Abstract
Modern high-throughput structural biology laboratories produce vast amounts of raw experimental data. The traditional method of data reduction is very simple—results are summarized in peer-reviewed publications, which are hopefully published in high-impact journals. By their nature, publications include only the most important results derived from experiments that may have been performed over the course of many years. The main content of the published paper is a concise compilation of these data, an interpretation of the experimental results, and a comparison of these results with those obtained by other scientists.
Due to an avalanche of structural biology manuscripts submitted to scientific journals, in many recent cases descriptions of experimental methodology (and sometimes even experimental results) are pushed to supplementary materials that are only published online and sometimes may not be reviewed as thoroughly as the main body of a manuscript. Trouble may arise when experimental results are contradicting the results obtained by other scientists, which requires (in the best case) the reexamination of the original raw data or independent repetition of the experiment according to the published description of the experiment. There are reports that a significant fraction of experiments obtained in academic laboratories cannot be repeated in an industrial environment (Begley CG & Ellis LM, Nature 483(7391):531–3, 2012). This is not an indication of scientific fraud but rather reflects the inadequate description of experiments performed on different equipment and on biological samples that were produced with disparate methods. For that reason the goal of a modern data management system is not only the simple replacement of the laboratory notebook by an electronic one but also the creation of a sophisticated, internally consistent, scalable data management system that will combine data obtained by a variety of experiments performed by various individuals on diverse equipment. All data should be stored in a core database that can be used by custom applications to prepare internal reports, statistics, and perform other functions that are specific to the research that is pursued in a particular laboratory.
This chapter presents a general overview of the methods of data management and analysis used by structural genomics (SG) programs. In addition to a review of the existing literature on the subject, also presented is experience in the development of two SG data management systems, UniTrack and LabDB. The description is targeted to a general audience, as some technical details have been (or will be) published elsewhere. The focus is on “data management,” meaning the process of gathering, organizing, and storing data, but also briefly discussed is “data mining,” the process of analysis ideally leading to an understanding of the data. In other words, data mining is the conversion of data into information. Clearly, effective data management is a precondition for any useful data mining. If done properly, gathering details on millions of experiments on thousands of proteins and making them publicly available for analysis—even after the projects themselves have ended—may turn out to be one of the most important benefits of SG programs.
Keywords: Databases, Data management, Structural biology, LIMS, PSI, CSGID
1 Introduction
1.1 Data in Structural Biology
Both structural genomics consortia and individual structural biology laboratories produce tremendous amounts of data, and having accurate, complete and consistent data is critical for reproducibility of biomedical research [1]. A single trip to a synchrotron for data collection by a productive crystallographic lab can generate hundreds of datasets totaling around 2 TB of raw data [2]. Modern data processing software can reduce, on the fly, a raw set of diffraction images into a single file that contains a description of every diffraction peak: Miller indices, intensity, and experimental uncertainty (sigma). These data are further reduced into one relatively small file that contains scaled and merged diffraction intensities. However, each file has to be associated with a particular sample (protein crystal) and the description of the experiment, which is usually written in the header of the diffraction image. These data are further used for structure determination and/or for function–structure relation studies.
To perform these studies the experimenter needs information about the protein (at a minimum, the protein sequence), crystallization conditions, and, for functional studies, protein production details. If this information is available, the process described above is simple to implement. Data harvesting from structure determination is relatively straightforward. The whole process following the placement of a crystal in the X-ray beam can be entirely controlled and captured by computer.
However, while this is very simple in theory, this simplicity has not yet been translated into practice. Analysis of the Protein Data Bank (PDB) [3, 4] shows that the number of data collection parameters marked as “NULL” in the header information (i.e., the detailed description of the experiment) is still significant [5, 6]. Moreover, data in the header are sometimes self-contradictory, contradictory to the experimental description in the paper citing the structure, or both [7, 8]. In that case, contacting the authors of the deposit and paper may be the only way to resolve the arising problems. Taking into account that only a small fraction, about 13 % [9], of structures determined by high-throughput consortia are converted (reduced) to peer-reviewed papers, the correctness of data uploaded to various databases like TargetTrack [10], TargetDB [11], and data banks like PDB is absolutely critical (see below).
1.2 Large-Scale Initiatives Create New Databases: TargetDB/PepcDB/TargetTrack
Since their inception, many structural genomics efforts have adopted policies that experimental data produced by member consortia should be made available to the community from the moment of target selection. This has been particularly true for the two large initiatives from the National Institutes of Health (NIH): the Protein Structure Initiative (PSI) established in 2000 by the National Institute of General Medical Sciences (NIGMS) and the SG centers focusing on infectious diseases established in 2007 by the National Institute of Allergy and Infectious Diseases (NIAID). Even some partially privately funded SG efforts like the Structural Genomics Consortium (SGC) have established policies to release some experimental data to the general public [12] (typically only after the structure is determined and deposited). In the specific case of the centers funded by NIGMS and NIAID, the NIH established the target registration database, TargetDB [11], and required that all member consortia deposit data on the progress of their targets. Subsequently many other SG centers world-wide have deposited some of their experimental data as well.
Initially, the main purpose of TargetDB was the prevention of duplication of effort between different SG centers and maximization of the structural coverage of the protein fold space. The scope of the data was very modest. It included protein identification information (sequence, organism) and the timeline of changes in experimental status for each target. Status events included target selection, cloning, expression, purification, as well as crystallization, diffraction, determination of crystal structure, and PDB deposition (for targets studied by X-ray crystallography) or obtaining the HSQC spectra, determination of NMR structure, and BMRB/PDB deposition (for targets studied by NMR).
However, even the modest amount of data available in TargetDB permitted interesting analyses of the overall SG structure determination pipeline [13, 14]. In particular, the overall efficiency of the pipeline—the ratio of solved structures to clones—was found to be below 10 % even in the most productive centers. The two steps that contributed most to the failure of a target in the pipeline were production of soluble protein and diffraction-quality crystals. Not surprisingly, the success ratio depended very strongly on the type of protein as well as the methodology used by particular centers. There was not a single overall bottleneck factor. In 2004, TargetDB was extended to the Protein Expression, Purification, and Crystallization Database (PepcDB) [15] which in addition to simple status history included multiple trials, tracking of failed as well as successful experiments, and more detailed descriptions of protocols.
In 2010, PepcDB and TargetDB were merged into a single new database, TargetTrack, part of the new PSI-Structural Biology Knowledge Base (PSI-SBKB) [10, 16]. The new repository extended the definition of a target to include protein–protein complexes and incorporated tracking of biological assays needed in the PSI:Biology phase. As of January 2013, TargetTrack contained data on over 300,000 targets and over 1,000 protocols.
1.3 Diverse Approaches to Data Management in SG Centers
Development of effective data management systems was a necessity for the large-scale SG centers, not only in order to provide the data to the scientific community but also particularly to effectively handle the huge amounts of experimental data, plan experiments, adjust experimental approaches (e.g., choice of cloning vectors, sequence truncation, crystallization conditions, structure determination procedures), and prioritize targets. These needs required gathering far more data than what was being required by TargetTrack.
In general, two levels of data management are needed in high-throughput, high-output structural biology programs: the target tracking level and the experiment tracking level. The target tracking level comprises target selection, overall experimental status of each target, center-wide efficiency statistics, and generation of reports to the public and to other databases such as TargetTrack. Almost all SG centers have a separate target-tracking database, though some functionality (e.g., target selection) can be “offloaded” to other specialized databases. The primary audience for the target-tracking level is everyone interested in a “high-level” view of the data produced by the center: the center’s scientists and administrators as well as members of the scientific community with interest in the targeted proteins. This level is typically not designed for uploading new data or providing all details of individual experiments; these tasks are better handled at the experimental tracking level.
The experimental tracking level comprises the tools used to collect the results of experiments performed in the laboratory. This type of tool is generally known as a “laboratory information management system” or LIMS. LIMSs are typically used day to day by the researchers conducting the experimental work of a laboratory and may be highly customized to the protocols and work flow of a particular laboratory. LIMSs may also provide tools to help design experiments, operate laboratory equipment, semi-automatically harvest data, track the use of resources, etc. As a result, the primary audience for the LIMS is composed of those interested in a “low-level” view of the data, the center researchers themselves. As compared to the target-tracking level, it is not uncommon to use more than one LIMS in a single SG center, as different systems may be used in different laboratories.
It should be noted that splitting the data management system of a typical SG center into two distinct levels, “high-level” target tracking and “low-level” experiment tracking, is somewhat arbitrary. Some data are natural candidates to be kept at the LIMS level only, for example, the location in the freezer where a particular clone is stored or the particular lot of a reagent or a crystallization buffer. Conversely, some data may only apply at the target-tracking level, for example, the number of publications referencing a given protein. In principle, it is possible for a single database and/or data management system to fully implement both levels. However, in practice, it seems that solutions where the two levels are implemented as separate systems/databases appear to be more common, especially for the larger scale projects.
There have been several “top-down” attempts to design a general framework for SG data management systems in the form of data dictionaries [17] or a protein production UML data model [18]. The latter has been implemented by several systems, such as HalX [19] or the Protein Information Management System (PiMS) [20] used by a number of European SG labs. However, most of the SG centers set up data management systems in a more ad hoc, “bottom-up” manner. Initially, some centers attempted to use commercial LIMS, but often these solutions were not flexible enough or even robust enough, and most SG centers developed their own solutions “in-house.” There are exceptions to this rule. For example, the Structural Genomics Consortium uses two commercially available software systems: the Beehive LIMS (Molsoft LLC; http://www.molsoft.com/beehive.html) and Electronic Laboratory Notebook (now iLabber; Contur Software; http://www.contur.com/home/). It should be noted however that unlike many SG consortia, SGC does not deposit the results of its experiments to PepcDB or TargetTrack. Several of the SG-developed data management systems have been described in the literature [21 – 23], but to our knowledge, none of these systems have been fully commercialized.
One comprehensive data SG management system that has gained wider use is Sesame, developed by Zsolt Zolnai at Center for Eukaryotic Structural Genomics (CESG) [22]. It has been adopted by a number of labs and specialized centers.
The data management system for the Joint Center for Structural Genomics (JCSG) was developed by the center’s programming team in parallel with the construction of the physical pipeline. The LIMS part of the system functions as a hub of information, recording all pipeline steps from target selection to deposition. The tracking database uses Oracle as its engine and tracks 424 experimental parameters, organized into 130 tables [24]. The tools and interfaces to the database contain approximately 360,000 lines of code, which illustrates the level of complexity of this and similar systems.
The Northeast Structural Genomics (NESG) consortium’s data management system is organized as a “federated database framework,” comprising a set of distributed, interconnecting databases [21]. The main target-tracking database, SPINE, serves as an analysis system, utilizing data mining and machine learning tools. In particular, decision trees are used for predicting chances for protein solubility, successful purification, and crystallization. These predictions are used in directing targets to X-ray crystallography or NMR studies [14].
The other two large-scale PSI:Biology centers—the Midwest Center for Structural Genomics (MCSG) and the New York Structural Genomics Research Consortium (NYSGRC)—use the data management system developed in the Minor Lab at the University of Virginia. In both cases, the system is based on a collection of customized LIMS in each site laboratory and a central database (UniTrack, described below) that curates and unifies data obtained by various laboratories. In the case of MCSG, several different LIMSs are used in different laboratories, including LabDB, Mnemosyne, and ANL-DB. In NYSGRC, two different instances of LabDB are used. Similar systems are also deployed in the Center for Structural Genomics of Infectious Diseases (CSGID) and the Enzyme Function Initiative (EFI).
2 A Centralized Target Management System: UniTrack
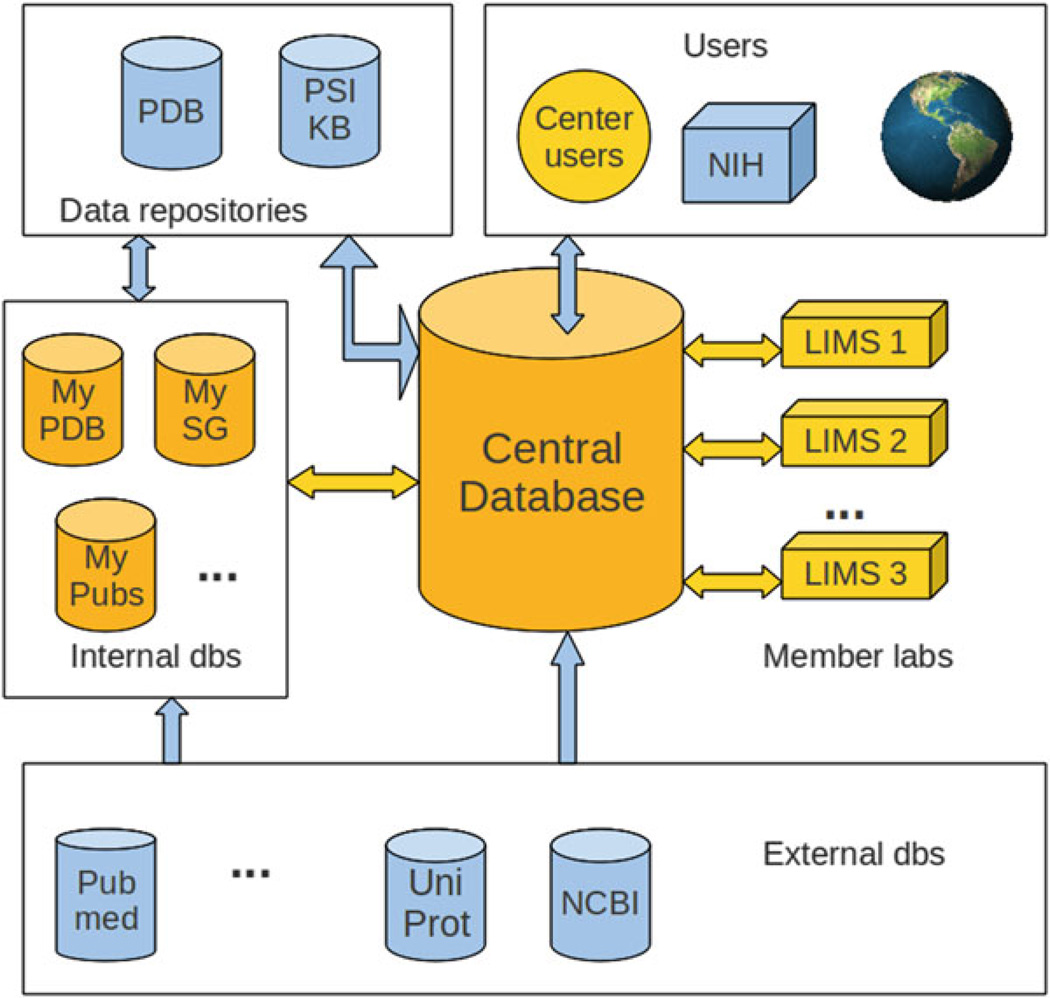
The central, public system comprising the target-tracking level of the SG management system developed by the Minor Lab at the University of Virginia is named UniTrack. As mentioned above, the MCSG, NYSGRC, CSGID, and EFI consortia are all driven by variants of the UniTrack system. The system comprises a core abstraction based on 10 years of experience in SG data management, with a common database architecture and set of tools for managing target and experimental data. Each site is based on the UniTrack core but is then highly customized for the needs of the particular center or consortium of research laboratories. In each case, the UniTrack-derived system comprises the central tracking database and a set of auxiliary databases and applications, which collect and integrate experimental data and are provided by distributed LIMSs deployed in participating laboratories (Fig. 1). Experimental data from different LIMSs are combined and incorporated into UniTrack via a standard protocol. In the most basic case, each LIMS generates XML files in a predefined format, which are parsed by UniTrack tools. An alternative (and more efficient) method, where a LIMS directly communicates with the tracking database, has also been developed. The LIMSs can be very diverse; however, they all must be able to provide the minimum set of required data for cloning, expression, purification, and crystallization experiments.
Fig. 1.
The architecture of the UniTrack data management system. The central database interacts with LIMSs distributed in member labs. A number of auxiliary databases are used to store data from the PDB, data from other SG centers, and SG publications. The central database is responsible for producing reports for external data repositories such as PSI-SBKB. UniTrack databases are synchronized with external data sources such as NCBI GenBank, UniProt, and PubMed via custom scripts. Users interact with the system via a web interface
The experimental pipeline starts with target selection and validation, which is specific for a particular center. The validation process is performed automatically and typically involves checking the accuracy of the amino acid and the nucleotide sequences as well as checking if the selected protein is homologous to proteins with structures in the PDB or to targets selected by other SG centers. Validated targets are inserted into the tracking database. Protein annotations and related data are automatically imported from external databases such as NCBI GenBank [25], Uniprot [26], PDB, and the PSI-SBKB. Depending on the needs of a particular center, between 30 and 80 attributes of any given protein target are stored in UniTrack.
UniTrack keeps a history and the results of the experiments for each target (Fig. 2). About 400 distinct data attributes are used to describe an experimental trial, from the cloning of a target through the determination of its structure. Almost all protein production and crystallization data can be automatically imported from the local LIMS or equipment database. However, smaller labs that do not have a LIMS deployed can still contribute data to UniTrack by entering it manually using the customized interface. Diffraction and structure determination data is currently imported automatically only from the LabDB instances that have the hkldb module enabled [27]. Researchers in other labs upload scaling logs and refinement files manually via the interface.
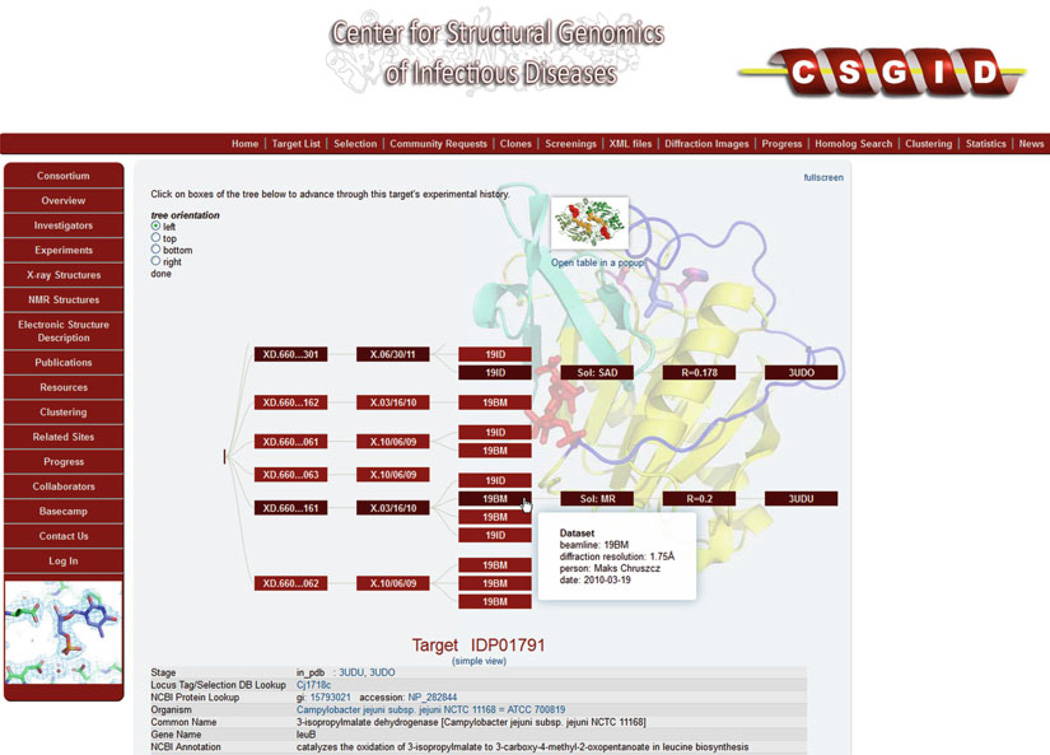
Fig. 2.
Fragment of an experiment tree displayed in the UniTrack-based CSGID interface. Boxes represent particular experiments: purification (P), crystallization drop (XD), crystal harvest (X), data collection (beamline name), structure solution (Sol), refinement (R factor), and PDB deposit (PDB id). Paths in the tree represent trials for a particular sample. The white box that appears when the cursor hovers over an item displays additional details about a particular step. In addition, clicking on any of the boxes display all the data known about this step stored in the database
The tracking database also generates real-time internal reports and statistics as well as the XML files that are being submitted to the TargetTrack repository. In addition, the periodic reports required by various bodies are generated in real time from the database and accessible to the general public. In some sense, all of the portions of UniTrack that generate publicly accessible web pages serve as reports.
The customized instances of UniTrack for each center drive dynamic parts of the centers’ corresponding web portals. The web interfaces are implemented using the Model–View–Controller (MVC) architecture, with separate layers for data retrieval (model), “business logic” (controller), and web page rendering (view). Even with the use of the CakePHP MVC framework (http://cakephp.org) the customized web interfaces for the centers are quite complicated; as an example, the implementation of the CSGID web interface contains over 50,000 lines of source code.
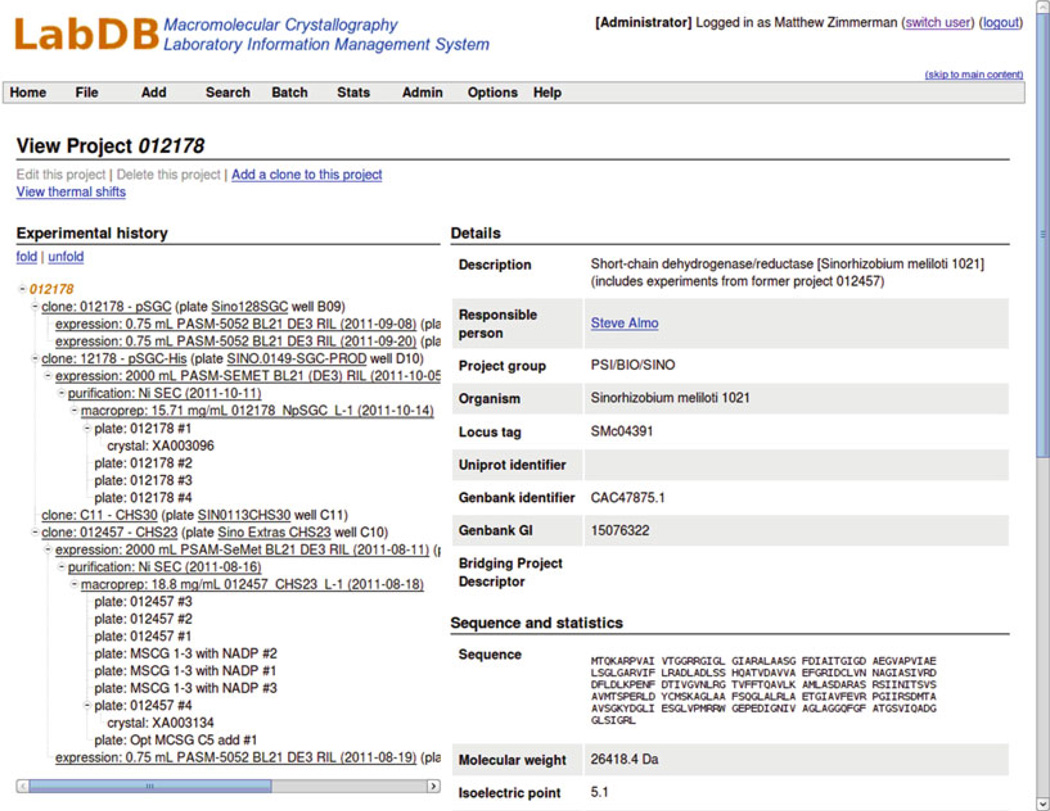
2.1 The LabDB “Super-LIMS”
LabDB is a modular “super-LIMS,” originally developed to track the structure determination pipeline from cloning to structure determination (Fig. 3). The central component of the system is a PostgreSQL database server coupled with a web-based framework, along with two specialized tools: Xtaldb, for designing and tracking crystallization experiments, and hkldb, a module of the HKL-2000/3000 system [27] for incorporating information from crystallographic data collection and structure determination. hkldb and Xtaldb can also be used with stand-alone databases.
Fig. 3.
A typical target overview page in the LabDB LIMS
One of the fundamental design goals of LabDB is to harvest data automatically or semiautomatically from laboratory equipment whenever possible. To that end, the system has modules to import data from a variety of different types of laboratory equipment, including chromatography systems (GE Healthcare AKTA systems), electrophoresis documentation and separation systems (Bio-Rad GelDoc, Caliper LabChip GX), crystallization observation robots (Rigaku Minstrel, Formulatrix Rock Imager), and others. The system provides tools to import data from groups of many similar experiments at once, for example from spreadsheet files, and to track shipments of purified protein and other samples from one laboratory to another.
A good example of how the LabDB system incorporates laboratory hardware to capture data automatically is the reagent tracking module. The system provides a tool to label bottles of chemical reagents with unique barcodes, which are tied to more detailed information about the chemicals in the database. When a researcher prepares a stock solution of a given reagent, he or she first scans the barcode of the reagent bottle before weighing out the chemical. LabDB uses this to track the particular lots and suppliers of chemicals and link them with the details of the stock solutions created (which are then also labeled with unique barcodes). These barcodes allow data to be carried along the pipeline, providing much more detailed information about the origin and history of given stock solutions than would be possible with hand-written labels. Furthermore, as this data is linked to later steps, it is possible to determine which reagent lots were used in successful vs. unsuccessful experiments, especially if complications arise in the replication of experimental results.
Two issues are critical for a LIMS to be widely adopted: the LIMS should facilitate experimental procedure whenever possible, and the system should harvest data accurately and efficiently (i.e., both quickly and easily). Automatic retrieval of data directly from lab equipment such as balances or solution formulation robots, along with efficient collection of experimental design parameters, minimizes manual data entry and facilitates a more complete and more accurate description of the experiment. Using barcode scanners and tablet computers, LabDB performs calculations on the fly based upon the information retrieved via the barcodes, such as calculating the amount of chemical needed to create a particular concentration given various volumes.
The most recent advances in LabDB are in the area of tracking other kinds of biomedical experiments beyond the traditional SG pipeline of clone to structure. These include spectrophotometric kinetic assays, fluorescence-based thermal shift assays, and isothermal titration calorimetry.
2.2 The Expansion of SG into Biomedical Research
The infectious disease centers funded by the NIAID were among the first to expand the traditional SG pipeline into biological and biomedical research. The CSGID and the Seattle Structural Genomics Center for Infectious Disease (SSGCID) are tasked to specifically characterize the structures of proteins with important biological roles in human pathogens, especially those on the NIAID Category A–C priority lists. A particular focus of these centers is screening purified proteins for binding to inhibitors, cofactors, substrates, and analogs. This screening is done both in silico and in vitro via a variety of techniques, including fluorescence-based thermal shift binding, spectrophotometry, isothermal titration calorimetry, and crystallography-based screening. Sometimes the results of computational experiments like model prediction or ligand binding are also included.
At its outset in 2000, the PSI was predominantly focused on developing new technologies and protocols for structure determination and, in its second phase, solving significant numbers of structures in part as an attempt to increase the structural coverage of the “fold space” of proteins [28, 29]. In its third phase, PSI:Biology, the initiative has expanded into large-scale biological and biomedical research. By focusing on targets of biological and medical significance, whether selected by PSI centers or nominated directly by the biological community, PSI:Biology centers can expand their impact by providing not only 3-D protein structures but also techniques for efficient protein production and purification and materials such as cloned expression vectors (made available through material repositories). In some cases, purified protein samples are even supplied directly to other laboratories. The determination of 3-D protein structures, in concert with advanced biomedical research, allows for more complete characterization of many significant proteins and presents the biochemical and biophysical data in the context of structural information. The ultimate goal is the creation of a powerful scientific and intellectual network to study even the most challenging biomedical problems.
The EFI, a U54 “Glue Grant” funded by NIGMS, is another example of the use of SG methods applied to a large-scale biological project. In this program, the traditional SG pipeline of clone to structure is only the first step in a broader program to develop a large-scale, multidisciplinary strategy to assign function to unknown enzymes identified by genome sequencing. Biological experiments performed by the EFI include enzymatic assays, binding assays, mass spectroscopy, metabolomics, and in silico binding studies.
2.3 Data Management Challenges in Collaborative Networks
One cannot overestimate the importance of target selection by the scientific community for such collaborative networks. For PSI:Biology the mechanism is twofold: (a) community members can submit targets through the community nomination target program and (b) the “high-throughput-enabled biology partnerships” supported by PSI:Biology can directly nominate targets relevant to their areas of functional study. These biological partnerships, where consortia of biological researchers from a variety of areas are paired with high-throughput structure determination consortia, focus on particular cellular organelles or protein complexes (such as mitochondrial proteins, nuclear receptors, tight junction membrane proteins) or particular systems (immune function complexes, natural product biosynthesis, cell–cell adhesion, etc.). As of February 2013, PSI programs had about 3,000 community requests and 6,500 targets selected by the high-throughput-enabled biological partnerships.
Collaborative networks provide special challenges in experimental data management, as biological research uses a very broad array of methods, including microscopy, enzymology, biophysical techniques, and whole-cell experiments to address projects of interest. The power of such a network can be dramatically enhanced when large centers provide not only structural information but also pure protein samples to the whole network. The protein samples can then be used for many different in vitro experiments. The importance of the ability to perform a large array of experiments using the same protein sample cannot be overemphasized, as inconsistent experimental results may be caused by the use of different protein samples, e.g., differences in affinity tags, cloning boundaries, and chemical incorporations [30–32].
Similarly, the NIAID centers also accept target nominations from the community. Targets directly requested by community and other “community-interest” targets constitute about a third of all targets for both the CSGID and SSGCID. As of February 2013, CSGID has accepted about 2,000 community targets from over 100 requesters—mostly academic researchers but also pharmaceutical companies such as Novartis and Merck. Close to one-half of all structures solved by the CSGID and about 40 % by SSGCID are community-nominated or community-interest proteins. Community collaborations impose specific demands on SG data management systems. They require establishing effective communication between the community researchers and the center, especially at the stages of selection, cloning, ligand binding, and functional studies. UniTrack contains tools that allow community requesters to monitor the progress of their targets.
In addition, the data management system for SG centers must interact with another component of the collaborative network—the material repositories. The two existing repositories, the PSI:Biology Materials Repository (http://psimr.asu.edu/) [33, 34] and BEI Resources (http://www.beiresources.org) [35], used by the infectious disease SG centers store tens of thousands of protein clones that are available to researchers worldwide. LabDB contains modules assisting the center researchers in tracking shipments of clones to the repositories, while the UniTrack interfaces allow checking the availability of particular constructs.
3 Tracking Biomedical Experiments with SG Data Management Systems
For the traditional structural biology pipeline, the experimental steps required to produce, for example, a structure by X-ray crystallography are well prescribed. A gene of interest is cloned and expressed, protein is purified and set up for crystallization, crystals are harvested, crystallographic data are collected, and the structure is determined (a similar pipeline can be described for structure solution by NMR). Despite differences in protocol, the basic data parameters of each type of experiment are well known. Data parameters comprise both the details of an experimental design and the measurable outcomes of the experiment. For example, design parameters for an expression experiment might include the strain of organism expressed, media used, temperature of expression, etc., and outcome parameters might include the rate and optical density of growth, estimates of expression yield, etc.
Furthermore, the “traditional” process is essentially linear; for each given step in the process, the prior step is a prerequisite. Thus, (1) the types of experiment steps needed (cloning, expression, etc.), (2) the data parameters to be collected at each step, and (3) the order in which steps are performed can all be defined a priori. This has made the design of the data management systems used to track high-throughput structural biology experiments somewhat straightforward. However, the process of target salvage or rescue, which involves returning to prior experimental steps once a target has “stalled” or otherwise failed in the pipeline, does add some complications.
Today, SG centers (and other programs that include high-throughput structural biology as a component) increasingly incorporate into their work flows other types of biomedical experiments spanning many other disciplines: biochemistry, biophysics, microbiology, cell biology, etc. This has raised significant challenges in data management, whether these biomedical experiments are performed in-house or by research partnerships. Unlike the traditional SG data pipeline, the number of different types of experiments that may be performed has expanded dramatically. Each of these experimental procedures differs significantly both in methodology and in parameters that are collected and thus require different types of tools to efficiently capture their data.
Additionally, the ways in which experiments are interrelated are more complex. Biomedical studies are generally not linear (i.e., they cannot be organized into a simple, step-by-step “pipeline”), and many experimental steps can be done in any order. For example, a ligand binding experiment can either be done before or after structure determination; one is not a prerequisite for the other. However, the two experiments can influence one another; the results of a ligand-binding screen can suggest potential soaking experiments, or conversely, unidentified density in a structure can suggest potential binding partners. Given the more complex interrelationships between experiments, the data structure required to track them is much more complicated.
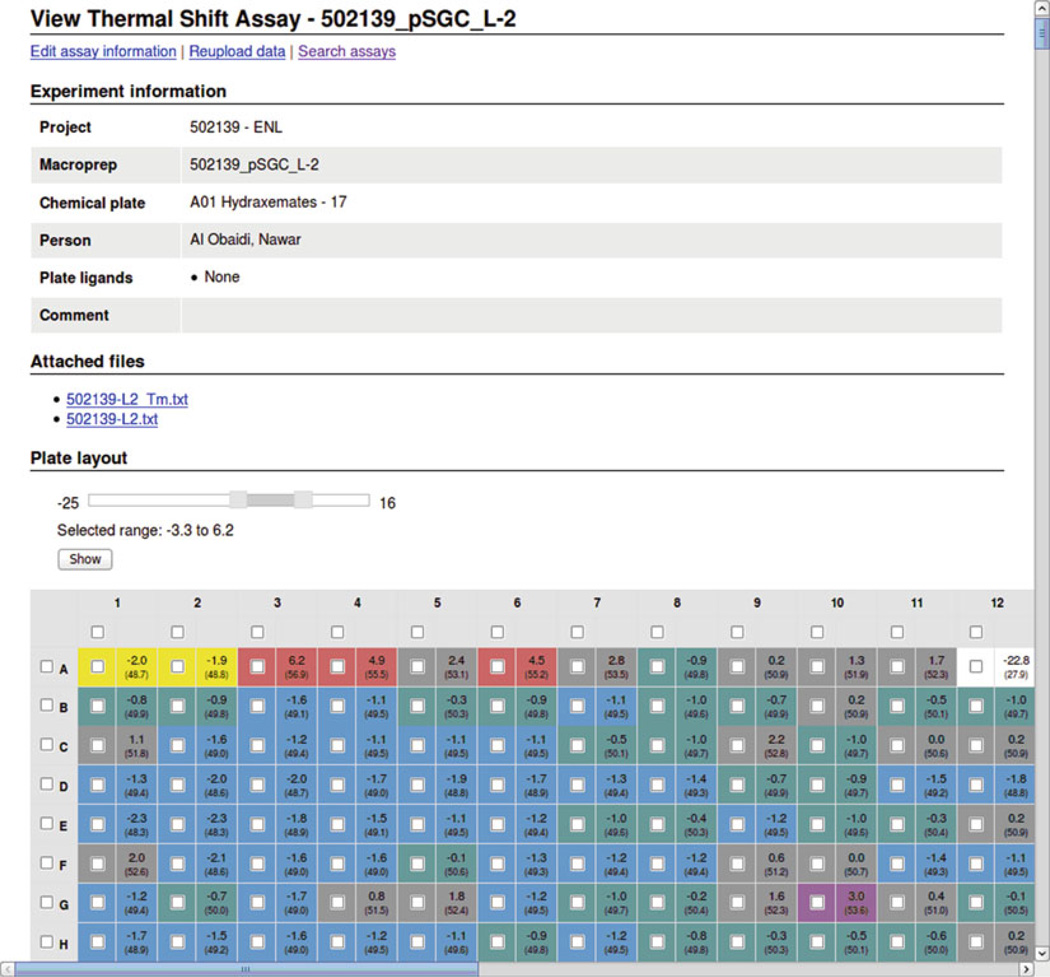
In an ideal world, individual components of a LIMS would be developed to track details of each kind of biological or functional experiment and track the appropriate data. The sheer diversity of techniques used makes this development slow and resource intensive. To some degree, such tools are in development. For example, the LabDB LIMS includes modules for tracking the results of spectrophotometric kinetic assays, fluorescence-based thermal shift assays, and protein and DNA electrophoresis. The Sesame LIMS includes modules for NMR and cryo-EM experiments as well as metabolomics. A key challenge for such LIMSs is that they should be able to automatically import detailed experimental information from laboratory equipment. For example, LabDB automatically parses data files from two different RT-PCR systems used for fluorescence-based thermal shift assays and converts the data into a common format for data comparison and analysis (Fig. 4).
Fig. 4.
The fluorescence-based thermal shift assay module of the LabDB LIMS, showing the graphical representation of the imported experimental data. Data were imported from an Applied Biosystems 7900-HT RT-PCR system
A somewhat complementary approach is to develop a more “generic” LIMS design, which allows the researcher to create a “protocol” describing a type of experiment and then provide data each time the protocol is used. Typically, the data provided for each experiment type is more general—for example, a textual description of the experiment or perhaps the names and values of parameters relevant to the experiment described. The TargetTrack specification allows experimenters to provide data in this format for “biological experiments” or “biophysical assays.” Another example of a LIMS that follows this model is PiMS, where most data input to the system is described in terms of protocols and samples. The advantage of such an approach is in its flexibility. New components of the LIMS are not needed to adapt to the new experimental types. This is at the expense of greater difficulty in data mining due to the relatively unstructured format of data imported into the system.
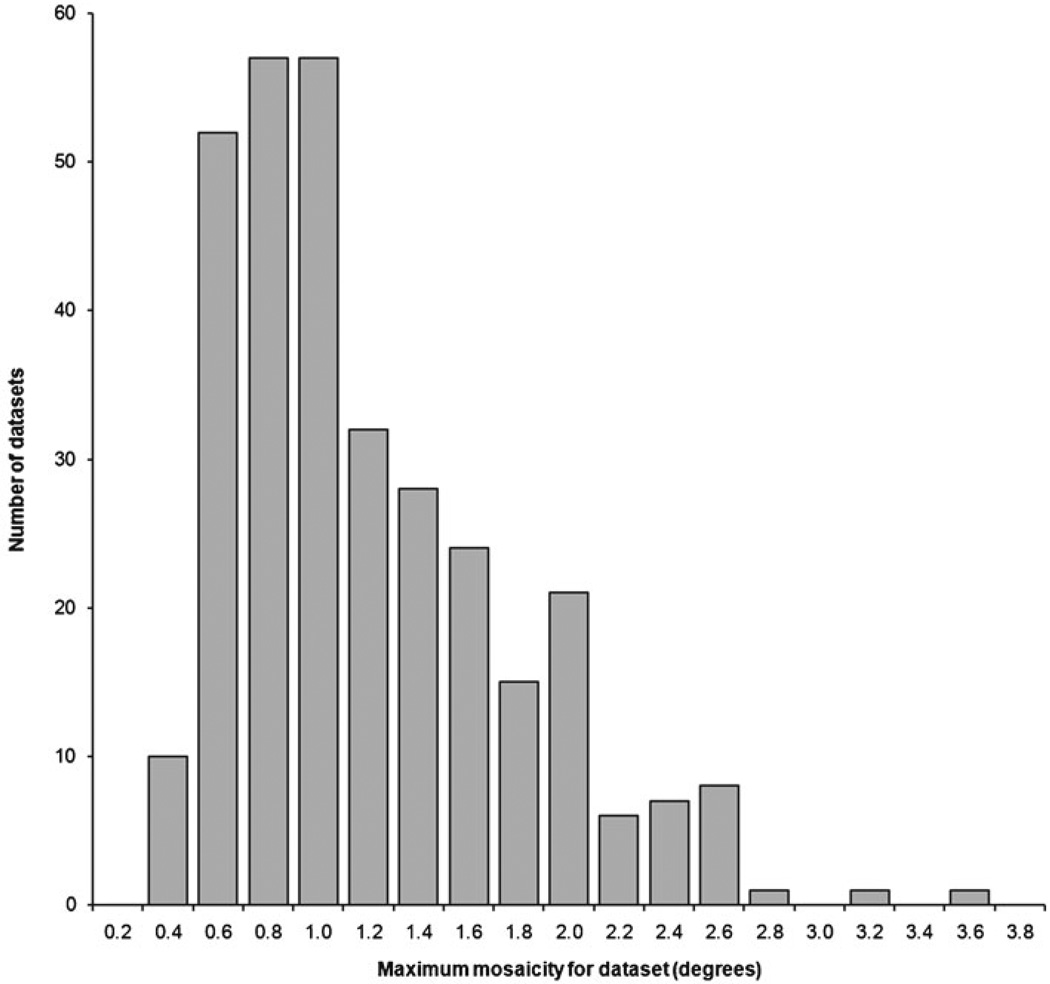
In order for a LIMS to be successful, the system must also provide tools that drive the design of new experiments. This is useful in multiple contexts, whether one is identifying targets for salvage/rescue or providing more immediate feedback while an experiment is still in progress. The tools for this purpose should make use of well-designed data mining mechanisms. For example, the new very-fast-pixel array detectors allow for data collection with narrow oscillation ranges, even below 0.01°. Tests of these detectors with high-quality crystals may show the advantages of using very narrow oscillations. In practice however, the mosaicity of typical macromolecular crystals used today for structure solution (for an example, see the distribution in Fig. 5) limits the advantages of narrow oscillations. For high-mosaicity crystals, experimenters should use larger oscillation ranges such as 0.5° rather than 0.05°. Unfortunately, there are no publicly available databases of experimental conditions used during diffraction experiments, and data collection protocols are based more on anecdotal evidence than on data mining. The large difference in productivity of similar synchrotron beamlines can be associated with differences in experimental protocols that synchrotron users are advised to adopt [36].
Fig. 5.
Histogram showing the distribution of maximum mosaicity value (as fit during integration) of diffraction datasets collected on MCSG targets processed at the University of Virginia, as tracked by the hkldb module of HKL-3000. Only datasets that resulted in both a scaled dataset and an initial model are counted in the distribution
3.1 Data Mining
The types of data mining that can be done with the data collected by SG centers can be divided into two broad categories. The first is real-time (or near-real-time) analyses, which provide not only overall summaries of the status of an experimental pipeline but also additional experimental guidance. The second is more detailed statistical analyses, which require more in-depth transformation and processing of the results.
Typically, real-time analyses can be done through the use of “dashboards” or “scoreboards,” which present a current (or nearly current) view of a particular type of data in a running database. These analyses can include such trivial measures as the overall success rate of a center, the success rate of individual experimental steps for particular labs or for particular organisms, and the mean time between target selection and deposition for various classes of proteins. It can also include some less trivial analyses that can be computed in real time, such as determination of phasing method—single-wavelength anomalous diffraction (SAD), multiple-wavelength anomalous diffraction (MAD), or molecular replacement (MR)—that would maximize the probability of success in the diffraction experiment. For structure validation the analysis of various parameters describing structure quality in the context of best similar structures is very important. There are a number of examples of such dashboards in the interfaces of the “unified” data management systems (Figs. 6 and 7).
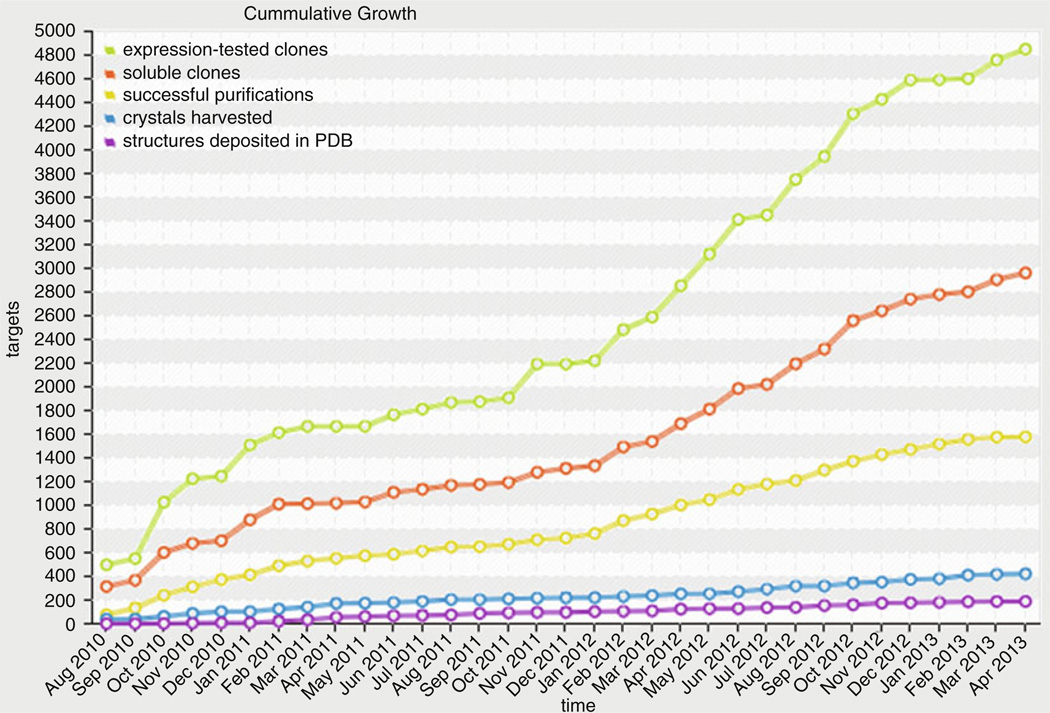
Fig. 6.
Example of a data dashboard: a plot of the cumulative progress for the MCSG center
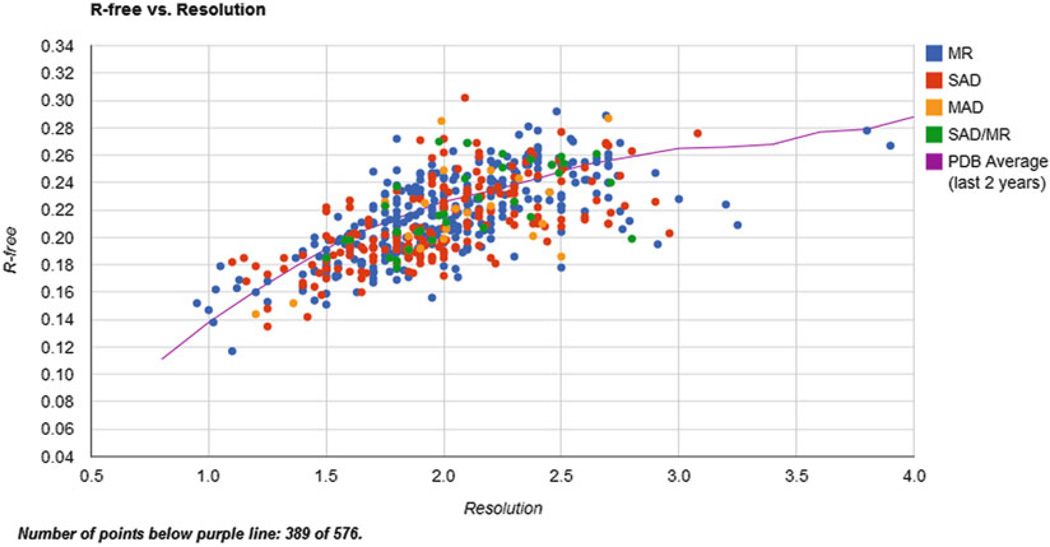
Fig. 7.
Example of a data mining “dashboard”: a plot of Rfree vs. resolution for structures determined by the CSGID
In particular, internal reports tracking the productivity of member labs (which tabulate the number of experimental steps performed at each lab overall as well as within the last 2 weeks or 2 months) have been very useful. These internal reports can aid in the early identification of bottlenecks arising in the experimental pipeline. Of course, this is only possible if the data in the database are current and not “censored” by experimenters. Censorship is defined in this case as an omission of unsuccessful experiments, mainly because the researcher did not see the value of a negative result. Other types of dashboards often used are the scatterplots representing the quality measures for deposited structures (such as R, Rfree, or the Molprobity clashscore vs. resolution; see Fig. 7). These plots can be filtered by various criteria, such as the project, organism, source of crystals, or name of the crystallographer. These reports make apparent which deposits are outliers with respect to the structure quality guidelines established by the NIH. The authors of such deposits are often subsequently asked to re-refine and redeposit them.
By contrast, more detailed analyses often require significant processing of the data, determination of data accuracy and completeness, calculation of statistical measures, etc. and thus require a more detailed (and off-line) processing of experimental data. These types of data mining studies have included in-depth measurements of the properties of peptides most likely to produce crystal structures [14, 37, 38] and the design of new formulations of crystallization screens [39, 40]. Ideally, such data mining studies should produce tools to help researchers design, validate, and optimize their experiments. For example, the Check My Metal server enables improved refinement of metal sites in protein structures [41].
3.2 Making Data and Information Available to the Public
A key goal of many SG programs is to make their results available and useful to the scientific community in forms other than publications or PDB deposits. This objective is addressed in part by the PSI Knowledgebase [10, 16], which provides a centralized web resource for searching SG structures, biological annotations, homology models, and experimental data and protocols. The ultimate purpose of the Knowledgebase is to convert SG data into useful information to be used by the biological community. Some individual centers also developed tools for dissemination of SG results. For example, JCSG developed Topsan [42], which is a wiki-type web resource that creates individual “pages” describing each PDB deposit to which the community can collaboratively add new information. This approach is also used by Proteopedia [43]. The SGC developed iSee interactive 3D presentations of structures solved by the consortium. These are generated using the ICM software developed by Molsoft LLC [44]. The UniTrack-based web portals have the ability to automatically generate a set of interactive 3D presentations for new protein structures using the ICM technology. Interactive content is embedded directly on the pages describing each structure and can be accessed using the freely available ActiveICM plug-in. Each structure presentation is accompanied by a short annotation written by the researcher who solved the structure. This includes a structure description and any potential functional information. Each automatically created presentation can be further expanded and/or highly customized by the annotator. An example of an extended and highly customized presentation can be seen using an ICM-enabled web browser on the CSGID website (http://csgid.org/csgid/deposits/view/3E4F). Within the presentation, users can rotate and manipulate structures to view structural units, ligands, oligomerization states, and B-factor distributions. Additionally, presentations can be downloaded and edited using ICM Browser, Browser Pro, or ICM Pro. ActiveICM is being used for scientific publishing [45] by journals such as PLoS ONE and Nature.
3.3 Unmet Challenges
A data management system is truly successful when the paradigm “data in, information out” is fully satisfied. Despite enormous progress, the major unmet challenge of high-throughput programs including structural genomics is an adequate rate of conversion of data into biomedically useful information, ideally as peer-reviewed papers. This is a general difficulty of modern science; one is swamped in experimental data, and extraction of useful information is quite often a Sisyphean task. Addressing this task effectively requires either very substantial manual labor or implementation of “knowledge-based systems,” with comprehensive tools for efficiently summarizing and mining experimental data, and in some cases implementation of machine learning methods. Ultimately, the only way to check the consistency and accuracy of a database is to examine reports generated by the database for internal and external users. The usefulness for external users, i.e., the scientific community, is the justification for the high costs related to the development and maintenance of databases. The scientific community is not limited to academic users but may also include commercial companies working on new drugs. Reliable information about the relationships between functional and structural data could potentially save millions of dollars in the drug discovery process [1].
Why is the development of data management systems so difficult? There may be no single, definitive answer to that question, but the problem is clearly widespread. The personal experience of one author shows that even a relatively simple database to track an airline’s checked baggage may fail when the baggage is lost and cannot be recovered for a number of days due to inadequate tools for checking data consistency. Similarly, the authors have received e-mails from an airline at (for example) 8:30 p.m. with a new late night departure time but also stating that they should still “be at the gate prior to 4:30 p.m.,” making one wonder if airline database programmers have mastered time travel. Unfortunately for database operators/developers, but fortunately for science, cutting-edge databases used in biomedical sciences appear to operate with fewer failures despite their tremendous complications. Keeping track of very diverse biological experiments performed in multiple labs, as well as tracking the shipments of constructs, proteins, crystals, and data between labs, is a problem of great complexity. In our opinion the main issue faced by data management systems in biological consortia is “creeping entropy,” the accumulation of inconsistent or plainly wrong data, causing users to lose confidence in the usefulness of the system. “Virtually all software systems today suffer to an unnecessary degree from the force of entropy” [46]. Correction of these issues requires data curation, which is very expensive in terms of time and resources. In fact, data curation should be considered a necessary part of the routine maintenance of any database to oppose its natural tendency toward disorder and inconsistency. This process cannot be (fully) automated; while tools can be developed to assist in the curation process, ultimately a human being must review the data to ensure its validity. In recent years, the needs of biology-related databases led to the formation of a new and growing profession, biocurator [47]. To illustrate the scope of this new field, scientists from over 250 different institutions worldwide are represented in the International Society for Biocuration [48].
A particular problem in designing and maintaining effective data management systems for large-scale biological programs is the interaction of two very different “cultures” involved with the system: the data management system developers and the biological researchers. People with training and experience in both software development and biological research are still relatively rare. Despite earnest efforts, the two groups often do not understand each other well. For example, addressing a request by a biologist, a system developer may propose a solution that is elegant, general, and yet fails completely to address the needs identified by the biologist. In turn, biologists are often bewildered when they are told by system developers that a supposedly minor modification of their experimental procedure would require an extensive redesign of the database schema taking several months of work. It is very important that project leaders try to bridge this cultural gap. This is especially crucial when designing new parts of the data management system. Development of an appropriate database abstraction is the single most important part of the design, requiring close collaboration of the two groups. At the testing and maintenance stage, it is crucial that real experiments leading to new structures and publications are performed by these two groups together. This approach is used in the development of LabDB and UniTrack, where both the people responsible for particular biomedical projects and the people who are writing the code are considered “developers” of the data management systems.
As mentioned above, one of the particular challenges of tracking biological data is the sheer diversity of potential experiments. When a chain of experiments is planned, one successful experiment in the chain can make others unnecessary. When data management systems were focused on tracking the “standard” structure determination pipeline, there was an implicit understanding of the scope of the methods that would be used, and thus most of the parameters that would need to be harvested could be determined or predicted a priori. The level of diversity increases even more when data from different consortia are brought together into a single database like TargetTrack.
Another particular challenge is in the sheer amount of experimental data to be collected. As the centers continue to become more efficient at producing greater numbers of experimental samples more quickly, the process of actually entering the results into the databases becomes a rate-limiting step, even when data are harvested semiautomatically. In particular, the process of protein crystallization, where each protein sample can potentially be used to produce thousands of individual crystallization trials, represents a virtual avalanche of data to be imported into the database. Further, given the comparatively large number of crystallization experiments typically required to yield useful results [49], the temptation to only include positive outcomes is strong, even though both positive and negative results are crucial for usefully data mining crystallization results. Some LIMSs, like LabDB, have partially addressed this issue by importing experimental data from the laboratory automatically or semiautomatically, but many systems still have challenges in ensuring that data entry and import are as simple as possible. Similarly, systems for importing data like the XML files used by TargetTrack will not be able to scale to the millions of data produced by the high-throughput centers.
Outside SG and other large projects, in many small-scale biological research laboratories, data are still primarily managed through written notebooks and spreadsheets. Such tools are not adequate to handle more complicated data. None of the available general-purpose commercial or open-source LIMSs have gained wide acceptance among small-scale laboratories. Some LIMS-like systems are in use; many pieces of scientific equipment come with specialized databases for automatically gathering and analyzing the data collected with that equipment. However, there is little incentive for equipment vendors to provide tools to integrate data from these databases with data from other databases, let alone data collected manually. Such tools are being created by the SG centers, and hopefully when they encompass a sufficiently broad range of experimental methods, they might be a decisive factor in encouraging adoption of modern data management systems in small-scale laboratories.
4 Conclusions
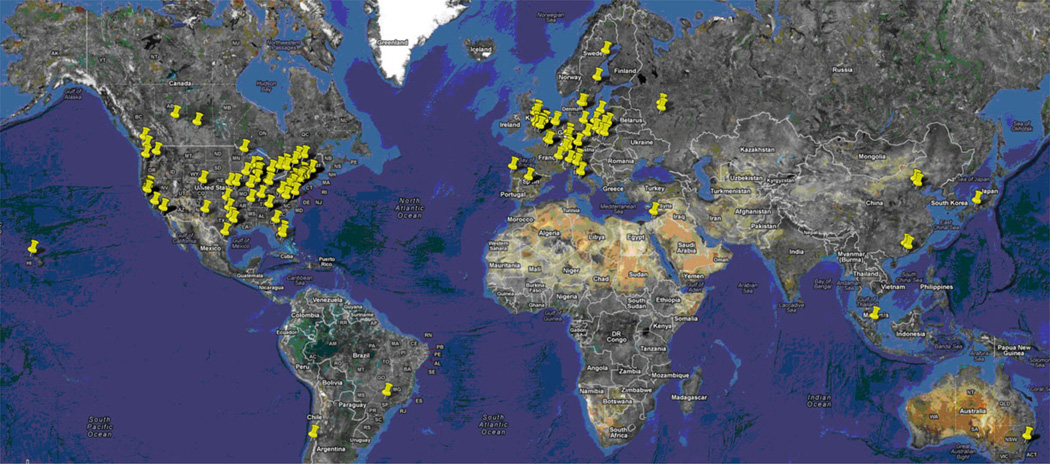
Data management in a large modern laboratory has become paramount for coordinating and tracking the vast amount of data generated across multiple experiments, time frames, and centers, not to mention the potential for data mining to extract even more useful and interesting information. Successful data management requires a system with a well-planned, cohesive, and flexible framework. How to best achieve this coordination and level of detail is currently being addressed in different ways, but the measure of success comes back to “data in, information out.” A coherent organizational structure using a “bottom-up” approach, along with mechanisms to connect these results into a unified system, has been working well for the SG centers, giving them the ability to adapt to new nonlinear and distributed experimental pipelines. In particular, the development of “super-LIMS” such as LabDB gives much needed flexibility as the frontier of the SG landscape continues to advance across organizations. The overall success of SG data management efforts should be measured not only in classical terms, i.e., the number of papers and/or number of citations, but most of all by the impact on the scientific community. There is no simple measure of that impact, but the number of papers published by an SG center jointly with other institutions is an indication of this impact. The map of collaborations for one SG center (Fig. 8) illustrates that the “big data” produced by the large-scale SG centers is also relevant to the biological research performed in small-scale laboratories around the world.
Fig. 8.
Map showing locations of collaborators of the MCSG (institutions of scientists who coauthored papers funded at least in part by the center)
Acknowledgments
The authors would like to thank Alex Wlodawer, Tom Terwilliger, Heidi Imker, Steve Almo, Wayne Anderson, Andrzej Joachimiak, Rachel Vigour, and Zbyszek Dauter for valuable comments on the manuscript. This work was supported by PSI:Biology grants U54 GM094585 and U54 GM094662 as well as grants R01 GM053163 and U54 GM093342. This work was also supported with federal funds from the NIAID, NIH, Department of Health and Human Services, under Contract Nos. HHSN272200700058C and HHSN272201200026C.
References
- 1.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 2.Minor W, et al. HKL-3000: the integration of data reduction and structure solution— from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 3.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10(12):980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 5.Peat TS, Christopher JA, Newman J. Tapping the Protein Data Bank for crystallization information. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 12):1662–1669. doi: 10.1107/S0907444905033202. [DOI] [PubMed] [Google Scholar]
- 6.Wlodawer A, et al. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008;275(1):1–21. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooft RW, et al. Errors in protein structures. Nature. 1996;381(6580):272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 8.Koclega KD, et al. ‘Hot’ macromolecular crystals. Cryst Growth Des. 2009;10(2):580. doi: 10.1021/cg900971h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.SBKB P-N PSI impact: ex-cited use of PSI structures [Google Scholar]
- 10.Gabanyi MJ, et al. The Structural Biology Knowledgebase: a portal to protein structures, sequences, functions, and methods. J Struct Funct Genomics. 2011;12(2):45–54. doi: 10.1007/s10969-011-9106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, et al. TargetDB: a target registration database for structural genomics projects. Bioinformatics. 2004;20(16):2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- 12.Edwards A. Open-source science to enable drug discovery. Drug Discov Today. 2008;13(17–18):731–733. doi: 10.1016/j.drudis.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 13.O’Toole N, et al. The structural genomics experimental pipeline: insights from global target lists. Proteins. 2004;56(2):201–210. doi: 10.1002/prot.20060. [DOI] [PubMed] [Google Scholar]
- 14.Goh CS, et al. Mining the structural genomics pipeline: identification of protein properties that affect high-throughput experimental analysis. J Mol Biol. 2004;336(1):115–130. doi: 10.1016/j.jmb.2003.11.053. [DOI] [PubMed] [Google Scholar]
- 15.Kouranov A, et al. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34(Database issue):D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman HM, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37(Database issue):D365–D368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westbrook J, et al. The Protein Data Bank and structural genomics. Nucleic Acids Res. 2003;31(1):489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pajon A, et al. Design of a data model for developing laboratory information management and analysis systems for protein production. Proteins. 2005;58(2):278–284. doi: 10.1002/prot.20303. [DOI] [PubMed] [Google Scholar]
- 19.Prilusky J, et al. HalX: an open-source LIMS (Laboratory Information Management System) for small-to large-scale laboratories. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 6):671–678. doi: 10.1107/S0907444905001290. [DOI] [PubMed] [Google Scholar]
- 20.Morris C, et al. The Protein Information Management System (PiMS): a generic tool for any structural biology research laboratory. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):249–260. doi: 10.1107/S0907444911007943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goh CS, et al. SPINE 2: a system for collaborative structural proteomics within a federated database framework. Nucleic Acids Res. 2003;31(11):2833–2838. doi: 10.1093/nar/gkg397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zolnai Z, et al. Project management system for structural and functional proteomics: sesame. J Struct Funct Genomics. 2003;4(1):11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 23.Raymond S, O’Toole N, Cygler M. A data management system for structural genomics. Proteome Sci. 2004;2(1):4. doi: 10.1186/1477-5956-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.JCSG web portal. [Accessed 4 Mar 2013]; http://www.jcsg.org/. [Google Scholar]
- 25.Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apweiler R, Bairoch A, Wu CH. Protein sequence databases. Curr Opin Chem Biol. 2004;8(1):76–80. doi: 10.1016/j.cbpa.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Cymborowski M, et al. To automate or not to automate: this is the question. J Struct Funct Genomics. 2010;11(3):211–221. doi: 10.1007/s10969-010-9092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair R, et al. Structural genomics is the largest contributor of novel structural leverage. J Struct Funct Genomics. 2009;10(2):181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Montelione GT, Rost B. Novel leverage of structural genomics. Nat Biotechnol. 2007;25(8):849–851. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
- 30.Bucher MH, Evdokimov AG, Waugh DS. Differential effects of short affinity tags on the crystallization of Pyrococcus furiosus maltodextrin-binding protein. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 3):392–397. doi: 10.1107/s0907444901021187. [DOI] [PubMed] [Google Scholar]
- 31.Koth CM, et al. Use of limited proteolysis to identify protein domains suitable for structural analysis. Methods Enzymol. 2003;368:77–84. doi: 10.1016/S0076-6879(03)68005-5. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, et al. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods. 2008;5(10):853–854. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cormier CY, et al. PSI:Biology-materials repository: a biologist’s resource for protein expression plasmids. J Struct Funct Genomics. 2011;12(2):55–62. doi: 10.1007/s10969-011-9100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cormier CY, et al. Protein structure initiative material repository: an open shared public resource of structural genomics plasmids for the biological community. Nucleic Acids Res. 2010;38(Database issue):D743–D749. doi: 10.1093/nar/gkp999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker R, Peacock S. BEI Resources: supporting antiviral research. Antiviral Res. 2008;80(2):102–106. doi: 10.1016/j.antiviral.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chruszcz M, Wlodawer A, Minor W. Determination of protein structures—a series of fortunate events. Biophys J. 2008;95(1):1–9. doi: 10.1529/biophysj.108.131789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page R, et al. Shotgun crystallization strategy for structural genomics: an optimized two-tiered crystallization screen against the Thermotoga maritima proteome. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 6):1028–1037. doi: 10.1107/s0907444903007790. [DOI] [PubMed] [Google Scholar]
- 38.Babnigg G, Joachimiak A. Predicting protein crystallization propensity from protein sequence. J Struct Funct Genomics. 2010;11(1):71–80. doi: 10.1007/s10969-010-9080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimber MS, et al. Data mining crystallization databases: knowledge-based approaches to optimize protein crystal screens. Proteins. 2003;51(4):562–568. doi: 10.1002/prot.10340. [DOI] [PubMed] [Google Scholar]
- 40.Newman J, et al. Towards rationalization of crystallization screening for small-to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 10):1426–1431. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 41.Zheng H, et al. Data mining of metal ion environments present in protein structures. J Inorg Biochem. 2008;102(9):1765–1776. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weekes D, et al. TOPSAN: a collaborative annotation environment for structural genomics. BMC Bioinforma. 2010;11:426. doi: 10.1186/1471-2105-11-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodis E, et al. Proteopedia—a scientific ‘wiki’ bridging the rift between three-dimensional structure and function of bio-macromolecules. Genome Biol. 2008;9(8):R121. doi: 10.1186/gb-2008-9-8-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee WH, et al. SGC—structural biology and human health: a new approach to publishing structural biology results. PLoS One. 2009;4(10):e7675. doi: 10.1371/journal.pone.0007675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raush E, et al. A new method for publishing three-dimensional content. PLoS One. 2009;4(10):e7394. doi: 10.1371/journal.pone.0007394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubert R. Convergent architecture: building model-driven J2EE systems with UML. New York: Wiley; 2001. [Google Scholar]
- 47.Howe D, et al. Big data: the future of biocuration. Nature. 2008;455(7209):47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bateman A. Curators of the world unite: the International Society of Biocuration. Bioinformatics. 2010;26(8):991. doi: 10.1093/bioinformatics/btq101. [DOI] [PubMed] [Google Scholar]
- 49.Chayen NE, Saridakis E. Protein crystallization: from purified protein to diffraction-quality crystal. Nat Methods. 2008;5(2):147–153. doi: 10.1038/nmeth.f.203. [DOI] [PubMed] [Google Scholar]