns within the microbiota, co-abundance associations of genera (CAGs) were suggested. Six CAGs were identified, and a distinct CAG dominance transition from the healthy community-dwelling subjects to the frail long-term care residents, most significantly in abundances of Prevotella and Ruminococcus CAGs (community-associated CAGs) and Alistipes and Oscillibacter CAGs (long-stay-associated CAGs). In addition, the long-stay subjects' microbiota was less diverse than that of the community-dwelling one; hence, the loss of community-associated microbiota was correlated with frailty (Claesson et al., 2012).
Other microbiome variations within an individual could be related to changes in diet, hormonal status, and exposure to external pathogens, drugs, and antibiotics (the latter is presented below as a case study).
Inter-individual variation: Gradient or clusters; Phylotypes or metabotypes?
In the pursuit of cataloguing the composition of a healthy gut microbiome, it was shown that more than 90% of resident gut microbes are members of only two phyla: Bacteroidetes and Firmicutes, and that the majority of human population have similar proportions of each (Zoetendal et al., 2008). However, in 2011, Arumugam and co-workers (Arumugam et al., 2011) identified three distinct clusters by the variation in the levels of one of three genera present in the gut. They labeled these clusters “enterotypes,” which they drove by assembling groups of species that tend to form a favored community structure (Table 1). The abundance of one genus was reported to correlate with those of the other two genera (i.e., discriminating genera either co-exist or annul each other) (Arumugam et al., 2011). Among the interesting findings of the former study is that enterotypes were reported to be neither continent- nor nation-specific.
Table 1.
Phylogenetic and Functional Variation Between the Three Suggested Human Enterotypes
| Phylogenetic variation | Functional variation | |||
|---|---|---|---|---|
| Main contributors | Co-occurring genus | Energy generation | Overrepresented vitamin | |
| Enterotype 1 | Bacteroides | Parabacteroides | Fermentation of carbohydrates and proteins | Biotin (vitamin B7) |
| Enterotype 2 | Prevotella | Desulfovibrio | Degradation of mucin glycoproteins in mucosal layer | Thiamine (vitamin B1) |
| Enterotype 3 | Ruminococcus (Sanger sequencing-based metagenomics) Clostridiales (Illumina sequencing-based metagenomics) | Akkermansia | Degradation of mucin | Heme (involved in vitamin B12 biosynthesis) |
From: Arumugam et al., 2011.
Shortly after the enterotype partitioning had emerged, Wu et al. (2011) showed that long-term diets correlated with enterotypes. Food rich in protein and animal fat were associated with the Bacteroides enterotype, while food rich in carbohydrate and simple sugars were associated with the Prevotella enterotype. Ruminococcus enterotype did not correlate with food types (Wu et al., 2011). The association between these enterotypes and food was paralleled in a previous comparative study (De Filippo et al., 2010). The impact of diet in children from Europe and rural Africa on the microbiome was investigated where fecal microbiota of 14 healthy children from Burkina Faso was compared to that of 15 healthy children from Italy of the same age. The European microbiome was enriched in taxa belonging to the Bacteroides enterotype, while the African microbiome was enriched in taxa belonging to the Prevotella enterotype, as expected based on western versus rural nutritional habits.
Huse and colleagues (2012) coined the term “biome types” after their data analysis showed that the biome types could be represented as gradients rather than clusters. Biome types were differentiated at the point where the ratio is greater than one, not when the relative abundance of one genus is greater than the other two discriminating genera. In agreement with Wu et al. (2011), biome types distinguish two rather than three distinct groups (Bacteroides/Clostridiales and Prevotella) based on the gradient combination of the three most dominant taxa in the human gut microbiome with an overlap between the Bacteroides and Clostridiales communities.
However, several technical factors influence the detection of enterotypes, including the clustering methodology, distance metrics, OTU-picking approaches, sequencing depth, data type (whole genome shotgun vs. 16S rRNA gene sequence data), and 16S rRNA region. A more recent study reported the impact of these factors on the interpretation of the results generated in different studies including data from the HMP and MetaHIT. This group concluded that there is a dire need for standardization of enterotyping methods to reach a consensus on how to define an enterotype (Koren et al., 2013).
Regardless of the technical factors listed above, enterotypes or biome types can both be described as phylotypes, as the distinction between microbiomes are made based on the community structure as identified by the microbial taxa. Another way of studying microbiomes puts less emphasis on their composition (“who the microbes are”) but focuses instead on their functional/metabolic attributes (“what they do”). Often, bacteria with identical phylogenetic assignment carry different functionally relevant, sometimes horizontally transferred, genes or genomic islands. A striking example is the identification of a digoxin-metabolizing operon in some but not all strains of Eggerthella lenta (Haiser et al., 2013) (Table 2). Thus, classifying microbiomes into different metabotypes might be more relevant to functional studies, including pharmacogenomic association studies (James, 2013; Kaddurah-Daouk and Weinshilboum, 2014).
Table 2.
Example Drug–Microbe Interactions Whose Molecular Mechanisms Have Been Delineated
| Drug (PubChemID) | Microbe (TaxID) | Pathway/ Reaction | Gene (s) | Interaction | Reference |
|---|---|---|---|---|---|
| Acetaminophen (1983) | Unknown | O-sulfonation | Unknown | Some gut microbes may increase acetaminophen toxicity by producing p-cresol, which competes with acetaminophen metabolism. | (Clayton et al., 2009), |
| Digoxin (2724385) | Eggerthella lenta (84112) | Cytochrome-encoding operon | Cardiac glycoside reductases: cgr1, cgr2 | cgr gene products, homologous to bacterial cytochromes, reduce digoxin by using it as an alternative electron acceptor. | (Haiser et al., 2013) |
| Cyclo-phosphamide (2907) | Firmicutes (1239) | Immune modulation | Unknown | Cyclophosphamide translocates firmicutes, thus altering the TH1 immune response, leading to synergistic anticancer activity. | (Viaud et al., 2013) |
Data compiled from: http://pharmacomicrobiomics.org.
The core issue
A fundamental question in the exploration of the human microbiome is the identification of a core of microbial taxa shared among all human populations across body habitats in the hope of establishing a baseline to contrast health versus disease state. This definition of a healthy core microbiome would enable scientists to discover new techniques and interventions for the restoration of the healthy homeostasis in cases of microbial imbalances, referred to as dysbiosis or dysbacteriosis (Pflughoeft and Versalovic, 2012).
To unravel what constitutes a core microbiome, Huse and co-workers (Huse et al., 2012) analyzed the HMP 16S tag sequencing data to search for a set of core OTUs common across individuals and body sites. When the core microbiome was defined as those OTUs present in 95% of the samples, oral sites were found to have the highest number of shared OTUs followed by stool and nares, then by skin and vaginal sites (Huse et al., 2012). Subsequently, researchers from the Craig Venter Institute sought a probabilistic interpretation of core taxa for individual body habitats and their collection into body regions. Similarly, 16S profiles generated by the HMP were analyzed, and conclusions were drawn by the analysis of graphical representations of the ubiquity (proportion of the cohort that a taxon of interest may be detected in) versus the abundance (the proportion that a taxon of interest exists in a specific donor's sample). This approach enabled the identification of a signature representing the underlying microbial community's structure. In addition, the concept of so-called minor core taxa was introduced and validated (Li et al., 2013). Through this two-parameter model, the numbers of core taxa detected across body sites were small, emphasizing the relatively high interpersonal variability within populations. The number of core genera increased across body regions in the following order: vagina, skin, stool, and oral cavity. When a subset of the cohort was resampled and analyzed to assess potential significant shifts in the microbiota, only the vaginal microbiome was stable between the two sampling visits, while skin, stool, and oral samples were significantly different in their taxonomical structure (Li et al., 2013).
Potential applications
The microbiome cloud model and the concept of intra-individual microbiome variability are likely to have several applications in personalizing therapeutic intervention. Below are examples on how different types of variability could be taken on consideration while planning a therapeutic regimen.
- Spatial and temporal variability: Within-individual variations of the skin microbiome have been recorded (Grice et al., 2009; Human Microbiome Project Consortium, 2012). Subsequently, the outcome of transdermal therapy may largely depend on where skin patches or topical products are applied, not just because of differences in skin thickness or humidity, but also because of microbiome variations and potential competition or interaction with applied drugs;
- Hormonal factors (menstrual cycle, pregnancy): Hormones largely control the population structure of the vaginal microbiome (Aagaard et al., 2012; Human Microbiome Project Consortium, 2012), which has direct implications on medicated vaginal inserts or pessaries;
- Temporal, dietary, or drug-dependent factors: The gut microbiota composition can change over time, and its diversity is highly sensitive to external factors such as diet, nutritional supplements, and health status. More dramatic factors, such as the use of antibiotics or chemotherapeutic agents, may radically disturb the balance of the gut microbiome. All these factors might have major impacts on drugs that are strongly modulated by gut bacteria;
- Health-related variability: Typically, several measures have to be implemented upon drug therapy of cancer, HIV, or organ transplant patients. Those measures take into consideration drug–drug interactions and the patients' immune status (often immunocompromised because of their condition or therapeutic regimen). Now, extra measures would be considered that are related to the microbiome alterations as a result of those diseases.
These examples highlight the potential need to monitor the microbiome composition [or at least some biomarkers of microbiome balance, such as enterotypes (Arumugam et al., 2011), bacteroieteds-to-firmicutes ratio in the gut (Turnbaugh et al., 2009), or markers of biome type (Turnbaugh et al., 2009)] within the same individual. Microbiome profiling of individuals may be added to high-density genotyping in future routine personalized medicine protocols (Rizkallah et al., 2010).
Practical implications
Personalized medicine is expensive, undoubtedly, and it is fully legitimate to argue that such approach would be considered a luxury in low or middle-income countries. The cost of adding more complexity to pharmacogenomics by taking the effect of an individual's microbiome into consideration might be prohibitive. However, in cases of life-threatening conditions, such as a drug with a narrow therapeutic index such as digoxin, which is modulated by the human microbiota (Dobkin et al., 1982; Mathan et al., 1989; Haiser et al., 2013), there is no alternative than adjusting the dosage based on the presence of digoxin-metabolizing genes/ biomarkers within the microbiome (Haiser et al., 2013). Sousa and co-workers reported that 18 cases of death because of microbial biotransformation were recorded in 1993 (Sousa et al., 2008). In addition, in cases of cancer chemotherapy where the patients are already closely monitored, it seems reasonable to add a microbiome assessment step to treatment protocols. Such addition is not only set to increase efficacy and safety of the treatment, but is likely to save costs in the long term (i.e., adjusting the dose of an expensive chemotherapeutic agent may save the healthcare system substantial amounts of money).
Focus Topics: Microbiome Variations and Personalized Medicine
The human intestinal microbiota is a diverse and complex community, which has an essential role in the maintenance of health state of its host. The link between imbalance in the composition of the members of gut microbiota (dysbiosis) and several diseases such as diabetes, obesity, and IBD has been frequently reported (e.g., Frank et al., 2007; Zhang et al., 2009).
Gut microbiome and colorectal cancer between association and causation
With 1.2 million new cases annually, colorectal cancer (CRC) is the most frequently diagnosed cancer in the developed world and the second most diagnosed worldwide (Jemal et al., 2011). CRC tumors consist of hyperproliferating cells that originate from mutated intestinal stem cells at the base of the colonic crypt. The genetic changes leading to epithelial hyperproliferation were described in the adenoma–carcinoma sequence, a series of common mutations occurring during the development of colorectal tumors, including tumor suppressor genes and oncogenes (Fearon and Vogelstein, 1990). Most of the common CRC-causing mutations are well documented, but the factors triggering them remain elusive. While cancer has long been considered a genetic disease, accumulating evidence implicates the intestinal microbiota in CRC development. First, the high bacterial density in the colon (∼1012 cells/mL) compared to the small intestine (∼102 cells/mL) is accompanied by a ∼12-fold increase in cancer incidence (Jemal et al., 2009). Second, IBD patients whose intestinal barrier function is reduced, and who are more exposed to microbes, have a ∼5-fold increased risk for CRC because of abnormal inflammatory reaction to commensal microbes (Rutter et al., 2004). Third, mice that are genetically susceptible to CRC develop fewer tumors under germ-free conditions (Sellon et al., 1998). Finally, several intestinal bacteria promote intestinal tumors by generating highly toxic compounds, including oxygen radicals (Huycke et al., 2002), carcinogenic byproducts of metabolism (Knasmuller et al., 2001), and substances that increase inflammation, cell proliferation, and induce DNA damage (Wu et al., 2009; Cuevas-Ramos et al., 2010; Arthur et al., 2012; Boleij et al., 2012; Schwabe and Wang, 2012; Boleij and Tjalsma, 2013; Kostic et al., 2013; Rubinstein et al., 2013).
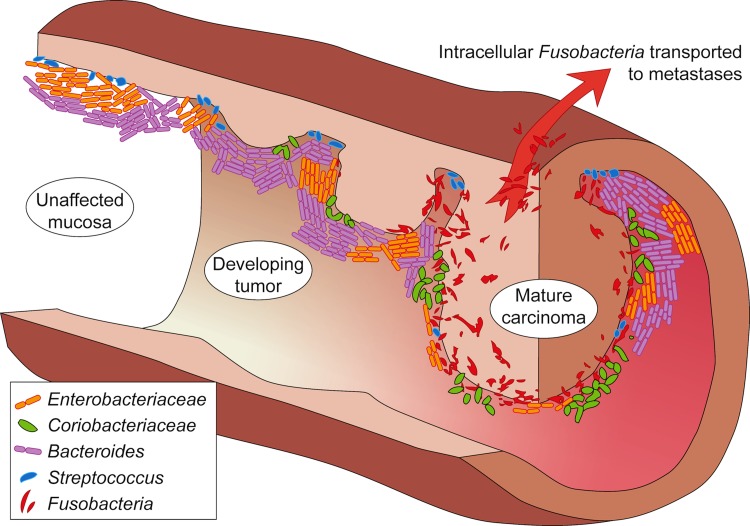
The lines of evidence above inspired a series of studies mapping the taxonomic distribution of the intestinal microbiota in CRC patients by using high-throughput sequencing of DNA isolated from clinical samples, including rRNA amplicon sequencing and shotgun metagenomics. Those studies found that a number of bacteria were enriched or depleted on tumors; although samples were analyzed at the genus level, these taxonomic associations remained difficult to interpret. For example, Fusobacterium, the most consistently tumor-enriched bacterium (Castellarin et al., 2012; Kostic et al., 2012), was originally expected to benefit the intestinal epithelium by producing butyrate, the preferred energy source for colonic cells. However, fusobacteria have recently been shown to possess pro-inflammatory and invasive features, and occur in CRC-metastases (Castellarin et al., 2012). Moreover, F. nucleatum promotes CRC by binding E-cadherin, activating β-catenin signaling (Rubinstein et al., 2013), and recruiting tumor-infiltrating myeloid cells (Kostic et al., 2013). Another example of functional ambiguity when analyzing taxonomic profiles is Bacteroides fragilis that includes both enterotoxigenic and nontoxigenic strains that are genetically highly similar but differ profoundly in their oncogenic activity (Wu et al., 2009). Finally, the bacteria enriched on tumors also include potentially probiotic species, such as Slackia, Collinsella, Roseburia, and Faecalibacterium (Marchesi et al., 2011; Geng et al., 2013), which convert dietary factors into human-beneficial catabolites, such as butyrate and the antioxidant equol. Thus, tumor promotion and inhibition are two possible outcomes of the species-specific colonization of CRC-tumors by passenger bacteria (Fig. 2).
FIG. 2.
A model for bacterial involvement in a progressing colorectal cancer (CRC) tumor. Several important driver and passenger bacteria associated with the different tumor stages are shown. For details and associated references, see text.
Besides the difficulty in inferring function from taxonomy, the recent metagenomic analyses also leave the cause-or-effect question unanswered. While providing detailed lists of taxa, these metagenomic associations of bacteria-enriched on/off CRC-tumors do not reveal a causal directionality, or even imply causality at all (Mokili et al., 2012). Interestingly, known CRC-promoting bacterial toxin genes were not more highly expressed on tumors (Dutilh et al., 2013), begging the question whether the enriched bacteria caused CRC, or were selected by the tumor-induced changes in the mucosal microenvironment. Addressing this, a driver-passenger model for bacterial involvement in CRC was postulated based on a meta-analysis of published studies (Tjalsma et al., 2012). This model states that the native microbiota of future CRC patients contains driver bacteria. These increase susceptibility to CRC by causing inflammation, increased cell proliferation, and/or production of genotoxins that contribute to the mutations compiling the adenoma–carcinoma sequence. Tumor formation is accompanied by tissue rupture and bleeding, which alters the microenvironment and the selective pressure on local microbiota. Subsequently, CRC-drivers are outcompeted by passenger bacteria that include tumor-foraging opportunistic pathogens, commensal or probiotic bacteria, and other bacteria with a competitive advantage in the tumor niche.
Addressing the question of cause and effect, a recent study in mice investigated the involvement of the gut microbiome in inflammation-associated colorectal cancer (Zackular et al., 2013). This study used a mouse line that could be triggered to develop inflammation-associated intestinal tumors. Under the laboratory experimental conditions, the intestinal microbiotas were homogeneous between individual mice, and showed a clear distinction between tumor-bearing mice and controls. Strikingly, when germ-free mice were exposed to fecal microbiota from tumor-bearing mice or from controls, the former showed significant increases in colonic tumorigenesis. Moreover, antibiotic treatment of the microbiota improved the disease outcome, decreasing both the size and number of observed tumors. Together, these results provide strong evidence that inflammation promoting intestinal microbiota contribute to colonic tumorigenesis. One important factor is probably the replacement of gut bacteria that maintain epithelial health and immune homeostasis, for example, by providing short-chain fatty acids including butyrate, by pro-inflammatory species. This leads to what Zackular and co-workers (2013) describe as “a pathogenic cascade between the gut microbiome and the host,” which decreases microbial diversity and leads to the generation of genotoxic reactive oxygen species and tumor-promoting inflammatory mediators, ultimately enhancing colorectal tumorigenesis.
Complexity of antibiotic–microbiome interactions
Dethlefsen and co-workers (2008) explored the effect of antibiotic administration on gut microbiota. The abundance of nearly 30% of bacterial taxa in the gut was affected by treatment with broad-spectrum ciprofloxacin, decreasing the taxonomic richness, diversity, and evenness of the community. By 4 weeks after treatment, the gut microbiota closely resembled its pretreatment state exhibiting a remarkable resilience, perhaps because the human appendix plays a role in repopulating the gut. However, the extent of recovery of pre-treatment community structure varied among individuals as some taxa failed to be restored within 6 months (Dethlefsen et al., 2008).
The gut microbiome may possess the ability to retain memory of past disturbance. In a study on healthy volunteers, two courses of ciprofloxacin were given over a 10 month period. Again, after each antibiotic course, there was a dramatic shift in the intestinal microbial community toward a stable state similar to, but distinct from, its pre-treatment state with no gastrointestinal symptoms, suggesting the presence of functional redundancy among the gut microbial taxa, a finding in agreement with the published HMP results (Human Microbiome Project Consortium, 2012). Interestingly, the speed and extent of recovery varied between the two ciprofloxacin treatments in the same individual. One potential explanation is the acquisition of antibiotic resistance genes within the gut community after the initial antibiotic perturbation (Dethlefsen and Relman, 2011), especially that antibiotic treatment has been reported to expand phage–bacterial interactions, increasing chances for gene exchange, enriching for multidrug resistance, and preserving functional robustness of bacterial communities in response to antibiotic stress (Modi et al., 2013).
Studies on the long-term impact of different antibiotic classes on the human gut microbiota showed the same resilience of the microbial communities to shift back to a stable yet distinct structure post the antibiotic perturbation period. For example, Bacteroides clonal diversity in the gut was significantly decreased for up to 2 years post clindamycin treatment with the emergence of clindamycin-resistant clones (Jernberg et al., 2007). Similarly, treatment with a combination of metronidazole, clarithromycin, and omeprazole led to a dramatic shift in the microbiota state that persisted for up to 4 years (Jakobsson et al., 2010).
Furthermore, the administration of antibiotics in early childhood has been associated with serious conditions such as Crohn's disease (Virta et al., 2012), asthma (Risnes et al., 2011), and obesity (Ajslev et al., 2011).
The Pharmacomicrobiomics Portal as a resource for drug–microbiome interactions
Reviewed elsewhere (Sousa et al., 2008; Rizkallah et al., 2010; Saad et al., 2012; Haiser and Turnbaugh, 2013), the study of drug–microbiome interactions gained momentum after the HMP was launched. Although over 60 such interactions have been documented, the underlying molecular mechanisms and genetic bases for those interactions remain largely unknown, and only very recently have some of those mechanisms unfolded for a few drugs, for example, acetaminophen (Clayton et al., 2009), digoxin (Haiser et al., 2013), and cyclophosphamide (Viaud et al., 2013)—reviewed in Table 2. With more microbiomes sequenced across the human population, it is expected that more interactions will be discovered and more mechanisms will be identified. The emerging interest in this branch of pharmacogenomics, which we have coined pharmacomicrobiomics (Rizkallah et al., 2010), necessitates the development of databases and bioinformatics tools to document and accelerate the discovery of drug–microbial gene, drug–microbe, and drug–microbiome interactions, and correlate them with drug interactions with the human host genes.
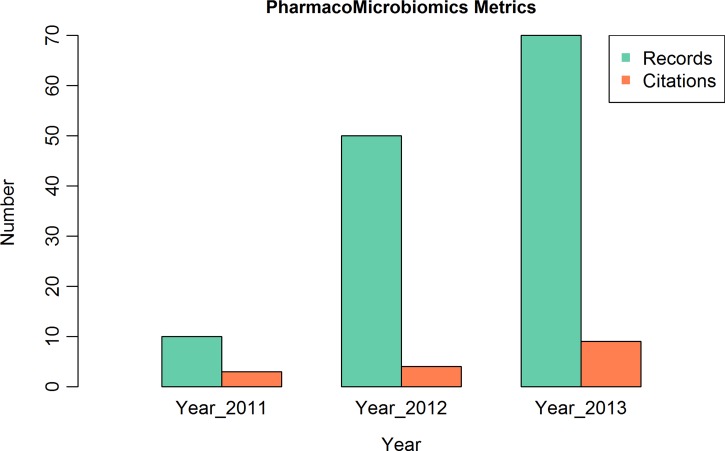
Having evolved from an educational initiative in 2010 aiming to mine the available literature for microbiome interactions and classify them (Aziz et al., 2011) into a publicly available web resource, the PharmacoMicrobiomics portal (http://www.pharmacomicrobiomics.org) has progressively grown since the time of its launch in 2011 (Rizkallah et al., 2012). In terms of data growth, PharmacoMicrobiomics started with ten data records in 2011 and expanded into a relational database that contained almost 70 interactions by the end of 2013. The data growth rate is limited by the number of curators and published drug–microbiome interactions (given that many of the studies in this area are performed at private institutions or companies). Steps towards crowdsourcing (i.e., allowing community submissions and revisions) are currently being undertaken to overcome the limited number of curators; on the other front, more awareness of the topic and more publicly funded research is needed to push forward the study of microbiome implications on pharmacotherapy. In addition to quantitative expansion of the database, efforts are being undertaken to include the information available on biochemical pathways involved in the drug–microbiome interactions from the SEED (Aziz et al., 2012) and KEGG (Kanehisa et al., 2012) databases. Future plans include directly linking current interactions to existing pharmacogenomics databases [e.g., PharmGKB (Owen et al., 2008), CTD (Davis et al., 2008), and PACdb (Gamazon et al., 2010)] and to human microbiome sequence databases (Gevers et al., 2012; Huang et al., 2014).
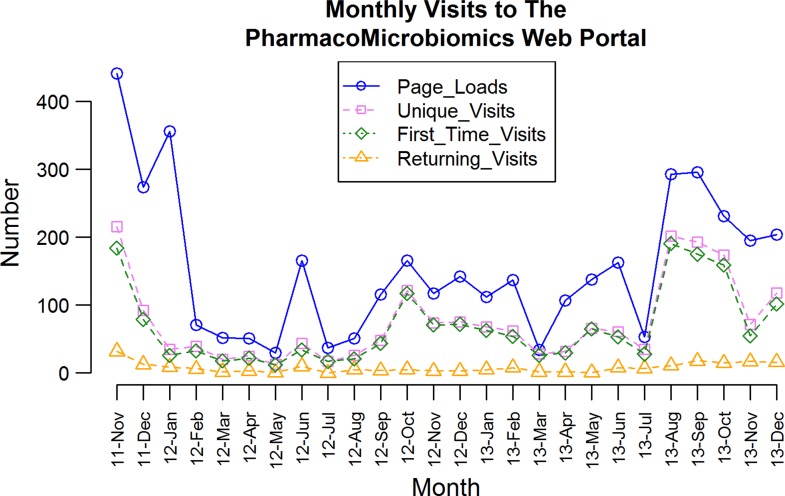
Given the recency of the subject and the short time elapsed since the Pharmacomicrobiomics portal was publicly released, it remains difficult to evaluate the usability, usage, and usefulness of this web resource. In terms of access statistics, not only the number of unique and returning users has increased (Fig. 3), but the number of publications citing the database and/or using the term pharmacomicrobiomics has been growing as well (Fig. 4). Analysis of Internet access logs provided by StatCounter (http://www.statcounter.com) estimates the number of unique visits to be 846 in the first year and 1,111 in the second year, with 96 and 110 returning visits, respectively (Fig. 3). Among the frequent visitors, pharmaceutical companies, and laboratories in the US, UK, and Germany show up in the visitors' Internet Protocol analysis (e.g., Glaxo, Pfizer, Harvard, Cambridge, the US National Institutes of Health, and the European Molecular Biology Laboratory EMBL).
FIG. 3.
Monthly visitor analysis of the PharmacoMicrobiomics web portal between November 2011 and December 2013. Data obtained from StatCounter and publicly available at the URL: http://statcounter.com/p6166637/summary/?account_id=436307.
FIG. 4.
Increase in number of citations and usage of the term “pharmacomicrobiomics” in the past 3 years based on data generated by Google Scholar (URL: http://scholar.google.com, accessed 20 Jan 2014) and the records in PharmacoMicrobiomics database from 2011–2013.
Conclusion
In summary, we are still at the early stages of envisaging the nascent fields that lie at the intersection of systems microbiology, genomics, systems pharmacology, and personalized medicine. The contribution of the human microbiome to phenotypic variability within and between humans is tremendous, and while the microbiome cloud model we suggest here enriches pharmacogenomics and systems pharmacology, it is certainly still too complex to be reduced into discrete microbiome states or a few biomarkers. Eventually, with more and deeper sequencing of humans from all around the globe and with more sequencing from same individuals over long periods of time, biomarkers will be developed that can be diagnostic for major microbiome-driven influences on health and therapeutics. Eventually, whole microbiome determination will be trivial as has become whole genome sequencing nowadays.
The possible beneficial applications of HMP on personalized health are countless and the challenges are not to be underestimated either. However, it is imperative to be prepared for those challenges ahead of time, and to start more intricate and systematic attempts to mine the human microbiome for all potential beneficial and detrimental drug–microbe interactions. In Africa, where financial resources are scarce, a computational biology project such as the development of large interconnected pharmacogenomics and pharmacomicrobiomics knowledge bases represents a great opportunity for advancing research through collaboration and crowdsourcing. Once those knowledge bases are well established, they offer ideal platforms to build smaller country-specific research projects and will be eventually used as tools for education, training, and recruitment of future genome scientists.
Abbreviations Used
- CAGs
co-abundance associations of genera
- CRC
colorectal cancer
- ENCODE
The ENCyclopedia Of DNA Elements
- HGP
Human Genome Project
- HMP
Human Microbiome Project
- HVP
Human Variome Project
- IBD
inflammatory bowel disease
- MetaHIT
Metagenomics of the Human Intestinal Tract
- OTU
operational taxonomic unit
Acknowledgments
BED is supported by the Dutch Virgo Consortium (FES0908, NGI 050-060-452) and CAPES/BRASIL.
Author contributions: JNC and RKA conceived and outlined the article; ME, BED, AB, MRR, and RKA collated literature and drafted the manuscript; AB and BED shared unpublished work about colorectal cancer; MRR analyzed usage data of the pharmacomicrobiomics web portal; ME, BED, JNC, and RKA wrote the manuscript in its final form.
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Aagaard K, Riehle K, Ma J, et al. (2012). A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE 7, e36466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlerberth I, Lindberg E, Aberg N, et al. (2006). Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: An effect of hygienic lifestyle? Pediatr Res 59, 96–101 [DOI] [PubMed] [Google Scholar]
- Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, and Jess T. (2011). Childhood overweight after establishment of the gut microbiota: The role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond) 35, 522–529 [DOI] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK. (2012). Rethinking pharmacogenomics in an ecosystem: Drug–microbiome interactions, pharmacomicrobiomics, and personalized medicine for the human supraorganism. Curr Pharmacogenomics Person Med 10, 258–261 [Google Scholar]
- Aziz RK, Devoid S, Disz T, et al. (2012). SEED Servers: High-performance access to the SEED genomes, annotations, and metabolic models. PLoS ONE 7, e48053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Saad R, and Rizkallah MR. (2011). PharmacoMicrobiomics or how bugs modulate drugs: An educational initiative to explore the effects of human microbiome on drugs. BMC Bioinformatics 12, A10 [Google Scholar]
- Bager P, Wohlfahrt J, and Westergaard T. (2008). Caesarean delivery and risk of atopy and allergic disease: Meta-analyses. Clin Exp Allergy 38, 634–642 [DOI] [PubMed] [Google Scholar]
- Boleij A, Dutilh BE, Kortman GA, et al. (2012). Bacterial responses to a simulated colon tumor microenvironment. Mol Cell Proteomics 11, 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A, and Tjalsma H. (2013). The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect Dis 13, 719–724 [DOI] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). (2006). Community-associated methicillin-resistant Staphylococcus aureus infection among healthy newborns—Chicago and Los Angeles County, 2004. MMWR Morb. Mortal. Wkly Rep. 55, 329–332 [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, et al. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 108, 4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184 [DOI] [PubMed] [Google Scholar]
- Clayton TA, Baker D, Lindon JC, Everett JR, and Nicholson JK. (2009). Pharmacometabonomic identification of a significant host–microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA 106, 14728–14733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, and Nougayrede JP. (2010). Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA 107, 11537–11542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. (2001). In a map for human life, count the microbes, too. Science 291, 2316. [DOI] [PubMed] [Google Scholar]
- Davis AP, Murphy CG, Rosenstein MC, Wiegers TC, and Mattingly CJ. (2008). The Comparative Toxicogenomics Database facilitates identification and understanding of chemical-gene-disease associations: Arsenic as a case study. BMC Med. Genomics 1, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107, 14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, and Bentley D. (2004). The HapMap project and its application to genetic studies of drug response. Pharmacogenomics J 4, 88–90 [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, and Relman DA. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6, e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, and Relman DA. (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108, 4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobkin JF, Saha JR, Butler VP, Jr, Neu HC, and Lindenbaum J. (1982). Inactivation of digoxin by Eubacterium lentum, an anaerobe of the human gut flora. Trans Assoc Am Physicians 95, 22–29 [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107, 11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh BE, Backus L, van Hijum SA, and Tjalsma H. (2013). Screening metatranscriptomes for toxin genes as functional drivers of human colorectal cancer. Best Pract Res Clin Gastroenterol 27, 85–99 [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. (2011). A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9, e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, and Vogelstein B. (1990). A genetic model for colorectal tumorigenesis. Cell 61, 759–767 [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, and Pace NR. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104, 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Duan S, Zhang W, et al. (2010). PACdb: A database for cell-based pharmacogenomics. Pharmacogenet Genomics 20, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Fan H, Tang X, Zhai H, and Zhang Z. (2013). Diversified pattern of the human colorectal cancer microbiome. Gut Pathog 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Knight R, Petrosino JF, et al. (2012). The Human Microbiome Project: A community resource for the healthy human microbiome. PLoS Biol 10, e1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F, and Malagelada JR. (2003). Gut flora in health and disease. Lancet 361, 512–519 [DOI] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, and Turnbaugh PJ. (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, and Turnbaugh PJ. (2013). Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res 69, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Brady A, Mahurkar A, et al. (2014). MetaRef: A pan-genomic database for comparative and community microbial genomics. Nucleic Acids Res. 42, D617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Ye Y, Zhou Y, and Fodor AA. (2012). A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE 7, e34242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke MM, Abrams V, and Moore DR. (2002). Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23, 529–536 [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. (2003). The International HapMap Project. Nature 426, 789–796 [DOI] [PubMed] [Google Scholar]
- Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, and Engstrand L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5, e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LP. (2013). Metabolomics: Integration of a new “omics” with clinical pharmacology. Clin Pharmacol Ther 94, 547–551 [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, and Forman D. (2011). Global cancer statistics. CA Cancer J Clin 61, 69–90 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, and Thun MJ. (2009). Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 [DOI] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, and Jansson JK. (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1, 56–66 [DOI] [PubMed] [Google Scholar]
- Johnson CH, Patterson AD, Idle JR, and Gonzalez FJ. (2012). Xenobiotic metabolomics: Major impact on the metabolome. Annu Rev Pharmacol Toxicol 52, 37–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, and Weinshilboum RM. (2014). Pharmacometabolomics: Implications for clinical pharmacology and systems pharmacology. Clin Pharmacol Ther 95, 154–167 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, and Tanabe M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40, D109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross JM, Darzi AW, and Nicholson JK. (2012). Gut microbiome–host interactions in health and disease. Genome Med 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross JM, von Roon AC, Holmes E, Darzi A, and Nicholson JK. (2008). The human gut microbiome: Implications for future health care. Curr Gastroenterol Rep 10, 396–403 [DOI] [PubMed] [Google Scholar]
- Knasmuller S, Steinkellner H, Hirschl AM, Rabot S, Nobis EC, and Kassie F. (2001). Impact of bacteria in dairy products and of the intestinal microflora on the genotoxic and carcinogenic effects of heterocyclic aromatic amines. Mutat Res 129–138, 480–481 [DOI] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, et al. (2013). A guide to enterotypes across the human body: Meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9, e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, et al. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22, 292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuitunen M, Kukkonen K, Juntunen-Backman K, et al. (2009). Probiotics prevent IgE-associated allergy until age 5 years in Cesarean-delivered children but not in the total cohort. J Allergy Clin Immunol 123, 335–341 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- Lederberg J. (2000). Infectious history. Science 288, 287–293 [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, and Gordon JI. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 [DOI] [PubMed] [Google Scholar]
- Li K, Bihan M, and Methe BA. (2013). Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS ONE 8, e63139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Bik EM, Costello EK, et al. (2013). Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS ONE 8, e53838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. (2012). ENCODE: The human encyclopaedia. Nature 489, 46–48 [DOI] [PubMed] [Google Scholar]
- Marchesi JR, Dutilh BE, Hall N, et al. (2011). Towards the human colorectal cancer microbiome. PLoS ONE 6, e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan VI, Wiederman J, Dobkin JF, and Lindenbaum J. (1989). Geographic differences in digoxin inactivation, a metabolic activity of the human anaerobic gut flora. Gut 30, 971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matto J, Maunuksela L, Kajander K, et al. (2005). Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome—A longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol 43, 213–222 [DOI] [PubMed] [Google Scholar]
- Medini D, Serruto D, Parkhill J, et al. (2008). Microbiology in the post-genomic era. Nature Rev 6, 419–430 [DOI] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, et al. (2011). The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res 21, 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi SR, Lee HH, Spina CS, and Collins JJ. (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili JL, Rohwer F, and Dutilh BE. (2012). Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony L, Feeney M, O'Halloran S, et al. (2001). Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment Pharmacol Ther 15, 1219–1225 [DOI] [PubMed] [Google Scholar]
- Owen RP, Altman RB, and Klein TE. (2008). PharmGKB and the International Warfarin Pharmacogenetics Consortium: The changing role for pharmacogenomic databases and single-drug pharmacogenetics. Hum Mutat 29, 456–460 [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Rosenblatt DS, Warnich L, et al. (2011). Towards an ecology of collective innovation: Human Variome Project (HVP), Rare Disease Consortium for Autosomal Loci (RaDiCAL) and Data-Enabled Life Sciences Alliance (DELSA). Curr Pharmacogenomics Person Med 9, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, and Brown PO. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5, e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. (2012). Genomics. ENCODE project writes eulogy for junk DNA. Science 337, 1159, 1161 [DOI] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, et al. (2009). The NIH Human Microbiome Project. Genome Res 19, 2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflughoeft KJ, and Versalovic J. (2012). Human microbiome in health and disease. Annu Rev Pathol 7, 99–122 [DOI] [PubMed] [Google Scholar]
- Proctor LM. (2011). The Human Microbiome Project in 2011 and beyond. Cell Host Microbe 10, 287–291 [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, et al. (2010). Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Bossdorf O, and Verhoeven KJ. (2010). Understanding natural epigenetic variation. New Phytol 187, 562–564 [DOI] [PubMed] [Google Scholar]
- Ring HZ, Kwok PY, and Cotton RG. (2006). Human Variome Project: An international collaboration to catalogue human genetic variation. Pharmacogenomics 7, 969–972 [DOI] [PubMed] [Google Scholar]
- Risnes KR, Belanger K, Murk W, and Bracken MB. (2011). Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. Am J Epidemiol 173, 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizkallah MR, Saad R, Aziz RK. (2010). The Human Microbiome Project, personalized medicine and the birth of pharmacomicrobiomics. Curr Pharmacogenomics Person Med 8, 182–193 [Google Scholar]
- Rizkallah MR, Saad R, and Aziz RK. (2012). The PharmacoMicrobiomics Portal: A database for drug-microbiome interactions. Curr Pharmacogenomics Person Med 10, 195–203 [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, and Han YW. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Saunders B, Wilkinson K, et al. (2004). Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology 126, 451–459 [DOI] [PubMed] [Google Scholar]
- Saad R, Rizkallah M, and Aziz R. (2012). Gut Pharmacomicrobiomics: The tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. (1977). Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31, 107–133 [DOI] [PubMed] [Google Scholar]
- Schwabe RF, and Wang TC. (2012). Cancer. Bacteria deliver a genotoxic hit. Science 338, 52–53 [DOI] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, et al. (1998). Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun 66, 5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleator RD. (2010). The human superorganism—Of microbes and men. Med Hypotheses 74, 214–215 [DOI] [PubMed] [Google Scholar]
- Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, and Basit AW. (2008). The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm 363, 1–25 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Campbell BJ, and Rhodes JM. (2006). Bacteria in the pathogenesis of inflammatory bowel disease. Curr Opin Infect Dis 19, 475–484 [DOI] [PubMed] [Google Scholar]
- Thiele I, Swainston N, Fleming RM, et al. (2013). A community-driven global reconstruction of human metabolism. Nat Biotechnol 31, 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H, Boleij A, Marchesi JR, and Dutilh BE. (2012). A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat Rev Microbiol 10, 575–582 [DOI] [PubMed] [Google Scholar]
- Tuohy KM, Gougoulias C, Shen Q, Walton G, Fava F, and Ramnani P. (2009). Studying the human gut microbiota in the trans-omics era—Focus on metagenomics and metabonomics. Curr Pharm Des 15, 1415–1427 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, and Gordon JI. (2009). The core gut microbiome, energy balance and obesity. J Physiol 587, 4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, and Gordon JI. (2007). The Human Microbiome Project. Nature 449, 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, and Gordon JI. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 [DOI] [PubMed] [Google Scholar]
- Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, and Knight R. (2012). The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol 129, 1204–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duynhoven J, Vaughan EE, Jacobs DM, et al. (2011). Metabolic fate of polyphenols in the human superorganism. Proc Natl Acad Sci USA 108, 4531–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. (2001). The sequence of the human genome. Science 291, 1304–1351 [DOI] [PubMed] [Google Scholar]
- Viaud S, Saccheri F, Mignot G, et al. (2013). The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta L, Auvinen A, Helenius H, Huovinen P, and Kolho KL. (2012). Association of repeated exposure to antibiotics with the development of pediatric Crohn's disease—S nationwide, register-based Finnish case-control study. Am J Epidemiol 175, 775–784 [DOI] [PubMed] [Google Scholar]
- Wilson ID, and Nicholson JK. (2009). The role of gut microbiota in drug response. Curr Pharm Des 15, 1519–1523 [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15, 1016–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie KM, Weinstock GM, and Storch GA. (2012). Emerging view of the human virome. Transl Res 160, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zackular JP, Baxter NT, Iverson KD, et al. (2013). The gut microbiome modulates colon tumorigenesis. MBio 4, e00692–00613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, et al. (2009). Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 106, 2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, and Dolan ME. (2008). Beyond the HapMap genotypic data: Prospects of deep resequencing projects. Curr Bioinform 3, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Rajilic-Stojanovic M, and de Vos WM. (2008). High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615 [DOI] [PubMed] [Google Scholar]