Abstract
Immune responses to some monoclonal antibodies (mAbs) and biologic proteins interfere with their efficacy due to the development of anti-drug antibodies (ADA). In the case of mAbs, most ADA target ‘foreign’ sequences present in the complementarity determining regions (CDRs). Humanization of the mAb sequence is one approach that has been used to render biologics less foreign to the human immune system. However, fully human mAbs can also drive immunogenicity. De-immunization (removing epitopes) has been used to reduce biologic protein immunogenicity. Here, we discuss a third approach to reducing the immunogenicity of biologics: introduction of Treg epitopes that stimulate Treg function and induce tolerance to the biologic protein. Supplementing humanization (replacing xeno-sequences with human) and de-immunization (reducing T effector epitopes) with tolerization (introducing Treg epitopes) where feasible, as a means of improving biologics ‘quality by design’, may lead to the development of ever more clinically effective, but less immunogenic, biologics.
Keywords: alemtuzumab, biologic, biosimilar, bio-better, Campath®, immunogenicity, mAb, monoclonal, quality by design, tolerance, tregitope
Fierce competition for market share among biologics manufacturers of monoclonal antibodies (mAbs) has contributed to the emergence of a range of technologies to improve performance in the clinic. Advances in protein engineering technologies, chemistry, manufacturing and control (CMC) considerations, and the development of completely new antibodies for established targets, such as humanized or fully human antibodies, may offer advantages in specificity, efficacy and cost. However, these advances only partially address the problem of immunogenicity, which has become a differentiating factor for biologics in clinical use.
Patients treated with mAbs and some biologic proteins occasionally develop neutralizing antibodies to the therapy, which reduce or eliminate the efficacy of the treatment. While numerous factors (such as aggregation, dose, route and target) can contribute to the immunogenicity of biologics, one of the key contributors to immunogenicity is T-cell epitope content. Ensuring that the primary sequence of the protein biologic is identical to ‘self’ and thus non-immunogenic should, in theory, reduce the potential for an immune response. The unexpected development of immune responses to fully human antibodies and proteins, when they are administered as drugs, has become one of the greatest puzzles of the protein therapeutics revolution.
Because immunogenicity can have dramatic effects on product safety and efficacy, regulatory agencies have drafted risk-based guidelines for immunogenicity screening that describe categories of protein biologics that would be subject to special scrutiny by the US FDA [101]. In February 2013, the FDA took a step further; posting a ‘Draft Guidance for Industry’ entitled ‘Immunogenicity Assessment for Therapeutic Protein Products’ [102]. Specifically, the guidance indicated that modifications to the protein backbone may be strategically introduced to remove stimulatory T-cell epitopes (T effector (Teff) epitopes) for reducing immunogenicity, but cautioned against the inadvertent removal of regulatory T-cell epitopes (also known as Tregitopes).
As is evident from the FDA document, a number of approaches to de-risking protein therapeutics are currently in use by biologics developers. Protein engineering methods for reducing the immunogenicity of mAbs include ‘humanization,’ which is accomplished by grafting fully human antibody sequences into regions of the antibody while retaining the complementarity determining regions (CDRs), and ‘de-immunization’ which involves removing T-cell epitopes where possible. De-immunization has also been applied to other protein biologics such as replacement enzymes or blood factors. The immunogenicity of biologic products has been somewhat reduced by de-immunization in some pre-clinical models [1] and products that are in clinical use [2] but other methods for reducing anti-drug antibody (ADA) responses currently prevail. These include co-administration of cytotoxic drugs (such as pre-treatment with methotrexate in children receiving recombinant human acid alpha-glucosidase for Pompe disease [3]) and pre-treatment with tolerizing doses of the biologic (such as FVIII) with or without other immune modulators, such as IVIG [4,5].
One of the earliest antibodies to be humanized is the Campath® 1G antibody, which was originally a chimeric (rat-human) mAb used to treat some B-cell leukemias. The antibody was humanized by grafting the anti-CD52 CDR regions onto a human IgG framework [6]. This antibody (Campath 1H) remains immunogenic in the clinic despite the humanization of its framework sequence. This is a well-known example of the somewhat unpredictable results of CDR grafting to humanize mAbs. Humanization was the favored approach to immunogenicity problems until recently [7,8]. The availability of several strains of mice expressing human antibody genes [9] quite naturally led to the development of what are called ‘fully human’ mAbs, which were believed to present a final solution to the immunogenicity problem, when they were first developed. Despite these advances, several well-known ‘humanized’ and fully human mAbs were subsequently shown to be just as immunogenic as their counterparts, for reasons that are partially explained by their residual differences from human germline [10] and in part due to their T-cell epitope content (regulatory and effector), as further described in this review.
An alternative strategy currently under consideration could be referred to as tolerization, which is the process of introducing tolerogenic sequences into the biologic that are known to trigger expansion of Treg cells to promote a tolerogenic immune response. In this review, we describe the contribution of T-cell epitopes to the immunogenicity of biologics, address some of the methods that biologics developers have used to identify these T-cell epitopes and use the Campath 1H and 1G to illustrate the effects of humanization, de-immunization and tolerization on biologic proteins, as a special case study.
Natural immune system mechanisms for controlling immunogenicity
T-cell epitopes: contributors to immunogenicity
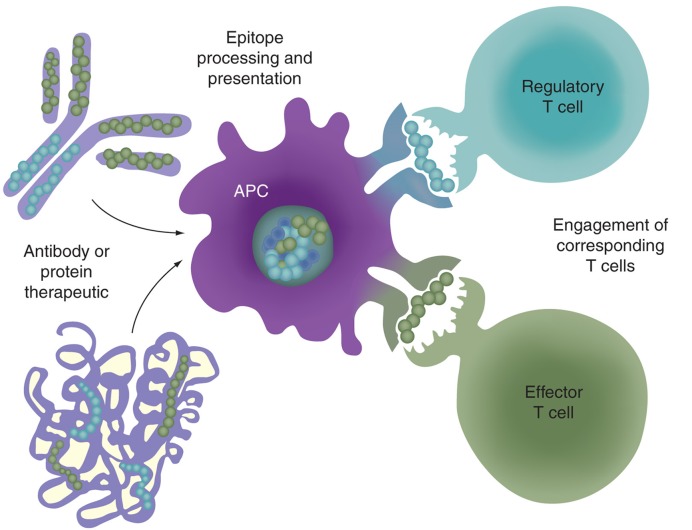
In studies designed to assist biologics developers with immunogenicity risk mitigation, the group headed by De Groot et al. has primarily focused on the role of T-cell epitopes in the primary amino acid sequence of biologics as drivers, or modulators, of immunogenicity. In the course of searching for Teff epitopes in biologic sequences, regulatory T-cell epitopes (Tregitopes) that are present in some mAbs were identified, and they proposed that these Tregitopes might modulate immune responses to immunoglobulins (Figure 1 & [11]). A retrospective review of the T- cell epitope (and Tregitope) content of mAb therapeutics published in 2009 revealed a close correlation between the presence of many highly conserved, highly promiscuous HLA class II Tregitopes and the absence of HLA-binding Teff epitopes, with lack of immunogenicity in published clinical studies [12]. These retrospective observations have been further validated by the group, through extensive prospective experience with mAb screening using an integrated on-line suite of immunoinformatics tools (the Interactive Screening and Protein Reengineering Interface (ISPRI) system) with commercial partners. Through these efforts, and in conjunction with validation by other groups [13,14], the contributions of T-cell epitopes (including Treg epitopes) to the immunogenicity of biologics has become apparent to biologics developers, many of whom have integrated immunogenicity screening using immunoinformatics tools that search for T-cell epitopes into their pre-clinical strategy.
Figure 1.
Processing and presentation of T-cell epitopes derived from monoclonal antibodies or biologic proteins to regulatory and effector T cells. T-cell epitopes present in an antibody or protein therapeutic will be processed and presented by APCs to either regulatory or effector T cells. Regulatory T-cell epitopes (such as Tregitopes) may serve to induce epitope-specific tolerance. It follows that Tregitopes could be combined with another immunogenic epitope, such as those present in a therapeutic antibody or protein, leading to epitope-specific tolerance induction.
APC: Antigen-presenting cell.
Tolerance to T-cell epitopes in biologic proteins
During immune system development, T cells that have high affinity receptors for autologous sequences are either deleted or anergized. Thus, immune responses to mAbs and non-mAb biologics are primarily directed to sequences that are foreign (non-human) or different from autologous proteins to which humans would normally be tolerant. In the case of mAbs, an immune response is driven by mouse- or rat-derived CDRs, although immune responses can also develop to fully or full humanized sequences in the CDR [15] as these re-ordered autologous sequences were likely not present during immune system development.
In the case of biologic proteins that are not antibodies, the problem of immunogenicity is more complex, as both residual tolerance (to epitope sequences present in the remaining, non-deleted gene sequence) and immune response to novel, foreign epitopes (not present during immunological development) play a role in the immune response. It may be the balance between these two opposing forces (further modulated by HLA-restriction), together with additional factors, that are integrated to trigger, or modulate, an immune response.
Monoclonal antibody immunogenicity
Following the US FDA approval of the first mAbs, administration to immune-competent patients was found to elicit ADA that compromised their clinical potential [16–19]. In hindsight, the immunogenicity was mainly associated with their non-human (murine- or rat-origin) sequences to which patients were sensitized during the required repeated dosing regimens. Initially, constant domains of antibody light and heavy chains were replaced by human constant regions to produce chimeric antibodies. This approach met with mixed success due to development of human anti-mouse antibodies against the mouse-derived variable regions. While an improvement over fully murine mAbs, chimeric antibodies were shown to raise immune responses that varied depending on their target, indication and the patient populations that were treated. For example, rituximab, a chimeric anti-CD20 antibody, elicited no immune response from B-cell chronic lymphocytic leukemia patients [20,21], but was immunogenic in 27% of Sjögren’s syndrome and 65% of systemic lupus erythomatosus patients [22,23].
To further reduce immunogenicity, mAb developers explored the grafting of CDR regions into fully human antibody frameworks (also known as humanization [24]) as a means to prevent the development of immune responses to the mAbs. These antibodies are fully human with the exception of mouse CDR regions. Antibody engineering technology has since advanced to develop completely human antibodies from humanized mice (mice expressing human antibody genes) [9]. Nonetheless, both humanized and fully human antibodies, remarkably, still may elicit immunogenic responses [12], leaving the problem of immunogenicity yet to be completely resolved.
Biologic protein (non-mAb) immunogenicity
Beyond mAbs, therapeutic products encompass diverse proteins such as human cytokines, cellular growth factors, hormones, clotting factors, enzymes and fusion proteins. Therapeutic proteins are attractive drug products, as they are generally considered safe, specific and non-toxic. However, their efficacy can be also dramatically compromised by the development of anti-therapeutic protein responses [25,26]. Like antibodies to mAbs, anti-therapeutic antibodies to biologic proteins (FVIII, erythropoietin) have the potential to neutralize their clinical effects [27,28] and can be associated with serious adverse events if cross-reaction occurs with endogenous protein antigens [29,30].
Biopharmaceuticals, such as fusion proteins, fall somewhere between mAbs and non-antibody biologics, as they are generally homologous with endogenous protein sequences (and carry Treg epitopes in the Fc region), yet they frequently incorporate point mutations intended to improve quality attributes of the final product such as stability, manufacturability or therapeutic activity. Even such small changes may present the risk of introducing new epitopes never before encountered by the host. The linkage between the Fc region and the fused protein may also introduce new T-cell epitopes.
Enzyme replacement therapies
Anti-therapeutic protein responses are not unexpected when the protein is foreign, either as the result of a different species of origin or a recipient in whom the natural analog of the therapeutic protein is deleted or modified. Examples of ‘foreign’ proteins include blood factors and enzymes that are provided to supplement or replace the same protein in patients who have genetic deficiencies (as with FVIII deficiency in hemophilia A and in acid alpha-glucosidase (GAA)-deficiency in Pompe disease). Immune responses to these products depend on several factors which include: i) the degree to which the endogenous protein has been deleted due to genetic mutations and ii) the prevalence of the protein in circulation [31].
Take, for example, the lyosomal storage disorder Pompe disease that is caused by a genetic defect in the GAA enzyme. Children affected by Pompe disease have protein expression defects ranging from complete lack of GAA protein expression, categorized as cross-reactive immunologic material (CRIM)-negative, or partial GAA protein expression, categorized as CRIM-positive. Treatment with fully human recombinant GAA can trigger high titer ADAs, and a clear correlation between the incidence of ADA and CRIM status in Pompe patients has been demonstrated. Thus, the less GAA expressed, the more severe the disease, the greater the dependence on the replacement protein, but the greater the risk and severity of ADA. Indeed, high ADA titers correlate with poor outcomes; thus many CRIM-negative Pompe infants with a complete GAA deficiency succumb quickly to disease [3,32,33].
Blood factors
De-immunization is one approach that has been used to address the immunogenicity of blood factors such as FVIII and other biologic proteins, however, modification of the primary sequence may result in reduced efficacy, particularly if the protein is large and de-immunization must be carried out in multiple sites. Furthermore, the relative importance of T-cell epitopes present in non-mAb proteins to tolerance induction (due to natural Treg responses to these epitopes) is unknown; therefore, de-immunization should be approached with caution. In the context of FVIII and GAA, residual circulating protein may contain T-cell epitopes to which subjects are tolerant or that actively induce regulatory T cells. Due to variability in the genetic mutations and in subject HLA, there may be no ‘one size fits all’ approach to de-immunizing these replacement proteins. Tolerization is an attractive alternative to de-immunization especially as it would augment Treg responses, suppressing ADA development. In the case of proteins that have patents expiring, engineering Treg epitopes within their framework is an additional strategy that could be used to develop ‘bio-better’ biologics that are more effective and highly competitive with biosimilars in the crowded biologics market.
Screening solutions for the immunogenicity problem
In vitro screening of biologics
Pre-clinical screening of biologic proteins for T-cell epitopes can be performed in vitro and in silico, providing an opportunity to improve the immunogenicity risk profile of a protein therapeutic at its very foundation. Wullner et al. have used T-cell assays to evaluate the immunogenicity of biologics [34]. Harding et al., using a time- and reagent-intensive overlapping peptide approach, have also confirmed the contribution of T-cell epitopes in the immunogenicity of biologics [15]. Screening overlapping peptides has generally been replaced by a combination approach, outlined in the next section.
In silico screening followed by in vitro validation
Another approach to immunogenicity screening is to evaluate T-cell epitope content in silico. The presence of T-cell epitopes is easily discernable using epitope-mapping immunoinformatics tools [12], many of which are available on the internet, although none of these freely available tools are specifically adapted for biologics. Several commercial companies currently offer comprehensive immunogenicity screening on a fee-for-service basis, the immunogenicity of these products is then evaluated in vitro, using peptides, or whole antigens. For example, Barbosa et al. confirmed the role of T cells on the immune response to Betaseron by linking ADA to HLA-DR type [35]. Of note, the in silico assessment of the same protein (using on-line tools) did not, in the authors’ view, correlate with observed immunogenicity in this instance; a separate reanalysis of the overlapping peptides by EpiMatrix provided better correlation [36]. Only a few comparisons between T-cell epitope predictors used in the context of immunogenicity screening have been published [12,37]. More often, drug developers have performed extensive (and expensive) in-house comparisons of services prior to selecting a single service.
Prospective evaluations of in silico screening
Several prospective studies have compared T-cell epitope mapping and immunogenicity screening side by side and found immunogenicity screening using selected immunoinformatics tools are validated by clinical outcomes for the biologic under study. For example, Koren et al. demonstrated the correlation between T-cell epitopes, HLA and immunogenicity in a double-blinded study of the FPX biologic; immunoinformatics tools were predictive and that immunogenicity was correlated with HLA-haplotype [38]. FPX is a recombinant fusion protein consisting of two identical, biologically active peptides linked to a human Fc fragment. Following a single administration of FPX in 76 healthy human subjects, 37% developed antibodies. A memory T-cell response against the carboxy-terminus of the peptide was observed in antibody-positive subjects, but not in antibody-negative subjects. The projected promiscuity of the predicted T-cell epitope(s) was confirmed by representation of all common HLA alleles in antibody-positive subjects. HLA-haplotype DRB1*0701/1501 was predicted to be associated with the highest T-cell and antibody response; subsequent detailed in silico studies confirmed the link between HLA and immunogenicity [39]. Further development of this product was abandoned due to clinical immunogenicity.
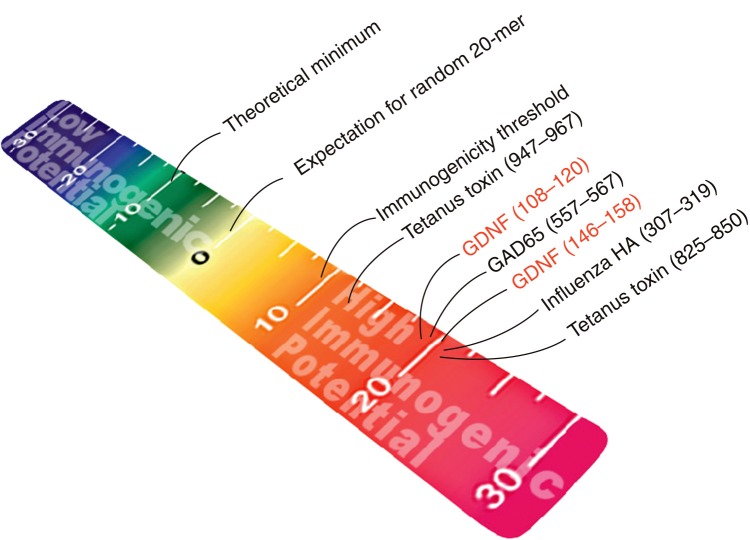
In a separate (also blinded) study, Tatarewicz et al. used EpiMatrix to screen GDNF, a protein therapeutic that was shown to be immunogenic in clinical trials [40]. The protein contains T-cell epitope clusters that rank as high as other well-known immunogenic epitopes on the EpiMatrix immunogenicity scale (Figure 2). Further clinical development of this product was cancelled due to concern about immunogenicity that emerged in clinical studies.
Figure 2.
EpiMatrix epitope cluster immunogenicity scale. Peptides are mapped onto the cluster immunogenicity scale according to their individual EpiMatrix scores. The EpiMatrix cluster immunogenicity score represents the deviation in putative epitope content from baseline expectation based on a random peptide standard. T-cell epitope clusters scoring above +10 are considered to be potentially immunogenic. Some well known positive control peptides, as well as some experimentally evaluated sequences, are arranged here by EpiMatrix score, from highest (red) to lowest (blue) (color figure can be found online at: www.expert-reviews.com/doi/full/10.1586/17512433.2013.835698).
Having observed the demonstrated serious consequences of immunogenicity and the published correlation with in silico results, researchers at selected pharmaceutical companies began to integrate in silico screening into the pre-clinical development phase of products. Most recently, Jawa et al. published an additional report on the prospective correlation between the EpiMatrix scores and immunogenicity for three additional biologic proteins that were predicted to be of low immunogenicity by EpiMatrix and proven to be of low immunogenicity in clinical trials [41].
Additional validation of immunogenicity predictions using selected immunoinformatics tools has been obtained from retrospective studies. In 2009, De Groot and Martin performed a detailed in silico analysis of mAbs in clinical use and described a strong correlation between T-cell epitope content and clinical immunogenicity (as published). The correlation was higher when the immunogenicity analysis was adjusted for Treg epitope content (see below for a discussion of Treg epitopes [12]). Immunoinformatics, when combined with in vitro and in vivo methods, provides an efficient alternative to conventional epitope mapping using overlapping peptides; reductions in time and effort up to 700-fold have been shown [42–46].
Development of web-based service centers for in silico immunogenicity screening
The team of De Groot and Martin has been using a full suite of T-cell epitope-based immunogenicity prediction tools since 2002. A self-serve secure-access ‘ISPRI’ website now enables users of these tools to screen biologics on demand, loading hundreds of candidates when, and as, needed. In 2008, subsequent to the discovery of Tregitopes (see below), the team integrated identification of validated Treg epitopes (Tregitopes) into the immunogenicity prediction, sharply improving the accuracy of the in silico analysis [12]. Several large biologics developers access this tool on a regular basis for their pre-clinical products. More than 1500 sequences are screened on average, per month, using this website for on-line immunogenicity screening tools.
Beyond screening: what to do when a biologic is immunogenic?
For products that are known to be immunogenic, de-immunization is an approach that has been used to reduce biologic protein immunogenicity for several decades (e.g., staphylokinase or SakSTAR [47]). De-immunization has been the focus of several previous reports and reviews [1, 48–51]. Here, we outline an emerging approach to reducing immunogenicity, by actively tolerizing immune responses to biologics, such as mAbs and protein therapeutics.
De-immunization
The inability of a specific HLA molecule to present epitopes from a given vaccine antigen is well known to be a cause of vaccine failure; by extension, the deletion of T-cell epitopes has been applied to reduce antibody responses to biologic proteins. For example, Celis et al. reported that a significant number of HBsAg-reactive T cells from various HBV-immune individuals recognize a determinant localized near the amino terminus of HBsAg, and individuals who cannot present the T-cell epitopes in this region are unable to mount a protective humoral response following vaccination [52]. Tumor cells [53] and pathogens [54,55] have also evolved to evade pro-inflammatory immune responses by accumulating mutations that alter T-cell epitope sequences. These mutations reduce the binding of their constituent epitopes to host HLA [56], rendering the host cell unable to alert T cells to the presence of the tumor or pathogen. The existence of viable ‘immune escape mutant’ viruses demonstrates that proteins, and indeed whole organisms, can tolerate certain immuno-modulatory mutations. Thus, it follows that deliberate removal of T-cell epitopes might also reduce the immunogenicity of biologic products [50].
A number of biologic proteins have been de-immunized by removing T-cell epitopes. One of the first attempts was the de-immunization of ‘SakSTAR’ or staphylokinase [47]. Modification or removal of the specific amino acids that contribute to HLA binding led to a reduction in the potential of the drug epitope to stimulate a T-cell response. Similarly, a number of epitope-abrogation studies have been performed using FVIII. Jones et al. identified a 15-mer sequence in human FVIII that bound strongly to DRB1*0401, *1101 and *1501, moderately to *0701, weakly to *0101, but not to *0301 and *1301 in HLA class II binding assays. Modification of the sequence of this epitope reduced its potential to bind to HLA. The modified peptide did not bind to any MHC class II molecule and was less immunogenic in vitro [57,58]. Epitope modification has also been applied to other proteins in studies performed by Hellendoorn et al., Tangri et al., Yeung et al. and others, using a variety of approaches [59–61]. For example, alanine substitutions to the MHC anchoring residues Y73, K74, R77, E80 and D82 of staphylokinase, alone or in combination, were shown to reduce or eliminate T-cell responses and clinical immunogenicity [62].
De Groot and collaborators have used a computational algorithm that iteratively searches for the optimal substitution for any given amino acid so as to reduce the impact of de-immunization on structure and function. This tool, OptiMatrix, used in concert with an established epitope mapping tool, EpiMatrix, can be tuned to minimize the number of sequence changes to one or two key amino acids per epitope, thus reducing the potential impact on protein structure and function, as described by Moise et al. for FVIII [1]. Using OptiMatrix, we have mapped and modified i) Botulinum neurotoxin type A [51], ii) lysostaphin [De Groot AS, Terry F, Cousens L, Martin W, Unpublished Data] and iii) a therapeutic mAb [De Groot AS, Terry F, Cousens L, Martin W, Unpublished Data]. De Groot and collaborators are currently working with Bailey-Kellogg and colleagues using a combined approach (EpiSweep, or Epi-3D) in which an algorithm is used to iteratively de-immunize epitope clusters while measuring the impact of the modifications on the stability of the protein structure. The set of modifications that are least likely to perturb stability are then tested in vitro (in HLA binding assays) and in vivo (immunization studies) [63].
Tolerization
Tolerization involves integration of previously identified Treg epitopes into the biologic protein sequence. While the concept of Tregitope-mediated tolerization is relatively novel, this method has emerged from studies carried out by Cousens et al., De Groot et al. and others, demonstrating that these specific, highly conserved and promiscuous T-cell epitopes derived from conserved regions of human immunoglobulins activate Treg cells, with the phenotypic properties of ‘natural’ Tregs [11] and suppress immune responses in vitro and in vivo [14]. While these Tregitope sequences do not contain any particular sequence that is unique to Treg epitopes, cross-conservation (at the T-cell receptor surface) with other highly conserved T-cell epitopes in autologous proteins has been described as a potential distinguishing feature [64]. The corresponding murine epitopes are also effective in murine models [14]. In vivo studies in autoimmune disease models have further validated the Tregitope discovery. Additional studies have demonstrated that co-administration of antigens with Tregitopes in vivo and in vitro leads to the induction of antigen-specific tolerance [11,65] and suppression of both humoral [13] and cellular immune responses [66,67] to co-administered antigens.
The discovery of the ‘Tregitope’ Treg epitopes in immunoglobulins (such as mAbs) is strengthened by published reports that immunoglobulin therapy (‘IVIG’) induces expansion of Tregs in vitro and in vivo [68–71], and IVIG experts generally agree that Treg epitopes such as the Tregitopes may be contributing to the tolerizing effects of IVIG [72]. The next step in the process of adapting Tregitopes to biologic therapy will be to actively introduce these epitopes into immunogenic biologics, thereby reducing potential immunogenicity; studies that support the effectiveness of this approach have been carried out by Cousens et al. [14,65,67] and are currently underway in the laboratories of a number of other research groups (e.g., Mingozzi and High [73] and Scott and collaborators [74]). In the following three sections, we review the role of T-cell epitopes in the immunogenicity problem faced by drug developers, discuss the discovery of regulatory T-cell epitopes known as Tregitopes and address their potential use as novel tolerizing agents for biologic proteins.
The tolerization solution to the immunogenicity problem
Natural tolerance
Immune responses to autologous proteins are controlled by a range of mechanisms, which have the potential to be exploited for the induction of tolerance to protein therapeutics. For T cells, self/non-self discrimination initially occurs in the thymus during T-cell maturation when medullary epithelial cells present tissue-specific self-protein epitopes, in the context of MHC, to immature T cells expressing antigen-recognition molecules (TCR). T cells whose receptors have a high affinity for self-peptide–MHC complexes, or whose receptors fail to bind at all, are deleted; T cells with low to moderate affinity escape deletion and may be converted to T cells with effector potential or ‘natural’ regulatory T cells [75].
Adaptive tolerance
Adaptive tolerance develops in the periphery where, in the presence of IL-10 and TGF-β, mature T cells are converted to the ‘adaptive’ Treg phenotype upon activation via their TCR. The mechanism of adaptive Treg induction is not well known; both bystander effects (via cytokines) and intracellular signaling (by antigen-presenting cells) have been evoked. The role of these ‘adaptive’ Treg cells may be to dampen effector immune responses (following the primary, vigorous immune reaction, as a means of controlling inflammation), or possibly to facilitate co-existence with some symbiotic bacteria and viruses. CTLA-4 (a T-cell surface molecule) may be involved, since the anti-CTLA-4 antibody ipilimumab (Yervoy) has been associated with adverse effects that parallel unregulated anti-self responses that might be expected in the absence of Tregs [76]. Adaptive Treg induction is associated with sustained tolerance (to grafts, to allergens and to autologous proteins) and probably requires the existence of Treg cells with the same antigen specificity as the self-reactive T cells.
Relevance of Tregitopes to IgG-mediated tolerance
The identification of Tregitopes contained in human IgG enables the integration of many independent observations of tolerance induction associated with mAb or mAb fragments, including tolerance following immunization with antigen-conjugated αDEC-205 [11,13,14]. Furthermore, murine T regulatory epitopes (mTregitopes) may explain earlier observations that Fc [77] and Fc-protein fusions [78] and IVIG [79–81] stimulate a tolerizing immune response.
Tolerization
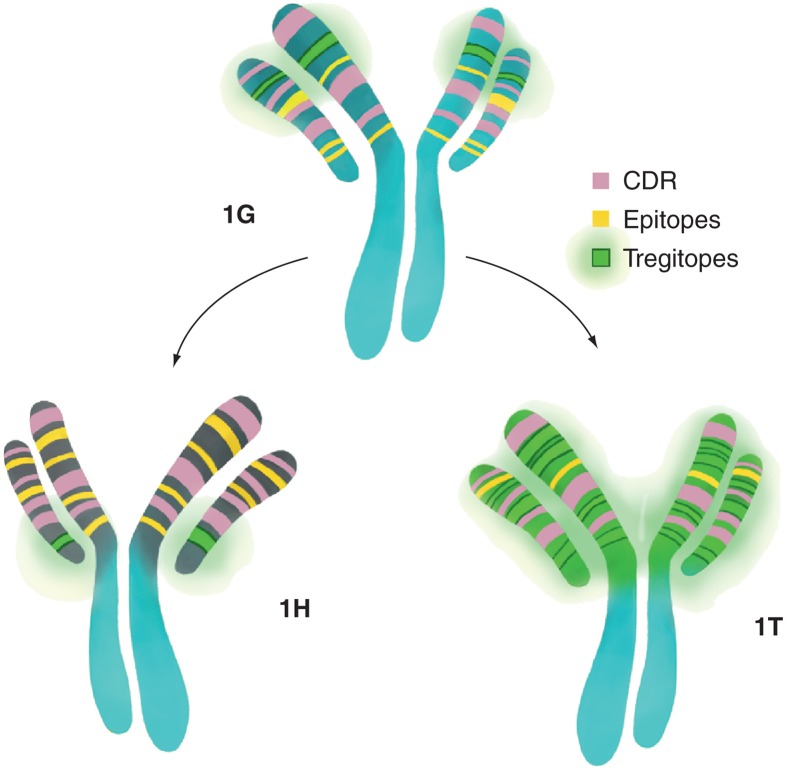
The discovery of Tregitopes and their close association with lack of immunogenicity to certain mAbs naturally led to the concept of actively integrating Tregitopes into biologics. Tolerization, as described here, could be considered to be an alternative to humanization of mAbs, it may also be applied to non-mAb biologic products. The approach is based on detailed studies showing Tregitopes lead to tolerance by Treg activation and epitope-specific tolerance induction. Using Campath (alemtuzumab) as an example, the tolerization approach is illustrated in Figure 3.
Figure 3.
The design of a less immunogenic Campath®. The Campath 1G to 1H transformation results in an almost equal gain/loss of Tregitopes and a net gain of potential Teff epitopes (based on Tregitope-adjusted EpiMatrix analysis, see [8]), while the 1G to 1T transformation results in a large gain of Tregitopes, a minimal gain of epitopes and a dramatically better EpiMatrix score.
CDR: Complementarity determining region.
The Campath example
To date, de-immunization and humanization approaches have provided a partial solution to the immunogenicity problem, and in the case of some fully human proteins, despite human sequence homologies, immunogenicity is still a problem in the clinic, for example, the alemtuzumab (Campath) mAbs that are directed against the antigen CD52 expressed on the surface of virtually all lymphocytes and monocytes. The humanized form, Campath 1H, is currently in use as a therapy for B-cell chronic lymphocytic leukemia, and has emerged as a potential therapeutic for multiple sclerosis in recent clinical studies [82]. The original rat-derived alemtuzumab (Campath 1G and Campath 1M) mAbs have limited clinical use due to the development of neutralizing antibodies in large numbers of subjects.
Three potential approaches have been considered for improving Campath 1H: i) administration of a tolerizing, non-binding (soluble) alemtuzumab [83], ii) de-immunization of immunogenic epitopes as performed in unpublished studies by De Groot et al. and iii) re-introduction of Tregitopes that may have already been present in the original mAb but perturbed in the process of humanization as proposed in greater detail in the next section. Initially, humanization appeared to have been a successful strategy for reducing immunogenicity. However, studies performed in immunocompetent patients have revealed that as many as 75% of patients develop antibody responses to the humanized alemtuzumab (Campath 1H) product, especially when several doses are given [83–86].
Assessing Campath 1G, 1H & ‘1T’ for T effector & regulatory T-cell epitopes
The process of humanization in the case of Campath involved the grafting of the CDR regions onto a new, human antibody framework. In Figure 3, we have compared Campath 1H with Campath 1G for T-cell epitope content and Treg (Tregitope) epitope content (defined using the ISPRI system) to illustrate that humanization did not reduce the number of Teff cell epitopes present in the derivative (1H) product, but rather removed one Tregitope. The unaltered Campath 1G molecule is predicted to be very immunogenic. The heavy and light chains have Tregitope-adjusted (TR-) EpiMatrix scores of 22.44 and 29.17, respectively. Modifying the Campath molecule from 1G to 1H involved grafting the CDR regions onto a new human antibody framework. This transformation resulted in a net change of 40 amino acids in the heavy chain, leading to an increase in the TR-EpiMatrix score from 22.44 to 42.36, meaning it became more immunogenic. One of the significant contributors to the increase in score was the introduction of 11 new epitopes. Another increase was due to the loss of one Tregitope, causing an 8-point increase. Humanization of the light chain (also through grafting CDR regions onto a new human antibody framework) resulted in a net 12 amino acid changes; the addition of three Tregitopes, and the removal of one, leaving a net gain of two Tregitopes. The TR-EpiMatrix score for the humanized light chain decreased from 29.17 to 0.77. The two major contributors to this decrease in score were the loss of six epitopes and the gain of three Tregitopes, causing a total decrease of nearly 50 points. This decrease was countered most aggressively by the loss of a Tregitope, leading to roughly a 20-point increase.
Tolerization of Campath 1G
Rather than humanizing, one might consider tolerizing 1G. The first step in tolerization is to identify where Treg epitopes may be present in a non-human version (rat or mouse version). Then amino acid changes are made to recover the human version of the Tregitope. In Figure 3, we have contrasted the humanization of Campath 1G to Campath 1H, with an alternative pathway creating Campath ‘1T’ from 1G through the incorporation of Tregitopes. A description of the approach follows: modifying the heavy chain from Campath 1G to introduce Tregitopes and reduce immunogenicity (called ‘1T’ in this case study) would require 21 amino acid point mutations, leading to a decrease in the EpiMatrix score from 22.44 to -66.21, These point mutations create 13 known Tregitopes and reduce or destroy five Teff epitopes. Tolerization of the light chain would require 10 amino acid point mutations, leading to a decrease in EpiMatrix score from 29.17 to -33.15. These point mutations create five known Tregitopes and reduce or destroy seven Teff epitopes. Changes to the CDR regions are avoided in this scenario.
Of course, this description ignores one of the most significant barriers to successful re-engineering: protein expression. Cell-culture production of the tolerized product may be impaired by the protein sequence modifications. Tools for predicting the impact of the T-cell epitope modification on the biologic protein stability have been developed [48] and have been applied to the de-immunization. Since the Tregitopes that were introduced are naturally located in these locations (in human framework regions), there is no expected perturbation of the structure. An analysis of the Campath 1G, 1H and 1T molecules for stability is in progress.
De-immunization may still be necessary in some cases. This is accomplished by identifying at which of the regions (where change is desired) one could make modifications without de-stabilizing the 3D structure of the protein. This approach requires protein-engineering experience, as amino acid changes in the primary sequence may have compensatory changes at a distal location within the protein. As previously mentioned, 3D modeling may improve attempts to de-immunize proteins without introducing destabilizing mutations. De-immunization of the Campath 1T heavy chain would require 1 amino acid change to the remaining Teff epitope, resulting in a decrease from −66.21 to −80.35, and 2 changes in the light chain with a resulting change in the EpiMatrix score from −33.15 to -75.92, further de-immunizing the tolerized Campath molecule. However, these changes may not be necessary as the tolerization described above might be sufficient. Clearly, each of these further modifications would have to be evaluated for de-stabilization, so this example is only provided as an illustration of an alternative approach to reducing the immunogenicity of a mAb.
Expert commentary
Anti-therapeutic antibodies can have a dramatic effect on the safety and efficacy of a protein therapeutic product. While several methods of reducing immunogenicity have been applied with a range of success (humanization and de-immunization), we propose a novel approach (tolerization) for engineering improving protein therapeutics by stimulating natural mechanisms of tolerance induction through the introduction of human regulatory T-cell epitopes into the biologic sequence.
One means of reducing immunogenicity is to develop products that have lower immunogenicity profiles. Over the past 10 years, immunoinformatics-driven immunogenicity screening has been fully integrated into the pipeline of companies developing biologicals. Immunogenicity screening has become de rigueur in most large companies. Unfortunately, on-line tools have been slow to adapt to the needs of the biologics community, and are not able to provide high-throughput screening in safe and secure interfaces, protected from ‘public’ viewing while products are still in development.
In response to increasing demand for an integrated immunoinformatics system for immunogenicity screening, De Groot and Martin developed a web-accessible toolbox called the ISPRI system that allows drug developers to accelerate the pre-clinical development of their protein products. Using this system, researchers can screen protein sequences of product candidates for the presence and immunogenic potential of putative T-cell epitopes (EpiMatrix) and epitope clusters (ClustiMer). Protein sequences can be ranked for immunogenic potential in comparison with known proteins on a normalized scale, and an interactive protein reengineering tool (OptiMatrix) allows researchers to modify, or de-immunize, T-cell epitope clusters in real time (in silico) by optimizing the amino acid sequence so that it is no longer able to interact with T cells.
The suite of tools developed by De Groot and Martin has been extensively validated internally and externally, with several key publications demonstrating the technology and rigorous testing procedures using known protein therapeutic targets [35,40,41]. In addition, this integrated system is the only available immunogenicity predictor that adjusts immunogenicity predictions for the presence of Tregitopes (see De Groot and Martin for additional information [12]).
If a protein therapeutic is known to be immunogenic but still has significant market value, it may be possible to create a ‘bio-better’ version. In this review article, we discuss two options for reducing protein immunogenicity (de-immunization and tolerization), and we illustrate the process of tolerization using Campath as an example. Based on available evidence, we expect that the integration of Tregitope sequences into biologics will stimulate regulatory cell expansion, cytokine production and suppress inflammatory cytokine levels and effectively prevent ADA production. Evaluation of Campath 1G revealed that there were a number of Tregitope-like sequences that could be converted to full Tregitopes by making single amino acid changes in the primary sequence with minimal change to the 3D structure, thus integration of new Tregitopes was not necessary. Clearly, these modified products would have to be produced in cell culture and tested in the appropriate laboratory assay, but it may be possible to identify changes that minimize the perturbation of the 3D structure, and 3D modeling tools can help with this process. This tolerization approach may accelerate development of a new generation of protein therapeutics, providing an effective solution to the problem of immunogenicity in this field.
Five-year view
Even though the potential for Tregitopes to regulate immunogenicity was recognized by De Groot and Martin in 2008, extensive validation studies were required before biologics developers were willing to accept the new concept and to begin to integrate Tregitopes into their biologics development plans. Extensive validation studies have now been performed in more than eight laboratories in six different regions of the world (Japan, Canada, USA, The Netherlands, France and Austria). Their potential for regulating immune responses to biologic proteins and for contributing to the development of improved ‘bio-betters’ is just beginning to mature. Thus, we expect that the Tregitopes described in this article will have a significant impact on biologics development over the next 5 years, leading to further differentiation of biologic products and improved competitiveness of Tregitope-containing products in the protein therapeutics field.
Acknowledgments
The authors would like to thank Genevieve De Groot for her beautiful illustrations created specially for this manuscript, Rebecca Martin for her analysis of Campath with the ISPRI toolkit and Kelsey Confreda for a thorough reading of this manuscript and editorial assistance.
Financial & competing interests disclosure
All of the authors are employees of EpiVax, and AS De Groot and W Martin are majority stockholders. These authors recognize the presence of a potential conflict of interest and affirm that the information represented in this paper is original and unbiased observations. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Writing assistance was provided by Kelsey Confreda, EpiVax, Inc.
Key issues
Immunogenicity is a differentiating factor in the market for biologics.
In silico screening has become the starting point for ‘quality by design’.
The discovery of Tregitopes has had a significant impact on immunogenicity screening, leading to higher accuracy predictions and better correlations with clinical outcomes.
Tolerization involves the introduction of Tregitope sequences into biologic proteins.
There is significant potential for this approach to accelerate the development of bio-better biologic products.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Moise L, Song C, Martin WD, Tassone R, De Groot AS, Scott DW. Effect of HLA DR epitope de-immunizaton of Factor VIII in vitro and in vivo . Clin. Immunol. 2012;142(3):320–331. doi: 10.1016/j.clim.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collen D, Lijnen HR. Thrombolytic agents . Thromb. Haemost. 2005;93(4):627–630. doi: 10.1160/TH04-11-0724. [DOI] [PubMed] [Google Scholar]
- 3.Joseph A, Munroe K, Housman M, Garman R, Richards S. Immune tolerance induction to enzyme-replacement therapy by co-administration of short-term, low-dose methotrexate in a murine Pompe disease model . Clin. Exp. Immunol. 2008;152(1):138–146. doi: 10.1111/j.1365-2249.2008.03602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messinger YH, Mendelsohn NJ, Rhead W et al. Successful immune tolerance induction to enzyme replacement therapy in CRIM-negative infantile Pompe disease . Genet. Med. 2012;14(1):135–142. doi: 10.1038/gim.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]; • One method that is currently used (in this context) for reducing immune responses to enzyme replacement therapy, that may not be broadly applicable to other biologics due to the systemic effects of methotrexate.
- 5.Kubisz P, Plamenová I, Hollý P, Stasko J. Successful immune tolerance induction with high-dose coagulation factor VIII and intravenous immunoglobulins in a patient with congenital hemophilia and high-titer inhibitor of coagulation factor VIII despite unfavorable prognosis for the therapy . Med. Sci. Monit. 2009;15(6):CS105–CS111. [PubMed] [Google Scholar]
- 6.Waldmann H. A personal history of the CAMPATH-1H antibody . Med. Oncol. 2002;19(Suppl.):S3–S9. doi: 10.1385/mo:19:2s:s03. [DOI] [PubMed] [Google Scholar]; • As described, an important description of the evolution of this antibody, from cloning to ‘humanization’.
- 7.Lefranc MP, Ehrenmann F, Ginestoux C, Giudicelli V, Duroux P. Use of IMGT(®) databases and tools for antibody engineering and humanization . Methods Mol. Biol. 2012;907:3–37. doi: 10.1007/978-1-61779-974-7_1. [DOI] [PubMed] [Google Scholar]
- 8.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics . Nat. Rev. Drug Discov. 2010;9(10):767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 9.Laffleur B, Pascal V, Sirac C, Cogné M. Production of human or humanized antibodies in mice . Methods Mol. Biol. 2012;901:149–159. doi: 10.1007/978-1-61779-931-0_9. [DOI] [PubMed] [Google Scholar]
- 10.Clark M. Antibody humanization: a case of the ‘Emperor’s new clothes’? . Immunol. Today. 2000;21(8):397–402. doi: 10.1016/s0167-5699(00)01680-7. [DOI] [PubMed] [Google Scholar]
- 11.De Groot AS, Moise L, McMurry JA et al. Activation of natural regulatory T cells by IgG Fc-derived peptide “Tregitopes” . Blood. 2008;112(8):3303–3311. doi: 10.1182/blood-2008-02-138073. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The first description of Tregitopes by De Groot; in this article, their ability to expand FoxP3-positive regulatory T cells (natural Tregs) and suppress immune response to co-administered antigens, in vitro and in vivo is described for the first time. The sequences of the human Fc-region Tregitopes are published provided, with their MHC binding affinities.
- 12.De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics . Clin. Immunol. 2009;131(2):189–201. doi: 10.1016/j.clim.2009.01.009. [DOI] [PubMed] [Google Scholar]; • Following the discovery of Tregitopes, this article describes a significant correlation between their presence in monoclonal antibodies and the absence of immunogenicity in clinical studies. A “Tregitope-adjusted” immunogenicity score is described.
- 13.Su Y, Rossi R, De Groot AS, Scott DW. Regulatory T cell epitopes (Tregitopes) in IgG induce tolerance in vivo and lack immunogenicity per se . J. Leukoc. Biol. 2013;94(2):377–383. doi: 10.1189/jlb.0912441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cousens LP, Najafian N, Mingozzi F et al. In vitro and in vivo studies of IgG-derived Treg epitopes (Tregitopes): A promising new tool for tolerance induction and treatment of autoimmunity . J. Clin. Immunol. 2013;33(1):43–49. doi: 10.1007/s10875-012-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Open-source review of Tregitope studies: their effects in autoimmune disease models are summarized and a more complete set of Tregitopes is provided, with their binding affinities. Potential applications of Tregitopes to autoimmunity are discussed.
- 15.Harding FA, Stickler MM, Razo J, DuBridge RB. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions . mAbs. 2010;2(3):256–265. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntyre JA, Kincade M, Higgins NG. Detection of IGA anti-OKT3 antibodies in OKT3-treated transplant recipients . Transplantation. 1996;61(10):1465–1469. doi: 10.1097/00007890-199605270-00009. [DOI] [PubMed] [Google Scholar]
- 17.Uckun FM, Messinger Y, Chen CL et al. Treatment of therapy-refractory B-lineage acute lymphoblastic leukemia with an apoptosis-inducing CD19-directed tyrosine kinase inhibitor . Clin. Cancer Res. 1999;5(12):3906–3913. [PubMed] [Google Scholar]
- 18.Kaminski MS, Zelenetz AD, Press OW et al. Pivotal study of iodine I 131 tositumomab for chemotherapy-refractory low-grade or transformed low-grade B-cell non-Hodgkin’s lymphomas . J. Clin. Oncol. 2001;19(19):3918–3928. doi: 10.1200/JCO.2001.19.19.3918. [DOI] [PubMed] [Google Scholar]
- 19.Stroomer JW, Roos JC, Sproll M et al. Safety and biodistribution of 99mTechnetium-labeled anti-CD44v6 monoclonal antibody BIWA 1 in head and neck cancer patients . Clin. Cancer Res. 2000;6(8):3046–3055. [PubMed] [Google Scholar]
- 20.Davis TA, Grillo-López AJ, White CA et al. Rituximab anti-CD20 monoclonal antibody therapy in non-Hodgkin’s lymphoma: safety and efficacy of re-treatment . J. Clin. Oncol. 2000;18(17):3135–3143. doi: 10.1200/JCO.2000.18.17.3135. [DOI] [PubMed] [Google Scholar]
- 21.Piro LD, White CA, Grillo-López AJ et al. Extended rituximab (anti-CD20 monoclonal antibody) therapy for relapsed or refractory low-grade or follicular non-Hodgkin’s lymphoma . Ann. Oncol. 1999;10(6):655–661. doi: 10.1023/a:1008389119525. [DOI] [PubMed] [Google Scholar]
- 22.Pijpe J, van Imhoff GW, Spijkervet FK et al. Rituximab treatment in patients with primary Sjögren’s syndrome: an open-label phase II study . Arthritis Rheum. 2005;52(9):2740–2750. doi: 10.1002/art.21260. [DOI] [PubMed] [Google Scholar]
- 23.Looney RJ, Anolik JH, Campbell D et al. B cell depletion as a novel treatment for systemic lupus erythematosus: a phase I/II dose-escalation trial of rituximab . Arthritis Rheum. 2004;50(8):2580–2589. doi: 10.1002/art.20430. [DOI] [PubMed] [Google Scholar]
- 24.M Kim JH, Hong HJ. Humanization by CDR grafting and specificity-determining residue grafting . Methods Mol. Biol. 2012;907:237–245. doi: 10.1007/978-1-61779-974-7_13. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg AS. Immunogenicity of biological therapeutics: a hierarchy of concerns . Dev. Biol. (Basel) 2003;112:15–21. [PubMed] [Google Scholar]
- 26.Barbosa MD. Immunogenicity of biotherapeutics in the context of developing biosimilars and biobetters . Drug Discov. Today. 2011;16:345–353. doi: 10.1016/j.drudis.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Eser A, Primas C, Reinisch W. Drug monitoring of biologics in inflammatory bowel disease . Curr. Opin. Gastroenterol. 2013;29(4):391–396. doi: 10.1097/MOG.0b013e328361f7f6. [DOI] [PubMed] [Google Scholar]
- 28.Shankar G, Pendley C, Stein KE. A risk-based bioanalytical strategy for the assessment of antibody immune responses against biological drugs . Nat. Biotechnol. 2007;25:555–561. doi: 10.1038/nbt1303. [DOI] [PubMed] [Google Scholar]; • One of the original ‘white papers’ describing a strategy for drug development that has been widely adapted by the biologics industry.
- 29.Haselbeck A. Epoetins: differences and their relevance to immunogenicity . Curr. Med. Res. Opin. 2003;19(5):430–432. doi: 10.1185/030079903125002063. [DOI] [PubMed] [Google Scholar]
- 30.Eckardt KU, Casadevall N. Pure red-cell aplasia due to anti-erythropoietin antibodies . Nephrol. Dial. Transplant. 2003;18:865–869. doi: 10.1093/ndt/gfg182. [DOI] [PubMed] [Google Scholar]
- 31.Haribhai D, Engle D, Meyer M et al. A threshold for central T cell tolerance to an inducible serum 870 protein . J. Immunol. 2003;170(6):3007–3014. doi: 10.4049/jimmunol.170.6.3007. [DOI] [PubMed] [Google Scholar]
- 32.Mendelsohn NJ, Messinger YH, Rosenberg AS, Kishnani PS. Elimination of antibodies to recombinant enzyme in pompe’s disease . N. Engl. J. Med. 2009;360(2):194–195. doi: 10.1056/NEJMc0806809. [DOI] [PubMed] [Google Scholar]
- 33.Garman RD, Munroe K, Richards SM. Methotrexate reduces antibody responses to recombinant human alpha-galactosidase A therapy in a mouse model of Fabry disease . Clin. Exp. Immunol. 2004;137(3):496–502. doi: 10.1111/j.1365-2249.2004.02567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wullner D, Zhou L, Bramhall E et al. Considerations for optimization and validation of an in vitro PBMC derived T cell assay for immunogenicity prediction of biotherapeutics . Clin. Immunol. 2010;137(1):5–14. doi: 10.1016/j.clim.2010.06.018. [DOI] [PubMed] [Google Scholar]; • A key article that describes in vitro immunogenicity screening using naïve blood donor cells. This approach has been adopted by many biologics developers, even though the predictive value of the approach is unknown and clinical validation is currently lacking.
- 35.Barbosa MD, Vielmetter J, Chu S, Smith DD, Jacinto J. Clinical link between MHC class II haplotype and interferon-beta (IFN-beta) immunogenicity . Clin. Immunol. 2006;118:42–50. doi: 10.1016/j.clim.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Jawa V, Cousens LP, Awwad M, Wakshull E, Kropshofer H, De Groot AS. T-cell dependent immunogenicity of protein therapeutics: preclinical assessment and mitigation. Clinical Immunology. doi: 10.1016/j.clim.2013.09.006. (In Press) [DOI] [PubMed] [Google Scholar]; • A biologics industry ‘White Paper’ describing the relevance and importance of T-cell epitopes as drivers of biologic protein immunogenicity.
- 37.Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach . PLoS Comput. Biol. 2008;4(4):e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koren E, De Groot AS, Jawa V et al. Clinical validation of the “in silico” prediction of immunogenicity of a human recombinant therapeutic protein . Clin. Immunol. 2007;124(1):25–32. doi: 10.1016/j.clim.2007.03.544. [DOI] [PubMed] [Google Scholar]; • One of the first prospective studies of in silico prediction; both T-cell epitope content and T-cell response in vitro were correlated with clinical immunogenicity.
- 39.Cohen T, Moise L, Ardito M, Martin W, De Groot AS. A method for individualizing the prediction of immunogenicity of protein vaccines and biologic therapeutics: individualized T cell epitope measure (iTEM) J. Biomed. Biotechnol. 2010;7 doi: 10.1155/2010/961752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatarewicz SM, Wei X, Gupta S, Masterman D, Swanson SJ, Moxness MS. Development of a maturing T-cell-mediated immune response in patients with idiopathic Parkinson’s disease receiving r-metHuGDNF via continuous intraputaminal infusion . J. Clin. Immunol. 2007;27(6):620–627. doi: 10.1007/s10875-007-9117-8. [DOI] [PubMed] [Google Scholar]
- 41.Jawa V, Cousens LP, De Groot AS. Schmidt SR. fusion protein technologies for biopharmaceuticals: Applications and Challenges. 1st. John Wiley & Sons, Inc; 2013. Immunogenicity of therapeutic fusion proteins: contributory factors and clinical experience. Edition. [Google Scholar]
- 42.Kast WM, Brandt RM, Sidney J et al. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins . J. Immunol. 1994;152:3904–3912. [PubMed] [Google Scholar]
- 43.Schafer JR, Jesdale BM, George JA, Kouttab NM, De Groot AS. Prediction of well-conserved HIV-1 ligands using a matrix-based algorithm, EpiMatrix . Vaccine. 1998;16(19):1880–1884. doi: 10.1016/s0264-410x(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 44.De Groot AS, Bosma A, Chinai N et al. From genome to vaccine: in silico predictions, ex vivo verification . Vaccine. 2001;19(31):4385–4395. doi: 10.1016/s0264-410x(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 45.De Groot AS, Saint-Aubin C, Bosma A, Sbai H, Rayner J, Martin W. Rapid determination of HLA B*07 ligands from the West Nile virus NY99 genome . Emerg. Infect. Dis. 2001;7(4):706–713. doi: 10.3201/eid0704.010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moutaftsi M, Peters B, Pasquetto V et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus . Nat. Biotechnol. 2006;24(7):817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 47.Collen D, Bernaerts R, Declerck P et al. Recombinant staphylokinase variants with altered immunoreactivity. I: Construction and characterization . Circulation. 1996;94(2):197–206. doi: 10.1161/01.cir.94.2.197. [DOI] [PubMed] [Google Scholar]
- 48.Parker AS, Choi Y, Griswold KE, Bailey-Kellogg C. Structure-guided deimmunization of therapeutic proteins . J. Comput. Biol. 2013;20(2):152–165. doi: 10.1089/cmb.2012.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Description of the Epi-Sweep approach to de-immunization. The structural modeling approach has recently been combined with EpiMatrix (EpiVax) to create ‘Epi-3D’.
- 49.Jones TD, Crompton LJ, Carr FJ, Baker MP. Deimmunization of monoclonal antibodies . Methods Mol. Biol. 2009;525:405–423. doi: 10.1007/978-1-59745-554-1_21. [DOI] [PubMed] [Google Scholar]
- 50.De Groot AS, Knopf PM, Martin W. De-immunization of therapeutic proteins by T-cell epitope modification . Dev. Biol. (Basel) 2005;122:171–194. [PubMed] [Google Scholar]
- 51.Cousens L, Moise L, Terry F, Martin W, De Groot AS. Immunogenic biologics: validation of screening, deimmunization and tolerization approaches (P3251) J. Immunol. 2013;190:192. [Google Scholar]
- 52.Celis E, Ou D, Otvos L. Recognition of hepatitis B surface antigen by human T lymphocytes. Proliferative and cytotoxic responses to a major antigenic determinant defined by synthetic peptides . J. Immunol. 1998;140:1808–1815. [PubMed] [Google Scholar]
- 53.Scanlan MJ, Jager D. Challenges to the development of antigen-specific breast cancer vaccines . Breast Cancer Res. 2001;3(2):95–98. doi: 10.1186/bcr278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullbacher A. Viral escape from immune recognition: multiple strategies of adenoviruses . Immunol. Cell Biol. 1992;70:59–63. doi: 10.1038/icb.1992.9. Pt 1. [DOI] [PubMed] [Google Scholar]
- 55.Hill AV, Jepson A, Plebanski M, Gilbert SC. Genetic analysis of host-parasite coevolution in human malaria . Philos. Trans. R. Soc. Lond. B Biol. Sci. 1997;352(1359):1317–1325. doi: 10.1098/rstb.1997.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vossen MT, Westerhout EM, Soderberg-Naucler C, Wiertz EJ. Viral immune evasion: a masterpiece of evolution . Immunogenetics. 2002;54(8):527–542. doi: 10.1007/s00251-002-0493-1. [DOI] [PubMed] [Google Scholar]
- 57.Jones TD, Phillips WJ, Smith BJ et al. Identification and removal of a promiscuous CD4+ T cell epitope from the C1 domain of factor VIII . J. Thromb. Haemost. 2005;3:991–1000. doi: 10.1111/j.1538-7836.2005.01309.x. [DOI] [PubMed] [Google Scholar]
- 58.Gilles JG, Lavend’homme R, Peerlinck K et al. Some factor VIII (FVIII) inhibitors recognise a FVIII epitope(s) that is present only on FVIII-vWF complexes . Thromb. Haemost. 1999;82:40–45. [PubMed] [Google Scholar]
- 59.Hellendoorn K, Jones T, Watkins J, Baker M, Hamilton A, Carr F. Limiting the risk of immunogenicity by identification and removal of T-cell epitopes (DeImmunisation™). Association for Immunotherapy of Cancer: Cancer Immunotherapy–2nd Annual Meeting Mainz, Germany. Cancer Cell Int. 2004;4(Suppl. 1):S20. doi: 10.1186/1475-2867-4-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tangri S, Mothe BR, Eisenbraun J et al. Rationally engineered therapeutic proteins with reduced immunogenicity . J. Immunol. 2005;174(6):3187–3196. doi: 10.4049/jimmunol.174.6.3187. [DOI] [PubMed] [Google Scholar]; • An excellent example of the de-immunization approach that includes successful production of variant proteins and demonstration that the variant proteins are still functional.
- 61.Yeung VP, Chang J, Miller J, Barnett C, Stickler M, Harding FA. Elimination of an immunodominant CD4+ T cell epitope in human IFN-beta does not result in an in vivo response directed at the subdominant epitope . J. Immunol. 2004;172(11):6658–6665. doi: 10.4049/jimmunol.172.11.6658. [DOI] [PubMed] [Google Scholar]
- 62.Warmerdam PA, Plaisance S, Vanderlick K et al. Elimination of a human T-cell region in staphylokinase by T-cell screening and computer modeling . Thromb. Haemost. 2002;87(4):666–673. [PubMed] [Google Scholar]
- 63.Parker AS, Choi Y, Griswold KE, Bailey-Kellogg C. Structure-guided de immunization of therapeutic proteins . J. Comput. Biol. 2013;20(2):152–165. doi: 10.1089/cmb.2012.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moise L, Gutierrez AH, Bailey-Kellogg C et al. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix . Hum. Vaccin. Immunother. 2012;9(7):1577–1586. doi: 10.4161/hv.24615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cousens LP, Su Y, McClaine E et al. Application of IgG-derived natural Treg epitopes (IgG Tregitopes) to antigen-specific tolerance induction in a murine model of type 1 diabetes . J. Diabetes Res. 2013:621693. doi: 10.1155/2013/621693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hui DJ, Basner-Tschakarjan E, Chen Y et al. Modulation of CD8+ T cell responses to AAV vectors with IgG-derived MHC class II epitopes. Mol. Ther. 2013 doi: 10.1038/mt.2013.166. doi:10.1038/mt.2013.166. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cousens LP, Tassone R, Mazer BD, Ramachandiran V, Scott DW, De Groot AS. Tregitope update: mechanism of action parallels IVIg . Autoimmun. Rev. 2012;12(3):436–443. doi: 10.1016/j.autrev.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 68.Schuster SJ, Neelapu SS, Gause BL et al. Vaccination with patient-specific tumor-derived antigen in first remission improves disease-free survival in follicular lymphoma . J. Clin. Oncol. 2011;29(20):2787–2794. doi: 10.1200/JCO.2010.33.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ephrem A, Chamat S, Miquel C et al. Expansion of CD4+CD25+ regulatory T cells by intravenous immunoglobulin: a critical factor in controlling experimental autoimmune encephalomyelitis . Blood. 2008;111(2):715–722. doi: 10.1182/blood-2007-03-079947. [DOI] [PubMed] [Google Scholar]
- 70.Tsurikisawa N, Saito H, Oshikata C, Tsuburai T, Akiyama K. High-dose intravenous immunoglobulin treatment increases regulatory T cells in patients with eosinophilic granulomatosis with polyangiitis . J. Rheumatol. 2012;39(5):1019–1025. doi: 10.3899/jrheum.110981. [DOI] [PubMed] [Google Scholar]
- 71.Lopez M, Clarkson MR, Albin M, Sayegh MH, Najafian N. A novel mechanism of action for anti-thymocyte globulin: induction of CD4+CD25+Foxp3+ regulatory T cells . J. Am. Soc. Nephrol. 2006;17(10):2844–2853. doi: 10.1681/ASN.2006050422. [DOI] [PubMed] [Google Scholar]
- 72.Schwab I, Nimmerjahn F. Intravenous immunoglobulin therapy: how does IgG modulate the immune system? . Nat. Rev. Immunol. 2013;13(3):176–89. doi: 10.1038/nri3401. [DOI] [PubMed] [Google Scholar]
- 73.Mingozzi F, High KA. Immune responses to AAV vectors: overcoming barriers to successful gene therapy . Blood. 2013;122(1):23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adair P, Su Y, Scott DW. Tolerance induction in hemophilia A animal models: battling inhibitors with antigen-specific immunotherapies . Discov. Med. 2013;15(84):275–282. [PubMed] [Google Scholar]
- 75.Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells . Nat. Rev. Immunol. 2003;3:253–257. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 76.Bakacs T, Mehrishi JN, Szabo M, Moss RW. Interesting possibilities to improve the safety and efficacy of ipilimumab (Yervoy) . Pharmacol. Res. 2012;66(2):192–197. doi: 10.1016/j.phrs.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 77.Baxevanis CN, Ioannides CG, Reclos GJ, Papamichail M. Evidence for distinct epitopes on human IgG with T cell proliferative and suppressor function . Eur. J. Immunol. 1986;16:1013–1016. doi: 10.1002/eji.1830160824. [DOI] [PubMed] [Google Scholar]
- 78.Zambidis ET, Scott DW. Epitope-specific tolerance induction with an engineered immunoglobulin . Proc. Natl Acad. Sci. U.S.A. 1996;93:5019–5024. doi: 10.1073/pnas.93.10.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vicente A, Barreto M, Demengeot J, Fesel C. Genetic factors in systemic lupus erythematosus. Instituto Gulbenkian de Ciência. 2005. Annual report Lisbon, Portugal.
- 80.Bystryn JC, Jiao D. IVIg selectively and rapidly decreases circulating pathogenic autoantibodies in pemphigus vulgaris . Autoimmunity. 2006;39(70):601–607. doi: 10.1080/08916930600972016. [DOI] [PubMed] [Google Scholar]
- 81.Jordan SC, Vo AA, Peng A, Toyoda M, Tyan D. Intravenous gammaglobulin (IVIG): a novel approach to improve transplant rates and outcomes in highly HLA-sensitized patients . Am. J. Transplant. 2006;6(3):459–466. doi: 10.1111/j.1600-6143.2005.01214.x. [DOI] [PubMed] [Google Scholar]
- 82.Coles AJ. Alemtuzumab treatment of multiple sclerosis . Semin. Neurol. 2013;33(1):66–73. doi: 10.1055/s-0033-1343797. [DOI] [PubMed] [Google Scholar]
- 83.Somerfield J, Hill-Cawthorne GA, Lin A et al. A novel strategy to reduce the immunogenicity of biological therapies . J. Immunol. 2010;185(1):763–768. doi: 10.4049/jimmunol.1000422. [DOI] [PubMed] [Google Scholar]
- 84.Isaacs JD, Watts RA, Hazleman BL et al. Humanised monoclonal antibody therapy for rheumatoid arthritis . Lancet. 1992;340(8822):748–752. doi: 10.1016/0140-6736(92)92294-p. [DOI] [PubMed] [Google Scholar]
- 85.Isaacs JD, Manna VK, Rapson N et al. CAMPATH-1H in rheumatoid arthritis: an intravenous dose-ranging study . Br. J. Rheumatol. 1996;35(3):231–240. doi: 10.1093/rheumatology/35.3.231. [DOI] [PubMed] [Google Scholar]
- 86.Matteson EL, Yocum DE, St Clair EW et al. Treatment of active refractory rheumatoid arthritis with humanized monoclonal antibody CAMPATH-1H administered by daily subcutaneous injection . Arthritis Rheum. 1995;38(9):1187–1193. doi: 10.1002/art.1780380903. [DOI] [PubMed] [Google Scholar]
Websites
- 101.Center for Drug Evaluation and Research (CDER) and the Center for Biologics Evaluation and Research (CBER) at the Food and Drug Administration. Guidance for Industry Immunogenicity Assessment for Therapeutic Protein Products, Assay Development for Immunogenicity Testing of Therapeutic Proteins. www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/default.htm. Accessed 1 December 2009.
- 102.Center for Drug Evaluation and Research (CDER) and the Center for Biologics Evaluation and Research (CBER) at the Food and Drug Administration. Guidance for Industry Immunogenicity Assessment for Therapeutic Protein Products, Guidance for Industry :Immunogenicity Assessment for Therapeutic Protein Products. www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM338856.pdf. Accessed 1 February 2013.