Abstract
There are two types of processive cellulases, exocellulases and processive endoglucanases. There are also two classes of exocellulases, ones that attack the reducing ends of cellulose chains and ones that attack the nonreducing ends. There are a number of ways of assaying processivity but none of them are ideal. It appears that exocellulases, all of which have their active sites in a tunnel, couple movement along a cellulose chain with cleavage of cellobiose from the end of the cellulose molecule. There are two sets of structures that suggest how an exocellulase might move along a cellulose chain. For family 48 exocellulases there are two different ways that a chain can be bound in the active site while for family 6 exocellulases there are several different ligand-bound structures. Site-directed mutagenesis of Thermobifida fusca exocellulases Cel48A and Cel6B and the processive endoglucanase Cel9A have identified some mutations that increase processivity and some that decrease processivity. In addition a mutation in Cel6B was identified that appears to allow the mutant enzyme to move along a cellulose chain in the absence of cleavage.
Keywords: Cellulose, Cellulase processivity, Exocellulases, Processive endoglucanases, Thermobifida fusca
1. Introduction
The first processive cellulases to be identified were exocellulases (also called cellobiohydrolases), which attack the end of a cellulase chain and cleave off cellobiose residues sequentially from one end of a cellulose chain until they dissociate or stall (1). There are two classes of exocellulases; one class which attacks the reducing ends of cellulose chains is found in either family GH-7 or GH-48 (2), while the other class of exocellulases attacks the nonreducing ends of cellulose chains and it is found in family GH-6 (2). All known exocellulases have their active sites in a tunnel, which is consistent with their processive activity (3–5). More recently, a new type of cellulase, processive endoglucanase, was discovered that contains a GH-9 catalytic domain with a family 3c carbohydrate binding module (CBM) bound at its C-terminus (6). The family 3c CBM was shown to be essential for processivity (7). Another type of processive endoglucanase was found recently that contains a family GH-5 catalytic domain (8). It appears that their processivity results from unusual subsite binding, as has been seen for some processive chitinases (9).
2. Processivity Assays
There is no perfect assay for determining cellulase processivity, which is defined as the average number of cleavages that an enzyme carries out on a cellulose chain before it dissociates from the chain. One assay that has been used, measures both the amount of soluble reducing sugars that an enzyme produces from filter paper or other insoluble substrate during an appropriate incubation, often overnight, and the amount of insoluble reducing ends it produces in the filter paper during the same incubation. This can be done by removing the filter paper at the end of the incubation and measuring the reducing sugars in the solution and in the rinsed filter paper disc using the DNS assay (10). Exocellulases produce more than 93% of the total reducing sugar in the solution while endocellulases produce 30–40% of the total reducing ends in the filter paper (insoluble). Thermobifida fusca Cel48A and Trichoderma reesei Cel7A showed the highest processivity in this assay with only 4% insoluble reducing ends while T. reesei Cel6A and T. fusca Cel6B gave 7% insoluble reducing ends. This may reflect the fact that family GH-6 cellulases contain only eight glucose-binding subsites while families GH-48 and GH-7 have more than ten such subsites (10). At this time it is not clear how the small number of insoluble ends are produced by an exocellulase, but it is unlikely that they result from endocellulolytic cleavages by the exocellulase. This is due to the fact that the buried surface area present in the loops that form the active site tunnel in exocellulases is large enough to prevent the tunnel from opening, especially in family GH-7 and GH-48 enzymes. The soluble-to-insoluble reducing sugar ratio assay is useful for distinguishing nonprocessive cellulases (most endoglucanases) from processive cellulases. A better assay for measuring the processivity of exocellulases is to determine the ratio of cellobiose to cellotriose that is produced by the exocellulase using HPLC (11). This assay is based on the assumption that during the first cleavage by an exocellulase there is an equal chance that it will produce either cellobiose or cellotriose depending on the stereochemistry of the chain end, as the glucose-binding subsites in an enzyme alternate in their binding specificity and cellulose chains are believed to have an equal number of each type of end. After the first cleavage, cellobiose will be the only product, as all the enzyme-bound ends have the same stereochemistry after the first cleavage. Thus, more processive enzymes will produce a higher ratio of cellobiose to cellotriose. This assay requires that the hydrolysis of cellotriose by the enzyme is slow and that cellobiose is not hydrolyzed by the enzyme, which is true for most processive cellulases Cel6B slowly hydrolizes cellotriose so the formula: G3−G1/G2+G1. This assay gives a value of 12–14 for the processivity of T. fusca WT Cel48A or Cel48cd on both bacterial cellulose and amorphous cellulose, (Kostylev M, Wilson DB, unpublished).
Using electron microscopy, it was shown that T. reesei Cel7A acts processively from the reducing end towards the nonreducing end of crystals of Valonia cellulose (12). A direct assay for processivity is single-molecule studies using either fluorescently labeled cellulases or atomic force microscopy (AFM). An AFM study of T. reesei Cel7A and Cel6A, which was published in 2009, provided clear evidence for processivity in T. reesei Cel7A (CBHI) but the movement of Cel6A (CBH II) on a cellulose chain was very limited (13). The Cel7A molecules moved a distance covering from 35 to 50 CB units on the cellulose showing a processivity of near 50. Cel7Acd did not bind unless its concentration was tenfold higher than the native enzyme, but the cd moved at the same rate as the intact enzyme showing that the CBM was important for binding the catalytic domain to cellulose but was not needed to allow cleavage or movement along the cellulose molecule. An inactive Cel7A mutant did not show any movement, supporting the idea that for the wild-type enzyme, cleavage is essential for movement along a cellulose chain. There are two studies of the cleavage of oligosaccharides by T. reesei Cel6A, which show that it acts processively, since this enzyme hydrolyzes cellohexose to cellobiose without releasing cellotetraose (14, 15).
Another assay of processivity is to label the reducing ends of cellulose by reacting them with a fluorescent group such as anthranilic acid or diaminopyridine. Then the release of labeled cellobiose under conditions that allow only one cycle of cellulase binding can be compared with the release of unlabelled cellobiose, which will be produced by all subsequent cleavages (16, 17). A similar assay is to reduce the reducing end to an alcohol, react the reduced cellulose with an exocellulase and measure the cellobiose produced along with the number of insoluble reducing groups that are produced when the alcohol group is cleaved off the end of a cellulose chain (16). By use of this assay, it was shown that the processivity of Cel7A from two different fungi was about three times higher on bacterial cellulose than on amorphous cellulose. The processivity values that have been determined seem fairly low relative to the length of the cellulose chains suggesting that release of the enzyme occurs easily, which is surprising given the large number of subsites in the active site tunnel of Cel7A. This may indicate that these assays have some undiscovered flaws.
A kinetic model of the initial burst phase of T. reesei Cel7A acting prosessively on cellulose was proposed and tested by a calorimetric assay using amorphous cellulose. It was proposed that the initial binding and processive cleavage is fast but that the enzyme gets stalled and dissociation of the stalled enzyme is slow (18).
3. Mechanistic Studies of Processivity
There are two structural studies that provide possible mechanisms for the processive movement of exocellulases. In one, two different binding modes for long oligosaccharides were seen in structures of two different mutants of CelF (Cel48A) from Clostridium cellulolyticum. One mode was identical to that seen in the WT enzyme, which is believed to be the catalytic site, while the other was in a site above the catalytic site and it was suggested that this site might be used during processive movement of the cellulose chain (19). In the other study a set of structures of Cel6A from Humicola insolens bound to different ligands suggests that the processive movement is possible due to the flexibility of the hydrophobic residues that bind the cellulose in the active site as well as the extensive hydration of the bound cellulose (20).
We have used the ratio of cellobiose to cellotriose assay to study the processivity of various site-directed mutants in the exocellulase: T. fusca Cel6B and the soluble/insoluble assay to study the processivity of mutants in T. fusca Cel9A. These experiments showed that certain mutations increase processivity while others decrease processivity and that both types of mutations were found in both enzymes. For Cel9A, it appeared that processivity depends on the balance between the binding affinity of the −4 to −1 subsites to the affinity of regions upstream of the cleavage site especially the family 3 CBM (19). Most mutations in potential substrate-binding residues in subsites −1 to −4 have decreased processivity and this seems reasonable, as the weaker binding would make dissociation of the chain from the CBM more likely than binding of the chain into the empty subsites after cleavage. It is less clear why most mutations in the family 3 CBM increase processivity since weaker binding to the CBM should increase both dissociation and movement of the chain into the subsites to about the same extent. A surprising finding is that a double mutant enzyme containing a cd mutation that increases productivity by itself and a CBM mutation that also increases processivity by itself, produces an enzyme with much lower than WT processivity and activity (21).
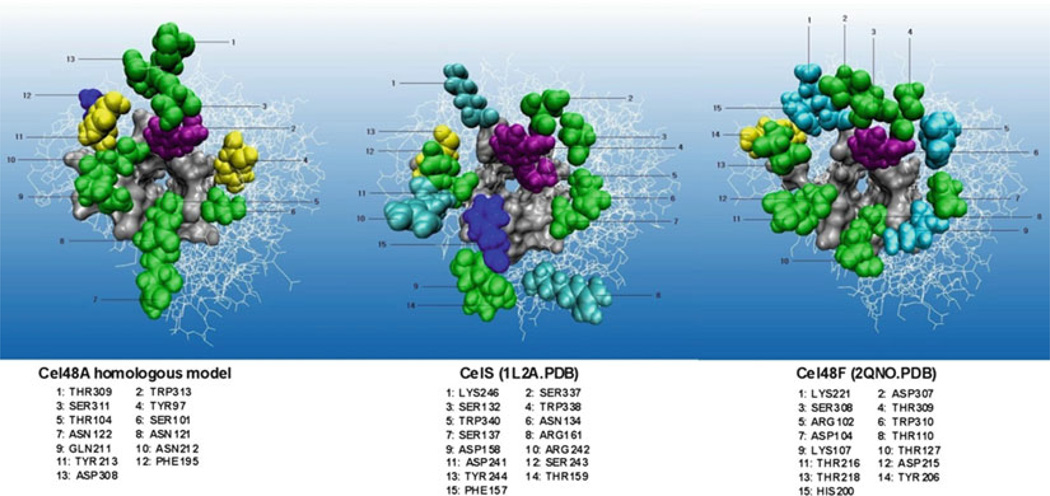
The studies of T. fusca Cel6B mutants showed that there was not a strong correlation between the activity of a mutant enzyme and its processivity (11). In addition, mutation of Asp226 to Ala appeared to allow the mutant enzyme to move along the substrate without cleavage. This is different from the WT enzyme and most mutant enzymes where movement along the substrate is coupled to cleavage (22). The evidence for this change was that the mutant enzyme had greatly reduced activity on swollen cellulose and bacterial cellulose but WT activity on carboxymethylcellulose [CMC]. Furthermore, it did not produce cellobiose from CMC but it did produce cellotriose, cellotetraose, cellopentose, and cellohexose suggesting that it made random cleavages along the CMC molecule. Further study is needed to explain how this mutation uncouples movement of the enzyme along the substrate from cleavage. For Cel48A, we have mutated surface residues that are close to the entrance of the active site tunnel (see Fig. 1).
Fig. 1.
Structural comparison of three GH family 48 exocellulases.
All such residues in the three GH-48 cellulases that we examined are potential cellulose-binding residues, and aromatic residues, which have the highest affinity for sugars, are about four times enriched relative to all the surface residues (19, 22). It is interesting that only 3 of the 13 tunnel entrance surface residues are conserved in all three enzymes. So far the three conserved residues have been mutated to Ala and the mutant enzymes have been characterized. Mutation of a highly conserved Trp residue(313) to Ala caused a decrease in activity on both amorphous and bacterial cellulose as does mutation of conserved Tyr213 while mutation of conserved Ser311 did not change the activity (Kostylev M, Wilson DB, unpublished). This seems reasonable, as the Tyr and Trp residues would be expected to bind cellulose more tightly than Ser. The Trp mutation decreased processivity while the other two mutations did not. A Trp residue is present at the entrance to the active site tunnel in all three exocellulase families, and it has been shown to be specifically required for crystalline cellulose hydrolysis in both family GH-6 and GH-48 exocellulases (Kostylev M, Wilson DB, unpublished) (24).
4. Conclusion
There is clearly a need to develop better assays to measure processivity. In addition, more research is needed to understand why the measured processivity of most exocellulases is quite low even though the processivity predicted based on the ratio of the on and off rates for Cel7A on cellulose is much higher (17).
Acknowledgments
This work was supported by the BioEnergy Science Center (BESC), which is a part of the U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science, Oak Ridge National Laboratory. We thank Mo Chen for preparing the figure.
References
- 1.Teeri TT, Koivula A, Linder M, Wohlfahrt G, Divne C, Jones TA. Trichoderma reesei cellobiohydrolases: why so efficient on crystalline cellulose? Biochem Soc Trans. 1998;26:173–178. doi: 10.1042/bst0260173. [DOI] [PubMed] [Google Scholar]
- 2.Barr BK, Hsieh YL, Ganem B, Wilson DB. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35:586–592. doi: 10.1021/bi9520388. [DOI] [PubMed] [Google Scholar]
- 3.Rouvinen J, Bergfors T, Teeri T, Knowles JK, Jones TA. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990;249:380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- 4.Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JKC, Teeri TT, Jones A. The three-dimensional structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265:524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- 5.Parsiegla G, Juy M, Reverbel-Leroy C, Tardif C, Belaich JP, Driguez H, Haser R. The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 Å resolution. EMBO J. 1998;17:5551–5562. doi: 10.1093/emboj/17.19.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakon J, Irwin D, Wilson DB, Karplus PA. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat Struct Biol. 1997;4:810–818. doi: 10.1038/nsb1097-810. [DOI] [PubMed] [Google Scholar]
- 7.Irwin D, Shin D-H, Zhang S, Barr BK, Sakon J, Karplus PA, Wilson DB. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J Bacteriol. 1998;180:1709–1714. doi: 10.1128/jb.180.7.1709-1714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson BJ, Zhang H, Longmire AG, Moon YH, Hutcheson SW. Processive endoglucanases mediate degradation of cellulose by Saccharophagus degradans. J Bacteriol. 2009;191:5697–5705. doi: 10.1128/JB.00481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zakariassen H, Aam BB, Horn SJ, Vårum KM, Sørlie M, Eijsink VG. Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J Biol Chem. 2009;284:10610–10617. doi: 10.1074/jbc.M900092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irwin DC, Spezio M, Walker LP, Wilson DB. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- 11.Vuong TV, Wilson DB. Processivity, synergism, and substrate specificity of Thermobifida fusca Cel6B. Appl Environ Microbiol. 2009;75:6655–6661. doi: 10.1128/AEM.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai T, Boisset C, Samejima M, Igarashi K, Sugiyama J. Unidirectional processive action of cellobiohydrolase Cel7A on Valonia cellulose microcrystals. FEBS Lett. 1998;432:113–116. doi: 10.1016/s0014-5793(98)00845-x. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi K, Koivula A, Wada M, Kimura S, Penttilä M, Samejima M. High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J Biol Chem. 2009;284:36186–36190. doi: 10.1074/jbc.M109.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harjunpää V, Teleman A, Koivula A, Ruohonen L, Teeri TT, Teleman O, Drakenberg T. Cello-oligosaccharide hydrolysis by cellobiohydrolase II from Trichoderma reesei Association and rate constants derived from an analysis of progress curves. Eur J Biochem. 1996;240:591. doi: 10.1111/j.1432-1033.1996.0584h.x. [DOI] [PubMed] [Google Scholar]
- 15.Nidetsky B, Zachariae W, Gercken G, Hayn M, Steiner W. Hydrolysis of cello-oligosaccharides by Trichoderma reesei cellobiohydrolases; experimental data and kinetic modeling. Enzyme Microb Technol. 1994;16:43–52. [Google Scholar]
- 16.Kurasin M, Väljamäe P. Processivity of cellobiohydrolases is limited by the substrate. J Biol Chem. 2011;286:169–177. doi: 10.1074/jbc.M110.161059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kipper K, Väljamäe P, Johansson G. Processive action of cellobiohydrolase Cel7A from Trichoderma reesei is revealed as ‘burst’ kinetics on fluorescent polymeric model substrates. Biochem J. 2005;385:527–535. doi: 10.1042/BJ20041144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Praestgaard E, Elmerdahl J, Murphy L, Nymand S, McFarland KC, Borch K, Westh P. A kinetic model for the burst phase of processive cellulases. FEBS J. 2011 doi: 10.1111/j.1742-4658.2011.08078.x. [DOI] [PubMed] [Google Scholar]
- 19.Parsiegla G, Reverbel C, Tardif C, Driguez H, Haser R. Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocello oligosaccharides give rise to a new view of the substrate pathway during processive action. J Mol Biol. 2008;375:499–510. doi: 10.1016/j.jmb.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 20.Varrot A, Frandsen TP, von Ossowski I, Boyer V, Cottaz S, Driguez H, Schülein M, Davies GJ. Structural basis for ligand binding and processivity in cellobiohydrolase Cel6A from Humicola insolens. Structure. 2003;11:855–864. doi: 10.1016/s0969-2126(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Irwin DC, Wilson DB. Processivity, substrate binding, and mechanism of cellulose hydrolysis by Thermobifida fusca Cel9A. Appl Environ Microbiol. 2007;73:3165–3172. doi: 10.1128/AEM.02960-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guimarães BG, Souchon H, Lytle BL, Wu D, Alzari PM. The crystal structure and catalytic mechanism of cellobiohydrolase CelS, major enzymatic component of the Clostridium thermocellum cellulosome. J Mol Biol. 2002;320:587–596. doi: 10.1016/s0022-2836(02)00497-7. [DOI] [PubMed] [Google Scholar]
- 23.Vuong TV, Wilson DB. The absence of a single identifiable catalytic base residue in Thermobifida fusca exocellulase Cel6B. FEBS J. 2009;276:3837–3845. doi: 10.1111/j.1742-4658.2009.07097.x. [DOI] [PubMed] [Google Scholar]
- 24.Koivula A, Kinnari T, Harjunpää V, Ruohonen L, Teleman A, Drakenberg T, Rouvinen J, Jones TA, Teeri TT. Tryptophan 272: an essential determinant of crystalline cellulose degradation by Trichoderma reesei cellobiohydrolase Cel6A. FEBS Lett. 1998;429:341–346. doi: 10.1016/s0014-5793(98)00596-1. [DOI] [PubMed] [Google Scholar]