Abstract
The formation of amyloid aggregates upon protein misfolding is related to several devastating degenerative diseases. The propensities of different protein sequences to aggregate into amyloids, how they are enhanced by pathogenic mutations, the presence of aggregation hot spots stabilizing pathological interactions, the establishing of cross-amyloid interactions between co-aggregating proteins, all rely at the molecular level on the stability of the amyloid cross-beta structure. Our redesigned server, PASTA 2.0, provides a versatile platform where all of these different features can be easily predicted on a genomic scale given input sequences. The server provides other pieces of information, such as intrinsic disorder and secondary structure predictions, that complement the aggregation data. The PASTA 2.0 energy function evaluates the stability of putative cross-beta pairings between different sequence stretches. It was re-derived on a larger dataset of globular protein domains. The resulting algorithm was benchmarked on comprehensive peptide and protein test sets, leading to improved, state-of-the-art results with more amyloid forming regions correctly detected at high specificity. The PASTA 2.0 server can be accessed at http://protein.bio.unipd.it/pasta2/.
INTRODUCTION
A broad range of human diseases arise from the failure of a specific peptide or protein to adopt, or remain in, its native functional conformational state. These pathological conditions are generally referred to as protein misfolding diseases (1). In many cases, misfolding of the wild type protein is associated with a late disease onset, whereas pathogenic familial variants, often single mutants, cause an early onset and more severe symptoms. The largest group of misfolding disease is associated with the conversion from a soluble functional form to highly organized fibrillar aggregates, generally described as amyloid fibrils. One hallmark of the amyloid structure is a specific supramolecular architecture called cross-beta structure, held together by hydrogen bonds extending repeatedly along the fibril axis. In recent years, it has been increasingly recognized that transient prefibrillar oligomeric species are in most cases responsible for cell toxicity (2). Toxic oligomers, however, often exhibit a cross-beta structure as well. Cross-amyloid interactions at a molecular level may also play a critical role in protein misfolding diseases, as evidenced by the co-aggregation of different disease-related proteins into heteromeric oligomer structures (3). Similarly, the inability of two homologous proteins to oligomerize together was hypothesized to be the molecular basis of the species barrier phenomenon, in the context of both mammals and yeast prions (4).
Amyloid and toxic oligomer formation is not restricted to those polypeptide chains that have recognized links to diseases. Several other proteins have been found to form both fibrillar and toxic oligomeric aggregates (5). This finding has led to the idea that the ability to form the cross-beta structure is an inherent property of polypeptide chains (6). The algorithm PASTA exploited this observation by assuming that the same universal mechanism is responsible for beta-sheet formation both in globular proteins and in cross-beta aggregates (7). PASTA predicts which interacting portions of a given protein are stabilizing the cross-beta structure by using an energy function. This is based on the propensities of two residues to be found within a beta-sheet facing one another on neighboring strands, as determined from a dataset of globular proteins of known native structure. Further proof of the effectiveness of this energy-based approach was shown in Cossio et al. and Sarti et al. (8,9), where a generalization of the PASTA energy function was used in the context of protein structure prediction to successfully discriminate native conformations among sets of alternative decoys. For PASTA, the predicted aggregation propensities rely on the assumption that the soluble form is natively unstructured. Predictions therefore need to be carefully gauged in the case of natively folded globular proteins. PASTA can discriminate the orientation between β-strands, either parallel or antiparallel. This distinction is rare among other methods, with (10) an early exception. Moreover, the algorithm can be quite easily extended to the case of two different co-aggregating sequences. The original PASTA server has been running since March 2007 and has received over 21 700 hits from over 60 different countries. In 2013 the web server has been used over 3500 times by over 520 different IP addresses. PASTA has become a milestone for benchmarking newer aggregation methods and has been sold to pharmaceutical companies. The new version PASTA 2.0 we present here extends the previous predictor in several ways. First of all, the underlying statistical potentials have been re-derived on a larger dataset of globular protein domains to improve accuracy, allowing a finer estimation of the expected true and false positive rate. Benchmarking was performed on a comprehensive dataset of 424 peptides with experimental information about their aggregating behavior (11–14) and on a second set of 33 proteins with experimental information about the location of aggregation hot spots (15). PASTA 2.0 improves performance over the previous version and compares very well with other state-of-the-art methods. For peptide discrimination, at a false positive rate of <5% it has a sensitivity of 40%, making it more specific than all other tested methods. When detecting the location of aggregating regions, at a false positive rate <10% it can recover regions with 30% sensitivity. Adjusting the energy threshold can increase sensitivity at the expense of specificity. The web server has also undergone a re-design, enhancing the output information with new graphs and stats (e.g. intrinsic disorder, secondary structure) and allowing the simultaneous execution of entire genomes in a single job. The energy cut-off for detection of cross-beta stretches and the resulting sensitivity and specificity can be directly manipulated by the user. Finally, it is now possible to calculate the difference in aggregation propensity after point-mutations and between different protein pairs, allowing the analysis of pathogenic mutations and of cross-amyloid interactions between protein heterodimers, as suggested e.g. by (16) and (17), respectively. To assess the effect of point-mutations on the aggregation profile, a free energy profile is now present in output, together with the probability profile already present in the old server.
MATERIALS AND METHODS
PASTA 2.0 predicts amyloid fibril regions from protein sequences using a pairwise energy potential at its core. In this version of the server we included methods for secondary structure and intrinsic disorder, which provide additional reinforcement to the fibril assignment. Briefly, a new machine learning algorithm was constructed to detect secondary structure while our previously developed disorder predictor ESpritz (18) was also included.
Energy pairing potential
The previous version of PASTA derived an energy function from the hydrogen bonding statistics on β-strands (7). Briefly, given a pair of residues i and j, whether they formed a parallel or antiparallel β-bridge within the DSSP algorithm (19), modified with a stricter threshold for hydrogen bond detection, was used to define potentials for pair (i,j). Thus, the aggregation potential of (i,j) can be related to its energy. The energy parameters were re-calibrated for PASTA 2.0 on a larger dataset derived from TESE (20) (see Supplementary Material for details).
Segment energy
Given two sequences, a segment can be allocated an energy by sliding two sequential regions of length L along the corresponding sequences. All possible pairings can be obtained by varying the region length L and relative orientations (antiparallel or parallel). The corresponding pairing aggregation scores are obtained by summing contributions for each of the L pairwise interactions using the energy pairing potential. Pairing aggregation scores are then combined together to compute aggregation probability profiles and aggregation free energy profiles, as a function of residue position along the protein chain. We also compute pairing probabilities and pairing free energies, as a function of the sequence positions of the paired residues. A more detailed mathematical formulation is given in Trovato et al. (7,21), and shortly recapitulated in the Supplementary Material. The sensitivity and specificity was calculated as a function of this newly tuned segment energy and implemented as a server option (see ‘Input’ and ‘Cut-off energy/top energies’ in Server description and Performance sections).
Secondary structure and intrinsic disorder
Sequence-based features may complement prediction of aggregation toward a better understanding of the sequence–structure relationship. Both intrinsic disorder and secondary structure predictors were trained using Bi-directional Recursive Neural Networks (BRNNs) (22). The only information supplied to the BRNNs was the amino acid sequence which proved accurate (see (18) for disorder and Supplementary Table S1 for secondary structure) while having an added speed advantage. Our speed/accuracy trade-off was in contrast to slightly more accurate predictors that used computationally challenging multiple sequence alignment calculations. While other sequence-based features may be envisaged, we chose to use secondary structure and intrinsic disorder as server output because they provide an easy way to interpret structural information that is orthogonal to the aggregation prediction. In fact, the presence of native structure plays a protective role against aggregation (23). Within this context, an intermediate partially disordered or flexible state was previously hypothesized in an aggregation model (24). Contradictory to this, highly disordered proteins were shown to be much lower in aggregation propensity than globular ones (25). Therefore, investigation is still needed to understand these conflicting views and offering aggregation, secondary structure and disorder in one web server should help.
Benchmark sets
Assessing the performance of aggregation is tricky, mainly due to the lack of experimental data. Despite this, over the last decade, small amounts of experimental data have been released in the literature. This allows performance to be assessed in two scenarios: (i) aggregation assignment to small peptides and (ii) aggregation assignment to a sequential stretch in a larger protein. Thus the server performance was measured on two sets.
Peptide detection (Pep424): this set collects all the available peptides annotated with experimental information. It contained 179 peptides from (11), 17 peptides from (12), 158 hexa-peptides from (13) and 70 peptides from human prion protein, human lysozyme, β2-microglobulin used in (14). In total, there were 424 peptides with 149 aggregating and 275 not. Thus, we measured the binary classification of each peptide as a whole.
Region detection (Reg33): this set annotates specific protein regions that are thought to aggregate; we took advantage of a dataset already constructed in (15). It contains 33 proteins with 1260 aggregating and 6472 regular residues annotated from the literature. For simplicity, the performance was measured on each residue in the 33 proteins.
PERFORMANCE
A comparison with other groups was only possible if their server allowed as input multiple sequences, or an easy to install stand-alone executable was available. This was particularly true for Pep424, as manually retrieving predictions became cumbersome. First, performance was assessed on small peptides classified as aggregating or not. Then, the ability of predictors to recover residues that are known aggregating hot spots was measured. Performance was assessed, in a leave-one-out validation, using sensitivity, specificity, Q2, Matthews correlation coefficient (MCC) and receiver operator characteristic curves (ROCs). For a more precise mathematical description of the performance measures, see Supplementary Material.
Peptide classification
Figure 1 shows the ROC curve that plots the true positive rate (sensitivity) versus the false positive rate (1-specificity) for PASTA 2.0 and other methods (11,14,26,27). PASTA 2.0 was well above random achieving a total area under the ROC (AUC) of 85.73 (random AUC is 0.5). In contrast, the next best curve FoldAmyloid (14) had AUC 2.42 worse. However, it is mostly the case that low false positive rate (high specificity) is desirable. PASTA 2.0 has a sensitivity of 42.95 and a high specificity of 94.85 when we select a strict energy threshold. Putting this into perspective, a hypothetical situation with 100 peptides and 90 known experimentally not to aggregate, PASTA 2.0 would return 9 candidate peptides. It would correctly predict 4 out of 10 positive and would incorrectly determine 5 out of the 90 negative peptides. With no a priori knowledge and using the web server to guide experiments, a laboratory test of these 9 peptides would reveal 4/9 were aggregating, a favorable scenario for most experimentalists. On the contrary, evaluating candidate peptides of low specificity algorithms would be rather time consuming for the experimentalist. Given this, Figure 1 also shows the AUC in the 0.0–0.1 false positive rate zone (i.e. >90% specificity). PASTA 2.0 clearly outperformed all other tested software (AUC 3.91) in this high specificity zone with the TANGO (11) method second to it (AUC 3.55). The two FoldAmyloid variants contact (26) and triple hybrid FoldAmyloid (14) and AGGRESCAN (27) were substantially lower with AUCs 1.18, 2.31 and 2.65 respectively.
Figure 1.
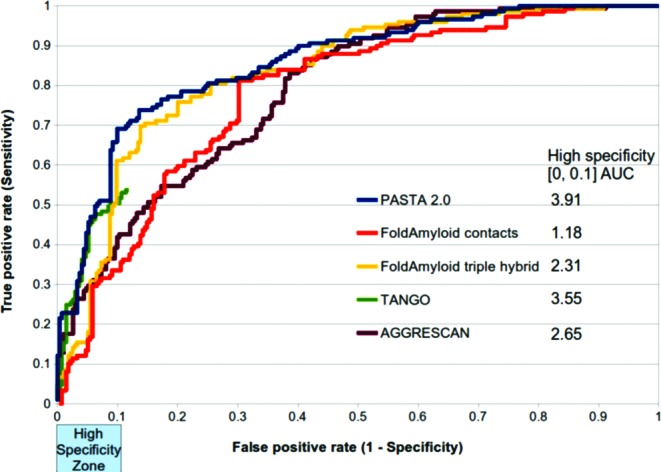
Receiver Operating Characteristic (ROC) curve for PASTA 2.0 and four other methods. Marked on the x-axis is an important area of the curve, the low false positive rate or high specificity zone. ROC calculated on Pep424 set. PASTA 2.0 is improved over all other comparisons in general. In the high specificity zone the area under the ROC (AUC) is measured and given in the legend. Tango stops at 12% FPR because all energies are 0 after this point and therefore no variation can be performed.
Region detection
A recent predictor, AMLYPRED2 (15), collected literature and annotated 33 proteins with aggregating hot spots. Given that AMLYPRED2 is a meta-predictor that was shown to improve over its 12 well-established constituent parts (11,26–36), we decided to compare against it and five other related methods (11,14,26,37,38). Table 1 shows the per residue sensitivity, specificity, Q2 and MCC for PASTA 2.0, in a leave-one-out test, when selecting thresholds defined for 90% and 85% specificity. The higher specificity option produced 90.00 specificity with 30.24% of the positive residues recovered (sensitivity). This is a conservative prediction, thus aggregation hot spots can be inferred with high confidence when selecting this option. To achieve the same specificity (∼85.0) as the other methods we needed to relax the selection of the top pairings and the energy cut-off. At this less stringent threshold, sensitivity increases (40.87) and specificity decreases (84.95) as expected and the PASTA MCC becomes superior to the other methods. The selection of the top pairings and the energy cut-off is described in the next section.
Table 1.
Performance on detecting aggregating residues from the Reg33 set
| Method | Sensitivity | Specificity | Q2 | MCC |
|---|---|---|---|---|
| Aggrescan | 35.37 | 79.26 | 57.32 | 0.13 |
| AMYLPRED2 | 39.27 | 84.48 | 61.88 | 0.22 |
| FoldAmyloid (contacts) | 20.71 | 86.97 | 76.17 | 0.08 |
| FoldAmyloid (triple hybrid) | 19.21 | 86.22 | 75.30 | 0.06 |
| Tango | 13.67 | 95.57 | 54.62 | 0.14 |
| MetAmyl (high specificity) | 39.05 | 83.14 | 77.24 | 0.19 |
| MetAmyl (global accuracy) | 52.46 | 70.73 | 68.29 | 0.17 |
| FishAmyloid | 13.73 | 93.68 | 82.98 | 0.10 |
| PASTA 2.0 (90% specificity) | 30.24 | 90.00 | 80.23 | 0.22 |
| PASTA 2.0 (85% specificity) | 40.87 | 84.95 | 77.77 | 0.24 |
Default thresholds used for FoldAmyloid, FishAmyloid and MetAmyl. Results for AMYLPRED2, Aggrescan and Tango are taken directly from (15).
Cut-off energy/top energies
The server predicts aggregation in energy units where 1 PASTA Energy Unit (PEU) is equivalent to 2 KBT at room temperature, that is 1.192 Kcal/mol (see Supplementary Material). The selection of an energy cut-off allows the user to alter the sensitivity and specificity of the server. In addition, the top X best energy pairings or combinations of energy cut-off and the top best can be chosen (see the Server description section). We envisage three prediction types: peptide discrimination, highly confident region detection and less confident region detection. The performances in Figure 1 and Table 1 allowed us to define optimal top X and energy cut-offs for the three cases. For peptide discrimination, only the best pairing is considered (top = 1) and an energy cut-off of −5 was found to produce 95% specificity (see Supplementary Figure S1 for sensitivity/specificity). For highly confident region detection, top = 22 and energy < −2.8 produced 90% specificity and 30% sensitivity. Finally, less confident regions were found with top = 44 and energy < −1.4 producing 85% specificity and 40% sensitivity. Supplementary Figure S2 shows an example of the three scenarios. These parameters are only recommendations and are available in a dropdown menu in the input page, however users are free to alter them as they see fit.
SERVER DESCRIPTION
The PASTA2 website is free and open to all users and there is no login requirement. The interface can process entire genomes and the sensitivity and specificity of the prediction can be suitably modified. In addition, version 2.0 of the server has increased functionality and other sequence-based predictions. Supplementary Table S2 shows what we believe to be the improvements over the PASTA 1.0 server (39). In the following, a description of the server, its improved functionality and other predictions are given in more detail.
Input interface
Single or multiple sequences in FASTA format are the only input required and can be either pasted or uploaded as a file. User email address and a query title are optional but recommended for user records on larger jobs. To facilitate navigation, help and example pages are available at the top of the interface. There are three modes of usage: self-aggregation (default), protein–protein aggregation and mutate one protein. Self-aggregation computes the aggregation by sliding each sequence over itself. The protein–protein option determines aggregation either on an all-against-all or one-against-all basis thus allowing aggregation to be determined between protein heterodimers. Finally, the mutate option allows the examination of many point mutations and their effects on the aggregation ability of one protein sequence. Large-scale processing is possible but it is recommended to turn on the ‘large-scale’ option since this will limit the protein–protein options and turn-off graph generation as both are computationally tough (recommended limits: without large-scale option <500 sequences and with it entire genome processing is possible). Importantly, the over/under prediction capabilities of the algorithm can be altered by a sliding bar that selects the energy cut-off and its measured sensitivity and specificity. Related to the energy selection the top best energy pairings can also be altered in a text-box. There are three recommended defaults for the top text-box and the energy cut-off (see ‘Cut-off energy/top energies’ in the Performance section).
Output layout
The PASTA 2.0 output is presented in two main pages. The first page, displays statistics, links to individual pages and a downloadable archive for all user supplied proteins. For self-aggregating sequences, the statistics include global information such as percentage α-helix, β-strand, coil, intrinsic disorder and most importantly the best aggregation pairing energy. Each statistic can be sorted by user preference, but by default all entries are sorted by lowest energy pair, thus ranking the most aggregation prone sequences. If the protein–protein option was selected, links to every possible pairing are provided at the bottom of the page (see Supplementary Figure S2 for a layout of the first output page).
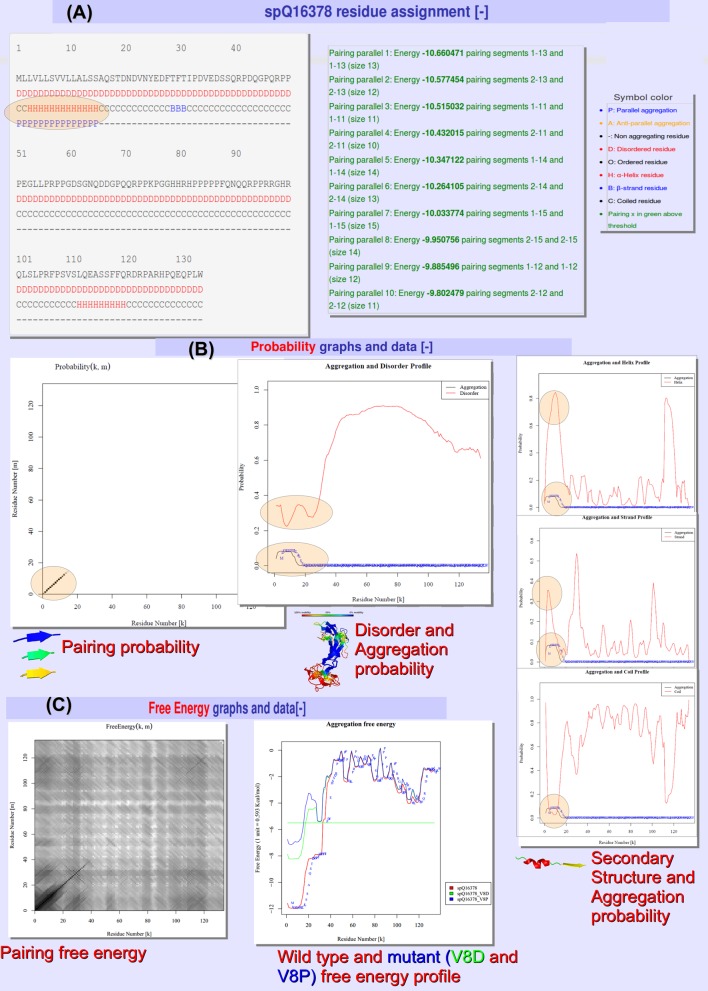
The second output pages display all the annotations at the residue level. In addition, graphical output of the aggregation free energies, aggregation probabilities, secondary structure and disorder probabilities are plotted and often combined. All of this information taken together can be a useful source of structural annotation. For example, using the web server we found nasopharyngeal carcinoma-associated proline-rich protein 4 (UniProt accession: Q16378) to be interesting because it was the most aggregation prone completely disordered protein in the human proteome. Figure 2 shows the output for this protein. The output is split into three main sections: the first residue assignment (Figure 2A) annotates each residue as disordered, helix, strand, coil and as parallel/antiparallel aggregating. Residues are only defined as aggregating if the energy is above the cut-off and inside the top pairings selected in the input page. In Figure 2A, all residues are predicted as disordered and the sole parallel aggregating region was predicted to be in a helix conformation with the rest of the protein mainly predicted in a coil arrangement. The second section shows the probability profiles and pairings (Figure 2B), in our example they reveal that both helix and strand probabilities are high, suggesting perhaps a conformational switch could be taking place in the aggregating region in conjunction with intermediate disordered states. In short, a global hypothesis can be made about this protein and moreover this interesting case was only found by scanning the human genome with the large-scale processing capabilities. Figure 2C shows the third output section, the free energy profiles and pairings. In Figure 2C, we mutated our example protein, in the aggregating region, at position 8 using the wildcard character (*) producing 19 mutants. The largest mutational effects were found to be proline and aspartic acid (V8P and V8D). All predictions and pairing matrices shown in Figure 2 are provided for download; an extensive description of each is available as part of the online help page.
Figure 2.
Sample output for nasopharyngeal carcinoma-associated proline-rich protein 4. (A) Residue assignment of disorder, α-helix, β-strand, coil and a parallel aggregation region marked with an oval, along with the energy of the aggregation pairings and legend. (B) Pairing and linear probability profiles as a function of the residue position. The probabilities show an interesting aggregation-prone region with large helix probability but also high strand probability. In addition, the protein is predicted to be completely disordered but tends to be less so in the aggregating region. The diagonal line in the pairing probability predicts a parallel in-register arrangement for the aggregation-prone stretch. (C) The free energy pairing matrix and the free energy profiles. In mutation mode the free energy profile can be used to visualize the changes in aggregation potential for the mutants. In this case the mutants are V8D and V8P, both decrease aggregation potential (higher energy) in green and blue, respectively.
Implementation and server run-time
The PASTA algorithm was developed in ANSI C, an executable is freely available for academic users on the server main page. The server is built on a Linux Debian 44 CPU cluster with each node having 8 GB RAM. Apache 2.2.16, Tomcat 7.1. web servers and JavaServer Pages (JSP) and Javascript scripting languages were used to build the server. Parallel execution is achieved by splitting multiple sequences into eight jobs, thus eight sequences are executed in the same time as one sequence without parallelization. The parallelization, efficient C code and other designed characteristics allow the processing of large amounts of data. To estimate the execution time on a real problem, we downloaded the human proteome from the National Center for Biotechnology Information FTP site, removed identical sequences, and found that PASTA 2.0 returned results in 28 h for the 31 641 proteins.
CONCLUSION
We have described PASTA 2.0, a novel web server for the prediction of protein aggregation from sequence. It allows the batch prediction of many sequences simultaneously, providing a rich structural overview. Each sequence is annotated not only with aggregation-prone regions but also α-helix, β-strand, coil and intrinsic disordered regions. All predictions concern structural characteristics of the sequence and we therefore believe their combination to be intuitively appealing. In addition, enhanced functionality such as protein dimer aggregation and mutational analysis is possible. Future work will concentrate on improving the functional description of the aggregating regions as well as integration with the MobiDB (40) database of disorder annotations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
I.W. and S.T. are grateful to members of the BioComputing UP lab for insightful discussions. F.S. and A.T. are grateful to Fabrizio Chiti and Amos Maritan for insightful discussions.
FUNDING
Padova University through Progetto di Ateneo [CPDA121890 to A.T.]; Italian Ministry for University and Research through FIRB Futuro in Ricerca [RBFR08ZSXY to S.T.]. Source of open access funding: FIRB Futuro in Ricerca [RBFR08ZSXY to S.T.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Fandrich M. Oligomeric intermediates in amyloid formation: structure determination and mechanisms of toxicity. J. Mol. Biol. 2012;421:427–440. doi: 10.1016/j.jmb.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg D., Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuite M.F., Serio T.R. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat. Rev. Mol. Cell Biol. 2010;11:823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 6.Hoang T.X., Marsella L., Trovato A., Seno F., Banavar J.R., Maritan A. Common attributes of native-state structures of proteins, disordered proteins, and amyloid. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6883–6888. doi: 10.1073/pnas.0601824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trovato A., Chiti F., Maritan A., Seno F. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PLoS Comput. Biol. 2006;2:e170. doi: 10.1371/journal.pcbi.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cossio P., Granata D., Laio A., Seno F., Trovato A. A simple and efficient statistical potential for scoring ensembles of protein structures. Sci. Rep. 2012;2:351. [Google Scholar]
- 9.Sarti E., Zamuner S., Cossio P., Laio A., Seno F., Trovato A. BACHSCORE. A tool for evaluating efficiently and reliably the quality of large sets of protein structures. Comput. Phys. Commun. 2013;184:2860–2865. [Google Scholar]
- 10.Tartaglia G.G., Cavalli A., Pellarin R., Caflisch A. Prediction of aggregation rate and aggregation-prone segments in polypeptide sequences. Protein Sci. 2005;14:2723–2734. doi: 10.1110/ps.051471205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotech. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 12.Roland B.P., Kodali R., Mishra R., Wetzel R. A serendipitous survey of prediction algorithms for amyloidogenicity. Biopolymers. 2013;100:780–789. doi: 10.1002/bip.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson M.J., Sievers S.A., Karanicolas J., Ivanova M.I., Baker D., Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garbuzynskiy S.O., Lobanov M.Y., Galzitskaya O.V. FoldAmyloid: a method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 2010;26:326–332. doi: 10.1093/bioinformatics/btp691. [DOI] [PubMed] [Google Scholar]
- 15.Tsolis A.C., Papandreou N.C., Iconomidou V.A., Hamodrakas S.J. A consensus method for the prediction of ‘aggregation-prone’ peptides in globular proteins. PloS One. 2013;8:e54175. doi: 10.1371/journal.pone.0054175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luheshi L.M., Tartaglia G.G., Brorsson A.C., Pawar A.P., Watson I.E., Chiti F., Vendruscolo M., Lomas D.A., Dobson C.M., Crowther D.C. Systematic in vivo analysis of the intrinsic determinants of amyloid Beta pathogenicity. PLoS Biol. 2007;5:e290. doi: 10.1371/journal.pbio.0050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraldo R. Amyloid assemblies: protein legos at a crossroads in bottom-up synthetic biology. Chembiochem. 2010;11:2347–2357. doi: 10.1002/cbic.201000412. [DOI] [PubMed] [Google Scholar]
- 18.Walsh I., Martin A.J., Di Domenico T., Tosatto S.C. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28:503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- 19.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 20.Sirocco F., Tosatto S.C. TESE: generating specific protein structure test set ensembles. Bioinformatics. 2008;24:2632–2633. doi: 10.1093/bioinformatics/btn488. [DOI] [PubMed] [Google Scholar]
- 21.Trovato A., Maritan A., Seno F. Aggregation of natively folded proteins: a theoretical approach. J. Phys.: Condens. Matter. 2007;19:285221. [Google Scholar]
- 22.Baldi P., Brunak S., Frasconi P., Soda G., Pollastri G. Exploiting the past and the future in protein secondary structure prediction. Bioinformatics. 1999;15:937–946. doi: 10.1093/bioinformatics/15.11.937. [DOI] [PubMed] [Google Scholar]
- 23.Chiti F., Dobson C.M. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 24.Uversky V.N., Fink A.L. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 26.Galzitskaya O.V., Garbuzynskiy S.O., Lobanov M.Y. Prediction of amyloidogenic and disordered regions in protein chains. PLoS Comput. Biol. 2006;2:e177. doi: 10.1371/journal.pcbi.0020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conchillo-Sole O., de Groot N.S., Aviles F.X., Vendrell J., Daura X., Ventura S. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Donnell C.W., Waldispuhl J., Lis M., Halfmann R., Devadas S., Lindquist S., Berger B. A method for probing the mutational landscape of amyloid structure. Bioinformatics. 2011;27:i34–42. doi: 10.1093/bioinformatics/btr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez de la Paz M., Serrano L. Sequence determinants of amyloid fibril formation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zibaee S., Makin O.S., Goedert M., Serpell L.C. A simple algorithm locates beta-strands in the amyloid fibril core of alpha-synuclein, Abeta, and tau using the amino acid sequence alone. Protein Sci. 2007;16:906–918. doi: 10.1110/ps.062624507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Chen H., Lai L. Identification of amyloid fibril-forming segments based on structure and residue-based statistical potential. Bioinformatics. 2007;23:2218–2225. doi: 10.1093/bioinformatics/btm325. [DOI] [PubMed] [Google Scholar]
- 32.Kim C., Choi J., Lee S.J., Welsh W.J., Yoon S. NetCSSP: web application for predicting chameleon sequences and amyloid fibril formation. Nucleic Acids Res. 2009;37:W469–W473. doi: 10.1093/nar/gkp351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian J., Wu N., Guo J., Fan Y. Prediction of amyloid fibril-forming segments based on a support vector machine. BMC Bioinformatics. 2009;10(Suppl. 1):S45. doi: 10.1186/1471-2105-10-S1-S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamodrakas S.J., Liappa C., Iconomidou V.A. Consensus prediction of amyloidogenic determinants in amyloid fibril-forming proteins. Int. J. Biol. Macromol. 2007;41:295–300. doi: 10.1016/j.ijbiomac.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Maurer-Stroh S., Debulpaep M., Kuemmerer N., Lopez de la Paz M., Martins I.C., Reumers J., Morris K.L., Copland A., Serpell L., Serrano L., et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 36.Frousios K.K., Iconomidou V.A., Karletidi C.-M., Hamodrakas S.J. Amyloidogenic determinants are usually not buried. BMC Struct. Biol. 2009;9:44. doi: 10.1186/1472-6807-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasior P., Kotulska M. FISH Amyloid—a new method for finding amyloidogenic segments in proteins based on site specific co-occurence of aminoacids. BMC Bioinformatics. 2014;15:54. doi: 10.1186/1471-2105-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emily M., Talvas A., Delamarche C. MetAmyl: a METa-predictor for AMYLoid proteins. PloS One. 2013;8:e79722. doi: 10.1371/journal.pone.0079722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trovato A., Seno F., Tosatto S.C. The PASTA server for protein aggregation prediction. Protein Eng. Des. Sel.: PEDS. 2007;20:521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- 40.Di Domenico T., Walsh I., Martin A.J., Tosatto S.C. MobiDB: a comprehensive database of intrinsic protein disorder annotations. Bioinformatics. 2012;28:2080–2081. doi: 10.1093/bioinformatics/bts327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.