Abstract
CTX-M enzymes, the plasmid-mediated cefotaximases, constitute a rapidly growing family of extended-spectrum β-lactamases (ESBLs) with significant clinical impact. CTX-Ms are found in at least 26 bacterial species, particularly in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. At least 109 members in CTX-M family are identified and can be divided into seven clusters based on their phylogeny. CTX-M-15 and CTX-M-14 are the most dominant variants. Chromosome-encoded intrinsic cefotaximases in Kluyvera spp. are proposed to be the progenitors of CTX-Ms, while ISEcp1, ISCR1 and plasmid are closely associated with their mobilization and dissemination.
Keywords: CTX-M, cefotaximase, extended-spectrum β-lactamase (ESBL), ISEcp1, ISCR1, plasmid
Introduction
Extended-spectrum β-lactamases (ESBLs) are the most influential mechanism for cephalosporin resistance in Enterobacteriaceae, particularly in Escherichia coli and Klebsiella pneumoniae. ESBLs confer resistance to penicillins, broad-spectrum cephalosporins with an oxyimino side chain (cefotaxime, ceftriaxone and ceftazidime) and the oxyimino-monobactam aztreonam, but can be inhibited by serine-type β-lactamase inhibitors as sulbactam, clavulanate and tazobactam (Philippon et al., 1989; Bradford, 2001). SHV-2 is the first ESBL, identified in a clinical isolate of Klebsiella ozaenae in Germany (Kliebe et al., 1985). To date, over 10 families have been documented to be associated with ESBLs, including CTX-M, SHV, TEM, PER, VEB, BES, GES, TLA, SFO and OXA (Paterson and Bonomo, 2005).
CTX-M enzymes, the plasmid-mediated acquired cefotaximases from a distinct phylogenetic lineage, constitute a rapidly growing family of ESBLs with significant clinical impact (Bonnet, 2004; Cantón and Coque, 2006; Livermore et al., 2007; Naseer and Sundsfjord, 2011). Chromosome-encoded genes of intrinsic cefotaximases in Kluyvera spp. are proposed to be the progenitors of CTX-M family (Humeniuk et al., 2002; Olson et al., 2005; Decousser et al., 2011). Most of CTX-Ms exhibit powerful activity against cefotaxime and ceftriaxone but not ceftazidime. However, some CTX-Ms, such as CTX-M-15 (Poirel et al., 2002a), CTX-M-16 (Bonnet et al., 2001) and CTX-M-19 (Poirel et al., 2001), exhibit enhanced catalytic efficiencies against ceftazidime.
This article summarizes the epidemiology of CTX-M-producing Gram-negative bacteria and the genetics of CTX-M ESBLs, with a focus on the phylogeny, origin and genetic platforms including ISEcp1, ISCR1 and plasmid.
Epidemiology of CTX-M ESBLs
Occurrence and bacterial hosts
A plasmid-mediated cefotaximase was identified from a clinical isolate of E. coli in Munich, Germany, and designated CTX-M in reference to its hydrolytic activity and the region where it was found (Bauernfeind et al., 1990). To date, the numbers of CTX-M variants and the recognized organisms harboring the genes have dramatically increased. At least 109 CTX-M variants, CTX-M-1 to CTX-M-124, have been identified (Table 1) and assigned in the Lahey database (Jacoby and Bush, 2012). The amino-acid sequences of CTX-M-14 and CTX-M-18 and of CTX-M-55 and CTX-M-57 are identical, and CTX-M-118 has been withdrawn. There is no detailed information available for the assigned members CTX-M-70, -73, -100, -103, -115, -119, -120 and -124 so far. In addition, CTX-M-76, -77, -78 and -95 are chromosome-encoded intrinsic cefotaximases in Kluyvera spp., and therefore, they are not counted into the CTX-M family. CTX-M-2, -3 and -37 are plasmid-mediated enzymes but also found on chromosomes in Kluyvera spp. To clarify the differences, the term c-CTX-M is used for such chromosome-encoded CTX-Ms in this article. Of the studied CTX-Ms, at least 19 variants display the enhanced catalytic efficiencies against ceftazidime (Table 1).
Table 1. .
CTX-M ESBLs and their bacterial hosts.
| CTX-M (alternate name) | Bacterial host | GenBank accession no. | Reference |
|---|---|---|---|
| CTX-M-1 (MEN-1) | Escherichia coli | X92506 | Bauernfeind et al., 1996 |
| Enterobacter cloacae | al Naiemi et al., 2006 | ||
| Klebsiella pneumoniae | Komatsu et al., 2001 | ||
| Proteus mirabilis | al Naiemi et al., 2006 | ||
| Pseudomonas aeruginosa | al Naiemi et al., 2006 | ||
| Salmonella enterica | Rodríguez et al., 2009 | ||
| Serratia marcescens | Choi et al., 2007 | ||
| Stenotrophomonas maltophilia | al Naiemi et al., 2006 | ||
| CTX-M-2 | Salmonella enterica | X92507 | Bauernfeind et al., 1996 |
| Acinetobacter baumannii | Nagano et al., 2004 | ||
| Citrobacter koseri | al Naiemi et al., 2006 | ||
| Escherichia coli | Arduino et al., 2003 | ||
| Enterobacter cloacae | Arduino et al., 2003 | ||
| Klebsiella pneumoniae | Arduino et al., 2003 | ||
| Morganella morganii | Power et al., 2005 | ||
| Proteus mirabilis | Bonnet et al., 2000 | ||
| Providencia stuartii | Minarini et al. 2009 | ||
| Pseudomonas aeruginosa | Arduino et al., 2003 | ||
| Serratia marcescens | Arduino et al., 2003 | ||
| Vibrio cholerae | Soler Bistué et al., 2006 | ||
| CTX-M-3 | Citrobacter freundii | Y10278 | Gniadkowski et al., 1998 |
| Aeromonas caviae | Ye et al., 2010 | ||
| Escherichia coli | Yan et al., 2000 | ||
| Enterobacter cloacae | De Champs et al., 2000 | ||
| Enterobacter aerogenes | Liu et al., 2009 | ||
| Klebsiella pneumoniae | Baraniak et al., 2002b | ||
| Klebsiella oxytoca | Baraniak et al., 2002b | ||
| Morganella morganii | Baraniak et al., 2002b | ||
| Proteus mirabilis | Eckert et al., 2006 | ||
| Salmonella enterica | Gierczyński et al., 2003 | ||
| Sarratia marcescens | Baraniak et al., 2002b | ||
| Shigella flexneri | Galimand et al., 2005 | ||
| Shigella sonnei | Acikgoz et al., 2003 | ||
| CTX-M-4 | Salmonella enterica | Y14156 | Gazouli et al., 1998b |
| CTX-M-5 | Salmonella enterica | U95364 | Bradford et al., 1998 |
| Acinetobacter baumannii | AF462635 | ||
| CTX-M-6 (renumbered) | Salmonella enterica | AJ005044 | Gazouli et al., 1998a |
| CTX-M-7 (renumbered) | Salmonella enterica | AJ005045 | Gazouli et al., 1998a |
| CTX-M-8 | Citrobacter amalonaticus | AF189721 | Bonnet et al., 2000 |
| Enterobacter cloacae | Bonnet et al., 2000 | ||
| Enterobacter aerogenes | Bonnet et al., 2000 | ||
| Escherichia coli | Minarini et al. 2009 | ||
| CTX-M-9 | Escherichia coli | AF174129 | Sabaté et al., 2000 |
| Citrobacter freundii | Minarini et al. 2009 | ||
| Enterobacter aerogenes | EF441350 | ||
| Enterobacter cloacae | Chanawong et al., 2002 | ||
| Enterobacter hormaechei | Ho et al., 2005b | ||
| Klebsiella pneumoniae | Chanawong et al., 2002 | ||
| Klebsiella oxytoca | Alobwede et al., 2003 | ||
| Salmonella enterica | García Fernández et al., 2007 | ||
| Serratia marcescens | Choi et al., 2007 | ||
| CTX-M-10 | Escherichia coli | AF255298 | Oliver et al., 2001 |
| Citrobacter freundii | Valverde et al., 2004 | ||
| Enterobacter cloacae | Cantón et al., 2002 | ||
| Enterobacter gergoviae | Cantón et al., 2002 | ||
| Klebsiella pneumoniae | Coque et al., 2002 | ||
| Salmonella enterica | Cartelle et al., 2006 | ||
| CTX-M-11 | Klebsiella pneumoniae | AY005110 | |
| CTX-M-12 | Klebsiella pneumoniae | AF305837 | Kariuki et al., 2001 |
| Escherichia coli | Bae et al., 2006b | ||
| Proteus mirabilis | Song et al., 2011 | ||
| CTX-M-13 | Klebsiella pneumoniae | AF252623 | Chanawong et al., 2002 |
| Escherichia coli | DQ058147 | ||
| Enterobacter cloacae | AF462399 | ||
| Enterobacter hormaechei | Ho et al., 2005b | ||
| Proteus mirabilis | Ho et al., 2005a | ||
| CTX-M-14 | Escherichia coli | AF252622 | Chanawong et al., 2002 |
| Citrobacter freundii | Kanamori et al., 2011 | ||
| Citrobacter koseri | Kanamori et al., 2011 | ||
| Enterobacter cloacae | Chanawong et al., 2002 | ||
| Enterobacter hormaechei | Ho et al. 2005b | ||
| Klebsiella pneumoniae | Chanawong et al., 2002 | ||
| Proteus mirabilis | Ho et al., 2005a | ||
| Providencia stuartii | Liu et al., 2009 | ||
| Salmonella enterica | Romero et al., 2004 | ||
| Serratia liquefaciens | AF462398 | ||
| Shigella flexneri | DQ350883 | ||
| Shigella sonnei | Pai et al., 2001 | ||
| CTX-M-15 (UOE-1) * | Escherichia coli | AY044436 | Karim et al., 2001 |
| Acinetobacter baumannii | Shakil & Khan, 2010 | ||
| Aeromonas hydrophila | Gómez-Garcés et al., 2011 | ||
| Citrobacter freundii | HQ214043 | ||
| Citrobacter koseri | Kanamori et al., 2011 | ||
| Enterobacter aerogenes | Kim et al., 2005 | ||
| Enterobacter cloacae | Moubareck et al., 2005 | ||
| Enterobacter gergoviae | EU118595 | ||
| Klebsiella pneumoniae | Lartigue et al., 2003 | ||
| Klebsiella oxytoca | Zhang et al., 2008 | ||
| Morganella morganii | al Naiemi et al., 2006 | ||
| Pantoea agglomerans | Aibinu et al., 2012 | ||
| Proteus mirabilis | Song et al., 2011 | ||
| Salmonella enterica | Weill et al., 2004 | ||
| Serratia marcescens | Baraniak et al., 2002a | ||
| Shigella flexneri | Zhang et al., 2011 | ||
| Shigella sonnei | Hrabák et al., 2008 | ||
| CTX-M-16 * | Escherichia coli | AY029068 | Bonnet et al., 2001 |
| CTX-M-17 | Klebsiella pneumoniae | AY033516 | Cao et al., 2002 |
| CTX-M-18§ | Klebsiella pneumoniae | AF325133 | Poirel et al., 2001 |
| CTX-M-19 * | Klebsiella pneumoniae | AF325134 | Poirel et al., 2001 |
| CTX-M-20 | Proteus mirabilis | AJ416344 | Saladin et al., 2002 |
| CTX-M-21 | Escherichia coli | AJ416346 | Saladin et al., 2002 |
| CTX-M-22 | Klebsiella pneumoniae | AY080894 | Yu et al., 2007 |
| Escherichia coli | Yu et al., 2007 | ||
| Enterobacter cloacae | Liu et al., 2007 | ||
| Serratia liquefaciens | HM470254 | ||
| Serratia marcescens | DQ309026 | ||
| CTX-M-23 * | Escherichia coli | AF488377 | Stürenburg et al., 2004 |
| Klebsiella pneumoniae | Stürenburg et al., 2004 | ||
| CTX-M-24 | Klebsiella pneumoniae | AY143430 | Yu et al., 2007 |
| Escherichia coli | Yu et al., 2007 | ||
| Enterobacter aerogenes | Ho et al., 2005b | ||
| Proteus mirabilis | Wu et al., 2008 | ||
| Shigella sonnei | FN594520 | ||
| CTX-M-25 * | Escherichia coli | AF518567 | Munday et al., 2004 |
| Klebsiella pneumoniae | Navon-Venezia et al., 2008 | ||
| Proteus mirabilis | Navon-Venezia et al., 2008 | ||
| CTX-M-26 | Klebsiella pneumoniae | AY157676 | Brenwald et al., 2003 |
| CTX-M-27 * | Escherichia coli | AY156923 | Bonnet et al., 2003 |
| Salmonella enterica | Bouallègue-Godet et al., 2005 | ||
| Shigella sonnei | HM595763 | ||
| CTX-M-28 | Escherichia coli | AJ549244 | Galimand et al., 2005 |
| Enterobacter sp. | EU531513 | ||
| Klebsiella pneumoniae | Yu et al., 2007 | ||
| Salmonella enterica | Hasman et al., 2005 | ||
| CTX-M-29 | Escherichia coli | AY267213 | Yu et al., 2007 |
| CTX-M-30 | Citrobacter freundii | AY292654 | Abdalhamid et al., 2004 |
| CTX-M-31 | Providencia stuartii | AJ567481 | Quinteros et al., 2003 |
| Escherichia coli | Quinteros et al., 2003 | ||
| CTX-M-32 * | Escherichia coli | AJ557142 | Cartelle et al., 2004 |
| Klebsiella pneumoniae | Mendonça et al., 2009 | ||
| Proteus mirabilis | Fernández et al., 2007 | ||
| CTX-M-33 | Escherichia coli | AY238472 | Galani et al., 2007 |
| CTX-M-34 | Escherichia coli | AY515297 | Miró et al., 2005 |
| CTX-M-35 * | Klebsiella pneumoniae | AB176532 | |
| Citrobacter koseri | Tian et al., 2010 | ||
| Escherichia coli | AB176533 | ||
| Klebsiella oxytoca | AB176534 | ||
| CTX-M-36 | Escherichia coli | AB177384 | |
| CTX-M-37 * | Enterobacter cloacae | AY649755 | |
| Salmonella enterica | Govinden et al., 2006 | ||
| CTX-M-38 | Klebsiella pneumoniae | AY822595 | |
| CTX-M-39 | Escherichia coli | AY954516 | Chmelnitsky et al., 2005 |
| Enterobacter cloacae | Navon-Venezia et al., 2008 | ||
| Klebsiella pneumoniae | Navon-Venezia et al., 2008 | ||
| CTX-M-40 * | Escherichia coli | AY750914 | Hopkins et al., 2006 |
| CTX-M-41 | Proteus mirabilis | DQ023162 | Navon-Venezia et al., 2008 |
| CTX-M-42 * | Escherichia coli | DQ061159 | Stepanova et al., 2008 |
| CTX-M-43 | Acinetobacter baumannii | DQ102702 | Celenza et al., 2006 |
| Enterobacter aerogenes | Celenza et al., 2006 | ||
| Enterobacter cloacae | Celenza et al., 2006 | ||
| Morganella morganii | Celenza et al., 2006 | ||
| Pseudomonas aeruginosa | Celenza et al., 2006 | ||
| CTX-M-44 (Toho-1) | Escherichia coli | D37830 | Ishii et al., 1995 |
| CTX-M-45 (Toho-2) | Escherichia coli | D89862 | Ma et al., 1998 |
| CTX-M-46 | Klebsiella pneumoniae | AY847147 | Cheng et al., 2008 |
| CTX-M-47 | Escherichia coli | AY847143 | Cheng et al., 2008 |
| Klebsiella pneumoniae | Cheng et al., 2008 | ||
| CTX-M-48 | Klebsiella pneumoniae | AY847144 | Cheng et al., 2008 |
| Escherichia coli | Cheng et al., 2008 | ||
| CTX-M-49 | Klebsiella pneumoniae | AY847145 | Cheng et al., 2008 |
| CTX-M-50 | Klebsiella pneumoniae | AY847146 | Cheng et al., 2008 |
| CTX-M-51 | Escherichia coli | DQ211987 | |
| CTX-M-52 | Klebsiella pneumoniae | DQ223685 | |
| CTX-M-53 * | Salmonella enterica | DQ268764 | Doublet et al., 2009 |
| CTX-M-54 * | Klebsiella pneumoniae | DQ303459 | Bae et al., 2006a |
| CTX-M-55 * | Escherichia coli | DQ885477 | Kiratisin et al., 2007 |
| Klebsiella pneumoniae | Kiratisin et al., 2007 | ||
| Shigella sonnei | Zhang et al., 2011 | ||
| CTX-M-56 | Escherichia coli | EF374097 | Pallecchi et al., 2007 |
| CTX-M-57§ | Salmonella enterica | DQ810789 | Hopkins et al., 2008 |
| Shigella sonnei | EU086736 | ||
| CTX-M-58 * | Escherichia coli | EF210159 | |
| CTX-M-59 | Klebsiella pneumoniae | DQ408762 | de Oliveira et al., 2008 |
| CTX-M-60 | Klebsiella pneumoniae | AM411407 | |
| CTX-M-61 | Salmonella enterica | EF219142 | Brasme et al., 2007 |
| Klebsiella pneumoniae | Mendonça et al., 2009 | ||
| CTX-M-62 * | Klebsiella pneumoniae | EF219134 | Zong et al., 2008 |
| CTX-M-63 | Klebsiella pneumoniae | AB205197 | |
| Morganella morganii | EU660216 | ||
| Salmonella enterica | Pornruangwong et al., 2011 | ||
| CTX-M-64 * | Shigella sonnei | AB284167 | Nagano et al., 2009 |
| Escherichia coli | Sun et al., 2010 | ||
| Enterobacter cloacae | GQ300937 | ||
| CTX-M-65 | Escherichia coli | EF418608 | Doi et al. 2008 |
| Citrobacter freundii | EF394372 | ||
| Salmonella enterica | FJ907380 | ||
| CTX-M-66 | Proteus mirabilis | EF576988 | Wu et al., 2008 |
| CTX-M-67 | Escherichia coli | EF581888 | Oteo et al., 2008 |
| CTX-M-68 | Klebsiella pneumoniae | EU177100 | Heffernan et al., 2009 |
| CTX-M-69 | Escherichia coli | EU402393 | |
| CTX-M-70† | Assigned | ||
| CTX-M-71 | Klebsiella pneumoniae | FJ815436 | Schneider et al., 2009 |
| CTX-M-72 | Klebsiella pneumoniae | AY847148 | Cheng et al., 2009 |
| CTX-M-73† | Assigned | ||
| CTX-M-74 | Enterobacter cloacae | GQ149243 | Minarini et al., 2009 |
| CTX-M-75 | Providencia stuartii | GQ149244 | Minarini et al., 2009 |
| c-CTX-M-76‡ | Kluyvera ascorbata | AM982520 | |
| c-CTX-M-77‡ | Kluyvera ascorbata | AM982521 | |
| c-CTX-M-78‡ | Kluyvera georgiana | AM982522 | Rodríguez et al., 2010 |
| CTX-M-79 | Escherichia coli | EF426798 | Tian et al., 2008 |
| CTX-M-80 | Klebsiella pneumoniae | EU202673 | Cheng et al., 2010 |
| CTX-M-81 | Klebsiella pneumoniae | EU136031 | Cheng et al., 2010 |
| CTX-M-82 * | Escherichia coli | DQ256091 | Liu et al., 2009 |
| CTX-M-83 | Salmonella enterica | FJ214366 | Cui et al., 2009 |
| CTX-M-84 | Salmonella enterica | FJ214367 | Cui et al., 2009 |
| CTX-M-85 | Salmonella enterica | FJ214368 | Cui et al., 2009 |
| CTX-M-86 | Salmonella enterica | FJ214369 | Cui et al., 2009 |
| CTX-M-87 (renumbered) | Escherichia coli | EU545409 | Yin et al., 2009 |
| CTX-M-88 | Salmonella enterica | FJ873739 | Ranjbar et al., 2010 |
| CTX-M-89 | Proteus mirabilis | FJ971899 | McGettigan et al., 2009 |
| Enterobacter cloacae | FJ966096 | ||
| CTX-M-90 | Salmonella enterica | FJ907381 | |
| Proteus mirabilis | Song et al., 2011 | ||
| CTX-M-91 | Proteus mirabilis | GQ870432 | |
| CTX-M-92 | Escherichia coli | GU127598 | Seputiene et al., 2010 |
| Klebsiella pneumoniae | Seputiene et al., 2010 | ||
| CTX-M-93 * | Escherichia coli | HQ166709 | Djamdjian et al., 2011 |
| CTX-M-94 | Escherichia coli | HM167760 | |
| c-CTX-M-95‡ | Kluyvera ascorbata | FN813245 | |
| CTX-M-96 (CTX-M-12a) | Klebsiella pneumoniae | AJ704396 | |
| CTX-M-97 | Escherichia coli | HM776707 | |
| CTX-M-98 | Escherichia coli | HM755448 | |
| CTX-M-99 | Klebsiella pneumoniae | HM803271 | |
| CTX-M-100† | Assigned | ||
| CTX-M-101 | Escherichia coli | HQ398214 | |
| CTX-M-102 | Escherichia coli | HQ398215 | |
| CTX-M-103† | Assigned | ||
| CTX-M-104 | Escherichia coli | HQ833652 | |
| CTX-M-105 | Escherichia coli | HQ833651 | |
| CTX-M-106 | Escherichia coli | HQ913565 | |
| CTX-M-107 | Shigella flexneri | JF274244 | Zhang et al., 2011 |
| CTX-M-108 | Shigella flexneri | JF274245 | Zhang et al., 2011 |
| CTX-M-109 | Shigella flexneri | JF274248 | Zhang et al., 2011 |
| CTX-M-110 | Shigella sonnei | JF274242 | Zhang et al., 2011 |
| CTX-M-111 | Shigella flexneri | JF274243 | Zhang et al., 2011 |
| CTX-M-112 | Shigella sonnei | JF274246 | Zhang et al., 2011 |
| CTX-M-113 | Shigella flexneri | JF274247 | Zhang et al., 2011 |
| CTX-M-114 | Providencia rettgeri | GQ351346 | |
| CTX-M-115† | Assigned | ||
| CTX-M-116 | Proteus mirabilis | JF966749 | |
| CTX-M-117 | Escherichia coli | JN227085 | |
| CTX-M-118 | Withdrawn | ||
| CTX-M-119† | Assigned | ||
| CTX-M-120† | Assigned | ||
| CTX-M-121 | Escherichia coli | JN790862 | |
| CTX-M-122 | Escherichia coli | JN790863 | |
| CTX-M-123 | Escherichia coli | JN790864 | |
| CTX-M-124† | Assigned |
with enhanced catalytic efficiencies against ceftazidime;
have been assigned in the Lahey database (Jacoby and Bush 2012);
chromosome-encoded intrinsic cefotaximase identified in Kluyvera spp.;
CTX-M-18 and CTX-M-14, CTX-M-57 and CTX-M-55 are identical in their amino acid sequences.
CTX-Ms have been detected in at least 26 bacterial species, including Acinetobacter baumannii, Aeromonas caviae, A. hydrophila, Citrobacter amalonaticus, C. freundii, C. koseri, E. coli, Enterobacter cloacae, E. aerogenes, E. gergoviae, E. hormaechei, K. pneumoniae, K. oxytoca, Morganella morganii, Proteus mirabilis, Pantoea agglomerans, Providencia rettgeri, P. stuartii, Pseudomonas aeruginosa, Salmonella enterica, Shigella flexneri, S. sonnei, Serratia marcescens, S. liquefaciens, Stenotrophomonas maltophilia and Vibrio cholera (Table 1).
CTX-M enzymes as the most prevalent ESBLs in E. coli, K. pneumoniae and P. mirabilis
The high prevalence of CTX-M ESBL genes in Enterobacteriaceae, particularly in E. coli, K. pneumoniae and P. mirabilis, has been documented worldwide (Bonnet, 2004; Cantón and Coque, 2006), while the CTX-Ms are not prominent in P. aeruginosa and A. baumannii (Zhao and Hu, 2010, 2012).
A study on the resistance of Enterobacteriaceae to third-generation cephalosporin was undertaken in 16 British hospitals over a 12-week period (Potz et al., 2006). Of 19,252 clinical isolates, CTX-M-producing strains accounted for 1.7%, higher than other ESBLs-producing strains (0.6%) and high-level AmpC-producing strains (0.4%). Particularly, of the resistance isolates of E. coli (n = 574) and Klebsiella spp. (n = 243), the CTX-M-producing strains accounted for 50.9% and 81.9%, respectively, by contrast with other ESBLs-producing strains (15.3% and 11.1%), high-level AmpC-producing strains (7.1% and 0.8%) and non-β-lactamase-producing strains (26.7% and 3.3%).
A rapid occurrence of CTX-M-producing strains in Enterobacteriaceae was documented by several longitudinal surveillances. Of 20,258 E. coli isolates studied in Italy, the prevalence of ESBL-producing strains increased from 0.2% in 1999 to 1.6% in 2003, of which CTX-M-positive strains increased from 12.5% to 38.2% (Brigante et al., 2005). Of 1574 P. mirabilis clinical isolates collected in a Taiwanese hospital during 1999–2005, 44 CTX-M-producing strains were detected at a rate of 0.7% in 1999 and approximately 6% after 2002 (Wu et al., 2008). Of 11,407 E. coli isolates from urine samples of outpatients in the USA, 107 CTX-M-producing strains were detected at a rate of 0.07% in 2003 and 1.66% in 2008 (Qi et al., 2010).
CTX-M-producing strains widespread not only in human but also in animals and in environments. Of 240 E. coli isolates from health and sick pets during 2007–2008 in China, 97 strains (40.4%) harbored ESBL-encoding genes, of which 96 strains were confirmed to be carriers of bla CTX-M genes (Sun et al., 2010). Of 16 multi-drug resistant E. coli isolates from river water during 2000–2001 in South Korea, 10 strains harbored CTX-M-14 gene (Kim et al., 2008). Of 79 food samples of animal origin in Tunisia, bla CTX-M-1-positive E. coli strains were isolated from 10 samples (Ben Slama et al., 2010).
A Japanese group surveyed the spread status of CTX-M genes in nosocomial Gram-negative bacteria collected from 132 geographically distant medical facilities during 2001–2003. Of the 1456 isolates resistant to oxyimino-cephalosporins, 21.8% were found to harbor bla CTX-M genes. The prevalent rates of CTX-Ms in ESBL-producing E. coli, K. pneumoniae and P. mirabilis were 77% (168/218), 56% (50/90) and 99% (71/72), respectively, while the rates of CTX-Ms in ESBL-producing A. baumannii and S. marcescens were 4.5% (4/89) and 7% (10/149), respectively (Shibata et al., 2006).
CTX-M-15 and CTX-M-14 as the most dominant variants in CTX-M family
Although the dominant variants of CTX-Ms are geographically different, CTX-M-15 and CTX-M-14 are the most common variants detected worldwide in clinically important pathogens, followed by CTX-M-2, CTX-M-3 and CTX-M-1 (Table 1). Conjugative plasmid-mediated horizontal transfer and clonal spread contributed to the increased prevalence.
Of 171 CTX-M-producing E. coli isolates from 11 Canadian medical centers in 2007, the positive rates for CTX-M-15, CTX-M-14, CTX-M-3 and CTX-M-27 were 86.5%, 9.9%, 2.9% and 0.6%, respectively (Peirano et al., 2010). Of 202 CTX-M-producing K. pneumoniae isolates from 41 medical centers in Hungary in 2005, 97% were CTX-M-15 producers derived from three genetically distinct clones (Damjanova et al., 2008). Of the CTX-M-producers (288 E. coli and 142 K. pneumoniae isolates) collected from 6 provinces in China during 1998–2002, CTX-M-14 was predominantly detected in 77.4% and 52.8% of the isolates, respectively, followed by CTX-M-3 (18.4% and 29.6%), CTX-M-24 (5.6% and 14.1%) and CTX-M-15 (0.7% and 1.4%) (Yu et al., 2007). An outbreak of CTX-M-producing S. enterica infection occurred in a university hospital in Algeria during 2008–2009, and all of 200 isolates from 138 patients were CTX-M-15 producers, identified to be a single clone (Naas et al., 2011).
Of 44 clinical isolates of CTX-M-producing P. mirabilis from a Taiwanese hospital, CTX-M-14 and CTX-M-3 positive strains accounted for 50% and 40.9%, respectively (Wu et al., 2008). Of 71 CTX-M-producing P. mirabilis isolates collected from 132 geographically distant hospitals in Japan, however, 100% of the strains carried the bla CTX-M-2-like genes (Shibata et al., 2006). CTX-M-2 was also predominant in C. koseri, accounting for 76.7% of ESBL-producing strains (n = 60) collected from 10 areas throughout Japan in a 5-month period between 2009 and 2010 (Kanamori et al., 2011).
Phylogeny, origin and evolution of CTX-M enzymes
Amino-acid identity and phylogeny
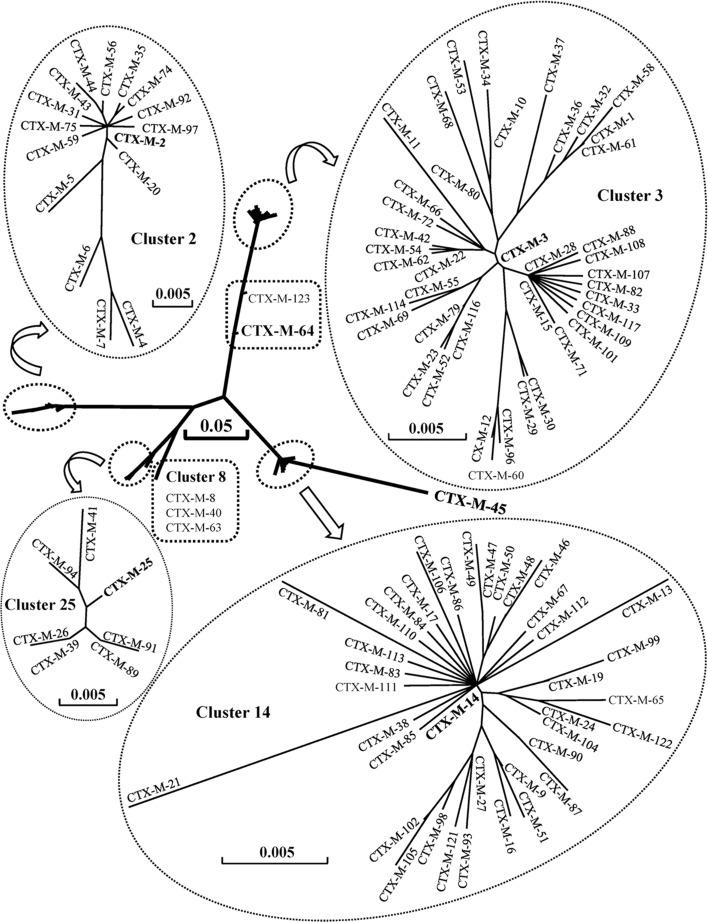
The deduced amino-acid sequences of CTX-Ms comprise 291 residues, with the exceptions of CTX-M-11 (282), CTX-M-107 and -108 (288), CTX-M-45 and -109 (289), CTX-M-40, -63 and -106 (290) and CTX-M-110 (292). Based on the phylogenetic tree of amino-acid sequences, CTX-M enzymes may be divided into seven clusters (Figure 1).
Figure 1. .
Phylogenetic tree of CTX-M family based on amino-acid sequences. DNASIS Pro v2.10 (Hitachi Software Engineering Co., Tokyo, Japan) was used to align the amino-acid sequences and construct the phylogenetic tree. The amino-acid sequences were downloaded from GenBank under the accession numbers cited in Table 1. The branch lengths are drawn to scale and are proportional to the number of different amino-acid residues. The scale bars of 0.05 and 0.005 represent 5% and 0.5% amino-acid difference, respectively.
CTX-M-3 cluster includes 42 members, sharing 97.6–99.7% identity in amino-acid sequences. The other clusters are as follows: CTX-M-14 cluster, 38 members, 97.3–99.7% identity; CTX-M-2 cluster, 16 members, 95.2–99.7% identity; CTX-M-25 cluster, 7 members, 98.6–99.7% identity; CTX-M-8 cluster, 3 members, 97.9–99.7% identity; CTX-M-64 cluster, 2 members, 95.9% identity. There is only one member in CTX-M-45 cluster. Among CTX-M variants, CTX-M-4 and CTX-M-45 are most divergent with 91 amino-acid substitutions.
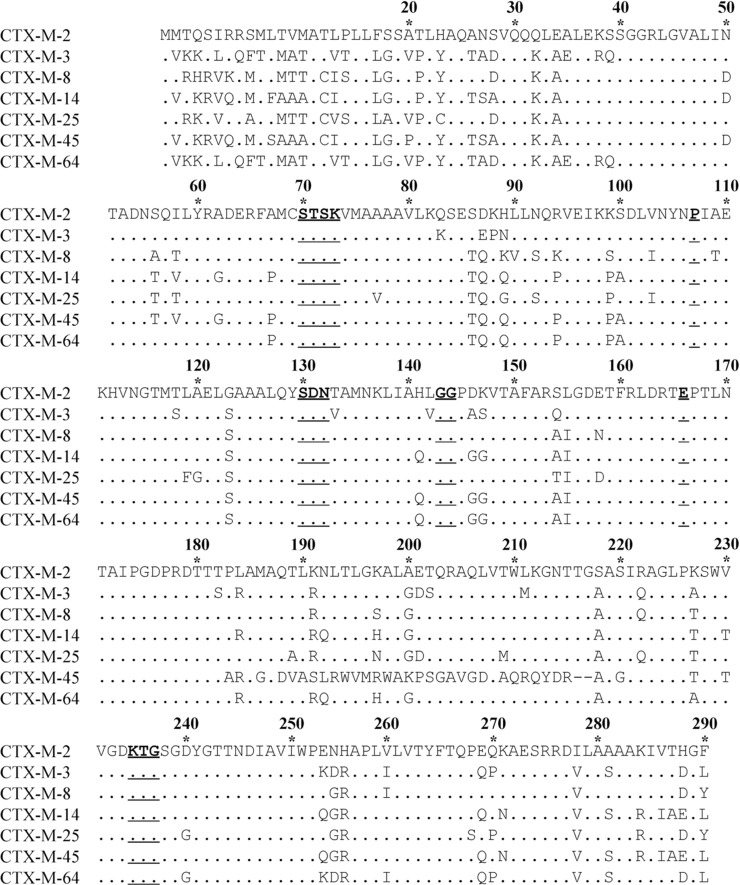
Variations of amino-acid sequences
Based on the central positions in phylogenetic tree (Figure 1), CTX-M-2, -3, -8, -14, -25, -45 and -64 are chosen as the representative enzymes in each cluster. The amino-acid sequences of the seven enzymes are aligned, and numbered according to the standard numbering scheme for the class A serine β-lactamases, giving the active site serine residue the Ambler number 70 (Ambler et al., 1991) (Figure 2). The sequences of CTX-M variants are then compared with their representative in each cluster (Table 2). In the CTX-M-3 cluster, for example, a single amino-acid is substituted between CTX-M-3 and CTX-M-15, -22, -42, -54, -62, -66, -72 or -80, while 5 amino-acids are substituted between CTX-M-3 and CTX-M-58.
Figure 2. .
Comparison of amino-acid sequences of seven representative enzymes in the CTX-M family. Amino-acids are numbered according to the standard numbering scheme for the class A serine β-lactamases, giving the active site serine residue the Ambler number 70. Dots indicate identical amino-acids compared to CTX-M-2. Deletion mutations are expressed with short lines. The underlined amino-acids, 70SXXK73, 107P, 130SDN132, 143GG144, 166E and 234KXG236, represent the conserved residues in typical class A serine β-lactamases.
Table 2. .
Amino acid substitutions of CTX-M variants compared to their representative enzymes.
| CTX-M | Amino acid substitution | CTX-M | Amino acid substitution |
|---|---|---|---|
| Cluster 2 | vs. CTX-M-2 | Cluster 8 | vs. CTX-M-8 |
| CTX-M-4 | L48Q, R61V, K98R, K99A, A125G, T171S, L225M, V230G | CTX-M-40 | K89N, T109A, N158D, N192H |
| CTX-M-5 | A26T, V230G, E253A, I278V | CTX-M-63 | K89N, T109A, N158D, N192H, S274N |
| CTX-M-6 | R61L, K99A, A125G, T171S, S228C, I278V | Cluster 14 | vs. CTX-M-14 |
| CTX-M-7 | R61V, K98R, K99A, E121Q, A125G, T171S, V230G, I278V | CTX-M-9 | V231A |
| CTX-M-20 | I278F | CTX-M-13 | V2M, A52K, A154E |
| CTX-M-31 | T159S | CTX-M-16 | V231A, D240G |
| CTX-M-35 | P167S | CTX-M-17 | E288K |
| CTX-M-43 | D240G, S274R | CTX-M-19 | P167S |
| CTX-M-44 | S274R | CTX-M-21 | A9G, A10G, C12G, L22F, V29G |
| CTX-M-56 | S274N | CTX-M-24 | S274R |
| CTX-M-59 | H89L | CTX-M-27 | D240G |
| CTX-M-74 | P167T | CTX-M-38 | S220R |
| Cluster 2 | vs. CTX-M-2 | Cluster 14 | vs. CTX-M-14 |
| CTX-M-75 | P14S | CTX-M-46 | S27N, A47P |
| CTX-M-92 | A205T | CTX-M-47 | G42R |
| CTX-M-97 | R3G | CTX-M-48 | S27N |
| Cluster 3 | vs. CTX-M-3 | CTX-M-49 | G42R, A47P |
| CTX-M-1 | A77V, N114D, A140S, D288N | CTX-M-50 | A47P |
| CTX-M-10 | A27V, R38Q | CTX-M-51 | A77V, V231A |
| CTX-M-11 | E35G, L119P, D277H, deletion of 282AAKIVTDGL290 | CTX-M-65 | A77V, S274R |
| CTX-M-12 | T12A, N89S, V278I | CTX-M-67 | N106S |
| CTX-M-15 | D240G | CTX-M-81 | K82E, K98Q, N132H |
| CTX-M-22 | D288N | CTX-M-83 | Q56H |
| CTX-M-23 | A77V, P167T, D288N | CTX-M-84 | T209A |
| CTX-M-28 | D240G, D288N | CTX-M-85 | L119P |
| CTX-M-29 | T12A, N114D, D240G, D288N | CTX-M-86 | I108F |
| CTX-M-30 | T12A, N114D | CTX-M-87 | A77V, P167L |
| CTX-M-32 | A77V, N114D, A140S, D240G, D288N | CTX-M-90 | A77V |
| CTX-M-33 | N106S, D240G | CTX-M-93 | L169Q, D240G |
| CTX-M-34 | A27V, R38Q, G238C | CTX-M-98 | A77V, D240G |
| CTX-M-36 | N114D, A140S, D288N | CTX-M-99 | P167S, S274R |
| CTX-M-37 | Y23H, R38Q, N114D | CTX-M-102 | A205E, D240G |
| CTX-M-42 | P167T | CTX-M-104 | S274N |
| CTX-M-52 | A77V, P167S | CTX-M-105 | A77V, A205E, D240G |
| CTX-M-53 | A27V, R38Q, A77V, D240G, T263I | CTX-M-106 | K234R, R276H, deletion of 290L |
| CTX-M-54 | P167Q | CTX-M-110 | K111E, insertion of N before 290L |
| CTX-M-55 | A77V, D240G | CTX-M-111 | P145Q |
| CTX-M-58 | A77V, N114D, A140S, P167T, D288N | CTX-M-112 | S123G |
| CTX-M-60 | T12A, N89S, V278I, A77V | CTX-M-113 | Q83R |
| CTX-M-61 | A77V, N114D, A140S | CTX-M-121 | A109T, D240G |
| CTX-M-62 | P167S | CTX-M-122 | A154S, S274R |
| CTX-M-66 | S19N | Cluster 25 | vs. CTX-M-25 |
| CTX-M-68 | Y23H, A27V, E158D | CTX-M-26 | V77A, Q222R, G240D |
| CTX-M-69 | A77V, D240G, K271N, D288N | CTX-M-39 | V77A, G240D |
| CTX-M-71 | G238C, D240G | CTX-M-41 | V77A, I103V, S123I |
| CTX-M-72 | R164G | CTX-M-89 | G240D |
| CTX-M-79 | A77V, D240G, D288N | CTX-M-91 | A189S, G240D |
| CTX-M-80 | A27V | CTX-M-94 | V77A, F119L |
| CTX-M-82 | A67P, D240G | Cluster 64 | vs. CTX-M-64 |
| CTX-M-88CTX-M-96 | D240G, R276HT12A, N89S, D240G, V278I | CTX-M-123 | P67A, Q83K, T86S, Q87E, K88P, Q89N, P94R, P99K, A100S, T118S, A227T, V230T |
| CTX-M-101 | S123I, D240G | T118S, A227T, V230T | |
| CTX-M-107 | K234R, D240G, deletion of 288DGL290 | ||
| CTX-M-108 | V95A, D240G, deletion of 288DGL290 | ||
| CTX-M-109 | Q56R, D240G, D288K, deletion of 289GL290 | ||
| CTX-M-114 | V74A, A77V, D240G | ||
| CTX-M-116 | A77V, D288N | ||
| CTX-M-117 | P174Q, D240G |
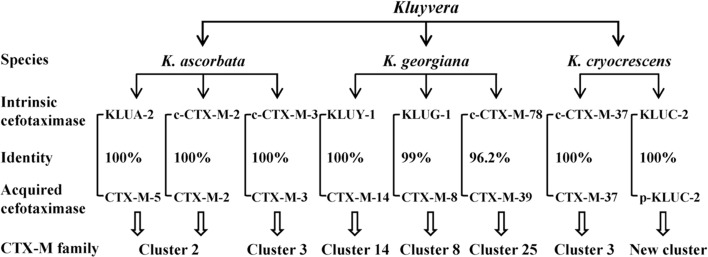
Origin of CTX-M family
In the family Enterobacteriaceae, the genus Kluyvera is a relatively new member, which has been isolated from various clinical specimens and regarded as a potentially virulent pathogen (Sarria et al., 2001). Some Kluyvera spp. harbor chromosome-encoded intrinsic genes of cefotaximases which are closely associated with CTX-Ms (Decousser, et al., 2001; Humeniuk et al., 2002; Rodríguez et al., 2004). Generally, Kluyvera spp. are susceptible to cefotaxime in despite of the presence of naturally occurring cefotaximases. However, the recombinant clones of E. coli with Kluyvera-derived cefotaximase genes exhibited a significant increase in resistance to cefotaxime (Decousser et al., 2001; Humeniuk et al., 2002; Rodríguez et al., 2004), suggesting that a proper genetic platform is necessary for the gene expression. The chromosome-encoded cefotaximases identified in Kluyvera spp. include KLUA, KLUG, KLUY, KLUC, c-CTX-M-2, c-CTX-M-3, c-CTX-M-37, c-CTX-M-76, c-CTX-M-77, c-CTX-M-78 and c-CTX-M-95. All of them comprise 291 amino-acid residues. An aspartate aminotransferase-encoding gene is found commonly upstream of these chromosomal bla genes, which is replaced by ISEcp1 or ISCR1 in the plasmid-harbored bla CTX-M genes (see the details under next section).
KLUA-1 to -5 and -8 to -12 (GenBank accession no. AJ272538, AJ251722, AJ427461, AJ427462, AJ427463, AJ427465, AJ427466, AJ427467, AJ427468, AJ427469) are a group of chromosomal cefotaximases identified in K. ascorbata, with minor variations (<5%) in their amino-acid sequences (Humeniuk et al., 2002). KLUA-2 shares 100% identity with plasmid-mediated CTX-M-5. CTX-M-2 and CTX-M-3 originally identified on plasmids were also found on the chromosomes of K. ascorbata (Rodríguez et al., 2004; Lartigue et al., 2006). The immediate upstream- and downstream-sequences of bla KLUA-1 and plasmid-mediated bla genes in CTX-M-2 cluster (bla CTX-M-2, -4, -5, -6, -7,-44) share 85 to 100% identities (Di Conza et al., 2002; Humeniuk et al., 2002). The architectures of the flanking regions corresponding to c-CTX-M-3 and plasmid-mediated CTX-M-3 are identical, including a 128 bp immediate upstream region and the first 373 bp of the downstream region of the bla gene (Rodríguez et al., 2004). The c-CTX-M-76, -77 and -95 (AM982520, AM982521, FN813245) identified in K. ascorbata also share high identities with the enzymes in CTX-M-2 cluster.
KLUY-1 to -4 (AY623932, AY623935, AY623934, AY623933) are a group of chromosomal cefotaximases identified in K. Georgiana (Olson et al., 2005). They share high homology with the enzymes in CTX-M-14 cluster. Typically, KLUY-1 exhibits 100% amino-acid identity with CTX-M-14. The upstream- and downstream-sequences of bla KLUY and bla CTX-M-9, -13, -14 also share consistent identity. A 42 bp upstream region of bla CTX-M-14 is identical to the corresponding region of bla KLUY genes. A 347 bp downstream region of bla CTX-M-9 and bla CTX-M-13 shares 95.7–98.6% identities with the corresponding region of bla KLUY genes (Olson et al., 2005).
KLUG-1 (AF501233) and c-CTX-M-78 (AM982522) are the chromosomal cefotaximases identified in K. Georgiana. KLUG-1 shares 99% amino-acid identity with the plasmid-mediated CTX-M-8 (Poirel et al., 2002b). The c-CTX-M-78 possesses high homology with the known members of CTX-M-25 cluster, sharing 95.2–96.2% identities (Rodríguez et al., 2010).
CTX-M-37 was also found on the chromosome of K. cryocrescens (FN813246), suggesting the c-CTX-M-37 as an origin of CTX-M-3 cluster. KLUC-1 (AY026417) and KLUC-2 (EF057432), with a single amino-acid substitution, are two chromosome-encoded cefotaximases identified in K. cryocrescens (Decousser et al., 2001). KLUC-1 and -2 are diverse from the known CTX-Ms, sharing only 87.6% identity with CTX-M-3. Notably, KLUC-2 was also identified on a plasmid carried by a clinical isolate of E. cloacae, indicating the transfer of bla KLUC from chromosome to the plasmid (Petrella et al., 2008). We would like to suggest the plasmid-mediated KLUC-2 as a novel cluster or member of CTX-M family.
CTX-M-64 shows a chimeric sequence of both CTX-M-14 (central portion) and CTX-M-15 (N- and C-terminal moieties), suggesting an origination owing to homologous recombination between the bla CTX-M-14 and -15 genes (Nagano et al., 2009).
Taken together, the origins of the acquired CTX-Ms in various clusters can be traced back to the intrinsic cefotaximase genes harbored by Kluyvera spp., of which the CTX-M-2 cluster appears to be derived from K. ascorbata, the CTX-M-14, CTX-M-8 and CTX-M-25 clusters from K. georgiana, while the CTX-M-3 cluster from both K. ascorbata and K. cryocrescens (Figure 3).
Figure 3. .
Identification of intrinsic cefotaximase genes in Kluyvera spp. as the original sources of acquired CTX-Ms based on their amino-acid identities and the homologies of neighboring sequences of the associated genes. c-CTX-M, CTX-M identified on chromosome of Kluyvera spp.; p-KLUC-2, KLUC-2 identified on plasmid in a clinical isolate of Enterobacter cloacae.
Genetic platforms of CTX-M enzymes
ISEcp1
Insertion sequences (ISs) are the smallest transposable elements (<2.5 kb) capable of independent transposition in an organism, thereby causing insertion mutations and genome rearrangements (Mahillon and Chandler, 1998). ISs play three basic roles in bacteria: encoding a transposase which makes a genetic element mobile; providing promoters to activate silent genes or enhance expression of downstream determinants; moving IS-mobilized genes among integrons, transposons, plasmids and chromosomes, thereby greatly increasing the opportunity a resistance determinant becomes transferable.
Of the genetic platforms associated with CTX-Ms, ISEcp1 is one of the most important elements (Table 3). ISEcp1 was first identified on the plasmid pST01 in E. coli strain 79 (AJ242809), hence its name (Stapleton, 1999). ISEcp1 is composed of an orf encoding a transposase with 420 amino-acids and two imperfect and inverted repeats. ISEcp1 can mobilize the downstream-located bla CTX-M gene and provide a promoter for its expression (Karim et al., 2001; Cao et al., 2002; Poirel et al., 2003, 2005; Dhanji et al., 2011b).
Table 3. .
Genetic platforms of CTX-M enzymes.
| CTX-M | Genetic platform | Bacterial host | Reference/GenBank accession no. |
|---|---|---|---|
| CTX-M-1 | ISEcp1–bla CTX-M-1 –orf477 | E. coli | Eckert et al., 2006 |
| ISEcp1Δ----IS26–ISEcp1Δ–bla CTX-M-1 | K. pneumoniae | Diestra et al., 2009 | |
| IS26–ISEcp1Δ–bla CTX-M-1 – orf477Δ | E. coli | Cullik et al., 2010 | |
| intI1–dfrA17–aadA5–qacEΔ1–sul1–ISCR1–bla CTX-M-1 –orf3–IS3000–qacEΔ1–sul1-like–orf5 | E. coli | Su et al., 2008 | |
| CTX-M-2 | intI1–aacA4–bla OXA-2 –orfD–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1 | P. mirabilis | Arduino et al., 2002 |
| intI1–aacA4–bla OXA-2 –orfD–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1–orf5 | V. cholera | Soler Bistué et al., 2006 | |
| intI1–aacA4–bla OXA-2 –orfD–qacEΔ1–sul1–ISCR1–dfrA10–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1–orf5–tniBΔ–IS1326 | S. enterica | AJ311891 | |
| intI1–dfrA12–orfF–aadA2–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1–orf5–IS1326 | K. pneumoniae | EU780013 | |
| intI1–estX–aadA1–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1–orf5–IS1326 | E. coli | Valverde et al., 2006 | |
| intI1–aac(6′)-Iq–aadA1–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1 | K. pneumoniae | EU622037 | |
| intI1–aadA1–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1 | K. pneumoniae | EU622040 | |
| intI1–aadA2–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1 | K. pneumoniae | EU622038 | |
| intI1–dhfrh1–aadA2–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1 | E. coli | Eckert et al., 2006 | |
| intI1–dfrA1–aadA1–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1 | S. enterica | EF592570 | |
| intI1–dfrA12–orfF–aadA2–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1 | S. enterica | EF592571 | |
| intI1–dfrA21–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1 | K. pneumoniae | EU622039 | |
| intI1–dfr22–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1 | K. pneumoniae | EU622041 | |
| intI1–orf1–cat–orf2–aadA1–qacEΔ1–sul1–ISCR1–bla CTX-M-2 –orf3Δ–qacEΔ1–sul1 | P. mirabilis | Song et al., 2011 | |
| ISEcp1–bla CTX-M-2 | P. mirabilis | Harada et al., 2012 | |
| CTX-M-3 | ISEcp1–bla CTX-M-3 –orf477 | K. pneumoniae | Eckert et al., 2006 |
| ISEcp1–bla CTX-M-3 –orf477–mucA | K. pneumoniae | Eckert et al., 2006 | |
| ISEcp1-like–bla CTX-M-3 –orf477-like | P. mirabilis | Wu et al., 2008 | |
| ISEcp1–bla CTX-M-3 | E. coli | Diestra et al., 2009 | |
| ISEcp1–IS1–bla CTX-M-3 –orf477–mucA | K. pneumoniae | Eckert et al., 2006 | |
| ISEcp1–bla CTX-M-3 –orf–mucA | C. freundii | Lartigue et al., 2004 | |
| IS26–ISEcp1Δ–bla CTX-M-3 | E. coli | Diestra et al., 2009 | |
| IS26–ISEcp1–bla CTX-M-3 –orf477–mucA | P. mirabilis | Eckert et al., 2006 | |
| CTX-M-9 | intI1–aadB–qacEΔ1–sul1–ISCR1–bla CTX-M-9 –orf3-like–IS3000 | E. cloacae | DQ108615 |
| intI1–dhfr12–orfX–aadA8–qacEΔ1–sul1–ISCR1–bla CTX-M-9 –orf3–orf339Δ | E. coli | Eckert et al., 2006 | |
| intI1–dfrA16–aadA2–qacEΔ1–sul1–ISCR1–bla CTX-M-9 –orf3-like–IS3000–qacEΔ1–sul1 | E. coli | Sabaté et al., 2002 | |
| ISCR1–bla CTX-M-9 | E. coli | Diestra et al., 2009 | |
| ISEcp1–bla CTX-M-9 | C. freundii | Minarini et al. 2009 | |
| CTX-M-10 | Tn1000-like–orf2–orf3–orf4–DNA-invertase-gene–bla CTX-M-10 –orf7–orf8–IS4321–orf10–orf11–IS5 | K. pneumoniae | Oliver et al., 2005 |
| ISEcp1–bla CTX-M-10 –orf–Tn5396 | E. coli | Lartigue et al., 2004 | |
| CTX-M-12 | ISEcp1–bla CTX-M-12 | P. mirabilis | Song et al., 2011 |
| CTX-M-13 | ISEcp1B–bla CTX-M-13 | E. coli | DQ058147 |
| CTX-M-14 | ISEcp1–bla CTX-M-14 –IS903 | E. coli | Lartigue et al., 2004 |
| ISEcp1-like–bla CTX-M-14 –IS903-like | P. mirabilis | Wu et al., 2008 | |
| ISEcp1–IS10–bla CTX-M-14 –IS903 | E. coli | Eckert et al., 2006 | |
| ISEcp1–IS10–bla CTX-M-14 –IS903D | E. coli | EU136400 | |
| IS26–ISEcp1–bla CTX-M-14 | K. pneumoniae | Eckert et al., 2006 | |
| IS26–ISEcp1–bla CTX-M-14 –IS903 | K. pneumoniae | GQ385317 | |
| IS26–bla CTX-M-14 –IS903D | S. enterica | Izumiya et al., 2005 | |
| ISEcp1B–bla CTX-M-14 | E. coli | Billard-Pomares et al., 2011 | |
| intI1–dfrA12–orfF–aadA2–qacEΔ1–sul1–ISCR1–bla CTX-M-14 –IS903-like | E. coli | Bae et al., 2007 | |
| intI1–dfrA12–orfF–aadA2–qacEΔ1–sul1–orf5–IS6100–ISCR1–ISEcp1Δ–bla CTX-M-14 –IS903D | E. coli | Bae et al., 2008 | |
| CTX-M-15 | ISEcp1–bla CTX-M-15 | A. hydrophila | Gómez-Garcés et al., 2011 |
| ISEcp1–bla CTX-M-15 –orf477 | E. coli | Eckert et al., 2006 | |
| ISEcp1–bla CTX-M-15 –orf477Δ–Tn3 | A. baumannii | JN788267 | |
| Tn3Δ–ISEcp1–bla CTX-M-15 –orf–Tn3Δ | E. coli | Lartigue et al., 2004 | |
| IS26–ISEcp1–bla CTX-M-15 –orf477 | E. coli | Eckert et al., 2006 | |
| IS26–ISEcp1–bla CTX-M-15 –orf477Δ | S. enterica | Fabre et al., 2009 | |
| bla TEM-1 –tnpR–tnpA–ISEcp1–bla CTX-M-15 –orf477 | E. coli | Eckert et al., 2006 | |
| CTX-M-16 | ISEcp1–bla CTX-M-16 –IS903 | E. coli | Brasme et al., 2007 |
| ISEcp1–bla CTX-M-16 –orf3–orf339–orf477 | E. coli | AM910790 | |
| CTX-M-17 | ISEcp1-like–bla CTX-M-17 –IS903C | K. pneumoniae | Cao et al., 2002 |
| CTX-M-19 | intI1-like–aacA4–cmlA1–qacEΔ1–sul1–Tn1721–ISEcp1B–bla CTX-M-19 –IS903D | K. pneumoniae | Poirel et al., 2003 |
| CTX-M-20 | ISEcp1–bla CTX-M-20 | P. mirabilis | AJ416344 |
| CTX-M-21 | ISEcp1–bla CTX-M-21 | E. coli | AJ416346 |
| CTX-M-22 | ISEcp1Δ–IS26–bla CTX-M-22 –orf477–ISEcp1Δ | S. liquefaciens | HM470254 |
| CTX-M-24 | ISEcp1–bla CTX-M-24 –IS903 | E. coli | Eckert et al., 2006 |
| ISEcp1-like–bla CTX-M-24 –IS903-like | P. mirabilis | Wu et al., 2008 | |
| CTX-M-25 | intI1–aacA4–bla OXA-2 –ISEcp1–bla CTX-M-25 –qacEΔ1–sul1 | P. mirabilis | Navon-Venezia et al. 2008 |
| ISEcp1Δ–IS50-A–ISEcp1Δ–bla CTX-M-25 –orfX | E. coli | Munday et al. 2004 | |
| CTX-M-26 | intI1–dhfr7–ISEcp1–bla CTX-M-26 –qacEΔ1–sul1 | K. pneumoniae | Navon-Venezia et al. 2008 |
| ISEcp1–bla CTX-M-26 –orfX | K. pneumoniae | Munday et al. 2004 | |
| CTX-M-27 | ISEcp1–bla CTX-M-27 | S. enterica | Bouallègue-Godet et al., 2005 |
| ISEcp1–bla CTX-M-27 –IS903 | E. coli | Sun et al., 2010 | |
| CTX-M-32 | ISEcp1Δ–IS5–IS1A–ISEcp1Δ–bla CTX-M-32 –orf477 | E. coli | Fernández et al., 2007 |
| ISEcp1Δ–IS5–ISEcp1Δ–bla CTX-M-32 | E. coli | Diestra et al., 2009 | |
| CTX-M-39 | intI1–dhfr7–ISEcp1–bla CTX-M-39 –qacEΔ1–sul1 | E. coli | Navon-Venezia et al. 2008 |
| intI1–aadA1–ISEcp1–bla CTX-M-39 –qacEΔ1–sul1 | E. coli | Navon-Venezia et al. 2008 | |
| CTX-M-40 | ISEcp1-like–bla CTX-M-40 | E. coli | Hopkins et al., 2006 |
| CTX-M-42 | ISEcp1–bla CTX-M-42 | E. coli | DQ061159 |
| CTX-M-53 | ISSen2---bla CTX-M-53 –orf477Δ–IS26 | S. enterica | Doublet et al., 2009 |
| CTX-M-54 | ISEcp1–bla CTX-M-54 –IS903-like | K. pneumoniae | Bae et al., 2006a |
| CTX-M-55 | ISEcp1–bla CTX-M-55 –orf477 | E. coli | Sun et al., 2010 |
| ISEcp1Δ–IS1294–bla CTX-M-55 –orf477 | E. coli | JN977127 | |
| CTX-M-59 | intI1–dfr15b–cmlA4-like–aadA2–qacEΔ1–sul1–ISCR1–bla CTX-M-59 –orf3Δ–qacEΔ1 | K. pneumoniae | EU622856 |
| CTX-M-62 | ISEcp1–bla CTX-M-62 –ISEcp1Δ1/Δ2 | K. pneumoniae | Zong et al., 2010 |
| CTX-M-64 | ISEcp1–bla CTX-M-64 –orf477 | S. sonnei | Nagano et al., 2009 |
| CTX-M-65 | ISEcp1–bla CTX-M-65 –IS903 | E. coli | Sun et al., 2010 |
| CTX-M-66 | ISEcp1-like–bla CTX-M-66 –orf477-like | P. mirabilis | Wu et al., 2008 |
| CTX-M-74 | ISCR1–bla CTX-M-74 –orf3Δ–qacEΔ1–sul1 | E. cloacae | Minarini et al. 2009 |
| CTX-M-75 | ISCR1–bla CTX-M-75 –orf3Δ–qacEΔ1–sul1 | P. stuartii | Minarini et al. 2009 |
| CTX-M-79 | ISEcp1–bla CTX-M-79 | E. coli | FJ169498 |
| CTX-M-82 | ISEcp1–bla CTX-M-82 | E. coli | GU477621 |
| CTX-M-89 | ISEcp1-like–bla CTX-M-89 –orf477-like | E. cloacae | FJ966096 |
| CTX-M-90 | ISEcp1–bla CTX-M-90 –IS903-like | P. mirabilis | Song et al., 2011 |
| ISEcp1–bla CTX-M-90 | P. mirabilis | Song et al., 2011 | |
| CTX-M-93 | ISEcp1–bla CTX-M-93 –IS903 | E. coli | Djamdjian et al., 2011 |
| CTX-M-98 | ISEcp1–bla CTX-M-98 –IS903 | E. coli | HM755448 |
| CTX-M-101 | ISEcp1–bla CTX-M-101 | E. coli | HQ398214 |
| CTX-M-102 | ISEcp1–bla CTX-M-102 –IS903 | E. coli | HQ398215 |
| CTX-M-104 | ISEcp1–bla CTX-M-104 –IS903 | E. coli | HQ833652 |
| CTX-M-105 | ISEcp1–bla CTX-M-105 –IS903 | E. coli | HQ833651 |
| CTX-M-116 | ISEcp1–bla CTX-M-116 | P. mirabilis | JF966749 |
| CTX-M-121 | ISEcp1–bla CTX-M-121 –IS903 | E. coli | JN790862 |
| CTX-M-122 | ISEcp1–bla CTX-M-122 –IS903 | E. coli | JN790863 |
| CTX-M-123 | ISEcp1–bla CTX-M-123 | E. coli | JN790864 |
Co-existence of ISEcp1 and bla CTX-M at a high rate in CTX-M-producing E. coli isolates is well documented. ISEcp1 was identified upstream of bla CTX-M genes in 86.9% of the isolates (93/107) recovered from health and sick pets in China, and no major clonal relatedness was observed (Sun et al., 2010). Similarly, ISEcp1 was identified upstream of bla CTX-M-14 in 91.4% of the clinical isolates (32/35) in Korea (Kim et al., 2011), and upstream of bla CTX-M-1 in 69.2% of the isolates (9/13) from food samples in Tunisia (Ben Slama et al., 2010). In addition, variations of ISEcp1 were also observed. ISEcp1B, originally identified upstream of a bla CTX-M-19 gene cassette (AF458080), differs from ISEcp1 by three nucleotide substitutions (Poirel, et al., 2003). Of the 174 ISEcp1-like and bla CTX-M-15 complex from E. coli isolates, the intact ISEcp1, truncated ISEcp1 with various lengths and a 24 bp remnant of ISEcp1 accounted for 62%, 33.3% and 4.6%, respectively (Dhanji et al., 2011b). Notably, ISEcp1 was also detected upstream of chromosomal bla CTX-M-2 genes in 4 P. mirabilis isolates in Japan (Harada et al., 2012), highlighting the ISEcp1-mediated movement of bla CTX-M genes between plasmids and chromosomes.
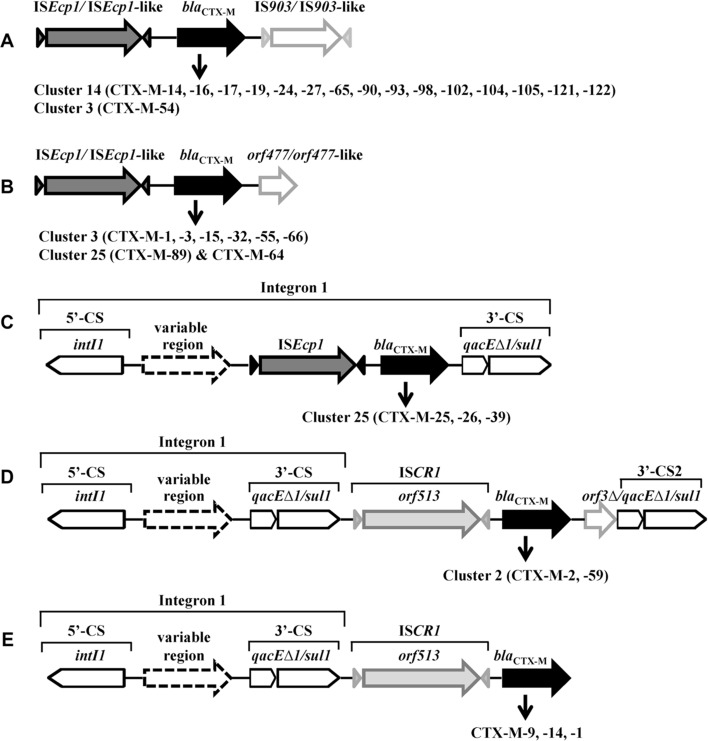
ISEcp1-bla CTX-M-IS903 (Figure 4A) and ISEcp1-bla CTX-M-orf477 (Figure 4B) are two major genetic platforms. In some cases, ISEcp1-mobilized bla CTX-M is inserted in a class 1 integron (Figure 4C). IS903 (V00359) encodes a transposase with 307 amino-acids and was originally found on a kanamycin resistance transposon Tn903 (Oka et al., 1981). IS903 and IS903-like elements, such as IS903C and IS903D, are located downstream of bla CTX-M genes (Table 3), including bla CTX-M-14-like genes (blaCTX-M-14, -16, -17, -19, -24, -27, -65, -90, -93, -98, -102, -104, -105, -121, -122) and bla CTX-M-3-like gene (bla CTX-M-54). orf477 encodes a protein of 158 amino-acids with unknown function and the orf477 and orf477-like elements were found downstream of plasmid-harbored bla CTX-M-3-like genes (bla CTX-M-1, -3, -15, -22, -32, -53, -55, -66), bla CTX-M-89 and bla CTX-M-64 (Table 3). The orf477 was also identified downstream of the chromosomal bla CTX-M-3 in K. ascorbata, of the chromosomal bla KLUY-1, -2, -3, -4 in K. georgiana, and of the chromosomal bla CTX-M-37 (FN813246) in K. cryocrescens (Rodriguez et al., 2004; Olson et al., 2005), footnoting the ISEcp1-mediated transfer of bla CTX-M genes together with the orf477 from the chromosomes of Kluyvera spp. to plasmids.
Figure 4. .
Typical genetic platforms of CTX-M enzymes. A & B: the bla CTX-M gene cassettes bracketed upstream by ISEcp1/ISEcp1-like and downstream by IS903/IS903-like (A) or orf477/orf477-like (B); C: bla CTX-M genes associated with class 1 integron-ISEcp1; D & E: bla CTX-M genes associated with class 1 integron-ISCR1 complex. CS, conserved segment; intI, integrase gene; qacE▵1, quaternary ammonium resistance gene; sul1, sulphonamide resistance gene; 3′-CS2, the second copy of 3′-conserved segment.
Class 1 integron-ISCR1 complex
Integrons are defined as mobile DNA elements that can capture genes by site-specific recombination (Stokes and Hall, 1989). A typical class 1 integron consists of a 5′ conserved segment (5′-CS), a variable region and a 3′ conserved segment (3′-CS). The 5′-CS consists of the gene encoding integrase (intI1), the site adjacent to intI1 for the insertion of captured genes (attI), and a promoter region (Pc). The 3′-CS often consists of a partially deleted qac gene (qacEΔ1) fused to a sul1 gene, and confers resistance to antiseptics and sulfonamide, respectively. Class 1 integrons play a critical role in acquiring and spreading metallo-β-lactamases (Mazel, 2006; Zhao and Hu, 2011a,b). The role of integrons in CTX-M gene acquisition and dissemination, however, is still unclear. The physical link of some bla CTX-M genes with class 1 integron-ISEcp1 complex (Figure 4C) and class 1 integron-ISCR1 complex (Figure 4D, 4E) indicates a possible association among the three genetic elements.
ISCR1 is another important element in the genetic platforms associated with the mobilization and dissemination of CTX-M genes (Rodriguez-Martinez et al. 2006; Toleman et al., 2006). Common region 1 (CR1) was first found as element associated with but distinct from class 1 integrons (Stokes et al., 1993). The CR1 element was renamed ISCR1 because it possesses the key motifs of IS91-like element and accommodates orf513 gene which codes a putative transposase of 513 amino-acids (Toleman et al., 2006). ISCR1 is particularly important for CTX-M-2 and CTX-M-9 genes (Table 3). In most instance, the ISCR1-bla CTX-M-2 is located between a typical class 1 integron and a fuse type of orf3Δ and qacEΔ1/sul1 (Table 3, Figure 4D). Notably, the genes harbored by class 1 integrons in their variable regions, such as bla OXA-2, aacA4, cmlA and dfr, are also associated with bacterial resistance to β-lactam, aminoglycoside, chloramphenicol and trimethoprim, respectively.
Molecular epidemiological study performed in Argentine during 1993–2000 showed that class 1 integron-ISCR1 complex was adjacent to bla CTX-M-2 in all the CTX-M-2 producers (n = 35), including Acinetobacter spp., E. cloacae, E. coli, K. pneumoniae, P. mirabilis, P. aeruginosa, S. enterica and S. marcescens, while only 1.5% of the bla CTX-M-2-negative isolates (n = 65) harbored ISCR1 (Arduino et al., 2003). These data strongly implicate the association of ISCR1 with the emergence and dissemination of bla CTX-M-2 gene. In addition, ISCR1 is also related to bla CTX-M-59, -74, -75 (members of CTX-M-2 cluster) and bla CTX-M-1, -9, -14 (Table 3).
Other IS and phage-related sequences
Besides ISEcp1, IS903 and ISCR1 described above, IS1, IS5, IS10, IS26, IS50A, IS1294, IS1326, IS3000, IS4321 and IS6100 were also found to be adjacent to bla CTX-M genes (Table 3). In some cases, several IS elements co-existed in a gene complex, for example, intI1-dfrA12-orfF-aadA2-qacEΔ1-sul1-ISCR1-IS6100-ISCR1-ISEcp1Δ-bla CTX-M-14-IS903D (Bae et al., 2008). Such heterogeneity may be explained by a continuously recombinatorial exchange of gene cassettes, denoting the sophisticated genetic rearrangement strategies that organisms acquire and dispense resistance genes.
A 12.2-kb DNA fragment containing bla CTX-M-10 gene in plasmid pRYCE21 was cloned from K. pneumoniae, and further detected in other bacterial species including E. coli, E. cloacae and E. gergoviae. Analysis of the sequence showed a phage-related 3.5-kb element immediately upstream of the bla CTX-M-10 gene cassettes. This phage-related fragment corresponds to four orfs, of which orf2, orf3 and orf4 display homology to the genes of conserved phage tail proteins (Oliver et al., 2005). Although there is a limited report on phage-related CTX-M genes, this finding indicates that phages may also function as a tool for bla CTX-M-associated genetic elements to become transferable.
Plasmids
The movement of IS-mobilized genes between chromosomes and plasmids greatly increase the opportunity a resistance determinant becomes transferable. Particularly, conjugative plasmid is one of the most important mechanisms for intra-species, inter-species and inter-genus gene transfers.
Plasmids are usually classified on their incompatibility (Inc), defined as the inability of two plasmids to be propagated stably in the same bacterial strain; thus, only compatible plasmids can be rescued in transconjugants (Novick et al., 1976). At least 29 Inc groups have been recognized among plasmids of enteric bacteria, including IncFI, IncFII, IncFIII, IncFIV, IncFV, IncFVI, IncI1, IncI2, IncIy, IncHI1, IncHI2, IncHI3, IncA/C, IncB, IncD, IncJ, IncK, IncL/M, IncN, IncO, IncP, IncS, IncT, IncU, IncV, IncW, IncX, IncY and com9 (Novick et al., 1976; Couturier et al., 1988). The IncFII, IncA/C, IncL/M, and IncI1 plasmids show the highest occurrence among the typed resistance plasmids (Carattoli, 2009).
Molecular epidemiological studies have revealed a close and significant linkage of bla CTX-M genes to plasmids, mainly belonged to IncF, IncI, IncN, IncHI2, IncL/M and IncK groups (Table 4). The IncF group (FIA, FIB and FII) is the most prevalent in transmitting bla CTX-M-15 genes, while IncF, IncK and IncI1 are closely related to the widespread of bla CTX-M-14 genes. In addition, the bla CTX-M-1 gene is dominantly harbored by IncN and IncI1, bla CTX-M-3 gene by IncL/M and IncI1, and bla CTX-M-9 gene by IncHI2.
Table 4. .
Plasmids associated with the spread of CTX-M genes.
| CTX-M gene (No. of isolates) | Inc group (No. of isolates) | Rate* | Resource | Reference |
|---|---|---|---|---|
| bla CTX-M-1 (119) | N (119) | 100% | E. coli from bovine on a dairy farm with high consumption of cephalosporins in Czech Republic, 2008 | Dolejska et al., 2011 |
| bla CTX-M-1 (10) | I1 (10) | 100% | S. enterica from poultry and humans in France, 2003–08 | Cloeckaert et al., 2010 |
| bla CTX-M-3 (14) | L/M (13) | 92.9% | Enterobacteriaceae from Bulgaria, Poland and France | Galimand et al., 2005 |
| bla CTX-M-9 (41) | HI2 (24) P1-α (10) FIB (4) HI2, F1 (2) I1 (1) |
58.5% 24.4% 9.8% 4.9% 2.4% |
Enterobacteriaceae from a university hospital in Spain, 1996–03 | Novais et al., 2006 |
| bla CTX-M-14 (40) | K (27) I1 (11) HI2 (2) |
67.5% 27.5% 5% |
E. coli from patients and healthy volunteers in Spain, 2000–05 | Valverde et al., 2009 |
| bla CTX-M-14 (25) | F (8) I1 (5) F, I1 (3) N (1) Q (1) |
32% 20% 12% 4% 4% |
E. coli from 20 hospitals in 15 provinces in China, 2007–08 | Cao et al., 2011 |
| bla CTX-M-14 (23) | FII (13) I1-Iγ (4) FIB (2) FII, I1-Iγ (1) K (1) |
56.5% 17.4% 8.7% 4.3% 4.3% |
E. coli from outpatients in Hong Kong, 2002–04 | Ho et al., 2011 |
| bla CTX-M-15 (18) | FII (17) FI (1) |
94.4% 5.6% |
E. coli from a hospital in Turkey, 2002–04 | Gonullu et al., 2008 |
| bla CTX-M-15 (36) | FI (36) | 100% | E. coli from a university hospital in Germany, 2006–07 | Mshana et al., 2009 |
| bla CTX-M-15 (55) | FIIA (41) A/C (3) FIIA, A/C (4) |
74.5% 5.5% 7.3% |
K. pneumoniae from patients in 9 Asian countries, 2008–09 | Lee et al., 2011 |
|
bla
CTX-M-1 (11) bla CTX-M-14 (15) bla CTX-M-15 (19) |
N (8) I1 (3) F (9) K (2) F (12) I1 (1) L/M (1) N (1) |
72.7% 27.3% 60% 13.3% 63.2% 5.3% 5.3% 5.3% |
E. coli from different areas in France, 1997–02 | Marcadé et al., 2009 |
|
bla
CTX-M-1 (7) bla CTX-M-9 (14) bla CTX-M-14 (13) bla CTX-M-15 (4) bla CTX-M-32 (3) |
N (5) FII (2) I1 (4) I1, P (3) HI2 (4) FIB (2) K (12) F (4) N (3) |
71.4% 28.6% 28.6% 21.4% 28.6% 14.3% 92.3% 100% 100% |
E. coli and K. pneumoniae from 11 hospitals in Spain, 2004 | Diestra et al., 2009 |
|
bla
CTX-M-2 (16) bla CTX-M-14 (8) |
A/C (11) FVII (1) I1 (1) I1 (6) |
68.8% 6.3% 6.3% 75% |
E. coli from a survey among 3193 healthy children in Peru & Bolivia, 2005 | Pallecchi et al., 2007 |
|
bla
CTX-M-3 (49) bla CTX-M-15 (11) |
I1 (36) FI (8) Y (3) N (2) FI (11) |
73.5% 16.3% 6.1% 4.1% 100% |
E. coli from faeces of residents in16 nursing homes in the UK, 2004–06 | Dhanji et al., 2011a |
Rate = (No. in the 2nd column/No. in the 1st column) × 100%.
Unlike the plasmids with broad host range, such as IncP, IncA/C and IncQ, IncF plasmids are limited by host range to the genera of Enterobacteriaceae (Toukdarian, 2004), footnoting the high prevalence and widespread of CTX-M genes in Enterobacteriaceae, but not in Acinetobacter and Pseudomonas.
Various resistance genes frequently co-exist on a plasmid, facilitating the dissemination of resistance determinants and the survival of bacteria under the pressure of various antibiotics. For example, plasmid pEK499 (a fusion of type FII and FIA replicons) identified in a UK variant of the internationally prevalent E. coli O25:H4-ST131 lineage is confirmed to harbor 10 resistance genes, conferring resistance to seven antibiotic classes, β-lactams (bla CTX-M-15, bla OXA-1, bla TEM-1), aminoglycoside (aac6′-Ib-cr, aadA5), macrolides (mph(A)), chloramphenicol (catB4), tetracycline (tet(A)), trimethoprim (dfrA7) and sulfonamide (sul1) (Woodford et al., 2009).
Secondary chromosomal integration
Most of the bla CTX-M genes are harbored by plasmids and the secondary chromosomal insertions of bla CTX-M genes are also confirmed, particularly in P. mirabilis. Of 25 clinical isolates of CTX-M-producing P. mirabilis collected in Korea, 21 strains harbored bla CTX-Ms on their chromosomes (Song et al., 2011). The genes of bla CTX-M-25 and-41 were also found on the chromosomes of P. mirabilis in Israel (Navon-Venezia et al., 2008).
In addition, chromosomal integration of bla CTX-M-15 gene was reported in E. coli, K. pneumoniae and S. enterica (Coque et al., 2008; Coelho et al., 2010; Fabre et al., 2009). Chromosomal bla CTX-M-9 was observed in one strain of 30 E. coli isolates collected in Barcelona during 1996–1999 (García et al., 2005).
Conclusion
Plasmid-mediated CTX-M enzymes are the most prevalent ESBLs, particularly in E. coli, K. pneumoniae and P. mirobilis. At least 109 members in CTX-M family are identified and can be divided into seven clusters based on their phylogeny. CTX-M-15 and CTX-M-14 are the most dominant variants in the family, followed by CTX-M-2, CTX-M-3 and CTX-M-1.
The CTX-M genes can be traced back to the chromosome-encoded cefotaximas genes in Kluyvera spp., strongly indicating that the plasmid-mediated CTX-M enzymes are originally from Kluyvera. Multiple genetic elements, especially ISEcp1 and ISCR1, are involved in the mobilization of bla CTX-M genes from the chromosomes to plasmids. Conjugative plasmids are responsible for the transfer of the bla CTX-M genes to new hosts, while the properties of plasmid incompatibility and host range are closely associated with the high prevalence and widespread of the CTX-M genes in Enterobacteriaceae, but not in Acinetobacter and Pseudomonas.
Footnotes
Declaration of interest: This work was supported by a grant (No. 24591489) from the Ministry of Education, Culture, Sports, Science and Technology, Japan and by a grant from Showa University Medical Foundation, Tokyo, Japan.
References
- Abdalhamid B, Pitout JD, Moland ES, Hanson ND. Community-onset disease caused by Citrobacter freundii producing a novel CTX-M beta-lactamase, CTX-M-30, in Canada. Antimicrob Agents Chemother. 2004;48:4435–4437. doi: 10.1128/AAC.48.11.4435-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acikgoz ZC, Gulay Z, Bicmen M, Gocer S, Gamberzade S. CTX-M-3 extended-spectrum beta-lactamase in a Shigella sonnei clinical isolate: first report from Turkey. Scand J Infect Dis. 2003;35:503–505. doi: 10.1080/00365540310013270. [DOI] [PubMed] [Google Scholar]
- Aibinu I, Pfeifer Y, Peters F, Ogunsola F, Adenipekun E, Odugbemi T, Koenig W. Emergence of bla CTX-M-15, qnrB1 and aac(6′)-Ib-cr resistance genes in Pantoea agglomerans and Enterobacter cloacae from Nigeria (sub-Saharan Africa) J Med Microbiol. 2012;61:165–167. doi: 10.1099/jmm.0.035238-0. [DOI] [PubMed] [Google Scholar]
- Alobwede I, M’Zali FH, Livermore DM, Heritage J, Todd N, Hawkey PM. CTX-M extended-spectrum beta-lactamase arrives in the UK. J Antimicrob Chemother. 2003;51:470–471. doi: 10.1093/jac/dkg096. [DOI] [PubMed] [Google Scholar]
- Ambler RP, Coulson AF, Frère JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276 (Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduino SM, Catalano M, Orman BE, Roy PH, Centrón D. Molecular epidemiology of orf513-bearing class 1 integrons in multiresistant clinical isolates from Argentinean hospitals. Antimicrob Agents Chemother. 2003;47:3945–3949. doi: 10.1128/AAC.47.12.3945-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduino SM, Roy PH, Jacoby GA, Orman BE, Pineiro SA, Centron D. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513 . Antimicrob Agents Chemother. 2002;46:2303–2306. doi: 10.1128/AAC.46.7.2303-2306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae IK, Lee BH, Hwang HY, Jeong SH, Hong SG, Chang CL, Kwak HS, Kim HJ, Youn H. A novel ceftazidime-hydrolysing extended-spectrum beta-lactamase, CTX-M-54, with a single amino acid substitution at position 167 in the omega loop. J Antimicrob Chemother. 2006a;58:315–319. doi: 10.1093/jac/dkl252. [DOI] [PubMed] [Google Scholar]
- Bae IK, Lee YH, Jeong HJ, Hong SG, Lee SH, Jeong SH. A novel blaCTX-M-14 gene-harboring complex class 1 integron with an In4-like backbone structure from a clinical isolate of Escherichia coli . Diagn Microbiol Infect Dis. 2008;62:340–342. doi: 10.1016/j.diagmicrobio.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Bae IK, Lee YN, Hwang HY, Jeong SH, Lee SJ, Kwak HS, Song W, Kim HJ, Youn H. Emergence of CTX-M-12 extended-spectrum beta-lactamase-producing Escherichia coli in Korea. J Antimicrob Chemother. 2006b;58:1257–1259. doi: 10.1093/jac/dkl397. [DOI] [PubMed] [Google Scholar]
- Bae IK, Lee YN, Lee WG, Lee SH, Jeong SH. Novel complex class 1 integron bearing an ISCR1 element in an Escherichia coli isolate carrying the blaCTX-M-14 gene. Antimicrob Agents Chemother. 2007;51:3017–3019. doi: 10.1128/AAC.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak A, Fiett J, Hryniewicz W, Nordmann P, Gniadkowski M. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum beta-lactamase (ESBL) in Poland. J Antimicrob Chemother. 2002a;50:393–396. doi: 10.1093/jac/dkf151. [DOI] [PubMed] [Google Scholar]
- Baraniak A, Fiett J, Sulikowska A, Hryniewicz W, Gniadkowski M. Countrywide spread of CTX-M-3 extended-spectrum beta-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob Agents Chemother. 2002b;46:151–159. doi: 10.1128/AAC.46.1.151-159.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli . Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas JM. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Slama K, Jouini A, Ben Sallem R, Somalo S, Sáenz Y, Estepa V, Boudabous A, Torres C. Prevalence of broad-spectrum cephalosporin-resistant Escherichia coli isolates in food samples in Tunisia, and characterization of integrons and antimicrobial resistance mechanisms implicated. Int J Food Microbiol. 2010;137:281–286. doi: 10.1016/j.ijfoodmicro.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Billard-Pomares T, Tenaillon O, Le Nagard H, Rouy Z, Cruveiller S, Médigue C, Arlet G, Denamur E, Branger C. Complete nucleotide sequence of plasmid pTN48, encoding the CTX-M-14 extended-spectrum ß-lactamase from an Escherichia coli O102-ST405 strain. Antimicrob Agents Chemother. 2011;55:1270–1273. doi: 10.1128/AAC.01108-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R, Dutour C, Sampaio JL, Chanal C, Sirot D, Labia R, De Champs C, Sirot J. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240–>Gly. Antimicrob Agents Chemother. 2001;45:2269–2275. doi: 10.1128/AAC.45.8.2269-2275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R, Recule C, Baraduc R, Chanal C, Sirot D, De Champs C, Sirot J. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother. 2003;52:29–35. doi: 10.1093/jac/dkg256. [DOI] [PubMed] [Google Scholar]
- Bonnet R, Sampaio JL, Labia R, De Champs C, Sirot D, Chanal C, Sirot J. A novel CTX-M beta-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob Agents Chemother. 2000;44:1936–1942. doi: 10.1128/aac.44.7.1936-1942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouallègue-Godet O, Ben Salem Y, Fabre L, Demartin M, Grimont PA, Mzoughi R, Weill FX. Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum beta-lactamase in a neonatal unit in Sousse, Tunisia. J Clin Microbiol. 2005;43:1037–1044. doi: 10.1128/JCM.43.3.1037-1044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford PA, Yang Y, Sahm D, Grope I, Gardovska D, Storch G. CTX-M-5, a novel cefotaxime-hydrolyzing beta-lactamase from an outbreak of Salmonella Typhimurium in Latvia. Antimicrob Agents Chemother. 1998;42:1980–1984. doi: 10.1128/aac.42.8.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–51, table of contents. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasme L, Nordmann P, Fidel F, Lartigue MF, Bajolet O, Poirel L, Forte D, Vernet-Garnier V, Madoux J, Reveil JC, Alba-Sauviat C, Baudinat I, Bineau P, Bouquigny-Saison C, Eloy C, Lafaurie C, Siméon D, Verquin JP, Noël F, Strady C, De Champs C. Incidence of class A extended-spectrum beta-lactamases in Champagne-Ardenne (France): a 1 year prospective study. J Antimicrob Chemother. 2007;60:956–964. doi: 10.1093/jac/dkm319. [DOI] [PubMed] [Google Scholar]
- Brenwald NP, Jevons G, Andrews JM, Xiong JH, Hawkey PM, Wise R. An outbreak of a CTX-M-type beta-lactamase-producing Klebsiella pneumoniae: the importance of using cefpodoxime to detect extended-spectrum beta-lactamases. J Antimicrob Chemother. 2003;51:195–196. doi: 10.1093/jac/dkg051. [DOI] [PubMed] [Google Scholar]
- Brigante G, Luzzaro F, Perilli M, Lombardi G, Colì A, Rossolini GM, Amicosante G, Toniolo A. Evolution of CTX-M-type beta-lactamases in isolates of Escherichia coli infecting hospital and community patients. Int J Antimicrob Agents. 2005;25:157–162. doi: 10.1016/j.ijantimicag.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Cantón R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Cantón R, Oliver A, Coque TM, Varela Mdel C, Pérez-Díaz JC, Baquero F. Epidemiology of extended-spectrum beta-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J Clin Microbiol. 2002;40:1237–1243. doi: 10.1128/JCM.40.4.1237-1243.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao V, Lambert T, Courvalin P. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum beta-lactamase CTX-M-17. Antimicrob Agents Chemother. 2002;46:1212–1217. doi: 10.1128/AAC.46.5.1212-1217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Cavaco LM, Lv Y, Li Y, Zheng B, Wang P, Hasman H, Liu Y, Aarestrup FM. Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J Clin Microbiol. 2011;49:2496–2501. doi: 10.1128/JCM.02503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartelle M, Canle D, Llarena FJ, Molina F, Villanueva R, Bou G. Characterisation of the first CTX-M-10-producing isolate of Salmonella enterica serotype Virchow. Clin Microbiol Infect. 2006;12:285–287. doi: 10.1111/j.1469-0691.2005.01300.x. [DOI] [PubMed] [Google Scholar]
- Cartelle M, del Mar Tomas M, Molina F, Moure R, Villanueva R, Bou G. High-level resistance to ceftazidime conferred by a novel enzyme, CTX-M-32, derived from CTX-M-1 through a single Asp240-Gly substitution. Antimicrob Agents Chemother. 2004;48:2308–2313. doi: 10.1128/AAC.48.6.2308-2313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G, Perilli M. Spread of bla CTX-M-type and bla PER-2 beta-lactamase genes in clinical isolates from Bolivian hospitals. J Antimicrob Chemother. 2006;57:975–978. doi: 10.1093/jac/dkl055. [DOI] [PubMed] [Google Scholar]
- Chanawong A, M’Zali FH, Heritage J, Xiong JH, Hawkey PM. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People’s Republic of China. Antimicrob Agents Chemother. 2002;46:630–637. doi: 10.1128/AAC.46.3.630-637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Gao W, Yin J, Sun Z, Ye Y, Gao YF, Li X, Li JB. Phenotypic and molecular characterization of two novel CTX-M enzymes carried by Klebsiella pneumoniae. Mol Biol Rep. 2010;37:1261–1267. doi: 10.1007/s11033-009-9499-1. [DOI] [PubMed] [Google Scholar]
- Cheng J, Wang Q, Chen Y, Ye Y, Li H, Li X, Li JB. Phenotypic and molecular characterization of a novel beta-lactamase carried by Klebsiella pneumoniae, CTX-M-72, derived from CTX-M-3. J Gen Appl Microbiol. 2009;55:207–216. doi: 10.2323/jgam.55.207. [DOI] [PubMed] [Google Scholar]
- Cheng J, Ye Y, Wang YY, Li H, Li X, Li JB. Phenotypic and molecular characterization of 5 novel CTX-M enzymes carried by Klebsiella pneumoniae and Escherichia coli. Acta Pharmacol Sin. 2008;29:217–225. doi: 10.1111/j.1745-7254.2008.00736.x. [DOI] [PubMed] [Google Scholar]
- Chmelnitsky I, Carmeli Y, Leavitt A, Schwaber MJ, Navon-Venezia S. CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob Agents Chemother. 2005;49:4745–4750. doi: 10.1128/AAC.49.11.4745-4750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Lee JE, Park SJ, Kim MN, Choo EJ, Kwak YG, Jeong JY, Woo JH, Kim NJ, Kim YS. Prevalence, microbiology, and clinical characteristics of extended-spectrum beta-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur J Clin Microbiol Infect Dis. 2007;26:557–561. doi: 10.1007/s10096-007-0308-2. [DOI] [PubMed] [Google Scholar]
- Cloeckaert A, Praud K, Lefevre M, Doublet B, Pardos M, Granier SA, Brisabois A, Weill FX. IncI1 plasmid carrying extended-spectrum-beta-lactamase gene bla CTX-M-1 in Salmonella enterica isolates from poultry and humans in France, 2003 to 2008. Antimicrob Agents Chemother. 2010;54:4484–4486. doi: 10.1128/AAC.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho A, González-López JJ, Miró E, Alonso-Tarrés C, Mirelis B, Larrosa MN, Bartolomé RM, Andreu A, Navarro F, Johnson JR, Prats G. Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int J Antimicrob Agents. 2010;36:73–78. doi: 10.1016/j.ijantimicag.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerging Infect Dis. 2008;14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque TM, Oliver A, Pérez-Díaz JC, Baquero F, Cantón R. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum beta-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000) Antimicrob Agents Chemother. 2002;46:500–510. doi: 10.1128/AAC.46.2.500-510.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M, Bex F, Bergquist PL, Maas WK. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Li J, Sun Z, Hu C, Jin S, Li F, Guo Y, Ran L, Ma Y. Characterization of Salmonella enterica isolates from infants and toddlers in Wuhan, China. J Antimicrob Chemother. 2009;63:87–94. doi: 10.1093/jac/dkn452. [DOI] [PubMed] [Google Scholar]
- Cullik A, Pfeifer Y, Prager R, von Baum H, Witte W. A novel IS26 structure surrounds bla CTX-M genes in different plasmids from German clinical Escherichia coli isolates. J Med Microbiol. 2010;59:580–587. doi: 10.1099/jmm.0.016188-0. [DOI] [PubMed] [Google Scholar]
- Damjanova I, Tóth A, Pászti J, Hajbel-Vékony G, Jakab M, Berta J, Milch H, Füzi M. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005–the new ‘MRSAs’? J Antimicrob Chemother. 2008;62:978–985. doi: 10.1093/jac/dkn287. [DOI] [PubMed] [Google Scholar]
- De Champs C, Sirot D, Chanal C, Bonnet R, Sirot J. A 1998 survey of extended-spectrum beta-lactamases in Enterobacteriaceae in France. The French Study Group. Antimicrob Agents Chemother. 2000;44:3177–3179. doi: 10.1128/aac.44.11.3177-3179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decousser JW, Poirel L, Nordmann P. Characterization of a chromosomally encoded extended-spectrum class A beta-lactamase from Kluyvera cryocrescens . Antimicrob Agents Chemother. 2001;45:3595–3598. doi: 10.1128/AAC.45.12.3595-3598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanji H, Doumith M, Rooney PJ, O’Leary MC, Loughrey AC, Hope R, Woodford N, Livermore DM. Molecular epidemiology of fluoroquinolone-resistant ST131 Escherichia coli producing CTX-M extended-spectrum beta-lactamases in nursing homes in Belfast, UK. J Antimicrob Chemother. 2011a;66:297–303. doi: 10.1093/jac/dkq463. [DOI] [PubMed] [Google Scholar]
- Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N. Variation in the genetic environments of bla CTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother. 2011b;66:1005–1012. doi: 10.1093/jac/dkr041. [DOI] [PubMed] [Google Scholar]
- Di Conza J, Ayala JA, Power P, Mollerach M, Gutkind G. Novel class 1 integron (InS21) carrying bla CTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob Agents Chemother. 2002;46:2257–2261. doi: 10.1128/AAC.46.7.2257-2261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diestra K, Juan C, Curiao T, Moyá B, Miró E, Oteo J, Coque TM, Pérez-Vázquez M, Campos J, Cantón R, Oliver A, Navarro F, Red Española de Investigación en Patología Infecciosa (REIPI), Spain Characterization of plasmids encoding bla ESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother. 2009;63:60–66. doi: 10.1093/jac/dkn453. [DOI] [PubMed] [Google Scholar]
- Djamdjian L, Naas T, Tandé D, Cuzon G, Hanrotel-Saliou C, Nordmann P. CTX-M-93, a CTX-M variant lacking penicillin hydrolytic activity. Antimicrob Agents Chemother. 2011;55:1861–1866. doi: 10.1128/AAC.01656-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y, Adams-Haduch JM, Paterson DL. Escherichia coli isolate coproducing 16S rRNA Methylase and CTX-M-type extended-spectrum beta-lactamase isolated from an outpatient in the United States. Antimicrob Agents Chemother. 2008;52:1204–1205. doi: 10.1128/AAC.01320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejska M, Jurcickova Z, Literak I, Pokludova L, Bures J, Hera A, Kohoutova L, Smola J, Cizek A. IncN plasmids carrying bla CTX-M-1 in Escherichia coli isolates on a dairy farm. Vet Microbiol. 2011;149:513–516. doi: 10.1016/j.vetmic.2010.11.032. [DOI] [PubMed] [Google Scholar]
- Doublet B, Granier SA, Robin F, Bonnet R, Fabre L, Brisabois A, Cloeckaert A, Weill FX. Novel plasmid-encoded ceftazidime-hydrolyzing CTX-M-53 extended-spectrum beta-lactamase from Salmonella enterica serotypes Westhampton and Senftenberg. Antimicrob Agents Chemother. 2009;53:1944–1951. doi: 10.1128/AAC.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various bla CTX-M genes. J Antimicrob Chemother. 2006;57:14–23. doi: 10.1093/jac/dki398. [DOI] [PubMed] [Google Scholar]
- Fabre L, Delauné A, Espié E, Nygard K, Pardos M, Polomack L, Guesnier F, Galimand M, Lassen J, Weill FX. Chromosomal integration of the extended-spectrum beta-lactamase gene bla CTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob Agents Chemother. 2009;53:1808–1816. doi: 10.1128/AAC.00451-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández A, Gil E, Cartelle M, Pérez A, Beceiro A, Mallo S, Tomás MM, Pérez-Llarena FJ, Villanueva R, Bou G. Interspecies spread of CTX-M-32 extended-spectrum beta-lactamase and the role of the insertion sequence IS1 in down-regulating bla CTX-M gene expression. J Antimicrob Chemother. 2007;59:841–847. doi: 10.1093/jac/dkm030. [DOI] [PubMed] [Google Scholar]
- Galani I, Souli M, Chryssouli Z, Giamarellou H. Detection of CTX-M-15 and CTX-M-33, a novel variant of CTX-M-15, in clinical Escherichia coli isolates in Greece. Int J Antimicrob Agents. 2007;29:598–600. doi: 10.1016/j.ijantimicag.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother. 2005;49:2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A, Navarro F, Miró E, Mirelis B, Campoy S, Coll P. Characterization of the highly variable region surrounding the bla CTX-M-9 gene in non-related Escherichia coli from Barcelona. J Antimicrob Chemother. 2005;56:819–826. doi: 10.1093/jac/dki345. [DOI] [PubMed] [Google Scholar]
- García Fernández A, Cloeckaert A, Bertini A, Praud K, Doublet B, Weill FX, Carattoli A. Comparative analysis of IncHI2 plasmids carrying bla CTX-M-2 or bla CTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob Agents Chemother. 2007;51:4177–4180. doi: 10.1128/AAC.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazouli M, Tzelepi E, Markogiannakis A, Legakis NJ, Tzouvelekis LS. Two novel plasmid-mediated cefotaxime-hydrolyzing beta-lactamases (CTX-M-5 and CTX-M-6) from Salmonella Typhimurium. FEMS Microbiol Lett. 1998a;165:289–293. doi: 10.1111/j.1574-6968.1998.tb13159.x. [DOI] [PubMed] [Google Scholar]
- Gazouli M, Tzelepi E, Sidorenko SV, Tzouvelekis LS. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A beta-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998b;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierczynski R, Szych J, Cieslik A, Rastawicki W, Jagielski M. The occurrence of the first two CTX-M-3 and TEM-1 producing isolates of Salmonella enterica serovar Oranienburg in Poland. Int J Antimicrob Agents. 2003;21:497–499. doi: 10.1016/s0924-8579(03)00044-x. [DOI] [PubMed] [Google Scholar]
- Gniadkowski M, Schneider I, Palucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing beta-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42:827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonullu N, Aktas Z, Kayacan CB, Salcioglu M, Carattoli A, Yong DE, Walsh TR. Dissemination of CTX-M-15 beta-lactamase genes carried on Inc FI and FII plasmids among clinical isolates of Escherichia coli in a university hospital in Istanbul, Turkey. J Clin Microbiol. 2008;46:1110–1112. doi: 10.1128/JCM.01974-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govinden U, Mocktar C, Moodley P, Sturm AW, Essack SY. CTX-M-37 in Salmonella enterica serotype Isangi from Durban, South Africa. Int J Antimicrob Agents. 2006;28:288–291. doi: 10.1016/j.ijantimicag.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Gómez-Garcés JL, Saéz D, Almagro M, Fernández-Romero S, Merino F, Campos J, Oteo J. Osteomyelitis associated to CTX-M-15-producing Aeromonas hydrophila: first description in the literature. Diagn Microbiol Infect Dis. 2011;70:420–422. doi: 10.1016/j.diagmicrobio.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Harada S, Ishii Y, Saga T, Kouyama Y, Tateda K, Yamaguchi K. Chromosomal integration and location on IncT plasmids of the bla CTX-M-2 gene in Proteus mirabilis clinical isolates. Antimicrob Agents Chemother. 2012;56:1093–1096. doi: 10.1128/AAC.00258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. beta-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother. 2005;56:115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- Heffernan HM, Woodhouse RE, Pope CE, Blackmore TK. Prevalence and types of extended-spectrum beta-lactamases among urinary Escherichia coli and Klebsiella spp. in New Zealand. Int J Antimicrob Agents. 2009;34:544–549. doi: 10.1016/j.ijantimicag.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Ho PL, Ho AY, Chow KH, Wong RC, Duan RS, Ho WL, Mak GC, Tsang KW, Yam WC, Yuen KY. Occurrence and molecular analysis of extended-spectrum beta-lactamase-producing Proteus mirabilis in Hong Kong, 1999-2002. J Antimicrob Chemother. 2005a;55:840–845. doi: 10.1093/jac/dki135. [DOI] [PubMed] [Google Scholar]
- Ho PL, Lo WU, Wong RC, Yeung MK, Chow KH, Que TL, Tong AH, Bao JY, Lok S, Wong SS. Complete sequencing of the FII plasmid pHK01, encoding CTX-M-14, and molecular analysis of its variants among Escherichia coli from Hong Kong. J Antimicrob Chemother. 2011;66:752–756. doi: 10.1093/jac/dkr010. [DOI] [PubMed] [Google Scholar]
- Ho PL, Shek RH, Chow KH, Duan RS, Mak GC, Lai EL, Yam WC, Tsang KW, Lai WM. Detection and characterization of extended-spectrum beta-lactamases among bloodstream isolates of Enterobacter spp. in Hong Kong, 2000-2002. J Antimicrob Chemother. 2005b;55:326–332. doi: 10.1093/jac/dki010. [DOI] [PubMed] [Google Scholar]
- Hopkins KL, Deheer-Graham A, Threlfall EJ, Batchelor MJ, Liebana E. Novel plasmid-mediated CTX-M-8 subgroup extended-spectrum beta-lactamase (CTX-M-40) isolated in the UK. Int J Antimicrob Agents. 2006;27:572–575. doi: 10.1016/j.ijantimicag.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Hopkins KL, Threlfall EJ, Karisik E, Wardle JK. Identification of novel plasmid-mediated extended-spectrum beta-lactamase CTX-M-57 in Salmonella enterica serovar Typhimurium. Int J Antimicrob Agents. 2008;31:85–86. doi: 10.1016/j.ijantimicag.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Hrabák J, Empel J, Gniadkowski M, Halbhuber Z, Rébl K, Urbásková P. CTX-M-15-producing Shigella sonnei strain from a Czech patient who traveled in Asia. J Clin Microbiol. 2008;46:2147–2148. doi: 10.1128/JCM.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeniuk C, Arlet G, Gautier V, Grimont P, Labia R, Philippon A. Beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob Agents Chemother. 2002;46:3045–3049. doi: 10.1128/AAC.46.9.3045-3049.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A beta-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya H, Mori K, Higashide M, Tamura K, Takai N, Hirose K, Terajima J, Watanabe H. Identification of CTX-M-14 beta-lactamase in a Salmonella enterica serovar Enteritidis isolate from Japan. Antimicrob Agents Chemother. 2005;49:2568–2570. doi: 10.1128/AAC.49.6.2568-2570.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G, Bush K. http://www.lahey.org/Studies/ β-Lactamase classification and amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant enzymes. 2012 Accessed on 1 March, 2012.
- Kanamori H, Yano H, Hirakata Y, Endo S, Arai K, Ogawa M, Shimojima M, Aoyagi T, Hatta M, Yamada M, Nishimaki K, Kitagawa M, Kunishima H, Kaku M. High prevalence of extended-spectrum ß-lactamases and qnr determinants in Citrobacter species from Japan: dissemination of CTX-M-2. J Antimicrob Chemother. 2011;66:2255–2262. doi: 10.1093/jac/dkr283. [DOI] [PubMed] [Google Scholar]
- Karim A, Poirel L, Nagarajan S, Nordmann P. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1 . FEMS Microbiol Lett. 2001;201:237–241. doi: 10.1111/j.1574-6968.2001.tb10762.x. [DOI] [PubMed] [Google Scholar]
- Kariuki S, Corkill JE, Revathi G, Musoke R, Hart CA. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob Agents Chemother. 2001;45:2141–2143. doi: 10.1128/AAC.45.7.2141-2143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K. Characterization of IncF plasmids carrying the bla CTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J Antimicrob Chemother. 2011;66:1263–1268. doi: 10.1093/jac/dkr106. [DOI] [PubMed] [Google Scholar]
- Kim J, Kang HY, Lee Y. The identification of CTX-M-14, TEM-52, and CMY-1 enzymes in Escherichia coli isolated from the Han River in Korea. J Microbiol. 2008;46:478–481. doi: 10.1007/s12275-008-0150-y. [DOI] [PubMed] [Google Scholar]
- Kim J, Lim YM, Jeong YS, Seol SY. Occurrence of CTX-M-3, CTX-M-15, CTX-M-14, and CTX-M-9 extended-spectrum beta-lactamases in Enterobacteriaceae clinical isolates in Korea. Antimicrob Agents Chemother. 2005;49:1572–1575. doi: 10.1128/AAC.49.4.1572-1575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiratisin P, Apisarnthanarak A, Saifon P, Laesripa C, Kitphati R, Mundy LM. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn Microbiol Infect Dis. 2007;58:349–355. doi: 10.1016/j.diagmicrobio.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kliebe C, Nies BA, Meyer JF, Tolxdorff-Neutzling RM, Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985;28:302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Ikeda N, Aihara M, Nakamachi Y, Kinoshita S, Yamasaki K, Shimakawa K. Hospital outbreak of MEN-1-derived extended spectrum beta-lactamase-producing Klebsiella pneumoniae. J Infect Chemother. 2001;7:94–101. doi: 10.1007/s101560100015. [DOI] [PubMed] [Google Scholar]
- Lartigue MF, Poirel L, Aubert D, Nordmann P. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene bla CTX-M of Kluyvera ascorbata . Antimicrob Agents Chemother. 2006;50:1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue MF, Poirel L, Heritier C, Tolun V, Nordmann P. First description of CTX-M-15-producing Klebsiella pneumoniae in Turkey. J Antimicrob Chemother. 2003;52:315–316. doi: 10.1093/jac/dkg335. [DOI] [PubMed] [Google Scholar]
- Lartigue MF, Poirel L, Nordmann P. Diversity of genetic environment of bla CTX-M genes. FEMS Microbiol Lett. 2004;234:201–207. doi: 10.1016/j.femsle.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents. 2011;38:160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Liu G, Ling BD, Xie YE, Lin L, Zeng Y, Zhang X, Lei J. Characterization of CTX-M-22 and TEM-141 encoded by a single plasmid from a clinical isolate of Enterobacter cloacae in China. Jpn J Infect Dis. 2007;60:295–297. [PubMed] [Google Scholar]
- Liu W, Chen L, Li H, Duan H, Zhang Y, Liang X, Li X, Zou M, Xu L, Hawkey PM. Novel CTX-M beta-lactamase genotype distribution and spread into multiple species of Enterobacteriaceae in Changsha, Southern China. J Antimicrob Chemother. 2009;63:895–900. doi: 10.1093/jac/dkp068. [DOI] [PubMed] [Google Scholar]
- Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- Ma L, Ishii Y, Ishiguro M, Matsuzawa H, Yamaguchi K. Cloning and sequencing of the gene encoding Toho-2, a class A beta-lactamase preferentially inhibited by tazobactam. Antimicrob Agents Chemother. 1998;42:1181–1186. doi: 10.1128/aac.42.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcadé G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J Antimicrob Chemother. 2009;63:67–71. doi: 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- McGettigan SE, Hu B, Andreacchio K, Nachamkin I, Edelstein PH. Prevalence of CTX-M beta-lactamases in Philadelphia, Pennsylvania. J Clin Microbiol. 2009;47:2970–2974. doi: 10.1128/JCM.00319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça N, Ferreira E, Louro D, Caniça M, ARSIP Participants Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int J Antimicrob Agents. 2009;34:29–37. doi: 10.1016/j.ijantimicag.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Minarini LA, Poirel L, Trevisani NA, Darini AL, Nordmann P. Predominance of CTX-M-type extended-spectrum beta-lactamase genes among enterobacterial isolates from outpatients in Brazil. Diagn Microbiol Infect Dis. 2009;65:202–206. doi: 10.1016/j.diagmicrobio.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Miró E, Mirelis B, Navarro F, Rivera A, Mesa RJ, Roig MC, Gómez L, Coll P. Surveillance of extended-spectrum beta-lactamases from clinical samples and faecal carriers in Barcelona, Spain. J Antimicrob Chemother. 2005;56:1152–1155. doi: 10.1093/jac/dki395. [DOI] [PubMed] [Google Scholar]
- Moubareck C, Daoud Z, Hakimé NI, Hamzé M, Mangeney N, Matta H, Mokhbat JE, Rohban R, Sarkis DK, Doucet-Populaire F. Countrywide spread of community- and hospital-acquired extended-spectrum beta-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J Clin Microbiol. 2005;43:3309–3313. doi: 10.1128/JCM.43.7.3309-3313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshana SE, Imirzalioglu C, Hossain H, Hain T, Domann E, Chakraborty T. Conjugative IncFI plasmids carrying CTX-M-15 among Escherichia coli ESBL producing isolates at a University hospital in Germany. BMC Infect Dis. 2009;9:97. doi: 10.1186/1471-2334-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday CJ, Boyd DA, Brenwald N, Miller M, Andrews JM, Wise R, Mulvey MR, Hawkey PM. Molecular and kinetic comparison of the novel extended-spectrum beta-lactamases CTX-M-25 and CTX-M-26. Antimicrob Agents Chemother. 2004;48:4829–4834. doi: 10.1128/AAC.48.12.4829-4834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T, Bentchouala C, Cuzon G, Yaou S, Lezzar A, Smati F, Nordmann P. Outbreak of Salmonella enterica serotype Infantis producing ArmA 16S RNA methylase and CTX-M-15 extended-spectrum ß-lactamase in a neonatology ward in Constantine, Algeria. Int J Antimicrob Agents. 2011;38:135–139. doi: 10.1016/j.ijantimicag.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Nagano N, Nagano Y, Cordevant C, Shibata N, Arakawa Y. Nosocomial transmission of CTX-M-2 beta-lactamase-producing Acinetobacter baumannii in a neurosurgery ward. J Clin Microbiol. 2004;42:3978–3984. doi: 10.1128/JCM.42.9.3978-3984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano Y, Nagano N, Wachino J, Ishikawa K, Arakawa Y. Novel chimeric beta-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 beta-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrob Agents Chemother. 2009;53:69–74. doi: 10.1128/AAC.00227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer U, Sundsfjord A. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist. 2011;17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- Navon-Venezia S, Chmelnitsky I, Leavitt A, Carmeli Y. Dissemination of the CTX-M-25 family beta-lactamases among Klebsiella pneumoniae, Escherichia coli and Enterobacter cloacae and identification of the novel enzyme CTX-M-41 in Proteus mirabilis in Israel. J Antimicrob Chemother. 2008;62:289–295. doi: 10.1093/jac/dkn182. [DOI] [PubMed] [Google Scholar]
- Novais A, Cantón R, Valverde A, Machado E, Galán JC, Peixe L, Carattoli A, Baquero F, Coque TM. Dissemination and persistence of bla CTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob Agents Chemother. 2006;50:2741–2750. doi: 10.1128/AAC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Clowes RC, Cohen SN, Curtiss R, 3rd, Datta N, Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976;40:168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Oliver A, Coque TM, Alonso D, Valverde A, Baquero F, Cantón R. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob Agents Chemother. 2005;49:1567–1571. doi: 10.1128/AAC.49.4.1567-1571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A, Pérez-Díaz JC, Coque TM, Baquero F, Cantón R. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing beta-lactamase (CTX-M-10) isolated in Spain. Antimicrob Agents Chemother. 2001;45:616–620. doi: 10.1128/AAC.45.2.616-620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson AB, Silverman M, Boyd DA, McGeer A, Willey BM, Pong-Porter V, Daneman N, Mulvey MR. Identification of a progenitor of the CTX-M-9 group of extended-spectrum beta-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob Agents Chemother. 2005;49:2112–2115. doi: 10.1128/AAC.49.5.2112-2115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteo J, Delgado-Iribarren A, Vega D, Bautista V, Rodríguez MC, Velasco M, Saavedra JM, Pérez-Vázquez M, García-Cobos S, Martínez-Martínez L, Campos J. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int J Antimicrob Agents. 2008;32:534–537. doi: 10.1016/j.ijantimicag.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Pai H, Choi EH, Lee HJ, Hong JY, Jacoby GA. Identification of CTX-M-14 extended-spectrum beta-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J Clin Microbiol. 2001;39:3747–3749. doi: 10.1128/JCM.39.10.3747-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallecchi L, Bartoloni A, Fiorelli C, Mantella A, Di Maggio T, Gamboa H, Gotuzzo E, Kronvall G, Paradisi F, Rossolini GM. Rapid dissemination and diversity of CTX-M extended-spectrum beta-lactamase genes in commensal Escherichia coli isolates from healthy children from low-resource settings in Latin America. Antimicrob Agents Chemother. 2007;51:2720–2725. doi: 10.1128/AAC.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano G, Richardson D, Nigrin J, McGeer A, Loo V, Toye B, Alfa M, Pienaar C, Kibsey P, Pitout JD. High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-beta-lactamase-producing Escherichia coli isolates from Canada. Antimicrob Agents Chemother. 2010;54:1327–1330. doi: 10.1128/AAC.01338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella S, Ziental-Gelus N, Mayer C, Renard M, Jarlier V, Sougakoff W. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob Agents Chemother. 2008;52:3725–3736. doi: 10.1128/AAC.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A, Labia R, Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Decousser JW, Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a bla CTX-M beta-lactamase gene. Antimicrob Agents Chemother. 2003;47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Gniadkowski M, Nordmann P. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J Antimicrob Chemother. 2002a;50:1031–1034. doi: 10.1093/jac/dkf240. [DOI] [PubMed] [Google Scholar]
- Poirel L, Kämpfer P, Nordmann P. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2002b;46:4038–4040. doi: 10.1128/AAC.46.12.4038-4040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Lartigue MF, Decousser JW, Nordmann P. ISEcp1B-mediated transposition of bla CTX-M in Escherichia coli. Antimicrob Agents Chemother. 2005;49:447–450. doi: 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Naas T, Le Thomas I, Karim A, Bingen E, Nordmann P. CTX-M-type extended-spectrum beta-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob Agents Chemother. 2001;45:3355–3361. doi: 10.1128/AAC.45.12.3355-3361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornruangwong S, Hendriksen RS, Pulsrikarn C, Bangstrakulnonth A, Mikoleit M, Davies RH, Aarestrup FM, Garcia-Migura L. Epidemiological investigation of Salmonella enterica serovar Kedougou in Thailand. Foodborne Pathog Dis. 2011;8:203–211. doi: 10.1089/fpd.2010.0626. [DOI] [PubMed] [Google Scholar]
- Potz NA, Hope R, Warner M, Johnson AP, Livermore DM, London & South East ESBL Project Group Prevalence and mechanisms of cephalosporin resistance in Enterobacteriaceae in London and South-East England. J Antimicrob Chemother. 2006;58:320–326. doi: 10.1093/jac/dkl217. [DOI] [PubMed] [Google Scholar]
- Power P, Galleni M, Di Conza J, Ayala JA, Gutkind G. Description of In116, the first bla CTX-M-2-containing complex class 1 integron found in Morganella morganii isolates from Buenos Aires, Argentina. J Antimicrob Chemother. 2005;55:461–465. doi: 10.1093/jac/dkh556. [DOI] [PubMed] [Google Scholar]
- Qi C, Pilla V, Yu JH, Reed K. Changing prevalence of Escherichia coli with CTX-M-type extended-spectrum beta-lactamases in outpatient urinary E. coli between 2003 and 2008. Diagn Microbiol Infect Dis. 2010;67:87–91. doi: 10.1016/j.diagmicrobio.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Quinteros M, Radice M, Gardella N, Rodriguez MM, Costa N, Korbenfeld D, Couto E, Gutkind G, Microbiology Study Group Extended-spectrum beta-lactamases in enterobacteriaceae in Buenos Aires, Argentina, public hospitals. Antimicrob Agents Chemother. 2003;47:2864–2867. doi: 10.1128/AAC.47.9.2864-2867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar R, Giammanco GM, Aleo A, Plano MR, Naghoni A, Owlia P, Mammina C. Characterization of the first extended-spectrum beta-lactamase-producing nontyphoidal Salmonella strains isolated in Tehran, Iran. Foodborne Pathog Dis. 2010;7:91–95. doi: 10.1089/fpd.2009.0382. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martinez JM, Poirel L, Canton R, Nordmann P. Common region CR1 for expression of antibiotic resistance genes. Antimicrob Agents Chemother. 2006;50:2544–2546. doi: 10.1128/AAC.00609-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, Schroeter A, Guerra B. Extended-spectrum beta-lactamases and AmpC beta-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J Antimicrob Chemother. 2009;64:301–309. doi: 10.1093/jac/dkp195. [DOI] [PubMed] [Google Scholar]
- Rodríguez MM, Power P, Radice M, Vay C, Famiglietti A, Galleni M, Ayala JA, Gutkind G. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob Agents Chemother. 2004;48:4895–4897. doi: 10.1128/AAC.48.12.4895-4897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez MM, Power P, Sader H, Galleni M, Gutkind G. Novel chromosome-encoded CTX-M-78 beta-lactamase from a Kluyvera georgiana clinical isolate as a putative origin of CTX-M-25 subgroup. Antimicrob Agents Chemother. 2010;54:3070–3071. doi: 10.1128/AAC.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero L, López L, Martínez-Martínez L, Guerra B, Hernández JR, Pascual A. Characterization of the first CTX-M-14-producing Salmonella enterica serotype Enteritidis isolate. J Antimicrob Chemother. 2004;53:1113–1114. doi: 10.1093/jac/dkh246. [DOI] [PubMed] [Google Scholar]
- Sabaté M, Navarro F, Miró E, Campoy S, Mirelis B, Barbé J, Prats G. Novel complex sul1-type integron in Escherichia coli carrying bla CTX-M-9 . Antimicrob Agents Chemother. 2002;46:2656–2661. doi: 10.1128/AAC.46.8.2656-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabaté M, Tarragó R, Navarro F, Miró E, Vergés C, Barbé J, Prats G. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing beta-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob Agents Chemother. 2000;44:1970–1973. doi: 10.1128/aac.44.7.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladin M, Cao VT, Lambert T, Donay JL, Herrmann JL, Ould-Hocine Z, Verdet C, Delisle F, Philippon A, Arlet G. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol Lett. 2002;209:161–168. doi: 10.1111/j.1574-6968.2002.tb11126.x. [DOI] [PubMed] [Google Scholar]
- Sarria JC, Vidal AM, Kimbrough RC., 3rd Infections caused by Kluyvera species in humans. Clin Infect Dis. 2001;33:E69–E74. doi: 10.1086/322686. [DOI] [PubMed] [Google Scholar]
- Schneider I, Queenan AM, Markovska R, Markova B, Keuleyan E, Bauernfeind A. New variant of CTX-M-type extended-spectrum beta-lactamases, CTX-M-71, with a Gly238Cys substitution in a Klebsiella pneumoniae isolate from Bulgaria. Antimicrob Agents Chemother. 2009;53:4518–4521. doi: 10.1128/AAC.00461-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seputiene V, Linkevicius M, Bogdaite A, Povilonis J, Planciuniene R, Giedraitiene A, Pavilonis A, Suziedeliene E. Molecular characterization of extended-spectrum ß-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from hospitals in Lithuania. J Med Microbiol. 2010;59:1263–1265. doi: 10.1099/jmm.0.021972-0. [DOI] [PubMed] [Google Scholar]
- Shakil S, Khan AU. Detection of CTX-M-15-producing and carbapenem-resistant Acinetobacter baumannii strains from urine from an Indian hospital. J Chemother. 2010;22:324–327. doi: 10.1179/joc.2010.22.5.324. [DOI] [PubMed] [Google Scholar]
- Shibata N, Kurokawa H, Doi Y, Yagi T, Yamane K, Wachino J, Suzuki S, Kimura K, Ishikawa S, Kato H, Ozawa Y, Shibayama K, Kai K, Konda T, Arakawa Y. PCR classification of CTX-M-type beta-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrob Agents Chemother. 2006;50:791–795. doi: 10.1128/AAC.50.2.791-795.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Bistué AJ, Martín FA, Petroni A, Faccone D, Galas M, Tolmasky ME, Zorreguieta A. Vibrio cholerae InV117, a class 1 integron harboring aac(6′)-Ib and bla CTX-M-2, is linked to transposition genes. Antimicrob Agents Chemother. 2006;50:1903–1907. doi: 10.1128/AAC.50.5.1903-1907.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Kim J, Bae IK, Jeong SH, Seo YH, Shin JH, Jang SJ, Uh Y, Shin JH, Lee MK, Lee K. Chromosome-encoded AmpC and CTX-M extended-spectrum ß-lactamases in clinical isolates of Proteus mirabilis from Korea. Antimicrob Agents Chemother. 2011;55:1414–1419. doi: 10.1128/AAC.01835-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton PD. Novel insertion sequence, ISEcp1, mobilizes the plasmid-mediated class C β-lactamase-coding gene, blaCMY-4. In Program and Abstracts of the Thirty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA. American Society for Microbiology; Washington, DC, USA: 1999. 132 pp. 1999. Abstract 1457, p. [Google Scholar]
- Stepanova MN, Pimkin M, Nikulin AA, Kozyreva VK, Agapova ED, Edelstein MV. Convergent in vivo and in vitro selection of ceftazidime resistance mutations at position 167 of CTX-M-3 beta-lactamase in hypermutable Escherichia coli strains. Antimicrob Agents Chemother. 2008;52:1297–1301. doi: 10.1128/AAC.01060-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Stokes HW, Tomaras C, Parsons Y, Hall RM. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid. 1993;30:39–50. doi: 10.1006/plas.1993.1032. [DOI] [PubMed] [Google Scholar]
- Stürenburg E, Kühn A, Mack D, Laufs R. A novel extended-spectrum beta-lactamase CTX-M-23 with a P167T substitution in the active-site omega loop associated with ceftazidime resistance. J Antimicrob Chemother. 2004;54:406–409. doi: 10.1093/jac/dkh334. [DOI] [PubMed] [Google Scholar]
- Su Z, Dai X, Chen J, Kong F, Wang H, Li Y, Peng S, Wang S, Shao Q, Lv L, Xu H. The bla CTX-M-1 gene located in a novel complex class I integron bearing an ISCR1 element in Escherichia coli isolates from Zhenjiang, China. J Antimicrob Chemother. 2008;62:1150–1151. doi: 10.1093/jac/dkn300. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zeng Z, Chen S, Ma J, He L, Liu Y, Deng Y, Lei T, Zhao J, Liu JH. High prevalence of bla CTX-M extended-spectrum ß-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin Microbiol Infect. 2010;16:1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x. [DOI] [PubMed] [Google Scholar]
- Tian GB, Adams-Haduch JM, Qureshi ZA, Wang HN, Doi Y. CTX-M-35 extended-spectrum beta-lactamase conferring ceftazidime resistance in Citrobacter koseri . Int J Antimicrob Agents. 2010;35:412–413. doi: 10.1016/j.ijantimicag.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian SF, Chen BY, Chu YZ, Wang S. Prevalence of rectal carriage of extended-spectrum beta-lactamase-producing Escherichia coli among elderly people in community settings in China. Can J Microbiol. 2008;54:781–785. doi: 10.1139/w08-059. [DOI] [PubMed] [Google Scholar]
- Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukdarian A. Plasmid strategies for broad-host-range replication in Gram-negative bacteria. In: Funnell B, Phillips G, editors. Plasmid Biology. Washington DC: ASM Press; 2004. pp. 259–270. [Google Scholar]
- Valverde A, Cantón R, Galán JC, Nordmann P, Baquero F, Coque TM. In117, an unusual In0-like class 1 integron containing CR1 and bla CTX-M-2 and associated with a Tn21-like element. Antimicrob Agents Chemother. 2006;50:799–802. doi: 10.1128/AAC.50.2.799-802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde A, Cantón R, Garcillán-Barcia MP, Novais A, Galán JC, Alvarado A, de la Cruz F, Baquero F, Coque TM. Spread of bla CTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother. 2009;53:5204–5212. doi: 10.1128/AAC.01706-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde A, Coque TM, Sánchez-Moreno MP, Rollán A, Baquero F, Cantón R. Dramatic increase in prevalence of fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J Clin Microbiol. 2004;42:4769–4775. doi: 10.1128/JCM.42.10.4769-4775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill FX, Perrier-Gros-Claude JD, Demartin M, Coignard S, Grimont PA. Characterization of extended-spectrum-beta-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol Lett. 2004;238:353–358. doi: 10.1016/j.femsle.2004.07.058. [DOI] [PubMed] [Google Scholar]
- Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother. 2009;53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Chen HM, Ko WC, Wu HM, Tsai SH, Yan JJ. Prevalence of extended-spectrum beta-lactamases in Proteus mirabilis in a Taiwanese university hospital, 1999 to 2005: identification of a novel CTX-M enzyme (CTX-M-66) Diagn Microbiol Infect Dis. 2008;60:169–175. doi: 10.1016/j.diagmicrobio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Yan JJ, Ko WC, Tsai SH, Wu HM, Jin YT, Wu JJ. Dissemination of CTX-M-3 and CMY-2 beta-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J Clin Microbiol. 2000;38:4320–4325. doi: 10.1128/jcm.38.12.4320-4325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Xu XH, Li JB. Emergence of CTX-M-3, TEM-1 and a new plasmid-mediated MOX-4 AmpC in a multiresistant Aeromonas caviae isolate from a patient with pneumonia. J Med Microbiol. 2010;59:843–847. doi: 10.1099/jmm.0.016337-0. [DOI] [PubMed] [Google Scholar]
- Yin J, Cheng J, Sun Z, Ye Y, Gao YF, Li JB, Zhang XJ. Characterization of two plasmid-encoded cefotaximases found in clinical Escherichia coli isolates: CTX-M-65 and a novel enzyme, CTX-M-87. J Med Microbiol. 2009;58:811–815. doi: 10.1099/jmm.0.006007-0. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ji S, Chen Y, Zhou W, Wei Z, Li L, Ma Y. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect. 2007;54:53–57. doi: 10.1016/j.jinf.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Zhang W, Luo Y, Li J, Lin L, Ma Y, Hu C, Jin S, Ran L, Cui S. Wide dissemination of multidrug-resistant Shigella isolates in China. J Antimicrob Chemother. 2011;66:2527–2535. doi: 10.1093/jac/dkr341. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou H, Shen XQ, Shen P, Yu YS, Li LJ. Plasmid-borne armA methylase gene, together with bla CTX-M-15 and bla TEM-1, in a Klebsiella oxytoca isolate from China. J Med Microbiol. 2008;57:1273–1276. doi: 10.1099/jmm.0.2008/001271-0. [DOI] [PubMed] [Google Scholar]
- Zhao WH, Hu ZQ. Beta-lactamases identified in clinical isolates of Pseudomonas aeruginosa . Crit Rev Microbiol. 2010;36:245–258. doi: 10.3109/1040841X.2010.481763. [DOI] [PubMed] [Google Scholar]
- Zhao WH, Hu ZQ. IMP-type metallo-ß-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011a;37:214–226. doi: 10.3109/1040841X.2011.559944. [DOI] [PubMed] [Google Scholar]
- Zhao WH, Hu ZQ. Epidemiology and genetics of VIM-type metallo-ß-lactamases in Gram-negative bacilli. Future Microbiol. 2011b;6:317–333. doi: 10.2217/fmb.11.13. [DOI] [PubMed] [Google Scholar]
- Zhao WH, Hu ZQ. Acinetobacter: a potential reservoir and dispenser for ß-lactamases. Crit Rev Microbiol. 2012;38:30–51. doi: 10.3109/1040841X.2011.621064. [DOI] [PubMed] [Google Scholar]
- Zong Z, Partridge SR, Iredell JR. ISEcp1-mediated transposition and homologous recombination can explain the context of bla CTX-M-62 linked to qnrB2 . Antimicrob Agents Chemother. 2010;54:3039–3042. doi: 10.1128/AAC.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Z, Partridge SR, Thomas L, Iredell JR. Dominance of bla CTX-M within an Australian extended-spectrum beta-lactamase gene pool. Antimicrob Agents Chemother. 2008;52:4198–4202. doi: 10.1128/AAC.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al Naiemi N, Bart A, de Jong MD, Vandenbroucke-Grauls CM, Rietra PJ, Debets-Ossenkopp YJ, Wever PC, Spanjaard L, Bos AJ, Duim B. Widely distributed and predominant CTX-M extended-spectrum beta-lactamases in Amsterdam, The Netherlands. J Clin Microbiol. 2006;44:3012–3014. doi: 10.1128/JCM.01112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Garcia D, Doi Y, Szabo D, Adams-Haduch JM, Vaz TM, Leite D, Padoveze MC, Freire MP, Silveira FP, Paterson DL. Multiclonal outbreak of Klebsiella pneumoniae producing extended-spectrum beta-lactamase CTX-M-2 and novel variant CTX-M-59 in a neonatal intensive care unit in Brazil. Antimicrob Agents Chemother. 2008;52:1790–1793. doi: 10.1128/AAC.01440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]