Abstract
Nonalcoholic fatty liver disease (NAFLD) involves significant changes in liver metabolism characterized by oxidative stress, lipid accumulation, and fibrogenesis. Mitochondrial dysfunction and bioenergetic defects also contribute to NAFLD. Herein, we examined whether differences in mtDNA influence NAFLD. To determine the role of mitochondrial and nuclear genomes in NAFLD, Mitochondrial-Nuclear eXchange (MNX) mice were fed an atherogenic diet. MNX mice have mtDNA from C57BL/6J mice on a C3H/HeN nuclear background and vice versa. Results from MNX mice were compared to wild-type C57BL/6J and C3H/HeN mice fed a control or atherogenic diet. Mice with the C57BL/6J nuclear genome developed more macrosteatosis, inflammation, and fibrosis compared with mice containing the C3H/HeN nuclear genome when fed the atherogenic diet. These changes were associated with parallel alterations in inflammation and fibrosis gene expression in wild-type mice, with intermediate responses in MNX mice. Mice with the C57BL/6J nuclear genome had increased State 4 respiration, whereas MNX mice had decreased State 3 respiration and RCR when fed the atherogenic diet. Complex IV activity and most mitochondrial biogenesis genes were increased in mice with the C57BL/6J nuclear or mitochondrial genome, or both fed the atherogenic diet. These results reveal new interactions between mitochondrial and nuclear genomes and support the concept that mtDNA influences mitochondrial function and metabolic pathways implicated in NAFLD.
Keywords: Liver, nonalcoholic fatty liver disease, steatosis, inflammation, bioenergetics, mtDNA
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease [1] with 10–30% prevalence rates in the general population and higher rates in metabolic syndrome patients [2]. The histopathological spectrum of NAFLD includes simple steatosis alone (i.e., fatty liver), steatosis with inflammation (nonalcoholic steatohepatitis or NASH), and NASH that includes necro-inflammation with or without fibrosis [3, 4]. NASH is a risk factor for cirrhosis and potentially hepatocellular carcinoma [5, 6]. Thus, a better understanding of NAFLD is needed for improved diagnosis, prevention, and treatment.
Experimental and clinical research supports the hypothesis that altered mitochondrial metabolism contributes to NAFLD [7]. While many of the molecular changes responsible for these mitochondrial changes remain poorly defined, they likely include alterations in mitochondrial bioenergetics, fatty acid oxidation, biogenesis, and mtDNA content. Sookoian and colleagues have shown that the hepatic mtDNA to nDNA ratio is significantly reduced in NAFLD patients compared to control subjects, and correlated with peripheral insulin resistance; a key risk factor for NAFLD/NASH [8]. In contrast, the liver mtDNA to nDNA ratio was significantly increased in mice fed a high fat diet for 20 wk, indicating a possible adaptive response to chronic metabolic stress [9]. Further, mtDNA deletions and mutations have long been associated with metabolic diseases like diabetes [10, 11]. Epigenetic modification of mtDNA and nuclear encoded mitochondrial genes may also contribute to NAFLD/NASH. For example, the mitochondrial-encoded Complex I subunit ND6 gene was found to be highly methylated in liver of NASH patients, but not in patients with simple steatosis [12]. This was associated with significantly decreased ND6 transcript and protein in liver of NASH patients [12], which would likely decrease Complex I function and mitochondrial energy production. Together, these results strongly implicate mitochondria and mtDNA alterations in NAFLD pathobiology.
While it is acknowledged that certain mtDNA haplotypes are associated with metabolic diseases [13, 14], it has been difficult to identify the precise molecular mechanisms that underpin how mtDNA differences influence disease susceptibility. This is especially the case for complex diseases like NAFLD. Therefore, we used the recently developed Mitochondrial-Nuclear eXchange (MNX) mice model [15] to test whether differences in the mtDNA genetic background influence mitochondrial function and NAFLD. MNX mice have interchanged nuclear and mitochondrial genomes from different mice strains, and therefore are different from conplastic [16] and xeno-mitochondrial animal models [17, 18]. MNX mice have 100% of the nDNA and mtDNA from respective donor strains (C57BL/6J and C3H/HeN) through nuclear transfer techniques [15]. For example, MNX mice have the nuclear genome of a C57BL/6J (C57n) or a C3H/HeN (C3Hn) mouse, and the mitochondrial genome of a C3H/HeN (C3Hmt) or C57BL/6J (C57mt) mouse, respectively. One previous study has shown that C57BL/6J mice are more, and C3H/HeN mice are less, susceptible to NAFLD [19]. Using the MNX model, Ballinger and colleagues [15] showed that mtDNA background modulated mitochondrial function, oxidant production, and response to injury in cardiac tissue. Herein, hepatic mitochondrial bioenergetics, histopathology, and gene expression were examined in liver of MNX mice and wild-type (WT) C57BL/6J and WT C3H/HeN mice fed an atherogenic diet. Our results show that the nuclear genome is a key determinant of inflammation and fibrosis; however, the mitochondrial genome influences certain aspects of lipid metabolism and mitochondrial bioenergetic function in liver.
EXPERIMENTAL
Mice and dietary treatments
Six wk old male C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME), C3H/HeNHsd mice (Harlan Research Models, Dublin, VA), and MNX mice (MNX C57 = C57n:C3Hmt and MNX C3H = C3Hn:C57mt) were housed in a temperature controlled environment (22 ± 2°C) with a 12 hr light-dark cycle. A detailed description of how MNX mice were generated is included in [15]. Mice were fed ad libitum a control diet (PicoLab Rodent Chow 20, 4.5% fat, 0.02% cholesterol) or an atherogenic diet (Teklad Atherogenic Rodent Diet TD.02028, 21% fat, 1.25% cholesterol, 0.5% cholic acid) for 12 wk. For experiments, mice were anesthetized by ketamine (60 mg/kg body weight) and xylazine (10 mg/kg body weight) injection (i.p.), and blood and liver were collected. Studies were approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham, USA (APN09319).
Serum chemistries and liver triglyceride measurement
Serum and liver triacylglycerol (TAG) and alanine aminotransferase (ALT) content were measured using TAG-GPO and ALT reagent kits (Pointe Scientific, Canton, MI). Serum total cholesterol was measured by fluorescence detection using the Amplex Red Cholesterol assay kit (Invitrogen, Grand Island, NY). Serum free cholesterol was measured with the Free Cholesterol E kit (Wako Diagnostics, Richmond, VA). Serum adiponectin was determined with the Quantikine® Adiponectin/Acrp30 kit (R&D Systems, Minneapolis, MN).
Histopathology
Hematoxylin & eosin (H&E) stained sections were labeled using a numeric code and evaluated by a pathologist blinded to the experimental design and status of individual animals. Sections were examined for steatosis and inflammatory activity. The presence or absence of steatosis was noted and the degree of macrosteatosis was expressed as the percentage of lobular parenchyma occupied by macrovesicular fat. Inflammatory activity was graded using the modified Knodell histology activity index for grading chronic hepatitis in humans [20]. The absence of intra-acinar inflammation was given a score of 0, one inflammatory focus or less per 10x field was scored 1, two to four foci was scored 2, five to ten foci was scored 3, and more than ten foci was scored 4. Sirius Red-stained sections were evaluated for fibrosis as described in [21]. Sirius Red (Direct Red 80, Sigma-Aldrich, St. Louis, MO) reacts with collagen and does not stain other matrix proteins. Four non-overlapping fields per slide were randomly selected under 400x magnification by an investigator blinded to sample identity. Fibrosis area was calculated using NIH Image J software (rsbweb.nih.gov). Images were converted to HSB stack with saturation set at 155 as MinThreshold and 255 as MaxThreshold to segment the collagen-stained area. The percentage of the surface area that fell in the threshold range within the whole image was taken as the fibrosis area.
Mitochondria isolation and respiration measurements
Mitochondria were prepared from fresh liver by differential centrifugation techniques [22]. Oxygen consumption of isolated liver mitochondria was monitored using a Clark-type oxygen electrode (Hansatech Instruments, Amesbury, MA). Respiratory capacity was assessed by measuring State 3 (i.e., ADP-dependent) and State 4 (i.e., ADP-independent) respiration using succinate as the oxidizable substrate. Succinate-driven respiration was done in the presence of rotenone (1 μM). The respiratory control ratio (RCR) was calculated as the ratio of State 3 to State 4 respiration rates. Mitochondrial complex (I, II–III, IV, and V) and citrate synthase activities were determined using standard spectrophotometric assays [23].
RNA Isolation and gene expression analysis
Total RNA was isolated from liver using TriReagent® (Sigma, St. Louis, MO). Reverse transcription was performed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA). Real-time PCR was performed using an Applied Biosystems 7900 HT instrument with Taqman Gene Expression assays containing gene specific primers and probes. Relative gene expression was determined using the comparative cycle threshold method. Data were normalized to GAPDH and expressed as a fold-change from C57BL/6J mice fed the control diet for statistical analysis. Gene expression data were transformed (log2) and plotted in a heatmap generated with Genespring ver12.6 (Agilent Technologies, Inc., Santa Clara, CA) to aid visualization of results (Figure 3).
Figure 3. Effect of diet, genotype, and haplotype on lipid metabolism, inflammation, and fibrosis genes.
Gene expression was determined for mice fed the control and atherogenic diets: WT C57, WT C3H, MNX C57, and MNX C3H. Results were normalized based on a change in expression from WT C57 mice fed the control diet. The sample size for each measurement was n=4 mice per group. Gene expression results were transformed (log2) and plotted in a heatmap generated using Genespring ver12.6 (Agilent Technologies, Inc., Santa Clara, CA). Down-regulated genes (relative to WT C57 Control Diet) are shown in green, and up-regulated genes (relative to WT C57 Control Diet) are shown in red. ANOVA results are provided in Supplemental Table 2 and p values for pair-wise comparisons are provided in Supplemental Table 3.
Statistical Analysis
Statistical differences were determined by ANOVA using SAS (SAS Institute Inc., Cary, NC) and SigmaStat (Systat Software, Inc. San Jose, CA). Three-way ANOVA was used to determine the significance of the main effect of diet, genotype, and haplotype, and interactions (diet X genotype, diet X haplotype, genotype X haplotype, and diet X genotype X haplotype). ANOVA results are compiled in Supplemental Tables 1 and 2. P values for key pair-wise comparisons between groups (post hoc tests) are compiled in Supplementary Table 3. Only results for the pair-wise comparisons within each group as a function of diet (control vs. atherogenic) are provided in Table 1 and Figures 1B, 1C, 2B, 4 and 5. Criterion for significance for all statistical analyses was set at p ≤ 0.05.
Table 1.
Effect of diet, genotype, and haplotype on body parameters, liver measurements, and select serum chemistries.
| WT C57
(C57n:C57mt) |
WT C3H
(C3Hn:C3Hmt) |
MNX C57
(C57n:C3Hmt) |
MNX C3H
(C3Hn:C57mt) |
|||||
|---|---|---|---|---|---|---|---|---|
| Control | Atherogenic | Control | Atherogenic | Control | Atherogenic | Control | Atherogenic | |
| Body weight (g) | 28.8 ± 1.3 | 24.9 ± 0.3* | 31.3 ± 0.8 | 42.4 ± 0.4** | 28.5 ± 0.8 | 24.5 ± 1.04* | 32.9 ± 0.8 | 42.4 ± 0.8** |
| Liver weight (g) | 1.4 ± 0.1 | 1.8 ± 0.01* | 1.7 ± 0.02 | 2.9 ± 0.1** | 1.3 ± 0.03 | 2.1 ± 0.1** | 1.5 ± 0.07 | 2.9 ± 0.1** |
| Liver weight/body weight | 4.6 ± 0.1 | 7.3 ± 0.2** | 5.6 ± 0.2 | 6.9 ± 0.2* | 4.7 ± 0.1 | 8.7 ± 0.3** | 4.6 ± 0.2 | 7.6 ± 0.5** |
| Liver TAG (mg/mg prot) | 3.1 ± 0.5 | 5.4 ± 1.4 | 3.9 ± 0.6 | 4.5 ± 0.4 | 2.7 ± 0.07 | 3.5 ± 1.5 | 2.1 ± 0.12 | 3.3 ± 0.05* |
| Serum TAG (mg/dL) | 31.0 ± 2.8 | 20.0 ± 3.8 | 87.4 ± 9.4 | 33.5 ± 4.2** | 21.4 ± 1.2 | 13.2 ± 2.00 | 93.8 ± 11.7 | 63.2 ± 12.4** |
| Total Serum Chol (mg/dL) | 56.5 ± 8.8 | 125.1 ± 9.9* | 148.1 ± 7.7 | 196.9 ± 29.8 | 55.8 ±12.7 | 113.4 ± 12.3* | 169.4 ± 9.1 | 298.4 ± 7.5** |
| Free Serum Chol (mg/dL) | 6.7 ± 1.3 | 16.6 ± 1.5* | 14.9 ± 0.8 | 16.8 ± 0.7 | 7.6 ± 0.9 | 15.5 ± 1.3* | 16.8 ± 1.07 | 18.2 ± 2.4 |
| Serum Adiponectin (μg/mL) | 6.1 ± 0.4 | 5.7 ± 0.7 | 2.8 ± 0.3 | 4.6 ± 0.4* | 8.0 ± 0.5 | 5.7 ± 0.5* | 5.0 ± 0.6 | 3.9 ± 0.4 |
| Serum ALT (IU/L) | 13.9 ± 3.5 | 51.2 ± 4.9* | 22.4 ± 5.2 | 35.5 ± 5.8 | 27.8 ± 4.6 | 88.2 ± 17.9* | 33.9 ± 9.1 | 95.8 ± 11.3** |
Mice were fed a control diet or an atherogenic diet for 12 wk. Data are presented as mean ± SE. The number of mice used for: 1) body weight, liver weight and liver/body weight ratio, n=5–15; 2) liver TAG, n=3–4; 3) serum TAG, n=4–9; 4) total serum cholesterol, n=3–5; 5) free serum cholesterol, n=5–11; 6) serum adiponectin, n=5; and 7) serum ALT, n=4–9. Statistical differences as compared with corresponding control counterparts:
p<0.05;
p< 0.0001 are for differences between dietary treatments (atherogenic diet compared to control diet). Other pair-wise comparisons are included in Supplemental Data Table 3.
Abbreviations: TAG, triacylglycerol; Chol, cholesterol; ALT, alanine aminotransferase.
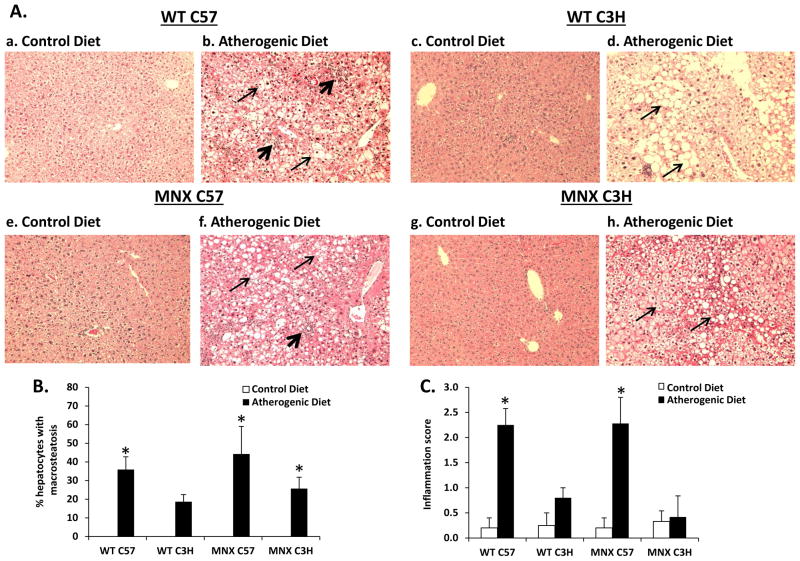
Figure 1. Effect of diet, genotype, and haplotype on liver histology.
Liver macrosteatosis and inflammatory foci were assessed in H&E stained sections for mice groups: WT C57, WT C3H, MNX C57, and MNX C3H. (A) Representative photomicrographs of H&E stained sections are shown from WT C57 (a, n=5), WT C3H (c, n=4), MNX C57 (e, n=5), and MNX C3H (g, n=6) mice fed the control diet; and WT C57 (b, n=16), WT C3H (d, n=15), MNX C57 (f, n=7), and MNX C3H (h, n=7) mice fed the atherogenic diet (magnification × 400). The narrow and wide arrows indicate hepatocyte steatosis and inflammatory infiltration, respectively. (B and C) Bar graphs representing % hepatocytes with macrosteatosis and inflammation score, respectively. Steatosis was not present in livers from mice fed the control diet and was scored as ‘0’; thus, no bars are given for control diet groups. Results are expressed as mean ± SE. *p<0.05, compared to corresponding control diet counterpart.
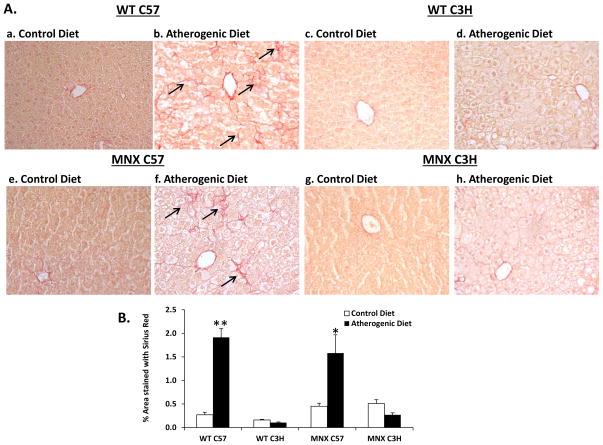
Figure 2. Effect of diet, genotype and haplotype on liver fibrosis.
Staining of collagen fibers was assessed for mice groups: WT C57, WT C3H, MNX C57, and MNX C3H. (A) Representative photomicrographs of Sirius Red-stained liver sections are shown WT C57 (a, n=5), WT C3H (c, n=3), MNX C57 (e, n=4), and MNX C3H (g, n=4) mice fed the control diet; and WT C57 (b, n=7), WT C3H (d, n=5), MNX C57 (f, n=6), and MNX C3H (h, n=5) mice fed the atherogenic diet (magnification × 400). The arrows indicate Sirius Red-stained collagen fibers. (B) Bar graph representing % area stained with Sirius Red. Results are expressed as mean ± SE. *p<0.05 or **p<0.0001, compared to corresponding control diet counterpart. ANOVA results are provided in Supplemental Table 1 and p values for pair-wise comparisons are provided in Supplemental Table 3.
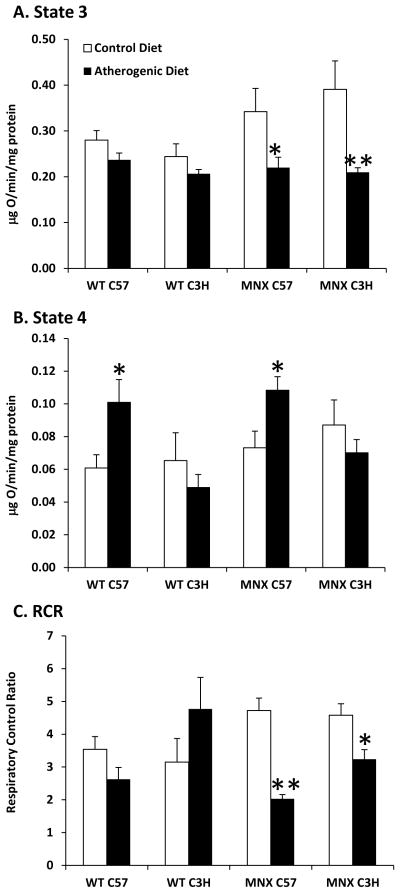
Figure 4. Effect of diet, genotype, and haplotype on mitochondrial function.
Mitochondrial respiration and respiratory control ratio (RCR) was measured for mice fed the control and atherogenic diets: WT C57, WT C3H, MNX C57, and MNX C3H. State 3 (A) and state 4 (B) respiration were measured and the respiratory control ratio (RCR, C) was determined. Succinate was used as the oxidizable substrate. Results are expressed as mean ± SE, n=4–8 for the three parameters measured. *p<0.05 or **p<0.0001, compared to corresponding control diet counterpart. ANOVA results are provided in Supplemental Table 1 and p values for pair-wise comparisons are provided in Supplemental Table 3.
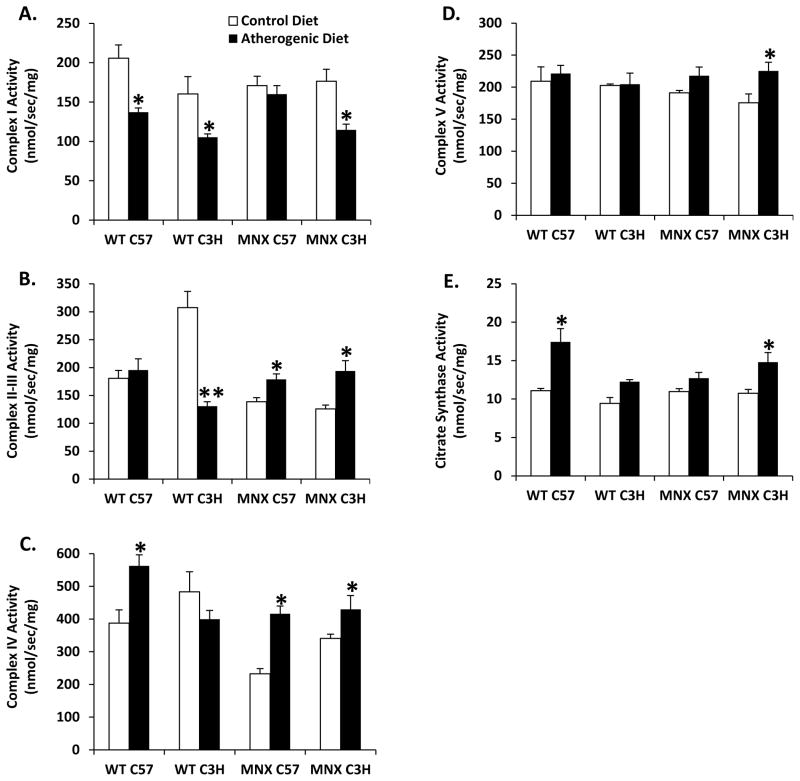
Figure 5. Effect of diet, genotype, and haplotype on respiratory complex activities.
Mitochondrial respiratory complexes activities were measured for (A) complex I, (B) complex II–III, (C) complex IV, (D) complex V, and (E) citrate synthase (CS) activity in liver mitochondria of WT and MNX mice fed control and atherogenic diets. Results are expressed as mean nmol/sec/mg protein ± SE, n=3–4 (Complex I), n=4–6 (Complex II–III), n=4–12 (Complex IV), n=4–6 (Complex V), and n=4–12 (CS). *p<0.05 or **p<0.0001, compared to corresponding control diet counterpart. ANOVA results are provided in Supplemental Table 1 and p values for pair-wise comparisons are provided in Supplemental Table 3.
RESULTS
The following abbreviations will be used to identify the four different strains of mice used: 1) WT C57 = C57n:C57mt; 2) WT C3H = C3Hn:C3Hmt; 3) MNX C57 = C57n:C3Hmt; and 4) MNX C3H = C3Hn:C57mt. WT C57 and MNX C57 mice contain the nuclear genome of C57 mice, and WT C3H and MNX C3H contain the nuclear genome of C3H mice. Further, WT C57 and MNX C3H contain the mitochondrial genome of C57 mice, and WT C3H and MNX C57 contain the mitochondrial genome of C3H mice.
Body parameters and serum chemistries
Wild-type C3H and MNX C3H mice weighed more when fed the atherogenic diet, whereas WT C57 and MNX C57 mice weighed less, compared to their control diet counterparts (Table 1). Liver weight and liver/body weight ratio were increased in all groups fed the atherogenic diet compared to mice fed the control diet (Table 1). Serum TAG and cholesterol (total and free) levels were significantly higher in control-fed WT C3H and MNX C3H mice than in WT C57 and MNX C57 mice (Table 1). The atherogenic diet decreased serum TAG levels in all groups (Table 1), whereas total cholesterol was increased in all groups fed the atherogenic diet, except WT C3H mice (Table 1). The atherogenic diet increased serum adiponectin in WT C3H mice, but not in WT C57 mice (Table 1). Serum adiponectin was decreased in MNX C57 mice fed the atherogenic diet, while no changes were observed in MNX C3H mice. The atherogenic diet significantly increased serum ALT in mice with the C57 genotype and haplotype (Table 1). Results for three-way ANOVA on serum chemistries are presented in Supplemental Table 1. A significant effect of diet was observed for all serum chemistries except adiponectin. A significant effect of genotype was observed for all serum chemistries except ALT. A significant effect of haplotype was observed for serum TAG and total cholesterol.
Histopathology
WT and MNX mice fed the atherogenic diet developed steatosis (Figure 1A); however, the degree of steatosis in WT C3H mice fed the atherogenic diet did not reach statistical significance (Figure 1B). Inflammation (Figure 1C) and pericellular fibrosis (Figure 2A) were present in livers of WT C57 and MNX C57 mice fed the atherogenic diet, whereas WT C3H and MNX C3H mice were resistant to inflammation and fibrosis. Collagen staining was 3–4-fold higher in WT C57 and MNX C57 mice fed the atherogenic diet compared to control diet counterparts (Figure 2B).
Expression of lipid metabolism, inflammation, and fibrosis genes
Having observed different effects of the atherogenic diet on liver pathology, we examined mRNA expression of well-established lipid metabolism, inflammation, and fibrosis mediators (see heatmap in Figure 3). All genes measured in this study were connected by network analysis (see analysis in Suppl. Figure 1). ANOVA and post hoc test results for gene expression are presented in Supplemental Tables 2 and 3, respectively.
Differences in the basal expression of proliferator-activated receptor gamma (PPARG), fatty acid binding protein 1 (FABP1), fatty acid synthase (FASN), patatin-like phospholipase domain-containing protein 3 (PNPLA3), and sterol regulatory element-binding protein 1c (SREBP1) were observed between WT C57 and WT C3H mice. Interestingly, basal expression of PPARG was markedly lower in WT C3H and MNX C3H mice compared with WT C57 and MNX C57 mice. WT and MNX mice fed the atherogenic diet had increased expression of the lipogenesis transcription factor SREBP1 and one of its downstream gene targets diacylglycerol O-acyltransferase 1 (DGAT1). FASN and PNPLA3 were increased only in MNX mice fed the atherogenic diet. Increased expression of the β-oxidation enzyme medium-chain acyl-CoA dehydrogenase (ACADM) and carnitine palmitoyltransferase 1A (CPT1A) were increased in mice fed the atherogenic diet. Peroxisome proliferator-activated receptor alpha (PPARA) was upregulated by the atherogenic diet in WT C57, MNX C57, and MNX C3H mice. The atherogenic diet had no effect FABP1 and PPARG.
Higher levels of inflammatory genes were observed in WT C57 and MNX C57 mice compared to WT C3H and MNX C3H mice fed the atherogenic diet. Importantly, significant main effects of diet and genotype were observed for all inflammation genes. It is important to note that C3H/HeN mice are toll-like receptor (TLR) competent, unlike C3H/HeJ mice that have non-functional TLR4 signaling and do not respond to lipopolysaccharide [24, 25]. Tumor necrosis factor alpha (TNFA) and nuclear factor kappa B (NFkB) were increased by the atherogenic diet in all four mice strains with the highest levels measured in WT C57 and MNX C57 mice. Heme oxygenase 1 (HMOX1) and inducible nitric oxide synthase (NOS2) were significantly increased in WT C57, MNX C57, and WT C3H mice fed the atherogenic diet compared to controls. WT C57 and MNX C57 mice also showed increased expression of TLR2, TLR4, and myeloid differentiation primary response gene 88 (MYD88) in response to the atherogenic diet. TLR9 expression was only up-regulated in WT C57 mice fed the atherogenic diet compared to control diet mice. A significant effect of haplotype was found for NOS2 and TLR9.
Higher levels of fibrosis genes were observed in WT C57 and MNX C57 mice in response to the atherogenic diet compared WT C3H and MNX C3H. Significant main effects of diet and genotype were observed for all three fibrosis genes (COL1A1, COL4A1, TGFB1).
Mitochondrial bioenergetics
Feeding the atherogenic diet caused a small decrease in State 3 respiration in WT mice. State 3 respiration was significantly decreased in MNX mice fed the atherogenic diet compared to control-fed mice (Figure 4A). In contrast, State 4 respiration was significantly increased in WT C57 and MNX C57 mice fed the atherogenic diet compared to controls (Figure 4B). The RCR was significantly decreased in MNX mice fed the atherogenic diet compared to controls (Figure 4C). A significant effect of diet was observed for State 3 respiration and RCR. A significant effect of genotype was observed for State 4 respiration. Additional ANOVA results are provided in Supplemental Table 1.
Higher and lower activities of Complex I and Complex II–III were measured in mitochondria from control-fed WT C57 mice compared to WT C3H mice (Figure 5A and B). Complex I activity was decreased in response to the atherogenic diet in WT C57, WT C3H, and MNX C3H mice (Figure 5A). Complex II–III activity was markedly decreased in WT C3H mice, but increased in MNX mice fed the atherogenic diet compared to controls (Figure 5B). Complex IV activity was significantly increased by the atherogenic diet in WT C57, MNX C57, and MNX C3H mice (Figure 5C). Complex V activity was increased by the atherogenic diet only in MNX C3H mice (Figure 5D). Citrate synthase activity was increased in WT C57 and MNX C57 mice fed the atherogenic diet (Figure 5E). A significant effect of diet was observed for all enzyme activities except Complex II–III. A significant effect of genotype was observed for Complex I. Significant diet X genotype interactions for Complexes II–III and IV, and diet X haplotype interactions for II–III, IV, and citrate synthase were observed. Other ANOVA results are in Supplemental Table 1.
To determine whether changes in bioenergetics might be associated with alterations in mitochondrial biogenesis genes, we measured the following: peroxisome proliferator-activated receptor gamma, coactivator 1-alpha (PPARGC1A) and -beta (PPARGC1B), nuclear respiratory factor 1 (NRF1), mitochondrial transcription factor B1 (TFB1M), sirtuin 1 (SIRT1), and hypoxia inducible factor 1-alpha (HIF1A). WT C57 and MNX C57 mice fed the atherogenic diet had higher levels of all mitochondrial biogenesis genes (Figure 3). The atherogenic diet increased NRF1 in WT C3H and MNX C3H mice, albeit at lower levels. A significant main effect of diet was observed for all biogenesis genes. A significant genotype effect was observed for all genes, except PPARGC1B. Additional ANOVA results are given in Supplemental Table 2.
DISCUSSION
Oxidative damage, ER stress, lipotoxicity, insulin resistance, and mitochondrial dysfunction have all been implicated as contributors to NAFLD/NASH [26, 27]. While genetic and dietary animal models have been very useful in studying NAFLD, most of these models fail to mimic many features of the human disease [28]. Most experimental animal studies have also not specifically considered the importance of mitochondrial genetic background in the disease process. To help address this question, we used a new experimental model, the MNX mouse, in combination with an atherogenic diet, to investigate whether differences in mitochondrial genetic background influence some of the established metabolic pathways and targets implicated in NAFLD/NASH.
MNX mice have the nuclear genome of one mouse strain and the mitochondrial genome of another mouse strain [15]. C57BL/6J and C3H/HeN mice were chosen as the parental strains for these studies as they are generally susceptible and resistant, respectively, to many metabolic syndrome disorders, including NAFLD [19]. C57BL/6J and C3H/HeN mice differ in mtDNA coding for the ND3 subunit of Complex I, subunit III of Complex IV, and position 9818 in the tRNAArg gene [29]. Thus, MNX mouse are a unique model to test the potential influence and/or interaction of the nuclear and mitochondrial genetic backgrounds in disease. Our results support the concept that the nuclear genome is a key determinant for inflammation and fibrosis because changing mtDNA had little effect on these pathologies. Specifically, mice with C57 nDNA (WT C57 and MNX C57) had higher inflammation and fibrosis scores when fed the atherogenic diet compared to mice with C3H nDNA (WT C3H and MNX C3H). These studies also show that mtDNA background affected serum TAG and cholesterol; two measurements linked to liver lipid metabolism and accumulation. Mitochondrial haplotype also influenced some mitochondrial bioenergetic parameters including State 3 respiration and Complex IV activity.
With regards to bioenergetics, we observed differences in mitochondrial function among the four distinct mice strains under both control and stress (atherogenic diet) conditions. For example, State 3 respiration and the RCR were decreased in MNX mice fed the atherogenic diet, whereas these parameters remained unchanged in WT mice. In contrast, State 4 respiration was significantly increased in WT C57 and MNX C57 mice fed the atherogenic diet. Increased State 4 respiration typically suggests an uncoupling effect; i.e., leakage of H+’s across the inner membrane. Uncoupling, if severe, can decrease efficiency for ATP synthesis [30] and lead to necrotic cell death [31]. Thus, this could partially explain increased liver injury in livers of WT C57 and MNX C57 mice. Alternatively, uncoupling can function as an adaptive response aimed to reduce mitochondrial superoxide anion production in response to chronic fatty acid overload in vivo [32]. While we did not measure oxidant production, oxidative stress/damage in WT C57 and MNX C57 mice fed the atherogenic diet is anticipated because hepatic inflammation and inflammatory gene expression (e.g., TNFA and NOS2) was much higher in these mice. Studies by Ballinger and colleagues showed increased reactive oxygen species (ROS) in cardiomyocytes isolated from WT C57 compared to WT C3H mice [15]. Future studies are needed to address the influence of diet and genetics (nuclear and/or mitochondrial) on ROS production in hepatocytes.
Chronic consumption of the atherogenic diet led to a significant increase in Complex IV activity in WT C57 mice, but not in WT C3H mice. Complex IV activity was also increased in both strains of MNX mice fed the atherogenic diet. This increase suggests an adaptive response to metabolic stress (e.g., mitochondrial biogenesis) in mice with C57 nDNA or mtDNA, or both. Further, it appears that the stress-mediated increase in Complex IV was influenced by C57 mtDNA because increased activity was observed in MNX C3H mice. As mentioned earlier, WT C57 and WT C3H mice differ in the coding sequence of subunit III of Complex IV. This mitochondrial-encoded subunit contains no redox centers and is thought to regulate complex activity and participate in proton uptake [33]. It is possible that the unique combination of C57 mtDNA and C3H nDNA altered retrograde signaling mechanisms (e.g., ROS production), which contributed to different adaptive responses in Complex IV. In contrast, different effects were observed for Complex I. Both WT C57 and WT C3H strains showed decreased Complex I activity in response to the atherogenic diet. These mice differ in the coding sequence of the mtDNA subunit ND3. Interestingly, the atherogenic diet-mediated decrease in Complex I activity did not occur in MNX C57 mice, again revealing a different response to stress in this novel mouse model. While the molecular signals responsible for these distinct changes are presently not known, our results show that the same stress (atherogenic diet) elicits different bioenergetic responses depending on the combination of mitochondrial and nuclear genetic backgrounds.
To complement these functional studies in mitochondria, we examined expression of genes known to be involved in mitochondrial maintenance and NAFLD, including PPARGC1A, PPARGC1B, SIRT1, NRF1, and TFB1M. PGC1α and PGC1β are transcriptional co-activators that serve as key regulators of metabolism, and function as mechanistic links between the mitochondrial and nuclear regulatory pathways of mitochondrial biogenesis [34]. We observed increased gene expression for both PPARGC1A and PPARGC1B in WT C57 and MNX C57 mice in response to the atherogenic diet. Similarly, the atherogenic diet increased hepatic SIRT1 gene expression only in WT C57 and MNX C57 mice. SIRT1, an NAD+-dependent deacetylase, promotes mitochondrial biogenesis by PGC1α activation [35]. Together, these data suggest a potential defect or block in mitochondrial biogenesis in mice with both C3H nDNA and mtDNA (i.e., WT C3H). Large increases in NRF1 were also measured in WT C57 and MNX C57 mice fed the atherogenic diet with a smaller increase detected in MNX C3H mice. NRF1 is a transcription factor that activates transcription of cytochrome c, respiratory chain complex subunits, and the transcriptional machinery of the mitochondrion including TFB1M [36]. Notably, NRF1 and TFB1M gene expression patterns positively correlated with Complex IV activities. This coordinated gene expression pattern supports the role of a mitochondrial biogenesis program that can be activated by metabolic stress (i.e., liver pathology) in mice with C57 nDNA (WT C57 and MNX C57) or C57 mtDNA only (MNX C3H).
Noteworthy in our study was that expression of the transcription factor PPARG was very low in WT C3H and MNX C3H mice compared to mice with the C57 nuclear genome. In contrast, mRNA levels of other classic mediators of lipid and triglyceride synthesis (SREBP1 and DGAT1) and fatty acid oxidation (PPARA, ACADM, and CPAT1A) were essentially the same in all control-fed mice and induced to the same levels in response to the atherogenic diet. PPARγ plays an important role in lipid metabolism by promoting lipid accumulation in adipose and liver [37]. For example, inactivation of liver PPARG reduced hepatic steatosis in lipoatrophic AZIP mice (a model of diabetes with insulin resistance) and wild-type mice fed a high fat diet [38]. Recently, Krek and colleagues reported that during stress, HIF1α activated PPARγ, which in turn induced cardiac hypertrophy and triglyceride accumulation [39]. Similarly, hypoxia and/or HIF1α has been implicated in the development of liver steatosis [9, 40–42] and fibrosis [43, 44]. Herein, we observed stress-mediated increase in HIF1A in WT C57 and MNX C57 mice, but not in WT C3H and MNX C3H mice. Further, HIF1A and PPARG expression positively correlated with the presence of fibrosis and fibrosis gene expression in WT C57 and MNX C57 mice fed the atherogenic diet. Together, these data support the mechanism proposed by Krek and colleagues [39] whereby a pathologic stress activates a HIF1α-PPARγ signaling axis resulting in tissue injury. Thus, the lack of activation of HIF1A and PPARG in WT C3H and MNX C3H mice may have contributed, in part, to a disease-resistant phenotype in these mice.
Along these same lines, we observed a similar pattern in the expression of inflammation genes. For example, TNFA, NOS2, TLR2, TLR4, and MYD88 were greatly increased in the liver of WT C57 and MNX C57 mice fed the atherogenic diet, with smaller increases detected in WT C3H and MNX C3H mice. These results demonstrate a blunted inflammatory response in WT C3H and MNX C3H mice even though C3H/HeN mice are TLR competent [24, 25]. We also observed a large increase in TLR9 in WT C57 fed the atherogenic diet, but not in MNX C57 mice. The reason for this difference is not known. Notably, TLR9 signaling has recently been shown to contribute to the development of NASH [45, 46]. Our finding regarding TLR9 may be of particular interest as mtDNA may act as a damage-associated molecular pattern (DAMP) molecule to activate TLR9 signaling [47]. Whether different mtDNA sequences or differences in mitochondria-nuclear interactions confer differential sensitivity or resistance to disease via activation of TLR signaling is an intriguing concept, and one that will require further investigation.
In summary, we show that the nuclear genome is an important determinant of liver inflammation and fibrosis. Utilization of the new MNX mice model [15], in combination with a NAFLD-inducing diet, indicates that the mitochondrial genome influences some aspects of lipid metabolism and mitochondrial respiratory function. Therefore, these studies support the hypothesis that differences in mtDNA contribute to the susceptibility and severity of NAFLD. These findings also provide a solid starting point for additional investigations focused on understanding how mitochondrial genetics influence cellular metabolism, redox signaling, and liver disease susceptibility.
Supplementary Material
SUMMARY STATEMENT.
The influence of mtDNA in nonalcoholic fatty liver disease (NAFLD) was tested using Mitochondrial-Nuclear eXchange (MNX) mice. Pathological features of NAFLD tracked with the nuclear genome of C57BL/6J mice, whereas mitochondrial functions were modulated in MNX mice.
Acknowledgments
FUNDING: This work was supported by National Institutes of Health grants to SMB [AA015172, AA018841] and SWB [HL094518, HL0103859]. In addition, work was supported by US Army Medical Research & Material Command award to SWB [W81XWH-07-1-0540d]. ALK was supported by a National Institutes of Health Research Supplement to Promote Diversity in Health-Related Research Award [linked to AA015172]. JLF was supported by an American Heart Association Fellowship [PRE2240046].
The authors would like to thank Dr. David Crossman, Department of Genetics and the Heflin Center for Genomic Science Core Laboratories, for preparation of the gene expression heatmap and network analysis.
ABBREVIATIONS USED
- ACADM
medium chain acyl-CoA dehydrogenase
- ALT
alanine aminotransferase
- ANOVA
analysis of variance
- C3H
C3H/HeN mouse
- C57
C57BL/6J mouse
- Chol
cholesterol
- COL1A1
collagen, type I, alpha 1
- COL4A1
collagen, type IV, alpha 1
- CPT1A
carnitine palmitoyltransferase 1A
- CS
citrate synthase
- DAMP
damage associated molecular pattern
- DGAT1
diacylglycerol O-acyltransferase 1
- FABP1
fatty acid binding protein 1
- FASN
fatty acid synthase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- H&E
hematoxylin & eosin
- HIF1A or HIF1α
hypoxia inducible factor 1 alpha
- HMOX1
heme oxygenase 1
- MNX
mitochondrial-nuclear exchange
- mtDNA
mitochondrial DNA
- MYD88
myeloid differentiation primary response gene 88
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- NFKB
nuclear factor kappa light chain enhancer of activated B cells
- NOS2
inducible nitric oxide synthase
- NRF1
nuclear respiratory factor 1
- PNPLA3
patanin-like phospholipase domain containing protein 3
- PPARA or PPARα
peroxisome proliferator-activated receptor alpha
- PPARG or PPARγ
peroxisome proliferator-activated receptor gamma
- PPARGC1A or PGC1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PPARGC1B
peroxisome proliferator-activated receptor gamma coactivator 1-beta
- RCR
respiratory control ratio
- ROS
reactive oxygen species
- SIRT1
sirtuin 1
- SREBP1 or SREBP1c
sterol regulatory element binding transcription factor 1c
- TAG
triacylglyercol
- TFB1M
transcription factor B1, mitochondrial
- TGFB1 or TGFβ1
transforming growth factor beta
- TLR2
toll-like receptor 2
- TLR4
toll-like receptor 4
- TLR9
toll-like receptor 9
- TNFA
tumor necrosis factor alpha
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION: Angela Betancourt, Adrienne King, and Jessica Fetterman performed the majority of the experimental work. Telisha Swain, Rachel Finley, and Claudia Oliva assisted in experiments. David Ralph Crowe analyzed liver histopathology (steatosis and inflammation) and Angela Betancourt analyzed Sirius Red staining (fibrosis). Jessica Fetterman, Scott Ballinger, and Shannon Bailey contributed to study design. Angela Betancourt and Shannon Bailey wrote the manuscript with helpful suggestions from co-authors.
DISCLOSURES: The authors have no conflicts of interest and no disclosures.
References
- 1.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 2.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34:274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 3.Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 1980;55:434–438. [PubMed] [Google Scholar]
- 4.Schaffner F, Thaler H. Nonalcoholic fatty liver disease. Prog Liver Dis. 1986;8:283–298. [PubMed] [Google Scholar]
- 5.Marrero JA, Fontana RJ, Fu S, Conjeevaram HS, Su GL, Lok AS. Alcohol, tobacco and obesity are synergistic risk factors for hepatocellular carcinoma. J Hepatol. 2005;42:218–224. doi: 10.1016/j.jhep.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Powell EE, Jonsson JR, Clouston AD. Steatosis: co-factor in other liver diseases. Hepatology. 2005;42:5–13. doi: 10.1002/hep.20750. [DOI] [PubMed] [Google Scholar]
- 7.Begriche K, Massart J, Robin MA, Bonnet F, Fromenty B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology. 2013;58:1497–1507. doi: 10.1002/hep.26226. [DOI] [PubMed] [Google Scholar]
- 8.Sookoian S, Rosselli MS, Gemma C, Burgueno AL, Fernandez Gianotti T, Castano GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 9.Carabelli J, Burgueno AL, Rosselli MS, Gianotti TF, Lago NR, Pirola CJ, Sookoian S. High fat diet-induced liver steatosis promotes an increase in liver mitochondrial biogenesis in response to hypoxia. J Cell Mol Med. 2011;15:1329–1338. doi: 10.1111/j.1582-4934.2010.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, Wallace DC. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- 11.van den Ouweland JM, Lemkes HH, Ruitenbeek W, Sandkuijl LA, de Vijlder MF, Struyvenberg PA, van de Kamp JJ, Maassen JA. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet. 1992;1:368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- 12.Pirola CJ, Gianotti TF, Burgueno AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castano GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 13.Dunham-Snary KJ, Ballinger SW. Mitochondrial genetics and obesity: evolutionary adaptation and contemporary disease susceptibility. Free Radic Biol Med. 2013;65:1229–1237. doi: 10.1016/j.freeradbiomed.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace DC. Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120267. doi: 10.1098/rstb.2012.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, Sammy MJ, Johnson M, Dunham-Snary KJ, Cao X, Bradley WE, Zhang J, Wei CC, Chacko B, Schurr TG, Kesterson RA, Dell’italia LJ, Darley-Usmar VM, Welch DR, Ballinger SW. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J. 2013;455:157–167. doi: 10.1042/BJ20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, Kunz M, Ibrahim SM. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res. 2009;19:159–165. doi: 10.1101/gr.078865.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenzie M, Trounce IA, Cassar CA, Pinkert CA. Production of homoplasmic xenomitochondrial mice. Proc Natl Acad Sci U S A. 2004;101:1685–1690. doi: 10.1073/pnas.0303184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trounce IA, Pinkert CA. Cybrid models of mtDNA disease and transmission, from cells to mice. Curr Top Dev Biol. 2007;77:157–183. doi: 10.1016/S0070-2153(06)77006-5. [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki Y, Kakizaki S, Takizawa D, Ichikawa T, Sato K, Takagi H, Mori M. Interstrain differences in susceptibility to non-alcoholic steatohepatitis. J Gastroenterol Hepatol. 2008;23:276–282. doi: 10.1111/j.1440-1746.2007.05150.x. [DOI] [PubMed] [Google Scholar]
- 20.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RN, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 21.Bailey SM, Mantena SK, Millender-Swain T, Cakir Y, Jhala NC, Chhieng D, Pinkerton KE, Ballinger SW. Ethanol and tobacco smoke increase hepatic steatosis and hypoxia in the hypercholesterolemic apoE(−/−) mouse: implications for a “multihit” hypothesis of fatty liver disease. Free Radic Biol Med. 2009;46:928–938. doi: 10.1016/j.freeradbiomed.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spach PI, Parce JW, Cunningham CC. Effect of chronic ethanol administration on energy metabolism and phospholipase A2 activity in rat liver. Biochem J. 1979;178:23–33. doi: 10.1042/bj1780023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darley-Usmar VM, Capaldi RA, Takamiya S, Millet F, Wilson MT, Malatesta F, Sarti P. In: In Mitochondria: a practical approach. Darley-Usmar VM, Rickwood D, Wilson MT, editors. IRL; Oxford: 1987. pp. 113–152. [Google Scholar]
- 24.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 25.Vazquez-Torres A, Vallance BA, Bergman MA, Finlay BB, Cookson BT, Jones-Carson J, Fang FC. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs M, Sanyal AJ. Lipotoxicity in NASH. J Hepatol. 2012;56:291–293. doi: 10.1016/j.jhep.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Feldstein AE, Bailey SM. Emerging role of redox dysregulation in alcoholic and nonalcoholic fatty liver disease. Antioxid Redox Signal. 2011;15:421–424. doi: 10.1089/ars.2011.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maher JJ. New insights from rodent models of fatty liver disease. Antioxid Redox Signal. 2011;15:535–550. doi: 10.1089/ars.2010.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham CC, Coleman WB, Spach PI. The effects of chronic ethanol consumption on hepatic mitochondrial energy metabolism. Alcohol Alcohol. 1990;25:127–136. doi: 10.1093/oxfordjournals.alcalc.a044987. [DOI] [PubMed] [Google Scholar]
- 31.Ivester P, Lide MJ, Cunningham CC. Effect of chronic ethanol consumption on the energy state and structural stability of periportal and perivenous hepatocytes. Arch Biochem Biophys. 1995;322:14–21. doi: 10.1006/abbi.1995.1430. [DOI] [PubMed] [Google Scholar]
- 32.Eccleston HB, Andringa KK, Betancourt AM, King AL, Mantena SK, Swain TM, Tinsley HN, Nolte RN, Nagy TR, Abrams GA, Bailey SM. Chronic exposure to a high-fat diet induces hepatic steatosis, impairs nitric oxide bioavailability, and modifies the mitochondrial proteome in mice. Antioxid Redox Signal. 2011;15:447–459. doi: 10.1089/ars.2010.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varanasi L, Hosler JP. Subunit III-depleted cytochrome c oxidase provides insight into the process of proton uptake by proteins. Biochim Biophys Acta. 2012;1817:545–551. doi: 10.1016/j.bbabio.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 36.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 37.Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, Evans RM. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, Vinson C, Gonzalez FJ, Reitman ML. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 39.Krishnan J, Suter M, Windak R, Krebs T, Felley A, Montessuit C, Tokarska-Schlattner M, Aasum E, Bogdanova A, Perriard E, Perriard JC, Larsen T, Pedrazzini T, Krek W. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9:512–524. doi: 10.1016/j.cmet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Mantena SK, Vaughn DP, Andringa KK, Eccleston HB, King AL, Abrams GA, Doeller JE, Kraus DW, Darley-Usmar VM, Bailey SM. High fat diet induces dysregulation of hepatic oxygen gradients and mitochondrial function in vivo. Biochem J. 2009;417:183–193. doi: 10.1042/BJ20080868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nath B, Levin I, Csak T, Petrasek J, Mueller C, Kodys K, Catalano D, Mandrekar P, Szabo G. Hepatocyte-specific hypoxia-inducible factor-1alpha is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology. 2011;53:1526–1537. doi: 10.1002/hep.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin J, Gao Z, He Q, Zhou D, Guo Z, Ye J. Role of hypoxia in obesity-induced disorders of glucose and lipid metabolism in adipose tissue. Am J Physiol Endocrinol Metab. 2009;296:E333–342. doi: 10.1152/ajpendo.90760.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copple BL, Bai S, Moon JO. Hypoxia-inducible factor-dependent production of profibrotic mediators by hypoxic Kupffer cells. Hepatol Res. 2010;40:530–539. doi: 10.1111/j.1872-034X.2010.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Copple BL, Bustamante JJ, Welch TP, Kim ND, Moon JO. Hypoxia-inducible factor-dependent production of profibrotic mediators by hypoxic hepatocytes. Liver Int. 2009;29:1010–1021. doi: 10.1111/j.1478-3231.2009.02015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miura K, Kodama Y, Inokuchi S, Schnabl B, Aoyama T, Ohnishi H, Olefsky JM, Brenner DA, Seki E. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology. 2010;139:323–334. e327. doi: 10.1053/j.gastro.2010.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roh YS, Seki E. Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. J Gastroenterol Hepatol. 2013;28(Suppl 1):38–42. doi: 10.1111/jgh.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.