Abstract
His-tag affinity purification is one of the most commonly used methods to purify recombinant proteins expressed in E. coli. One drawback of using the His-tag is the co-purification of contaminating histidine-rich E. coli proteins. We engineered a new E. coli expression strain, LOBSTR (low background strain), which eliminates the most abundant contaminants. LOBSTR is derived from the E. coli BL21(DE3) strain and carries genomically modified copies of arnA and slyD, whose protein products exhibit reduced affinities to Ni and Co resins, resulting in a much higher purity of the target protein. The use of LOBSTR enables the pursuit of challenging low-expressing protein targets by reducing background contamination with no additional purification steps, materials, or costs, and thus pushes the limits of standard His-tag purifications.
Keywords: BL21(DE3), E. coli protein expression strain, His-tag affinity purification, LOBSTR
INTRODUCTION
Many methods of recombinant protein purification have been developed. One of the most widely used techniques is the His-tag affinity purification1. A small His-tag (usually 6 or 10 histidines) is fused to either the N or C terminus of the target protein, enabling capture by nickel or cobalt ions coordinated on a variety of commercially available resins. The small size of the His-tag, low cost, and ease of use have made it the most popular affinity-tag available. Expression of recombinant His-tagged proteins is largely carried out in E. coli because it is easy to culture and it allows for the production of target proteins with high yield. However, one major drawback of His-tag affinity purification of proteins expressed in E. coli is the presence of naturally histine-rich host proteins, resulting in co-purification of these contaminants2,3. The two most common E. coli contaminants are ArnA, a bifunctional enzyme involved in the modification of lipid A phosphates with aminoarabinose4, and SlyD, a peptidyl-prolyl cis/trans-isomerase1,5. ArnA has several non-consecutive histidine residues, which are surface exposed and form clusters within the hexameric structure2,3,6. In contrast, SlyD is characterized by a 48 amino acid unstructured C-terminal tail containing 15 histidines4,7. Because the Ni-binding mechanism of ArnA and SlyD mimics that of the His-tag, both proteins are co-purified along with the target protein in His-tag affinity purifications. For well expressing recombinant proteins, these endogenous proteins are a small problem because they are out-competed by the sheer amount of the protein of interest. However, many proteins, including human proteins, large, multi-domain proteins and co-expressed protein complexes are ignored as viable targets for in vitro studies because they express poorly and consequently cannot be isolated in sufficient amounts or with high purity. When protein expression is low, host proteins, especially ArnA and SlyD, have a similar abundance and compete for binding on Ni or Co resins. As a result, ArnA and SlyD are purified in nearly equal amounts when compared to the target protein. The most effective means to increase the purity of the target protein is to use additional affinity tags or multiple purification steps, however this lowers the yield and increases the purification time and cost. Because both arnA and slyD knockout strains suffer growth defects, these strains are not viable options for recombinant protein expression8,9. To address these problems, we designed a new E. coli expression strain named LOBSTR (low-background-strain), which features genomic modifications in arnA and slyD based on surface engineering. LOBSTR maintains normal cell growth but significantly reduces the Ni- and Co-binding affinities of both host proteins. LOBSTR drastically reduces ArnA and SlyD contamination, thus enabling the purification even of poorly expressing target proteins.
MATERIALS AND METHODS
Wild type arnA was PCR amplified from E. coli genomic DNA with NdeI and XhoI restriction site overhangs on the 5’ and 3’ ends, respectively, using primers 1F and 1R (See all primer details in Table S1), and cloned into the bacterial expression vector pColaDuet (EMD Millipore). Two serine point mutations were introduced at site 1 (H359S and H361S) using primers 2F and 2R. Two additional serine point mutations were introduced at site 2 (H592S and H593S) using primers 3F and 3R to generate the final arnA mutant containing a total of four histidine to serine mutations.
The arnA knockout strain was generated with the E. coli recombineering technique10, using the pKD4 plasmid as a template for the selectable marker and BL21(DE3) as the parental strain. The forward and reverse primers, 4F and 4R, were designed to maintain the reading frame of arnB, which shares its start codon with the stop codon of arnA within the arn operon11 (also called pmrHFIJKLM operon12). A slightly modified scheme was used to introduce the arnA mutant back into the arnA knockout strain at the original locus (Fig. S1). First, mutant arnA was amplified and combined with the amplified selectable marker in a second PCR step. The resulting PCR product containing mutated arnA and the selectable marker was transformed into the arnA knockout strain for recombination using the λ Red recombinase plasmid (pKD46). The selectable marker was eliminated using the FLP plasmid (pCP20). For the modification in slyD, the arnA mutant strain was transformed with a PCR product (generated using primers 5F and 5R) containing a selectable marker flanked by homologous overhangs that, after recombination, result in the elimination of the 46-residue C-terminal, histidine-rich segment of SlyD. Again, the selectable marker was later removed using pCP20. Proper genomic integration was confirmed by PCR and sequencing. The RIL plasmid (Agilent Technologies) encoding rare tRNAs was transformed into the final expression strain to improve the expression of our eukaryotic target proteins.
The binding affinity of wild type and mutant ArnA were assessed by immobilizing purified protein onto a 1 ml His-Trap FF column (GE Healthcare) equilibrated in 50 mM potassium phosphate pH 8.0, 300 mM NaCl, and 5 mM beta-mercaptoethanol. Protein was eluted with a linear gradient of 0–150 mM imidazole. The imidazole concentration at the elution peak of each protein was recorded and compared.
Growth analysis was performed at 18, 25 and 37°C for both LOBSTR and the BL21(DE3) strains carrying the same test expression plasmid (See table S2 for a list of all test constructs). Cultures of 1L were grown in LB medium supplemented with 0.4% (w/v) glucose and antibiotic selection at 37°C to OD600 ~0.7. Protein expression was induced with 0.2 mM IPTG 20 minutes after the cultures were shifted to the desired expression temperature. OD600 was measured from the initial synchronization time and until the cells were harvested ~20–22 hours after induction.
To test protein purification, BL21(DE3) and LOBSTR cultures were started at 37°C in LB medium supplemented with 0.4% (w/v) glucose and appropriate antibiotic selection. At OD600 ~0.7, cultures were shifted to 18°C and induced with 0.2 mM IPTG ~20 min later. Cultures were harvested after 18–20 hours. For each strain and construct tested, a total of ~3.5g of cells were resuspended in 50 mL of resuspension buffer (40 mM potassium phosphate pH 8.0, 150 mM NaCl, 40 mM imidazole, and 3mM beta-mercaptoethanol) and lysed with a cell disrupter (Constant Systems). Lysates were cleared for 25 min at 9500×g and the soluble fraction was incubated with 400 µl bed volume of Ni Sepharose 6 Fast Flow (GE Healthcare) resin for 1 hour while stirring at 4°C. The resin was collected and washed with 6 mL of resuspension buffer and eluted with 2 mL of elution buffer (40 mM potassium phosphate pH 8.0, 150 mM NaCl, 250 mM imidazole, and 3 mM beta-mercaptoethanol). Elution fractions were analyzed on a 4–15 % SDS-PAGE gradient gel (Bio-RAD) and stained with Coomassie Blue R250. Purifications using Ni-NTA (Qiagen) and Talon (Clontech) resins were performed using resuspension buffer containing 20 mM or 5 mM imidazole, respectively, following manufacturer’s recommendations.
RESULTS
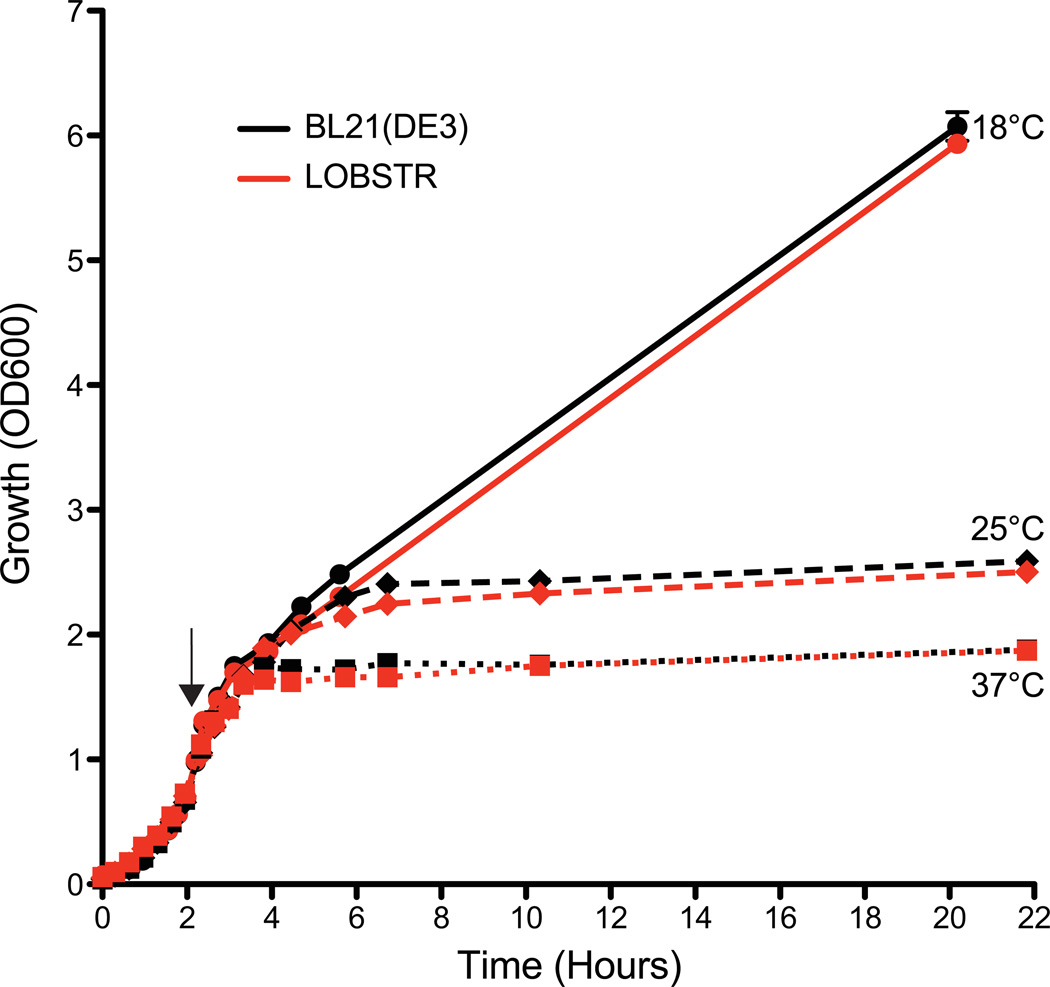
We designed surface engineered forms of E. coli ArnA and SlyD based on their crystal and NMR structures, respectively6,7 (Fig. S2). Both proteins have exposed histidine-rich surfaces that result in binding to immobilized metal-affinity resins. ArnA is a hexamer, formed by a dimer of trimers. The structure revealed two prominent surface-exposed patches of histidine residues. One of the patches is at a trimer interface and results in a cluster of 9 histidines per trimer (Fig. S2, site 1). We mutated histidine residues 359 and 361 to serines to abolish this histidine-rich surface. The second cluster of surface-exposed histidines was removed by mutating histidines 592 and 593 to serines (Fig. S2, Site 2). To determine whether the histidine to serine mutations resulted in weaker Ni-binding affinity, both recombinant wild type and mutant ArnA were first purified in batch. Subsequently, pure protein was loaded onto a His-trap Ni-column and eluted with a linear imidazole gradient. Wild type ArnA eluted at a concentration of ~60 mM imidazole, while mutant ArnA showed significantly weaker binding affinity, eluting at ~30 mM imidazole (Fig. S3). Thus, mutating four histidine residues to serines in ArnA (24 per hexamer) lowers the Ni-affinity to a level comparable to non-specific binding. Similarly, analysis of the SlyD NMR structure showed that all of the clustered histidine residues reside in an unstructured tail at the very C terminus of the protein (Fig. S2). A previous study suggested that deleting this tail has little effect on cell growth13,14. Thus, we truncated SlyD at residue 150, thereby maintaining the structural integrity of the catalytic N-terminal domain while removing the entire unstructured tail. Using a modified recombineering10 approach, we then replaced the genomic copies of arnA and slyD in the host strain BL21(DE3) with our mutant versions to create LOBSTR (overview Fig. S1). To confirm that the combined genetic modifications in LOBSTR also maintain normal growth, we monitored and compared its growth rate to the parental BL21(DE3) strain at 18, 25 and 37°C. A test construct (See table S2 for a list of all test constructs) was expressed over the duration of the growth analysis. No significant difference in growth rate at any of the induction temperatures was observed between LOBSTR and BL21(DE3), and the final OD600 of the cultures after overnight induction were very similar (Fig. 1).
Figure 1. LOBSTR and the parental BL21(DE3) strain show comparable growth.
The growth (OD600) of both LOBSTR and the parental BL21(DE3) strain was measured from initial synchronization at 0 hours until the final harvest. Both strains carried the same expression plasmid and were grown at 37°C until an OD600 ~0.7, at which point protein expression was induced at 18, 25 and 37°C (black arrow). The growth curves for LOBSTR and BL21(DE3) are shown in red and black, respectively. Cell growth during log phase and final cell density is similar for both strains.
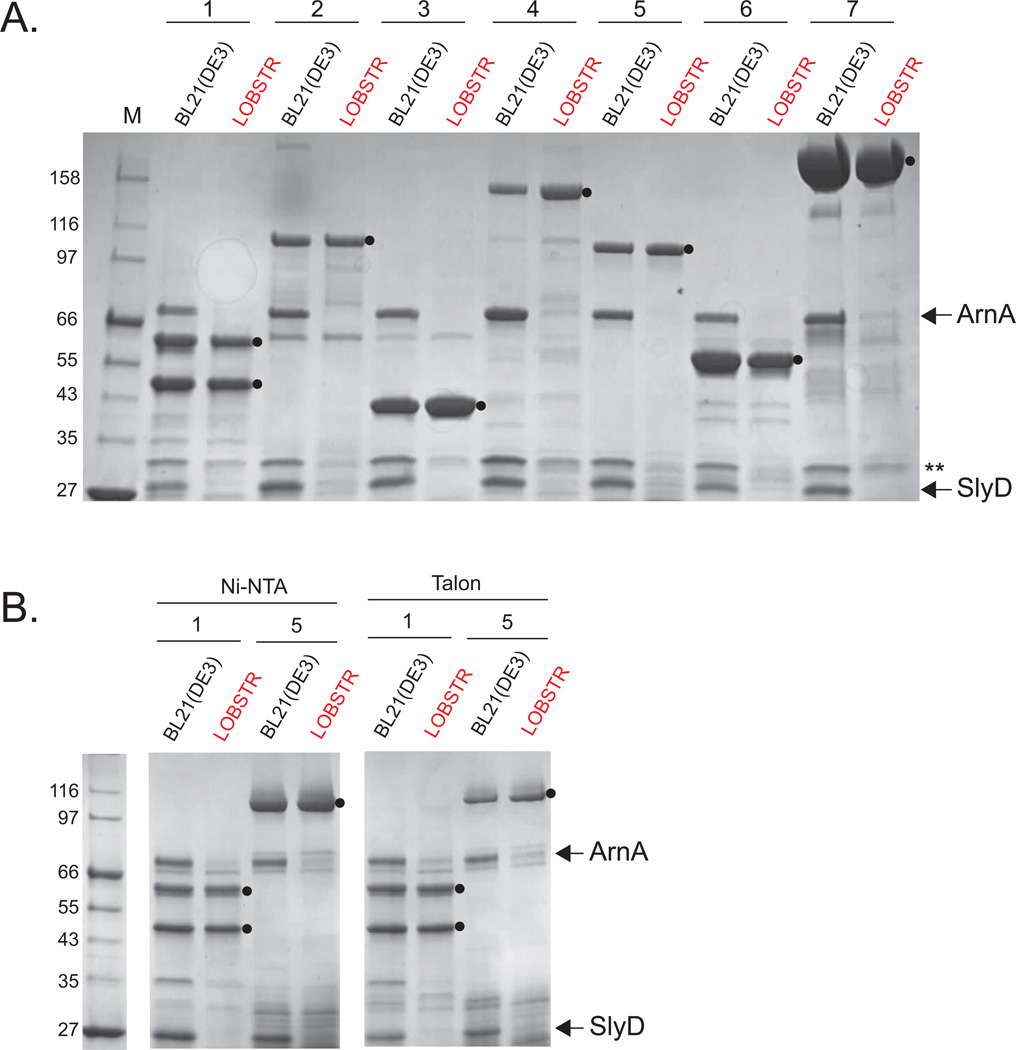
To verify that LOBSTR reduces ArnA and SlyD contamination, we performed small-scale purifications of seven different protein constructs in the parental BL21(DE3) strain and in LOBSTR. The seven constructs (Table S2) were chosen to represent a wide range of potential targets, including low- and higher-expressing constructs, monomeric proteins, dimeric complexes, 6×- and 10×His-tagged proteins. Most of our test constructs contain a SUMO-tag fused to the N terminus to increases protein solubility. In the BL21(DE3) strain background, high levels of contamination by both ArnA and SlyD can be seen in the elutions (Fig. 2). Illustrating the low expressions levels of target proteins, ArnA and SlyD are purified in amounts nearly equivalent to that of the target protein, as seen in constructs 2, 4, and 5. However, in LOBSTR, the vast majority of contaminants are eliminated, with the target protein now being the most prominent protein. Purification of construct 1, a heterodimeric complex with one binding partner carrying a 6×His-tag, is also greatly enhanced in LOBSTR. Furthermore, the amounts of all target proteins purified are similar between the BL21(DE3) strain and LOBSTR. Since the initial purity is much greater, fewer subsequent purification steps are required to obtain pure protein, resulting in equivalent, if not greater, final yields from LOBSTR. Curiously, a secondary contaminant, indicated by a double asterisk (**) in Fig. 2, is also reduced in LOBSTR. This protein, identified by mass spectrometry as Hsp15, is reported to bind nucleic acids15. While no modifications have been made to this protein in LOBSTR, we speculate that it may have non-specific binding affinity to SlyD, which is highly negatively charged. To ensure that the results seen here are reproducible on a variety of commercially available resins, we purified constructs 1 and 5 on two additional commonly used resins, Ni-NTA (Qiagen) and Talon (Clontech) (Fig, 2B). Both resin manufacturers recommend lower imidazole concentrations in the binding and washing buffers compared to the Ni Sepharose 6 FF resin (GE Healthcare), which was used for the purifications above. Still, nearly complete elimination of ArnA and SlyD contamination is observed on these resins as well (Figure 2B).
Figure 2. ArnA and SlyD are eliminated from His-tag purifications from LOBSTR.
Elution samples of test purifications from BL21(DE3) and LOBSTR using common metal affinity resins are shown. A. Seven protein constructs were purified from both the parental BL21(DE3) strain and LOBSTR using Ni Sepharose 6FF resin (GE Healthcare). The constructs are numbered 1–7, and contain either a 6×His-tag (1 and 4) or a 10×His-tag (2,3,5–7). See table S2 for a list of all test constructs. The elution samples were run on an SDS-PAGE gel and stained with Coomassie Blue R250. ArnA and SlyD are indicated by arrows and target proteins indicated with a black circle (●). The double asterisk (**) indicates Hsp15, another protein showing reduced Ni-binding affinity in LOBSTR. B. Purifications of constructs 1 and 5 from BL21(DE3) and LOBSTR were also carried out on two additional commonly used resins, Ni-NTA (Qiagen) and Talon (Clontech). In each case, ArnA and SlyD are successfully eliminated in LOBSTR.
DISCUSSION
LOBSTR enables the pursuit of poorly expressed protein targets in E. coli by lowering the background contamination of ArnA and SlyD. Previously, constructs yielding only 0.1–1 mg of target protein per liter of culture could be considered inadequate for in vitro studies. At such low levels of expression, ArnA and SlyD compete for the binding capacity of the metal affinity resin and are co-purified in equivalent or even greater amounts. LOBSTR enables a significantly higher yield and purity of poorly expressed target protein eluted from Ni or Co resins. Protein purity is of key importance for most downstream purposes, whether the protein is used in medical applications, binding studies, functional assays, or structural studies (EM, SAXS, NMR, and crystallography). An alternate approach to eliminate E. coli host contaminants has been developed previously9. Here, ArnA, SlyD, and Can were genomically tagged with a chitin-binding domain and eliminated over chitin beads, pulling out the contaminants and leaving the target protein in the flow-through. In addition, GlmS is mutated to reduce binding to Ni and Co. While this method is successful in removing the contaminants, it requires an additional purification step as well as an additional resin, increasing both the time and cost of each purification. However, LOBSTR only requires a one-step purification to eliminate the major E. coli contaminants ArnA and SlyD with no additional costs and is specifically designed for low-expressing proteins. An alternate purification strategy is to simply perform a second IMAC step after cleaving off the His-tag from the protein of interest so that contaminants are rebound while the cleaved protein remains in the flow-through. While this method is successful when the contaminants make up only a small fraction of the total immobilized protein in the first IMAC step, it is highly inefficient if the contaminants are abundant and thus substantially reduce the initial yield. LOBSTR instead, incorporates genomic modifications to arnA and slyD in order to reduce the affinity of their gene products for metal affinity resins, eliminating them from co-purification with recombinant proteins of interest. Thus, proteins that were previously ignored as targets for recombinant expression and purification are now accessible.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anna Kaertner and Marta Carrara for initial help on the project. Simon Ringgaard for advice on recombineering and Greg Kabachinski for reading the manuscript. K.R.A. was supported by the Lundbeck Foundation and a Sapere Aude DFF postdoc grant. This work was supported by NIH Grant GM077537 (T.U.S.) and a Pew Scholar Award (T.U.S.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare potential competing financial interests.
REFERENCES
- 1.Hochuli E, Bannwarth W, Döbeli H, Gentz R. Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. Nature Biotechnology. 1988;6(11):1321–1325. [Google Scholar]
- 2.Wülfing C, Lombardero J, Plückthun A. An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J. Biol. Chem. 1994;269:2895–2901. [PubMed] [Google Scholar]
- 3.Bolanos-Garcia VM, Davies OR. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim. Biophys. Acta. 2006;1760:1304–1313. doi: 10.1016/j.bbagen.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Williams GJ, Breazeale SD, Raetz CRH, Naismith JH. Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis. J. Biol. Chem. 2005;280:23000–23008. doi: 10.1074/jbc.M501534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roof WD, Horne SM, Young KD, Young R. slyD, a host gene required for phi X174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J. Biol. Chem. 1994;269:2902–2910. [PubMed] [Google Scholar]
- 6.Gatzeva-Topalova PZ, May AP, Sousa MC. Structure and mechanism of ArnA: conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance. Structure. 2005;13:929–942. doi: 10.1016/j.str.2005.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weininger U, Haupt C, Schweimer K, Graubner W, Kovermann M, Brüser T, Scholz C, Schaarschmidt P, Zoldak G, Schmid FX, Balbach J. NMR Solution Structure of SlyD from Escherichia coli: Spatial Separation of Prolyl Isomerase and Chaperone Function. J. Mol. Biol. 2009;387:295–305. doi: 10.1016/j.jmb.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 8.Roof WD, Fang HQ, Young KD, Sun J, Young R. Mutational analysis of slyD, an Escherichia coli gene encoding a protein of the FKBP immunophilin family. Mol. Microbiol. 1997;25:1031–1046. doi: 10.1046/j.1365-2958.1997.5201884.x. [DOI] [PubMed] [Google Scholar]
- 9.Robichon C, Luo J, Causey TB, Benner JS, Samuelson JC. Engineering Escherichia coli BL21(DE3) Derivative Strains To Minimize E. coli Protein Contamination after Purification by Immobilized Metal Affinity Chromatography. Applied and Environmental Microbiology. 2011;77:4634–4646. doi: 10.1128/AEM.00119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross J, Englesberg E. Determination of the order of mutational sites governing Larabinose utilization in Escherichia coli B/r bv transduction with phage Plbt. Virology. 1959;9:314–331. doi: 10.1016/0042-6822(59)90125-4. [DOI] [PubMed] [Google Scholar]
- 12.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect. Immun. 2000;68:6139–6146. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leach MR, Zhang JW, Zamble DB. The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD. J. Biol. Chem. 2007;282:16177–16186. doi: 10.1074/jbc.M610834200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JWJ, Butland GG, Greenblatt JFJ, Emili AA, Zamble DBD. A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J. Biol. Chem. 2005;280:4360–4366. doi: 10.1074/jbc.M411799200. [DOI] [PubMed] [Google Scholar]
- 15.Korber P, Zander T, Herschlag D, Bardwell JC. A new heat shock protein that binds nucleic acids. J. Biol. Chem. 1999;274:249–256. doi: 10.1074/jbc.274.1.249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.