Abstract
Purpose
To identify genes whose depletion is detrimental to Pim1-overexpressing prostate cancer cells and to validate this finding in vitro and in vivo.
Experimental Design
RNAi screening was employed to identify genes whose depletion is detrimental to Pim1-overexpressing cells. Our finding was validated using shRNA or PLK1 specific inhibitor BI 2536. Xenograft studies were performed using both PLK1 knockdown cells and BI 2536 to investigate the effects of PLK1 inhibition on tumorigenesis in Pim1-overexpressing cells. Finally, PLK1 and PIM1 expression patterns in human prostate tumors were examined by immunohistochemistry using tissue microarrays.
Results
We identified the mitotic regulator polo-like kinase (PLK1) as a gene whose depletion is particularly detrimental to the viability of Pim1-overexpressing prostate cancer. Inhibition of PLK1 by shRNA or BI 2536 in Pim1-overexpressing prostate cancer xenograft models resulted in a dramatic inhibition of tumor progression. Notably, Pim1-overexpressing cells were more prone to mitotic arrest followed by apoptosis due to PLK1 inhibition than control cells. Furthermore, inhibition of PLK1 led to the reduction of MYC protein levels both in vitro and in vivo. Our data also suggest that PIM1 and PLK1 physically interact and PIM1 might phosphorylate PLK1. Finally, PLK1 and PIM1 are frequently co-expressed in human prostate tumors, and co-expression of PLK1 and PIM1 was significantly correlated to higher Gleason grades.
Conclusions
Our findings demonstrate that PIM1-overexpressing cancer cells are particularly sensitive to PLK1 inhibition, suggesting that PIM1 might be used as a marker for identifying patients that will benefit from PLK1 inhibitor treatment.
Keywords: PIM1, PLK1, siRNA, Prostate Cancer, Tumorigenesis
Introduction
Members of the PIM family of serine-threonine kinases (PIM1, PIM2 and PIM3) are overexpressed in a variety of malignancies, including leukemias, lymphomas, prostate and pancreatic cancers (1, 2). The PIM kinases increase cell survival, proliferation and tumorigenicity and appear to play these roles by phosphorylating multiple substrates including Cdc25A, NuMA, p21, Bad, C-TAK1, Cdc25C, and p27 (3–9). A highly notable feature of PIM kinase-driven tumorigenesis is the dramatic cooperativity between PIM kinases and MYC. In prostate cancers, PIM1 and MYC are frequently co-expressed (10, 11) and recent work in animal models has shown that PIM1 synergizes with c-MYC to induce advanced prostate cancer in a kinase-dependent manner (11). PIM1 can stabilize MYC protein levels and enhance MYC transcriptional activity (12). Importantly, PIM1 is required to maintain the tumorigenicity of MYC/PIM1-expressing tumor cells (13), supporting the notion that PIM1 could be a valid therapeutic target. Accordingly, there are many ongoing efforts aimed at developing small molecule inhibitors of the PIM kinases as anti-cancer therapeutic agents. PIM1 inhibition is potentially an attractive strategy for treating prostate cancer as Pim kinase-deficiency in mice is generally well tolerated, suggesting that PIM kinases are not required for essential cellular functions. Furthermore, the presence of a unique hinge region in the ATP-binding site of PIM1 facilitates the development of specific small molecule kinase inhibitors.
Conceptually, another approach to targeting PIM kinase-expressing tumor cells is to identify the specific vulnerabilities of these cells. In this study, we employed RNAi screening to systematically identify genes whose expression is required for the viability of PIM1-expressing prostate epithelial cells. RNAi screening has been used to identify synthetic lethal interaction between genes with relevance to cancer treatment (14, 15).
Here we used a collection of siRNAs that target genes encoding selected serine/threonine kinases, tyrosine kinases, cell cycle protein and apoptosis proteins to identify genes that may be potential targets for inhibiting PIM1-expressing cells, leading to the identification of PLK1. PLK1 is a mitotic regulator that plays a crucial role at various steps of mitosis and is overexpressed in many tumor types including prostate cancer, where PLK1 overexpression was found to correlate with Gleason grade (16). The inhibition of PLK1 has been shown to have potent antitumor effects in experimental in vitro or in vivo models (17–20). The fact that PLK1 is required for normal mitotic progression has raised some concerns about the potential toxicity of anti-PLK1 therapeutic agents. However, in principle, the identification of molecular changes that make tumor cells more sensitive to the effects of PLK1 inhibition will lead to an increase in therapeutic index and better tolerability. Our findings demonstrate that molecular changes induced by oncogenes such as PIM1 can make cancer cells particularly sensitive to the inhibition of PLK1, a feature that can be exploited for therapeutic purposes.
Materials and Methods
RNAi Screen
RWPE-diploid and polyploid cells (21) were screen for siRNA libraries targeting a total of 570 genes involved in key cancer relevant pathways (111 cell cycle, 318 apoptosis, 87 serine-threonine kinase and 54 tyrosine kinase genes) (siRNA library, Dharmacon). Before the screening, transfection condition and reagents were optimized and validated using reverse-transfection (Dharmacon). The data from both cell lines were combined and hits were determined by Z scores ≥2 or ≤−2 (22). For validation, we selected 10 genes including some of the top hits that reduced cell viability. The siRNAs against these 10 genes were custom ordered and tested using RWPE1-Pim1 and control RWPE1-Neo to identify genes whose knock-down specifically affect cell viability in Pim1 overexpressing cells. Cellular viability was determined after 72 hrs of reverse transfection by using the CellTiter-Glo Luminescent cell viability assay (Promega).
Cell Culture
All the cell lines were authenticated. RWPE-1, LNCaP and PC3 cells were purchased from ATCC and were maintained in keratinocyte serum–free medium (KSFM for RWPE1) or RPMI medium (for LNCaP, PC3) with 10 % FBS in a humidified 37°C incubator with 5% CO2. RWPE-Neo, Pim1, diploid and polyploid cells have been described in previous papers (21, 23, 24). LNCaP-Neo/Pim1, PC3-Neo/Pim1 cells has also been described previously (23). NHPrE cells were maintained in F12/DMEM medium as described (25). To establish PLK1 knock-down cells, lentiviral PLK1 shRNAmir and control shRNAmir (Open Biosystems) were transduced into LNCaP-Neo/Pim1 and PC3-Neo/Pim1 cells and stable clones were selected by using 1–2 ug/ml puromycin. For BI 2536 (Selleckchem) treatment, different dose of BI 2536 (10 nM −100 nM) were added to the cells and cell lysates were prepared 24 hrs later.
Western Blotting and Immunoprecipitation
Western blotting was performed as described previously (23). The following antibodies were used: PIM1 (Santa Cruz, sc-13513), PLK1 (Santa Cruz, sc-17783), phospho-PLK1 (Thr 210; Cell Signaling, #5472), phosphor-histone H3 (Upstate, #06-570), cleaved PARP (Cell Signaling, # 9541), MYC (Abcam, ab32072) and phospho-serine 62-MYC (Abcam, ab51156). For immunoprecipitation (Ipt), lysates were prepared with the lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.2% NP-40, 1X protease inhibitor cocktail (Roche)) were incubated with mouse α-PIM1 or α-PLK1 overnight at 4°C, followed by incubation with the protein A/G agarose beads for 2 h at 4°C. The lysates were washed with the Ipt/washing buffer three times, then the proteins bound to the beads were eluted in 2X SDS sample buffer, separated by SDS-PAGE, and blotted with the antibodies.
Immunofluorescence
Cells were processed as reported previously (23). Briefly, cells on coverslips were fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with PBS three times, the cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min. Following washing and blocking, cells were incubated with the appropriate primary antibodies overnight and incubated with fluorescent secondary antibodies (molecular probe). After washing and staining with 4,6-diamidino-2-phenylindole (DAPI), slides were mounted, sealed, and examined.
In vitro Kinase Assay
The kinase assay was carried out in the kinase buffer (20 mM Pipes, pH 7.0, 5 mM MnCl2, 0.25 mM β-glycerophosphate, 0.4 mM spermidine and 7 mM β-mercaptoethnol) containing 10 μM nonradioactive γATP, 10 μCi [32P]ATP, 2 μM aprotinin and 0.1–2μg PIM1 or PLK1. The reactions were then incubated at room temperature for 30 min, stopped by SDS sample buffer, separated by SDS-PAGE, fixed, amplified, dried and the film was developed.
In vivo Xenograft
For xenograft studies, cells (107 for both LNCaP and PC3) modified to overexpress Pim1 with or without stable PLK1 knockdown were mixed with 200 μl of matrigel (Beckton Dickinson Labware) and injected subcutaneously into the flanks of 8 week old male nu/nu mice and tumor size was measured once a week for 5 weeks. For BI 2536 treatment, LNCaP-Pim1/Neo cells (106 cells per graft) were grafted subcutaneously in nu/nu mice and one week later BI 2536 were injected intravenously (i.v) at a dose of 25 mg/kg twice a week on two consecutive days for three weeks. Tumor size was measured once a week. Nude mice bearing PC3-Pim1 and Neo were also treated with BI 2536 i.v. for 6 cycles at a dose of 25 mg/kg twice a week starting from 5.5 weeks after grafting. All samples were processed for H&E and immunostaing with the following antibodies; phospho-histone H3 (Upstate, #06-570), activated caspase 3 (Cell Signaling, #9661), cleaved PARP (Cell Signaling, #9541), MYC (Abcam, #ab32072).
Results
siRNA screening identifies genes required for the viability of Pim1-ovexpressing prostate cells
To identify genes selectively required for the viability of Pim1-expressing cells, we used RWPE-Pim1 cells. These cells stably overexpress Pim1 at expression levels previously determined to be within the range seen in human prostate tumors (23). With additional passaging, RWPE-Pim1 cells become polyploid and tumorigenic (21, 24). We used early passage, diploid, and non-tumorigenic RWPE-Pim1 and late passage, polyploid, and tumorigenic RWPE-Pim1 cells in our screen to capture potential differences in genetic vulnerability between the diploid and polyploid cells. We screened a panel of siRNAs targeting 111 cell cycle, 318 apoptosis, 87 serine-threonine kinase and 54 tyrosine kinase genes in both cell lines. Each gene was targeted by four different siRNAs arrayed in a 96-plate format that facilitates reverse transfection. Cellular viability was determined after 72 hrs of reverse transfection by using the CellTiter-Glo Luminescent cell viability assay and the results were normalized to RISC-free siRNA as a control. We did not observe any consistent differences in viability between the diploid and the polyploid RWPE1-Pim1 cells upon knockdown of any of the genes examined.
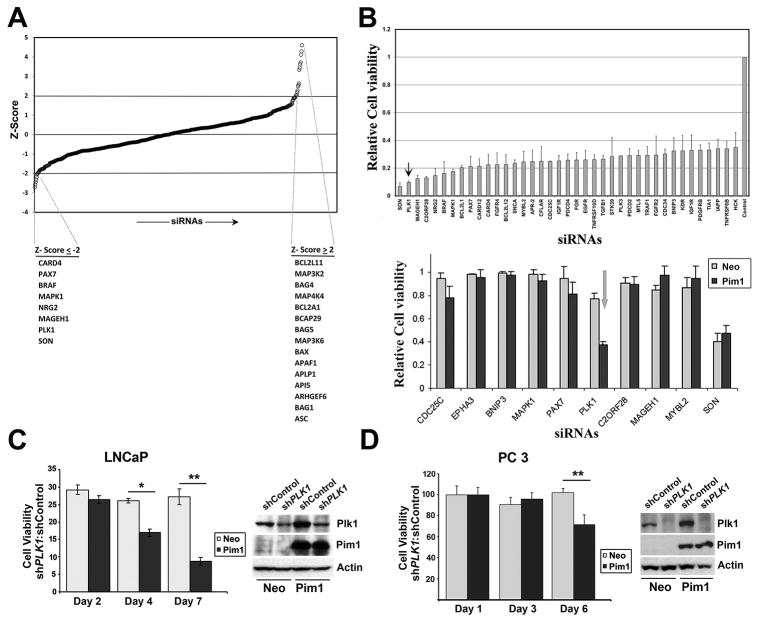
We therefore combined the data from both cell lines to determine the effect of siRNAs on cell viability of RWPE-Pim1 cells regardless of ploidy status. Figure 1A shows a ranking of individual siRNA effects on cell viability relative to control set by Z scores. Notably, the siRNAs that resulted in increased cell viability (Z ≥ 2) in this initial screen are known pro-apoptotic proteins, including BAX, APAF1, APLP1, API5, ARHGEF6, BAG1 and ASC. Conversely, among the genes whose siRNAs reduced viability (Z ≤ −2) are MAPK1 and BRAF, which are part of the growth factor receptor/MAPK signaling pathway. Since RWPE1 cells are grown in media supplemented with mitogens including EGFR and bovine pituitary extract (which contains basic FGF and PDGF), these results might simply reflect the fact that pathways mediated by these growth factors are important for the viability for RWPE1 cells.
Figure 1. siRNA screening identifies genes required for the viability of Pim1-ovexpressing prostate cells.
A, Graph of normalized cell viability in RWPE-Pim1 cells transfected with siRNAs targeting 570 genes (111 cell cycle, 318 apoptosis, 87 serine-threonine kinase and 54 tyrosine kinase genes) by Z score. The top hits that decreased (Z ≤ −2) or increased (Z ≥ 2) cell viability are indicated.
B (top), Graph of genes whose knockdown led to less than 30 % cell viability in RWPE1-Pim1 cells; (bottom) Relative cell viability of custom ordered 10 genes in RWPE1-Pim1 and control RWPE-Neo cells. Note the Pim1- specific effect of PLK1 knockdown.
C (left), Relative cell viability of PLK1 knockdowned LNCaP cells. Cell viability of shPLK1 cells was normalized with that of shControl cells; (right) Western blot for the indicated proteins in PLK1 knockdowned cells by shRNA in LNCaP are shown. shPLK1; shRNA targeting PLK1, shControl: control shRNA.
D (left), Relative cell viability of PLK1 knockdowned PC3 cells. Cell viability of shPLK1 cells was normalized with that of shControl cells; (right) Western blot for the indicated proteins in PLK1 knockdowned cells by shRNA in PC3 cells are shown.
The most significant reduction in cell viability was seen for siRNAs targeting SON, a DNA binding protein (also known as Bax antagonist selected in Saccharomyces 1) and PLK1 (Fig. 1B). SON is a Ser/Arg (SR)-related protein that functions as a splicing cofactor. Depletion of SON has been shown to result in severe impairment of proper spindle pole and microtubule function leading to genomic instability (26). Interestingly, these molecular defects result from inefficient RNA splicing of a specific set of cell cycle-related genes that possess weak splice sites in SON-depleted cells (26). Inhibition of PLK1 has been shown to inhibit proliferation of prostate cancer cells preferentially compared to non-transformed prostate epithelial cells (27).
To determine whether any of these siRNAs selectively impairs the viability of Pim1-overexpressing cells, we performed a secondary screen including 10 genes in RWPE-Neo and RWPE-Pim1 cells. As shown in Figure 1B (bottom), in this assay, SON depletion affected RWPE-Neo and RWPE-Pim1 viability equally, while PLK1 siRNA showed a selective effect on RWPE-Pim1 cells.
To confirm and extend this observation to additional cell lines, we decided to knockdown PLK1 in other prostate tumor cell lines using shRNA targeting PLK1 (shPLK1). We first examined PLK1 levels and found that PLK1 is robustly expressed in all three prostate cancer cell lines tested (DU145, PC3 and LNCaP) but not in the non-transformed cell line NHPrE (Supplementary Fig. S1A). We established stable PLK1 knockdown cells by shRNA in LNCaP and PC3 cells with and without Pim1 overexpression (Fig. 1D). Cell lines with severe PLK1 depletion were not viable, thus cells with modest amounts of PLK1 knockdown were selected and used in this study. Analysis of cell viability of these cells indicates that Pim1-overexpressing LNCaP and PC3 cells are more sensitive to the inhibitory effect of PLK1 knockdown than control Neo cells (Fig. 1D). To examine whether Pim1 affected PLK1 expression levels, Pim1 overexpressing cells as well as control Neo cells were synchronized in mitotic phase using Nocodazole and PLK1 expression levels were examined. Results showed that there was no difference in PLK1 expression levels between Pim1 overexpressing cells and control Neo cells, indicating that increased sensitivity of Pim1-overexpressing cells to PLK1 knockdown is not due to Pim1’s ability to affect PLK1 expression levels (Supplementary Fig. S1C).
PIM1 and PLK1 interact and co-localize in the centrosome and midbody
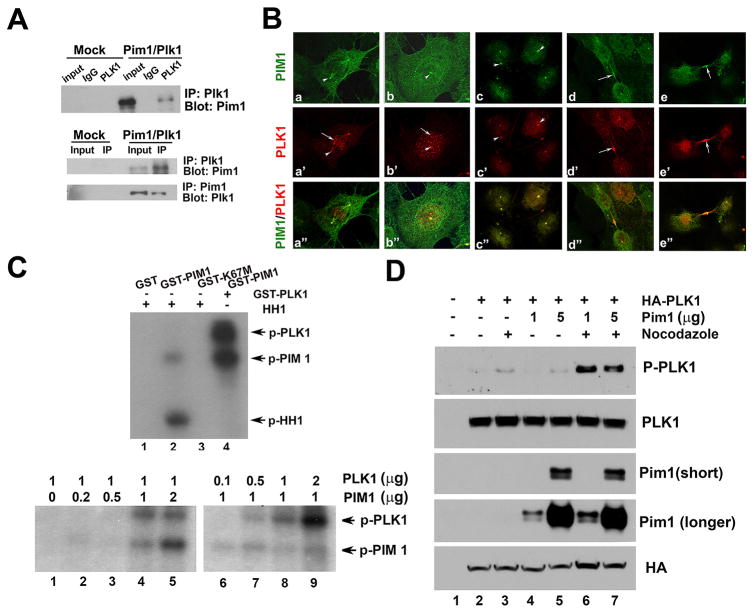
To explore possible mechanisms for the observed dependency of Pim1-overexpressing cells on PLK1, we examined for a possible interaction between Pim1 and PLK1. For this, Pim1 and PLK1 were co-transfected into 293T cells and samples were processed for co-immunoprecipitation using either Pim1 or PLK1 antibodies. Our result indicated that Pim1 and PLK1 do interact (Fig. 2A). Next, immunofluorescence experiments were performed to determine the co-localization of PIM1 and PLK1. PLK1 was detected in centrosome, kinetochore and midbody during mitosis (Fig. 2B), consistent with its multiple mitotic functions as reported (28). Interestingly, we detected co-localization of PIM1 and PLK1 in the centrosome and midbody, but not in the kinetochore (Fig. 2B).
Figure 2. PIM1 and PLK1 interact and co-localize in the centrosome and midbody.
A, PIM1 and PLK1 interaction in 293T cells. cDNA of PIM1 and PLK1 were transfected into 293T cells, immunoprecipitated by Pim1 or Plk1 antibodies and probed with reciprocal antibodies.
B, Immunofluorescence images showing co-localization of endogenous PIM1 and PLK1 at centrosome (a–c) and midbody (d–e). a–c: PIM1 and PLK1 are co-localized at centrosomes (arrowheads), but not at kinetochores (arrows). d–e: midbodies are shown by arrows.
C (left), Kinase assay showing phosphorylation of PLK1 and PIM1 upon incubation of wild type recombinant PIM1 with PLK1. GST-PIM1 but not GST-K67M or GST phosphorylates histone H1 (lanes 1–3). Phosphorylated PLK1 and PIM1 were detected in sample which contains both PIM1 and PLK1 (lane 4); (right) Kinase assay indicating that PIM1 phosphorylates PLK1. For this, different amount of PIM1 or PLK1 were mixed with fixed amount of PLK1 or PIM1 with 32P ATP in kinase buffer.
D, Pim1 phosphorylates PLK1. HA-PLK1 and Pim1 were transfected into 293 T cells, cells were treated with 100 nM nocozadole for 12 hr to induce mitotic phase and samples were processed for immnoblotting. Notice strong phosphorylation of PLK1 (Thr 210) in Pim1 transfected cells after nocodazole treatment (lane 7 & 8).
Since both Pim1 and PLK1 are serine-threonine kinases, we next examined if one can phosphorylate the other. We tested the activity of PIM1 by incubating it with histone H1, a known Pim1 substrate. Wild type recombinant PIM1 was active as shown by PIM1 autophosphorylation and by phosphorylation of histone H1, but histone H1 was not phosphorylated by kinase dead mutant form of PIM1, K67M or GST only recombinant protein (Fig. 2C, left, lane 1–3). When we mixed wild type PIM1 with PLK1, phosphorylated forms of both PIM1 and PLK1 were detected (Fig. 2C, left, lane 4). To distinguish which molecule phosphorylates which molecule, we performed a kinase assay with fixed amounts of PLK1 and increasing amounts of PIM1. PLK1 phosphorylation was not observed in the absence of PIM1 but became apparent with increasing amounts of PIM1 (Fig. 2C, right, lanes 1–5). Incubation of a fixed amount of PIM1 with increasing amounts of PLK1 revealed a dose-dependent increase in phosphorylated PLK1 but not in phosphorylated PIM1 (Fig. 2C, right, lanes 6–9). PIM1 is known to autophosphorylate itself and since the phosphorylated PIM1 band intensity remained constant with increasing amounts of PLK1 (Fig. 2C, right, lanes 6–9), these data indicate that PIM1 is not phosphorylated by PLK1 under these condition. Overall, these results indicate that PLK1 is a PIM1 substrate. To further investigate phosphorylation of PLK1 by Pim1, we overexpressed PLK1 and Pim1 and then examined phosphorylation of PLK1 using phospho-specific antibody (p-Thr210). Specifically, HA-PLK1 and Pim1 were transfected into 293 T cells and cells were treated with nocozadole to induce mitotic phase since PLK1 is reported to be phosphorylated in vivo in mitosis (29). Phosphorylation of PLK1 was increased after nocodozole treatment without Pim1 overexpression as expected (Fig. 2D, lane 2 & 3) and Pim1 overexpression further increased phosphorylation of PLK1 after nocodazole treatment (Fig. 2D, lane 6 & 7). This data strongly indicates that Pim1 phosphorylates PLK1 at threonine 210, a site previously reported to be phosphorylated by aurora A kinase during mitosis (30).
Targeting PLK1 inhibits tumor progression in Pim1 overexpressing prostate tumors
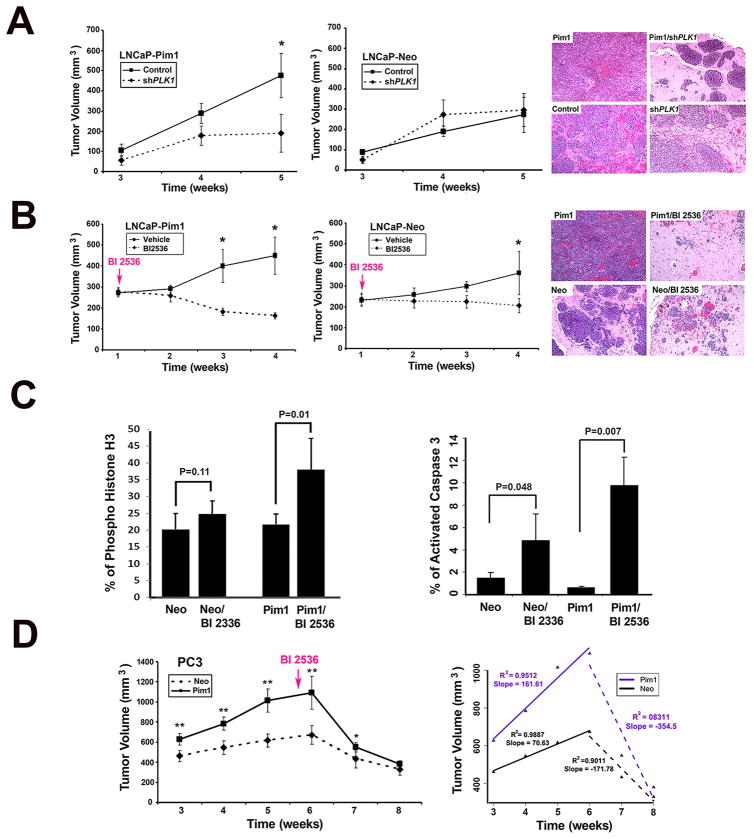
We next sought to determine if PLK1 depletion will impair the in vivo tumorigenicity of Pim1-overexpressing cells. We generated xenograft tumors using LNCaP-Pim1 cells or LNCaP-Neo cells with and without stable shPLK1 expression (Fig. 3A). As noted earlier, we selected cells with modest reduction in PLK1 because cells with drastic knockdown of PLK1 were not viable. In this assay, we found that Pim1-overexpressing LNCaP cells with PLK1 knockdown (LNCaP-Pim1/shPLK1 cells) formed significantly smaller tumors than the LNCaP-Pim1 with control shRNA (LNCaP-Pim1/Control cells) (Fig. 3A, left and right). By contrast, no differences were observed between LNCaP-Neo/Control and LNCaP-Neo/shPLK1 cells (Fig. 3A, middle and right).
Figure 3. Targeting PLK1 inhibits tumor progression in Pim1 overexpressing prostate tumors.
A (left), shPLK1 and shControl LNCaP-Pim1/Neo cells were injected into flanks of nude mice and tumor volumes measured over time. Mean tumor volume ± SD are shown. N = 10 per group; (right) H&E images of representative grafts from each group. Magnification, x 20.
B (left), LNCaP-Pim1/ Neo cells were grafted subcutaneously onto nude mice and one week later BI 2536 were injected intravenously. Tumor sizes were measured once a week. Mean tumor volume ± SD are shown. N= 10 per group. Tumor volume was dramatically reduced in BI 2536-treated Pim1 cells; (right) Representative H&E images of each group. Notice massive tumor in Pim1 cells and dramatic reduction of tumors in BI 2536-treated Pim1 cells (Pim1/BI 2536). Neo cells also formed tumor and BI 2536 treatment led to the reduction of tumor, but Pim1 cells were more sensitive to BI 2536 treatment. Magnification, x 20.
C (left), The mitotic index as determined by quantitation of phospho-histone H3 positive cells in BI 2536-treated LNCaP-Neo/Pim1 cells; (right) Quantitation of active caspase 3 positive cells in each group. N = 4 (150–200 cells for each) per group.
D (left), Nude mice bearing PC3-Pim1 and Neo were treated with BI 2536 intravenously for 6 cycles starting from 5.5 weeks after grafting. N =10 per group. Mean tumor volume ± SD are shown; (right) Mean tumor volume of PC3-Pim1 and Neo cells with slope showing kinetics of tumor size before and after BI 2536 treatment. N = 10 per group. *, P < 0.05; **, P < 0.01.
To confirm this finding, we employed the PLK1 specific small molecule inhibitor, BI 2536. In the first set of experiments, BI 2536 was given early during tumor development (one week post-grafting). We found the dramatic inhibition of LNCaP-Pim1 tumor growth by BI 2536 treatment (Fig. 3B, left and right). In contrast, control LNCaP-Neo tumors treated with BI 2536 demonstrated the modest inhibition of tumor growth (Fig. 3B, middle and right). PLK1 inhibition is reported to arrest cells in mitotic phase (M phase) followed by apoptosis (28). Accordingly, BI 2536-treated LNCaP tumors showed evidence of arrest in M phase as determined by histone H3 phosphorylation (Fig. 3C, left and supplementary Fig. S2A) as well as apoptosis determined by active caspase 3 staining (Fig. 3C, right and supplementary Fig. S2B). Interestingly, LNCaP-Pim1 tumors treated with BI 2536 showed much higher rates of M phase arrest and apoptosis compared to BI 2536-treated control LNCaP-Neo tumors.
Next, we examined the efficacy of BI 2536 treatment in established tumors. For this experiment, we used the more aggressive PC3 cell line. BI 2536 was administered after tumors have developed (5.5 weeks post-grafting). The results showed that the PC3-Pim1 tumors regressed at a faster rate than the PC3-Neo tumors (Fig. 3D, left). Calculation of the slopes of tumor growth and regression showed that PC3-Pim1 tumors regressed at twice the rate of PC3-Neo tumors upon BI 2536 treatment (slope 354.5 vs 171.8) (Fig 3D, right).
Pim1-overexpressign cells are hypersensitive to the molecular effects of PLK1 inhibition
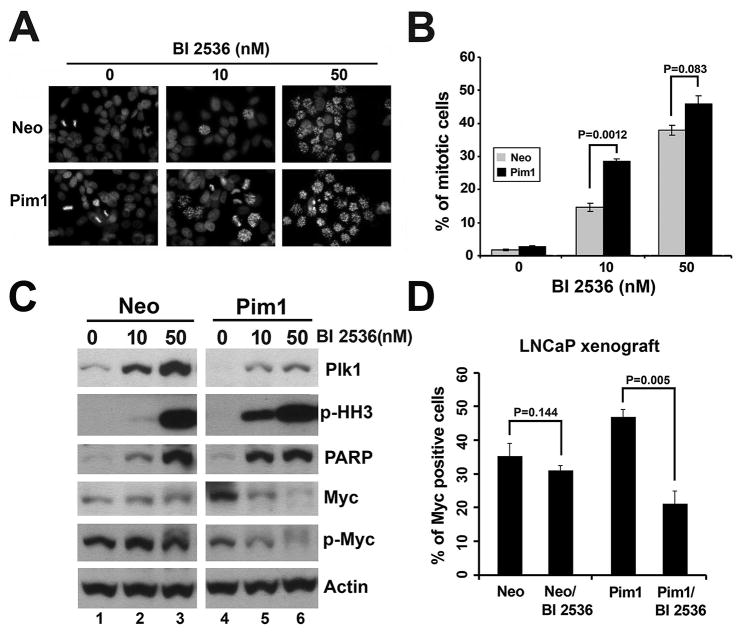
To further understand the underlying mechanism responsible for the increased sensitivity of Pim1-expressing cells to PLK1 inhibition, we first checked expression levels of PLK1 in Neo and Pim1 cells. Since our data showed that Pim1 does not consistently modulate PLK1 expression levels even in nocodazole synchronized cells (Supplementary Fig. S1C), we then treated LNCaP-Neo and-Pim1 cells with different dose of BI 2536 to find the differential effects of Neo and Pim1 cells to PLK1 inhibition. Results showed that there are more mitotic arrested cells in Pim1 cells than in Neo cells as shown by phospho-specific histone H3 (p-HH3) immunofluorescence (Figs. 4A and B) and by western blotting of p-HH3 (Fig. 4C). The difference is more obvious in lower dose of BI 2536, most likely due to saturation effects of BI 2536 in higher dose. In addition to the increased p-HH3, the apoptotic marker PARP, another characteristic of PLK1 inhibition, also increased more in BI 2536 treated Pim1-expressing cells than control cells, especially at a lower dose (Fig. 4C). These data suggest that Pim1 cells are more sensitive to the inhibitory effects of BI 2536 as shown by increased mitotic arrest and apoptosis.
Figure 4. Pim1-overexpressign cells are hypersensitive to the molecular effects of PLK1 inhibition.
A, Immunofluorescence of phospho-specific histone H3 in BI 2536-treated LNCaP-Neo/Pim1 cells.
B, Quantitation of phospho-specific histone H3 positive cells in ‘A’. There are more mitotic arrested cells in Pim1 cells than Neo cells upon BI 2536 treatment.
C, Western blotting showing more mitotic arrested (shown by phospho-histone H3) and apoptotic (shown by PARP) cells as well as lower MYC levels in LNCaP-Pim1 cells after BI 2536 treatment compared with LNCaP-Neo cells. The difference is obvious at lower dose of BI 2536 (10 nM). Both total MYC expression and phosphor-MYC levels were lower in Pim1 cells than Neo cells after BI 2536 treatment.
D, Quantitation of MYC positive cells in BI 2536-treated LNCaP-Neo/Pim1 xenograft cells. N = 5 (150–200 cells for each) per group.
A recent study suggested a link between PLK1 and the regulation of MYC protein levels in the G2 phase of the cell cycle (31). The ubiquitin ligase β-TrCP was found to ubiquitylate MYC, leading to increased MYC stability. Phosphorylation of MYC by PLK1 increases its association with β-TrCP, thereby enhancing MYC stability. Thus PLK1 inhibition might be expected to lead to a reduction in MYC protein levels. We tested this idea by examining MYC protein levels in cells following PLK1 inhibition by BI 2536. Interestingly, MYC levels were much lower in Pim1 cells than in control Neo cells after BI 2536 treatment (Fig. 4C), and this is correlated with phosphorylation of MYC (Fig. 4C). Furthermore, immunostaining showed reduced levels of MYC expression in both LNCaP-shPLK1 cells and BI 2536 treated Pim1 xenografts compared to control Neo cells (Fig. 4D and supplementary Figs. S2C and D). MYC expression levels were also lower in BI 2536-treated PC3-Pim1 cells compared to control Neo cells (Supplementary Fig. S3B). Decreased MYC expression levels were also observed in parental LNCaP, DU145 and PC3 cells after BI 2536 treatment (Supplementary Fig. S3C). The reduced MYC expression seen in cells in which PLK1 is inhibited is particularly interesting in light of the known cooperativity between MYC and PIM1. Together our results suggest that PIM1-expressing cells are hypersensitive to PLK1 inhibition most likely due to increased mitotic arrest followed by apoptosis as well as reduced MYC protein levels upon PLK1 inhibition.
Co-expression of PLK1 and PIM1 in human prostate tumors
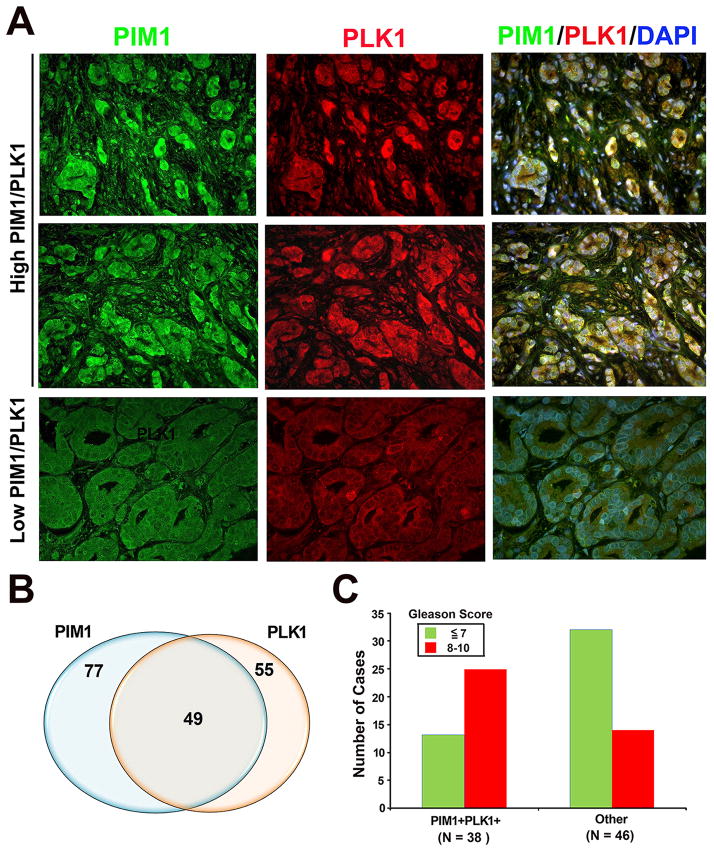
Based on the data presented so far, we decided to examine whether there is a relationship between PIM1 and PLK1 in human prostate tumor tissue microarrays. Expression levels of PIM1 or PLK1 were much higher in tumor samples compared with normal prostate tissues, and they were frequently co-localized (Fig. 5A). Out of 162 specimens examined, PIM1 staining was observed in 77 (48%) cases and PLK1 staining was present in 55 (34 %) cases (Fig. 5B). There was considerable overlap between samples that overexpress both PLK1 and PIM1; 49 cases (30. 2%) (Fig. 5B). In addition, co-expression of PLK1 and PIM1 was significantly correlated to higher Gleason scores. The majority (66.8%) of PLK1/PIM1 high expressing samples are of high Gleason score 8–10, while the majority (69.6%) of the PLK1/PIM1 low expressing samples are of low Gleason score less than 7 (p = 0.0012; Chi-square test) (Fig. 5C). These results support the idea that PLK1 and PIM1 are frequently co-expressed in human prostate tumors.
Figure 5. Co-expression of PLK1 and PIM1 in human prostate tumors.
A, Representative images showing high expression levels of PIM1 and PLK1 as well as co-localization in human prostate tumors. Normal prostate glands show low expression levels of both PIM1 and PLK1.
B, Venn diagram showing a significant percentage of human prostate tumor samples co-expressing PIM1 and PLK1. N = 162.
C, Co-expression of PLK1 and PIM1 was significantly correlated to higher Gleason scores (P = 0.00012, Chi-square). Human microarray tissues were immunostained with both PIM1 and PLK1 antibodies.
Discussion
In these studies, we used a focused RNAi screen to identify PLK1 as a target whose inhibition impairs the viability of PIM1-expressing prostate tumor cells. PIM1 is overexpressed in numerous solid tumors including prostate cancers (1, 2, 32) and has been known to play significant roles in tumorigenesis and chemoresistance in various cancers cells (33–36). As such, a number of PIM1 inhibitors have been developed (37), however most of these have not been tested yet either in in vitro or in vivo. In this study, we employed an alternative strategy to target PIM1; we reasoned that the molecular changes induced by overexpression of PIM1 might render cancer cells particularly sensitive to the knockdown of certain genes. Our studies led to the finding that PIM1-expressing cells are particularly sensitive to PLK1 inhibition.
PLK1 is overexpressed in a wide variety of malignancies including prostate cancer and its expression frequently correlates with poor patient prognosis (16, 28, 38, 39). It plays a key role in cell division and its activity is elevated in cells with a high mitotic index including cancer cells (27, 28). Notably, a genome-wide RNAi screens have identified PLK1 as the kinase selectively required for the viability of activated Ras (14). All these data coupled with the unique structure of PLK1 have made PLK1 an attractive anti-cancer drug target. Several inhibitors targeting PLK1 have been developed so far and they have been under investigation in multiple clinical trials (28, 40).
In our study, we chose to use PLK1-specific, small molecule inhibitor, BI 2536. BI 2536 is an ATP-competitive PLK1 inhibitor identified through high-throughput screening. BI 2536 showed high efficacy in vivo at well-tolerated doses and caused tumor regression in several xenograft models (19). Several clinical trials that include hormone-refractory prostate cancer have also revealed that BI 2536 exhibits some anti-tumor activity in patients (41, 42). A clinical trial with BI 2536 as a single agent administered to prostate cancer patients showed some sign of antitumor activity measured by prostate specific antigen (PSA) (43). However, this study was performed on a small scale in patients with undefined genetic backgrounds. It is well known that not all patients respond to the same drugs and the extent of PSA decline and measurable tumor regression are variable. One explanation for this could be different genetic backgrounds of each individual as well as their different prior treatment options. In this regard, our finding that PIM1 overexpressing prostate cells show better response to PLK1 inhibition is intriguing.
One of the major concerns in drug targeting for cancer therapy is the potential toxicity in normal tissues. Inhibition of PLK1 induces apoptosis and cancer cells seem to be more sensitive to PLK1 inhibition than normal cells (14, 44–46). In this study, we found that even in tumor cell lines such as LNCaP and PC3 cells, there exists differential sensitivity to PLK1 inhibition because Pim1 overexpressing LNCaP or PC3 cells, which are more tumorigenic than their control cells, are much more sensitive to PLK1 inhibitory effects than control cells. Thus, identifying genetic changes such as PIM1 overexpression in individual tumors might be of value in selecting patients to be put on PLK1 inhibitor therapy regimens.
Mitotic arrest and apoptosis after PLK1 inhibition are well documented in previously published literatures (19, 27, 47). In this study, we observed more mitotic arrested cells and apoptotic cells in Pim1 overexpressing cells after PLK1 inhibition than in control cells. Previously, we reported that Pim1 overexpression induces genomic instability characterized by polyploidy, abnormal tubulin and defects in mitotic checkpoint and cytokinesis (23). As mentioned earlier, PLK1 plays key roles in regulating cell cycle-related events such as bipolar spindle formation, centrosome maturation, chromosome segregation, activation of the anaphase-promoting complex/cyclosome (APC/C) and cytokinesis (28). Thus, the mitotic stress Pim1-overexpressing cells are experiencing might be exacerbated when the function of key cell cycle-related molecules such as PLK1 is disturbed. Previous reports that cells with activated Ras or p53 mutation-bearing cancer cells depend more on PLK1 for their viability than their isogenic cells support this idea (14, 45). It is possible that PIM1 and PLK1 have common downstream effectors that are required for cell cycle progression. Our data showing the co-localization of PIM1 and PLK1 in centrosome and midbody are consistent with this notion.
A possible common effector for both PIM1 and PLK1 is MYC. PIM1 interacts with MYC and increases its transcriptional activity (48). On the other hand, PLK1 stabilizes MYC as BI 2536 treatment decreases MYC levels in cells in culture (31). We observed reduced MYC expression in Pim1 xenografts and cells after BI 2536 treatment (Fig. 4D and supplementary Figs. S2C and D). Thus one of the potential mechanisms by which inhibition of PLK1 impairs the tumorigenicity of PIM1-expressing cells is by de-stabilizing MYC.
In summary, this study identified PLK1 as a synthetic partner of PIM1 and provides a rationale for the potential clinical utility of PLK1 inhibition in PIM1-overexpressing prostate cancer. Our study represents an attempt to assess the impact of PLK1 inhibition in genetically defined tumor model systems with Pim1 overexpression. It would be interesting to test PLK1 inhibition in additional model systems that faithfully reflect human prostate cancer.
Supplementary Material
Translational Relevance.
PIM1 kinase is overexpressed in many tumor types including lymphomas and prostate cancer, where it is known to cooperate with the MYC oncogene in promoting tumorigenicity. PIM1 has emerged as an attractive target for drug discovery but few PIM1 inhibitors have been tested in vitro or in vivo. In this regard, knowledge of the vulnerabilities of PIM1-overexpressing tumor cells will be of great value in efforts to develop novel anti-cancer therapeutics. In this study, we show that PLK1 inhibition is particularly effective against PIM1-overexpressing prostate tumors, possibly due to interaction between PIM1 and PLK1. Furthermore, PIM1 and PLK1 are frequently co-expressed in human prostate tumors. These data suggest that targeting PLK1 could be exploited for therapeutic purposes specifically in prostate cancer patients with PIM1 overexpression and show the usefulness of RNAi screen-based approach for identification of tumor biomarkers as well as therapeutic targets.
Acknowledgments
Grant Support
This study was supported by the Department Of Defense (grant no. W81XWH-10-1-0246) and the National Cancer Institute (grant no. CA123484).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Contributions
Conception and design: M. Roh, S. Abdulkadir
Development of methodology: M. Roh, S. Abdulkadir
Acquisition of data (provided animals, provided facilities, etc.): M. Roh, R. Van Der Meer
Analysis and interpretation of data (e.g., statistical analysis, biostatistics): M. Roh
Writing, review, and/or revision of the manuscript: M. Roh
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Roh
Study supervision: M. Roh
References
- 1.Magnuson NS, Wang Z, Ding G, Reeves R. Why target PIM1 for cancer diagnosis and treatment? Future Oncol. 2010;6:1461–78. doi: 10.2217/fon.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11:23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 3.Mochizuki T, Kitanaka C, Noguchi K, Muramatsu T, Asai A, Kuchino Y. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999;274:18659–66. doi: 10.1074/jbc.274.26.18659. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya N, Wang Z, Davitt C, McKenzie IF, Xing PX, Magnuson NS. Pim-1 associates with protein complexes necessary for mitosis. Chromosoma. 2002;111:80–95. doi: 10.1007/s00412-002-0192-6. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Bhattacharya N, Mixter PF, Wei W, Sedivy J, Magnuson NS. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim Biophys Acta. 2002;1593:45–55. doi: 10.1016/s0167-4889(02)00347-6. [DOI] [PubMed] [Google Scholar]
- 6.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571:43–9. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 7.Bachmann M, Hennemann H, Xing PX, Hoffmann I, Moroy T. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J Biol Chem. 2004;279:48319–28. doi: 10.1074/jbc.M404440200. [DOI] [PubMed] [Google Scholar]
- 8.Bachmann M, Kosan C, Xing PX, Montenarh M, Hoffmann I, Moroy T. The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int J Biochem Cell Biol. 2006;38:430–43. doi: 10.1016/j.biocel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Morishita D, Takami M, Yoshikawa S, Katayama R, Sato S, Kukimoto-Niino M, et al. Cell-permeable carboxyl-terminal p27(Kip1) peptide exhibits anti-tumor activity by inhibiting Pim-1 kinase. J Biol Chem. 2011;286:2681–8. doi: 10.1074/jbc.M109.092452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Kim J, Roh M, Franco OE, Hayward SW, Wills ML, et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010;29:2477–87. doi: 10.1038/onc.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Roh M, Abdulkadir SA. Pim1 promotes human prostate cancer cell tumorigenicity and c-MYC transcriptional activity. BMC Cancer. 2010;10:248. doi: 10.1186/1471-2407-10-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Anderson PD, Luo W, Gius D, Roh M, Abdulkadir SA. Pim1 kinase is required to maintain tumorigenicity in MYC-expressing prostate cancer cells. Oncogene. 2012;31:1794–803. doi: 10.1038/onc.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–96. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weichert W, Schmidt M, Gekeler V, Denkert C, Stephan C, Jung K, et al. Polo-like kinase 1 is overexpressed in prostate cancer and linked to higher tumor grades. Prostate. 2004;60:240–5. doi: 10.1002/pros.20050. [DOI] [PubMed] [Google Scholar]
- 17.Spankuch-Schmitt B, Wolf G, Solbach C, Loibl S, Knecht R, Stegmuller M, et al. Downregulation of human polo-like kinase activity by antisense oligonucleotides induces growth inhibition in cancer cells. Oncogene. 2002;21:3162–71. doi: 10.1038/sj.onc.1205412. [DOI] [PubMed] [Google Scholar]
- 18.Elez R, Piiper A, Kronenberger B, Kock M, Brendel M, Hermann E, et al. Tumor regression by combination antisense therapy against Plk1 and Bcl-2. Oncogene. 2003;22:69–80. doi: 10.1038/sj.onc.1206038. [DOI] [PubMed] [Google Scholar]
- 19.Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–22. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 20.McInnes C, Wyatt MD. PLK1 as an oncology target: current status and future potential. Drug Discov Today. 2011;16:619–25. doi: 10.1016/j.drudis.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Roh M, Franco OE, Hayward SW, van der Meer R, Abdulkadir SA. A role for polyploidy in the tumorigenicity of Pim-1-expressing human prostate and mammary epithelial cells. PLoS ONE. 2008;3:e2572. doi: 10.1371/journal.pone.0002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin H, Bernal A, Amato FA, Pinhasov A, Kauffman J, Brenneman DE, et al. High-throughput siRNA-based functional target validation. Journal of biomolecular screening. 2004;9:286–93. doi: 10.1177/1087057104263533. [DOI] [PubMed] [Google Scholar]
- 23.Roh M, Gary B, Song C, Said-Al-Naief N, Tousson A, Kraft A, et al. Overexpression of the oncogenic kinase Pim-1 leads to genomic instability. Cancer Res. 2003;63:8079–84. [PubMed] [Google Scholar]
- 24.Roh M, Song C, Kim J, Abdulkadir SA. Chromosomal instability induced by Pim-1 is passage-dependent and associated with dysregulation of cyclin B1. J Biol Chem. 2005;280:40568–77. doi: 10.1074/jbc.M509369200. [DOI] [PubMed] [Google Scholar]
- 25.Jiang M, Strand DW, Fernandez S, He Y, Yi Y, Birbach A, et al. Functional remodeling of benign human prostatic tissues in vivo by spontaneously immortalized progenitor and intermediate cells. Stem Cells. 2010;28:344–56. doi: 10.1002/stem.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, et al. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell. 2011;42:185–98. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reagan-Shaw S, Ahmad N. Silencing of polo-like kinase (Plk) 1 via siRNA causes induction of apoptosis and impairment of mitosis machinery in human prostate cancer cells: implications for the treatment of prostate cancer. Faseb J. 2005;19:611–3. doi: 10.1096/fj.04-2910fje. [DOI] [PubMed] [Google Scholar]
- 28.Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–60. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- 29.Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem. 2002;277:44115–20. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- 30.Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–23. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 31.Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12:973–81. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 32.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–6. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 33.Hammerman PS, Fox CJ, Cinalli RM, Xu A, Wagner JD, Lindsten T, et al. Lymphocyte transformation by Pim-2 is dependent on nuclear factor-kappaB activation. Cancer Res. 2004;64:8341–8. doi: 10.1158/0008-5472.CAN-04-2284. [DOI] [PubMed] [Google Scholar]
- 34.Xie Y, Xu K, Dai B, Guo Z, Jiang T, Chen H, et al. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene. 2006;25:70–8. doi: 10.1038/sj.onc.1209058. [DOI] [PubMed] [Google Scholar]
- 35.Xie Y, Xu K, Linn DE, Yang X, Guo Z, Shimelis H, et al. The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. J Biol Chem. 2008;283:3349–56. doi: 10.1074/jbc.M707773200. [DOI] [PubMed] [Google Scholar]
- 36.Walpen T, Kalus I, Schwaller J, Peier MA, Battegay EJ, Humar R. Nuclear PIM1 confers resistance to rapamycin-impaired endothelial proliferation. Biochem Biophys Res Commun. 2012;429:24–30. doi: 10.1016/j.bbrc.2012.10.106. [DOI] [PubMed] [Google Scholar]
- 37.Morwick T. Pim kinase inhibitors: a survey of the patent literature. Expert opinion on therapeutic patents. 2010;20:193–212. doi: 10.1517/13543770903496442. [DOI] [PubMed] [Google Scholar]
- 38.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24:267–76. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 39.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–30. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 40.Murugan RN, Park JE, Kim EH, Shin SY, Cheong C, Lee KS, et al. Plk1-targeted small molecule inhibitors: molecular basis for their potency and specificity. Molecules and cells. 2011;32:209–20. doi: 10.1007/s10059-011-0126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofheinz RD, Al-Batran SE, Hochhaus A, Jager E, Reichardt VL, Fritsch H, et al. An open-label, phase I study of the polo-like kinase-1 inhibitor, BI 2536, in patients with advanced solid tumors. Clin Cancer Res. 2010;16:4666–74. doi: 10.1158/1078-0432.CCR-10-0318. [DOI] [PubMed] [Google Scholar]
- 42.Schoffski P, Awada A, Dumez H, Gil T, Bartholomeus S, Wolter P, et al. A phase I, dose-escalation study of the novel Polo-like kinase inhibitor volasertib (BI 6727) in patients with advanced solid tumours. Eur J Cancer. 2012;48:179–86. doi: 10.1016/j.ejca.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Pandha HSea. An open label phase II trial of BI 2536, a novel Plk1 inhibitor, in patients with metastatic hormone refractory prostate cancer (HRPC) J Clin Oncol. 2008:26. [Google Scholar]
- 44.Liu X, Lei M, Erikson RL. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol. 2006;26:2093–108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sur S, Pagliarini R, Bunz F, Rago C, Diaz LA, Jr, Kinzler KW, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;106:3964–9. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu B, Mahmud H, Maass AH, Yu B, van Gilst WH, de Boer RA, et al. The Plk1 inhibitor BI 2536 temporarily arrests primary cardiac fibroblasts in mitosis and generates aneuploidy in vitro. PLoS ONE. 2010;5:e12963. doi: 10.1371/journal.pone.0012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–15. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 48.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9:932–44. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.