Abstract
Mannose binding lectin (MBL) is a pathogen pattern recognition protein involved in antimicrobial activities. Variation in MBL2 gene has been extensively implicated in differential outcomes of infectious diseases in studies conducted outside Africa, but virtually very little is known on the role of this candidate gene in the African continent. We investigated human genetic variations in MBL2 in a Zimbabwean pediatric population and their putative associations with HIV infection in perinatally exposed children. One hundred and four children aged 7 to 9 years comprising 68 perinatally exposed to HIV (32 who were born infected and 36 who were uninfected) and 36 unexposed controls were recruited. DNA samples were genotyped for MBL2 polymorphisms using PCR-RFLP and sequencing. HIV infected children had markedly variable and significantly lower mean height (p=0.03) and weight (p=0.005) when compared to the uninfected children. Using all samples, frequencies for MBL2 genetic variants for the Zimbabwean population were calculated. Twelve single nucleotide polymorphisms were observed and minor alleles occurred with the following frequencies: −550C>G (G: 0.02), −435G>A (A: 0.08), −428A>C (C: 0.39), −394A>G (A: 0.39), −328AGAGAA ins/del (AGAGAA ins: 0.44), −245G>A (A: 0.05), −221C>G (C: 0.12), −111A>T (T: 0.10), −70C>T (C: 0.46), +4C>T (C: 0.45), novel −595G>A (A: 0.02), and 170G>A (0.24). We found that the MBL2 +4T variant displayed a trend for association with reduced risk of HIV transmission from mother-to-child but the remaining vast majority of the genetic markers did not show a significant association. We conclude (1) the MBL2 gene is highly polymorphic in the Zimbabwean population, and (2) MBL2 genetic variation does not appear to play a major role in influencing the risk of mother-to-child HIV transmission in our study sample. These observations contest the hitherto significant role of this candidate gene for HIV transmission from mother-to-child in non-African populations and thus, further speak to the limits of extrapolating genomic association studies directly to the African populations from studies conducted elsewhere. It is hoped that more OMICS research in a diverse set of African countries can shed further light on the putative role (or the lack thereof ) of this candidate gene in HIV transmission in the continent, a major global health burden in Africa.
Introduction
Mannose binding lectin (MBL), a protein encoded by the MBL2 gene, is a member of the collectins family which has affinity for carbohydrates such as D-mannose and N-acetyl-D-glucosamine on the surface of pathogens (Holmskov et al., 1994; Sastry and Ezekowitz, 1993). MBL binds to polysaccharide chains on pathogen surfaces, leading to destruction of the micro-organism by activation of the lectin pathway of the complement system, opsonization, or direct microbial killing (Holmskov et al., 1994). Thus, MBL recognizes and binds to mannose residues on HIV gp120 (Ezekowitz et al., 1989). By binding gp120, MBL blocks the attachment of HIV to host cells bearing gp120 receptors, thus inhibiting infection of the host cell (Saifuddin et al., 2000). MBL polypeptide chains assemble into trimers that in turn oligomerize to form structures of up to 18 polypeptide chains (Garred et al., 2006; Kishore et al., 1997). Multimerization enhances the overall affinity of MBL towards carbohydrates on pathogen surfaces, thus failure to assemble into these structures results in reduced immunity (Clark et al., 2000).
Genetic variation in the promoter and exon 1 of the MBL2 gene affects expression and oligomerization of MBL, leading to differential susceptibility to HIV infection and disease progression (Mangano et al., 2008; Singh et al., 2008). Distribution of MBL2 allelic variants differs among populations, for example, of the exon 1 SNPs implicated in HIV infection and disease progression, African populations predominantly carry the C allele, whilst Caucasian populations mainly present with the B allele (Lipscombe et al., 1992; Madsen et al., 1998). However, the distribution of MBL2 variants and their possible role in HIV infection and neurocognitive function is not well described among African populations. Identifying genetic variants that influence HIV transmission and disease progression may help predict disease course, guide therapy, and provide potential therapeutic targets (Singh and Spector, 2009). This study therefore describes the variations in the MBL2 gene and its possible role in risk of HIV infection in children born to HIV-infected mothers. We further discuss the extent to which MBL2 genetic variation might play a role (or not) in HIV transmission from mother-to-child in the African continent.
Methods
Study participants
Participants were recruited from the Better Health for the African Mother and Child (BHAMC) cohort, a longitudinal study of mother–child pairs followed up since 2002 at three Harare peri-urban clinics in Epworth, St Mary′s Chitungwiza, and Seke North. One hundred and four (N=104) unrelated children aged 7 to 9 years were of bantu African origin (Kurewa et al., 2010). Participants included 32 children perinatally-infected with HIV (EI) and 72 healthy HIV-uninfected children comprising of 36 exposed to HIV in utero but not infected (EU), and 36 unexposed and uninfected (UEUI) were recruited as controls. All HIV-infected mothers of children included in the study were administered a single dose of 200 mg nevirapine at delivery of the children to prevent mother-to-child transmission of HIV. A 5 mL blood sample was collected from each child for CD4+ T-cell count and genotyping purposes. The demographic characteristics of the recruited children were captured from their medical records. Written informed consent was obtained from each child's parent or legal guardian before the samples were collected. The study received ethics approval from the Medical Research Council of Zimbabwe and the University of Cape Town, Faculty of Health Sciences Research ethics committee.
Genotyping for MBL2 genetic variants
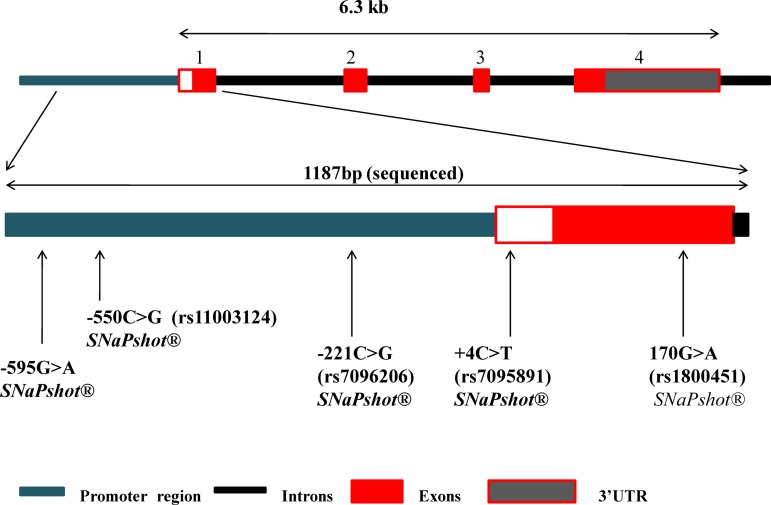
Genomic DNA was extracted from blood samples using the Nucleospin® Blood L kit (Macherey-Nagel, Germany) according to the manufacturer's instructions. Primers for amplification of genomic DNA regions were designed using Primer 3 online package http://frodo.wi.mit.edu/, NCBI Primer Blast and OligoAnalyzer SciTool (Integrated DNA Technologies®; http://eu.idtdna.com) bioinformatics tools. To investigate genetic variation in the promoter and exon 1 of MBL2 gene, a 1187 bp fragment was amplified by polymerase chain reaction (PCR) (Fig. 1), and subsequently sequenced. The PCR contained 10 picomoles of each of the sense (5′-AGGCTGCTGAGGTTTCTTAGG-3′) and antisense (5′-ATGCCAGAGAATGAGAGCTGA-3′) primers, 200 μM dNTPs (Bioline, UK), 1X GoTaq Flexi Green buffer (Promega, USA), 1.5 mM MgCl2 (Fermentas, Canada), 1U Taq polymerase (Fermentas), and 50 ng genomic DNA in a total volume of 25 μL. The cycling conditions were as follows; initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C, primer annealing at 64°C, and primer extension at 72°C for 30 sec at each step. The reaction was completed by an extension step at 72°C for 5 min. The PCR products were further purified using alkaline phosphatase (FastAP™) (Fermentas) and exonuclease I, and then sequenced including capillary electrophoresis on ABI® PRISM 3130 Genetic Analyser (Life Technologies, Grand Island, NY). Alignment of sequences with a reference sequence was performed using the Lasergene® 10 software (DNASTAR®, Madison, WI).
FIG. 1.
Schematic representation of the MBL2 gene. The top panel shows the full gene structure whilst the bottom panel zooms in on the region that was investigated in this study. The arrows indicate the positions of the SNPs that were genotyped and the genotyping methods employed.
Statistical analysis
Genotype and allele frequencies in the HIV exposed infected (EI), exposed uninfected (EU), and unexposed (UE) children cases and controls were calculated using Stata 11.2 (StataCorp LP, College Station, TX) and/or SHEsis online version (Yong and Lin, 2005). All the samples were combined to calculate the frequencies of the MBL2 variants in the Zimbabwean population. Conformation to Hardy Weinberg Equilibrium (HWE) was determined. Chi-square and Fisher's exact tests were also used to evaluate the effects of MBL2 genotypes and alleles on HIV and disease status. p<0.05 was considered statistically significant. Pairwise Linkage Disequilibrium (LD) analyses between MBL2 SNPs was carried out using SHEsis (Li et al., 2009). Lewontin's D′ value and r2 were used to quantify the level of LD. Haplotypes were inferred using the expected maximisation algorithm in SHEsis.
Results
Demographic characteristics
HIV infected (EI) children presented with significantly lower mean height (cm) of 117.11±6.2 SD (p=0.03) and lower mean weight (kg) of 20.31±1.99 SD (p=0.005) when compared to uninfected children (exposed uninfected+unexposed and uninfected) with 120.37±6 SD and 22.35±3.01 SD, respectively, as shown in Table 1. This indicated possible slowed growth due to HIV/AIDS-related challenges.
Table 1.
Demographic and Clinical Characteristics of Study Participants
| Characteristics | HIV-infected (EI) n=32 | HIV-uninfected (EU+UEUI) n=72 | P value |
|---|---|---|---|
| Mean age in years±SD (range) | 8.22±0.57 (7.25–9.08) | 8.43±0.55 (7.5–9.08) | 0.28 |
| Mean height in cm±SD (range) | 117.11±6.22 (109–131) | 120.37±6.00 (108–134) | 0.03 |
| Mean weight in kg±SD (range) | 20.31±1.99 (17–24) | 22.35±3.01 (15–31) | 0.005 |
| Mean head circum in cm±SD (range) | 51.14±1.57 (49–54) | 51.37±1.2 (48–54) | 0.48 |
| Sex | |||
| Female | 19 (0.59) | 36 (0.50) | 0.37 |
| Male | 13 (0.41) | 36 (0.50) | |
EI-HIV, exposed and infected; EU, exposed but uninfected; N/A, non-applicable; UEUI, unexposed and uninfected.
MBL2 genetic polymorphism distribution and HIV infection
All 104 participants were used to calculate frequencies for MBL2 genetic variants to impute the likely frequencies in the Zimbabwean population in general. 12 single nucleotide polymorphisms (SNPs) were observed and the minor alleles occurred with the following frequencies; −595G>A (A: 0.02), −550C>G (G: 0.02), −435G>A (A: 0.08), −428A>C (C: 0.39), −394A>G (A: 0.39), −328AGAGAA ins/del (AGAGAA ins: 0.44), −245G>A (A: 0.05), −221C>G (C: 0.12), −111A>T (T: 0.10), −70C>T (C: 0.46), +4C>T (C: 0.45), novel −595G>A, and 170G>A (0.24). Two previously reported SNPs, 154C>T (rs5030737) and 161G>A (rs1800451), were monomorphic in the Zimbabwean population. However, none of the SNPs investigated was associated with differential susceptibility to HIV infection, although the MBL2 +4T variant appeared to show a trend towards association with reduced risk. Table 2 shows the genotype and minor allele frequencies of the SNPs detected. All SNPs conformed to the Hardy-Weinberg Equilibrium.
Table 2.
SNPs Detected by DNA Sequencing of the MBL2 Gene Promoter and Exon 1 Regions
| SNP | n | F | wt/wt* | wt/mt** | mt/mt* | MA (Freq) |
|---|---|---|---|---|---|---|
| −595 G>A (Novel) | 104a | 99 (0.93) | 5 (0.07) | 0 | A (0.02) | |
| −550C>G (rs11003125) | 28 | 27 (0.96) | 1 (0.04) | 0 | G (0.02) | |
| −435G>A (rs7100749) | 26 | 2 | 22 (0.85) | 4 (0.15) | 0 | A (0.08) |
| −428A>C (rs11003124) | 28 | 6 (0.21) | 10 (0.36) | 12 (0.43) | C (0.39) | |
| −394A>G (rs7084554) | 23 | 5 | 6 (0.26) | 6 (0.26) | 11 (0.48) | A (0.39) |
| −328AGAGAA (rs45560739) | 23 | 5 | 8 (0.33) | 5 (0.21) | 11 (0.46) | AGAGAA (0.44) |
| −245G>A (rs35236971) | 21 | 7 | 19 (0.90) | 2 (0.10) | 0 | A (0.05) |
| −221C>G (rs7096206) | 104a | 0 | 26 (0.25) | 78 (0.75) | C (0.12) | |
| −111A>T (rs67990116) | 21 | 7 | 19 (0.90) | 2 (0.10) | 0 | T (0.10) |
| −70C>T (rs11003123) | 23 | 5 | 7 (0.31) | 7 (0.30) | 9 (0.39) | C (0.46) |
| +4C>T (rs7095891) | 104a | 24 (0.23) | 45 (0.43) | 35 (0.34) | C (0.45) | |
| 154C>T (rs5030737) | 26 | 2 | 26 (1.00) | 0 | 0 | T (0) |
| 161G>A (rs1800450) | 26 | 2 | 26 (1.00) | 0 | 0 | A (0) |
| 170G>A (rs1800451) | 104a | 60 (0.58) | 38 (0.36) | 6 (0.06) | A (0.24) |
F, failed samples; MA (Freq), minor allele frequency; N, total number of samples genotyped; N.t, nucleotide; wt, wild type allele; mt, mutant allele.
SNPs were further genotyped in the samples remaining after sequencing; *wt=refers to the starting allele as indicated in the nucleotide base substitution column, and **mt=to the second allele. This designation has nothing to do with functional significance.
We report a novel G>A variation at position −595 upstream of the exon 1 start site (MBL2 −595G>A) which occurred with a frequency of 2% for the −595A allele (Fig. 1). No homozygous −595A/A genotype was observed. Functional analysis of −595G>A variants done using a predictive bioinformatics software called TFSearch (Heinemeyer et al., 1998) showed that the MBL2 −595G>A position does not carry any transcription factor binding site regardless of the presence of G or A allele.
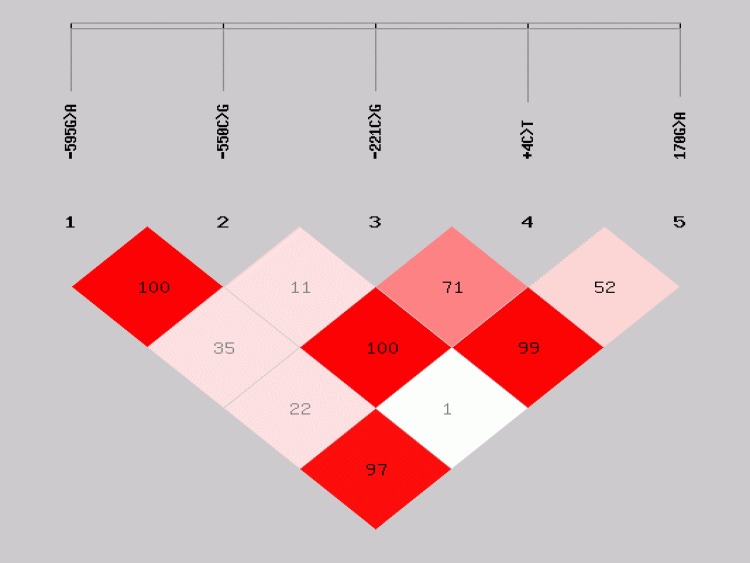
In addition to the MBL2 −595G>A, distribution of genotypes of four SNPs previously implicated in HIV infection and disease progression in the literature (−550C>G, 221C>G, +4C>T, and 170G>A) were also compared between HIV EI and EU children (Table 3). None of the MBL2 genotypes showed a significant difference in distribution between HIV EI and EU children. Pairwise LD calculation for the five MBL2 SNPs; −595G>A, −550C>G, −221C>G,+4C>T, and 170G>A in the HIV uninfected (EU+UEUI) children showed even distribution of strong and weak pairwise LD (Fig. 2). Strong LD limited the number of MBL2 haplotypes inferred from the SNPs −550C>G, −221C>G, +4C>T, and 170G>A. The mutant alleles of MBL2 exon 1 nonsynonymous SNPs 170G>A (G57E), 154C>T (R52C), and 161G>A (G54D) are alternatively termed “C,” “D,” and “B,” respectively, whilst the promoter and 5′-UTR variants −221C/G and +4C>T are referred to as; −221X/Y and +4P/Q, respectively (Lipscombe et al., 1992; Madsen et al., 1998; Sumiya et al., 1991). This alternative nomenclature is commonly used to describe MBL2 haplotypes (Fig. 3). Seven haplotypes with respect to the −550C>G, −221C>G, +4C>T, and 170G>A SNPs were observed, three being reported for the first time (HYQA, HYPC, and HXPA) (Fig. 3). None of the haplotypes' distribution was significantly different between the HIV groups.
Table 3.
Frequency and Distribution MBL2 Genotypes and Their Association with HIV Status
| Genotype | HIV EI n=32 | HIV EU n=36 | UEUI n=36 | P value EI v EU |
|---|---|---|---|---|
| MBL2 −595G>A (novel) | ||||
| −595G/G | 30 (0.94) | 34 (0.94) | 35 (0.97) | |
| −595G/A | 2 (0.07) | 2 (0.06) | 1 (0.03) | 0.90 |
| MBL2 −550C>G (rs11003124) | ||||
| −550C/C | 27 (0.84) | 26 (0.72) | 34 (0.94) | |
| 11−550C/G | 5 (0.16) | 10 (0.28) | 2 (0.06) | 0.23 |
| MBL2 −221C>G (rs7096206) | ||||
| −221G/G | 23 (0.73) | 27 (0.75) | 28 (0.78) | |
| −221C/G | 9 (0.27) | 9 (0.25) | 8 (0.22) | 0.77 |
| MBL2 170G>A (rs1800451) | ||||
| 170G/G | 19 (0.59) | 21 (0.58) | 20 (0.56) | |
| 170G/A | 9 (0.28) | 13 (0.36) | 16 (0.44) | 0.48 |
| 170A/A | 4 (0.13) | 2 (0.06) | 0 | 0.31 |
| MBL2 +4C>T (rs7095891) | ||||
| +4T/T | 11 (0.34) | 11 (0.31) | 13 (0.36) | |
| +4C/T | 12 (0.38) | 21 (0.58) | 12 (0.33) | 0.09 |
| +4C/C | 9 (0.28) | 4 (0.11) | 11 (0.36) | 0.08 |
EI-HIV, exposed infected; EU-HIV, exposed uninfected; N/A, non-applicable; UEUI- HIV, unexposed; OR (95%CI), odds ratio (95% confidence interval).
FIG. 2.
Linkage disequilibrium plot of MBL2 SNPs. The plot shows pairwise LD of MBL2 variants in the promoter and exon1. The numbers in the boxes are percentage D′ values indicating strength of LD.
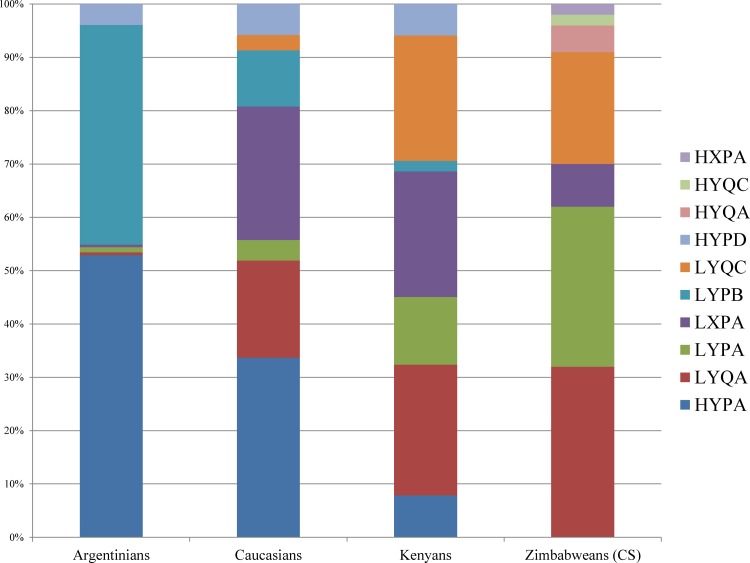
FIG. 3.
Frequency of MBL2 haplotypes in the Zimbabwean population and how they compare to other populations (Madsen et al., 1998).
Discussion
Deficiency in MBL protein, which is mainly due to reduced gene expression and poor oligomerization, has been linked with increased susceptibility to HIV (Boniotto et al., 2000; Garred et al., 2006). We describe MBL2 genetic variation among Zimbabweans and discuss their possible role in differential susceptibility to HIV infection in utero.
The frequency of MBL2 +4C (P) variant (45%) observed in our study population is comparable to what has been reported among other African populations (38%–50%) but higher than the frequency observed in Caucasian (17%) and Asian populations (12%) (Mangano et al., 2008; Ou et al., 2011; Thye et al., 2011). The difference in the frequency of the +4C allelic variant in different populations may be a pointer to possible molecular causes of the observed geographical differences in HIV prevalence. However, this is somewhat contradicted by the lower frequency of the +4C variant among Asians (12%) compared to Caucasians (17%), yet HIV prevalence is much higher in the former group (Ou et al., 2011; Singh et al., 2008). This could be due to the fact that HIV susceptibility is influenced by a multitude of factors of which MBL2 variation could be a constituent player. Moreover, presence of the +4C allelic variant has been associated with mild downregulation of MBL2 gene expression and is often overshadowed in the presence of the MBL2 −221C allele (Israëls et al., 2012). MBL2 +4C and −221C are in strong LD, thus, presence of +4C allele is unlikely to cause a detrimental reduction in antimicrobial activity of MBL protein. MBL2 −221C allele has been reported to significantly reduce serum MBL. However, frequency of the −221C allele (12%) in Zimbabweans fell in the same range with what has been reported among other African populations (12%–17%), Caucasians (17%), and Asians (15%) (Ou et al., 2011; Singh et al., 2008; Thye et al., 2011), casting doubt on its importance in the interindividual differences in susceptibility to HIV infection and distribution.
The frequency of MBL2 170A allele (C) observed in the Zimbabwean population (24%) is comparable what has been reported in other African populations (20%–30%) (Lipscombe et al., 1992; Thye et al., 2011), but higher than in both Asian (Ou et al., 2011) and Caucasian populations (Lipscombe et al., 1992). Neither D nor B alleles were observed in the Zimbabweans, yet the D allele occurs in 15% of Caucasian and 25% Asian populations whilst the B is rare in most populations (<5%) (Chen et al., 2009; Lipscombe et al., 1992). The opposing frequency distribution between MBL2 C and B variants between Caucasians and Africans is unlikely to explain the differences in HIV prevalence because the effects of the B and C alleles on MBL oligomerization is thought to be almost similar (Israëls et al., 2012). The resulting amino acid changes disrupt the α-helical structure of the MBL polypeptide chain, thus interfering with the formation of functional oligomers (Eisen and Minchinton, 2003). This reduces the antiviral activity of MBL and may increase susceptibility to a wide range of diseases, nonetheless, on a similar scale for both B and C alleles. We therefore speculate that MBL2 genetic variation may not solely explain differences in HIV distribution in world populations. This is supported by the findings on BST-2 (tetherin) differential genetic variation in Southern African populations, which is thought to affect or mirror the HIV-1 infection prevalence (Skelton et al., personal communication).
Our observations are contradicted by earlier reports where MBL deficiency variants have been identified as strong risk factors for HIV infection when compared to MBL sufficiency variants (Boniotto et al., 2003; Kuhn et al., 2006). Others have reported accelerated disease progression in individuals carrying MBL deficiency variants (Mangano et al., 2008; Singh et al., 2008). However, there are several explanations for the failure to observe any association between MBL2 variation and HIV infection. Distribution of MBL2 genetic variants is likely to have been influenced by other micro-organisms more ancient than HIV. For example, the MBL2 170A (C) allele, which results in MBL deficiency, may have been selected for in African populations because it protects the host against MBL-mediated Mycobacterium africanum (M. africanum) infection (Søborg et al., 2007; Thye et al., 2011). MBL-mediated opsonization is used by some intracellular organisms such as M. africanum to enter host cells. Moreover, the lectin (MBL) pathway of the complement system can also be activated by two other pathways (Endo et al., 2006); hence reduction in MBL-induced complement activation may not be detrimental to life (Mangano et al., 2008), thus may have a negligible effect on HIV outcomes.
The distribution of MBL2 haplotypes, like that of the individual SNPs differs among world populations. Frequencies of MBL2 haplotypes in the Zimbabwean population studied were comparable to those reported among Kenyans but differed from Caucasians and Argentinians (Fig. 3) (Madsen et al., 1998). The HYA containing haplotypes, which have been linked with high secretion of MBL (Jensen et al., 2005), were present at very low frequencies in both Zimbabwean and Kenyan populations whilst they are present at high frequencies in Caucasians and Argentinians (Madsen et al., 1998). The differences in haplotypes may account for the higher prevalence of infectious diseases such as HIV, hepatitis, and TB among Africans when compared to other populations.
Our study makes a contribution towards understanding host genetic variation with regard to HIV infection among Africans, particularly, Zimbabweans. However, these observations must be interpreted cautiously as the study was carried out on a limited sample size whose participants may also have suffered “survivor bias”, since only those children who were alive at the time the cross-sectional study was undertaken (7–9 years after birth). The study population was limited to individuals alive after close to a decade of follow-up; information on the genetic make-up of those children who died during follow-up period was not available.
Conclusions
We demonstrate high variability in the promoter and exon 1 of the MBL2 gene among Zimbabwean children, with differences in SNP and haplotype distribution when compared to other populations. Discovery of a novel −595G>A polymorphism suggests that there may be more undetected gene variants in African populations as most have not been studied at genome level. The novel SNPs may hold the key to our understanding of contribution of most of the so-called susceptibility genes. Our observations collectively contest the hitherto significant role of this candidate gene for HIV transmission mother-to-child in non-African populations and thus further speak to the limits of extrapolating genomic association studies directly to the African populations from studies conducted elsewhere. It is hoped that more OMICS research in a diverse set of African countries can shed further light on the putative role (or the lack thereof ) of this candidate gene in HIV transmission in the continent, a major global health burden in Africa.
Acknowledgments
We would like to thank Letten Foundation, Norway; University of Cape Town; National Research Foundation (NRF) of South Africa, Medical Research Council of South Africa and, the University of Cape Town for funding both the student and research project costs. We are also indebted to the National Institute of Health Research and University of Zimbabwe for facilitating some of the research work. None of this would have been possible without the voluntary participation of members of the BHMAC cohort for which we are thankful. We also want to thank the late Dr. EN Kurewa for facilitating this study.
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Boniotto M, Crovella S, Pirulli D, et al. (2000). Polymorphisms in the MBL2 promoter correlated with risk of HIV-1 vertical transmission and AIDS progression. Genes Immun 1, 346–348 [DOI] [PubMed] [Google Scholar]
- Boniotto M, Braida L, Pirulli D, Arraes L, Amoroso A, and Crovella S. (2003). MBL2 polymorphisms are involved in HIV-1 infection in Brazilian perinatally infected children. AIDS 17, 779–780 [DOI] [PubMed] [Google Scholar]
- Chen J, Xu Z, Ou X, Wang M, Yang X, and Li Q. (2009). Mannose-binding lectin polymorphisms and recurrent respiratory tract infection in Chinese children. Eur J Pediatr 168, 1305–1313 [DOI] [PubMed] [Google Scholar]
- Clark HW, Reid K, and Sim RB. (2000). Collectins and innate immunity in the lung. Microbes Infect 2, 273–278 [DOI] [PubMed] [Google Scholar]
- Eisen DP, and Minchinton RM. (2003). Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin Infect Dis 37, 1496–1505 [DOI] [PubMed] [Google Scholar]
- Endo Y, Takahashi M, and Fujita T. (2006). Lectin complement system and pattern recognition. Immunobiology 211, 283–293 [DOI] [PubMed] [Google Scholar]
- Ezekowitz RA, Kuhlman M, Groopman JE, and Byrn RA. (1989). A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J Exp Med 169, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garred P, Larsen F, Seyfarth J, Fujita R, and Madsen HO. (2006). Mannose-binding lectin and its genetic variants. Genes Immun 7, 85–94 [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, et al. (1998). Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmskov U, Malhotra R, Sim RB, and Jensenius JC. (1994). Collectins: Collagenous C-type lectins of the innate immune defense system. Immunol Today 15, 67–74 [DOI] [PubMed] [Google Scholar]
- Israëls Jl, Scherpbier HJ, Frakking FN, van de Wetering MD, Kremer LC, and Kuijpers TW. (2012). Mannose-binding lectin and the risk of HIV transmission and disease progression in children: A systematic review. Pediatr Infect Dis J 12, 1272–1278 [DOI] [PubMed] [Google Scholar]
- Jensen PH, Weilguny D, Matthiesen F, McGuire KA, Shi L, and Højrup P. (2005). Characterization of the oligomer structure of recombinant human mannan-binding lectin. J Biol Chem 280, 11043–11051 [DOI] [PubMed] [Google Scholar]
- Kishore U, Eggleton P, and Reid K. (1997). Modular organization of carbohydrate recognition domains in animal lectins. Matrix Biol 15, 583–592 [DOI] [PubMed] [Google Scholar]
- Kuhn L, Coutsoudis A, Trabattoni D, et al. (2006). Synergy between mannose-binding lectin gene polymorphisms and supplementation with vitamin A influences susceptibility to HIV infection in infants born to HIV-positive mothers. Am J Clin Nutr 84, 610–615 [DOI] [PubMed] [Google Scholar]
- Kurewa NE, Mapingure MP, Munjoma MW, Chirenje MZ, Rusakaniko S, and Stray-Pedersen B. (2010). The burden and risk factors of sexually transmitted infections and reproductive tract infections among pregnant women in Zimbabwe. BMC Infect Dis 10, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Z, He Z, et al. (2009). A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis (http://analysis.bio-x.cn). Cell Res 19, 519–523 [DOI] [PubMed] [Google Scholar]
- Lipscombe RJ, Sumiya M, Hill AV, et al. (1992). High frequencies in African and non-African populations of independent mutations in the mannose binding protein gene. Hum Mol Genet 1, 709–715 [DOI] [PubMed] [Google Scholar]
- Madsen HO, Satz ML, Hogh B, Svejgaard A, and Garred P. (1998). Different molecular events result in low protein levels of mannan-binding lectin in populations from southeast Africa and South America. J Immunol 161, 3169–3175 [PubMed] [Google Scholar]
- Mangano A, Rocco C, Marino SM, et al. (2008). Detrimental effects of mannose-binding lectin (MBL2) promoter genotype XA/XA on HIV-1 vertical transmission and AIDS progression. J Infect Dis 198, 694–700 [DOI] [PubMed] [Google Scholar]
- Ou XT, Wu JQ, Zhu LP, et al. (2011). Genotypes coding for mannose-binding lectin deficiency correlated with cryptococcal meningitis in HIV-uninfected Chinese patients. J Infect Dis 203, 1686–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søborg C, Andersen AB, Range N, et al. (2007). Influence of candidate susceptibility genes on tuberculosis in a high endemic region. Mol Immunol 44, 2213–2220 [DOI] [PubMed] [Google Scholar]
- Saifuddin M, Hart ML, Gewurz H, Zhang Y, and Spear GT. (2000). Interaction of mannose-binding lectin with primary isolates of human immunodeficiency virus type 1. J Gen Virol 81, 949–955 [DOI] [PubMed] [Google Scholar]
- Sastry K, and Alan Ezekowitz R. (1993). Collectins: Pattern recognition molecules involved in first line host defense. Curr Opin Immunol 5, 59–66 [DOI] [PubMed] [Google Scholar]
- Singh KK, Lieser A, Ruan PK, Fenton T, and Spector SA. (2008). An age-dependent association of mannose-binding lectin-2 genetic variants on HIV-1-related disease in children. J Allergy Clin Immunol 122, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, and Spector SA. (2009). Host genetic determinants of human immunodeficiency virus infection and disease progression in children. Pediatr Res 65, 55R–63R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiya M, Tabona P, Arai T, et al. (1991). Molecular basis of opsonic defect in immunodeficient children. Lancet 337, 1569–1570 [DOI] [PubMed] [Google Scholar]
- Thye T, Niemann S, Walter K, et al. (2011). Variant G57E of mannose binding lectin associated with protection against tuberculosis caused by Mycobacterium africanum but not by M. tuberculosis. PloS One 6, e20908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong YONG, and Lin HE. (2005). SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15, 97–98 [DOI] [PubMed] [Google Scholar]