Abstract
Control of estrogenic activity within the uterus is evident as unopposed estrogen action is associated with endometrial pathologies such as endometriosis and endometrial carcinoma. MicroRNAs have emerged as important post-transcriptional regulators which are postulated to fine tune the actions of steroids in many systems including the uterus. The objective of the current study was to examine uterine expression of microRNAs in response to estrogen treatment within the mouse uterus using an ovariectomized, steroid reconstituted mouse model. MicroRNA microarray analysis and subsequent qRT-PCR verification revealed that expression of mirn155, mirn429 and mirn451 was significantly increased by estrogen administration while mirn181b and mirn204 expression was significantly reduced. Pretreatment with the estrogen receptor antagonist ICI 182,780 confirmed that estrogen regulation was mediated via the classical estrogen receptor pathway. This study demonstrates that estrogen regulates specific miRNAs within the murine uterus which may participate in post-transcriptional regulation of estrogen-regulated genes.
Keywords: Uterus, estrogen, microRNA
Introduction
The female sex hormone estrogen regulates diverse biological processes which include bone metabolism, cardiovascular functions, developmental processes and reproductive events.1 Estrogen action upon the uterus is biphasic and can be divided into early and late responses. Early response events include increased RNA transcription, hyperemia and water imbibition while late events include DNA synthesis and mitosis of epithelium.2 Estrogen-regulated genes involved in growth factor signaling, Wnt signaling, cell cycle progression and apoptosis have been associated with these early and late estrogenic responses within the uterus. Regulation of these estrogenic events has been shown to occur via both estrogen receptors (ER)-α and ER-β3 as well as through alternative estrogen signaling pathways.2,4–6
Understanding the molecular mechanisms that contribute to the development and subsequent function of the uterus are absolutely essential for successful reproduction and/or adequate treatment of uterine disorders that impede the reproductive process. As suggested above, it is well established that numerous estrogen regulated factors (cytokines, growth factors, steroids, etc.) dictate the normal pattern of uterine development and that disruption of the normal action of these factors plays a causative role in uterine abnormalities and infertility. It is becoming increasingly clear that post-transcriptional regulation of such factors may also impact organ development. As such, developing a thorough understanding of the factors which regulate uterine development and function is of paramount importance within the context of reproductive biology.
MicroRNAs (miRNAs) are a family of small (19 to 22 nucleotides) non-coding RNAs that are involved in post-transcriptional gene regulation by affecting mRNA translation and/or stability.7,8 miRNA post-transcriptional regulation of specific gene products has been proposed to play a role in the normal development of the lung,9 limbs,10 and skeletal muscle11 as well as the female germline12 and female reproductive tract.13–15 Recently, post-transcriptional regulation of gene expression by miRNAs has been proposed to play a role in the normal physiology of the human endometrium.16–18 Further, studies in rodents have identified specific miRNAs within the uterus which are proposed to participate in the preparation of the uterus for embryo implantation.19–21 While there is limited information on the role of miRNAs in normal uterine physiology, there has been a greater emphasis on studying the role of miRNAs in endometrial diseases which are associated with enhanced or unopposed estrogen responsiveness such as endometriosis22–26 and endometrial cancer.27–29 Collectively, these studies suggest that miRNAs play a pivotal role in the molecular regulation of multiple organ systems, including reproductive function and specifically uterine implantation and uterine pathophysiological conditions.
Unfortunately, beyond these limited studies there is no information on the expression and regulation of miRNAs in the uterus and more importantly the potential role of miRNA control of early and late phase estrogen responses in vivo. As such, the objective of the current study was to examine uterine expression of miRNAs in response to estrogen treatment during the early and late response phases within the mouse uterus.
Materials and Methods
Animals and treatments
Mature female mice (CD1; Harlan) were ovariectomized, rested two weeks and treated s.c. with either estradiol 17-β (E2; 10 μg/kg BW) or sesame seed oil vehicle and sacrificed 2, 4, 8, 16, 24, or 30 h after treatment administration. Uteri were removed and stored in RNalater (Ambion) until processed for RNA isolation.
A second study was conducted to examine whether the E2 regulation of uterine miRNAs was mediated via its cognate nuclear receptor. To do so, mice were injected s.c. with estrogen receptor antagonist ICI-182,780 (ICI; Tocris Cookson Inc., Ellisville, MO) 30 min before steroidal treatment. Estradiol was again administered, mice were sacrificed at 8 h post steroid administration, and tissues were processed as described above. We selected the 8h time point as this was shown to be a time after estradiol administration for maximal change in the level of expression for each examined miRNA.
miRNA microarray analysis
Uterine tissue was pooled from 3 mice per 0, 2, 4 and 8h time points and from 2 mice per 16, 24 and 30h time points. A total of 3 pools for each of the seven time points (N=3/time point) were analyzed by miRNA microarray analysis (a total of 21 arrays). Total RNA was isolated from uterine tissue pools using miRNEasy columns according to the recommendations of the manufacturer (Qiagen; Valencia, CA). Small RNA isolation and hybridization were performed by LC Sciences (Houston, TX) using 5 to 10 μg of total RNA. Briefly, small RNA species were isolated from total RNA by column exclusion. The concentrated small RNAs were 3′-polyadenylated with poly(A) polymerase. A nucleotide tag was then ligated to the poly(A) tails. The tagged RNAs were hybridized to a μParaflo superfluidic array chip (miRBase version 10.0) containing probes for 568 murine microRNA sequences as well as controls. Tagged RNA was subsequently labeled in a second hybridization reaction with Cy5 dendrimer dye. After overnight hybridization, arrays were stringently washed and scanned on an Axon GenePix 4000B laser scanner (Molecular Devices, Sunnyvale, CA). Data extraction, imaging and statistical analysis were performed by LC Sciences using ArrayPro software (Media Cybernetics; Bethesda, MD). After signal amplification, the background was subtracted and normalized using LOWESS (locally weighted regression) method. For a transcript to be listed as detectable, it had to meet the following criteria: signal intensity higher than 3× (background SD), spot coefficient of variation <0.5 (coefficient of variation = SD/signal intensity), and signals from at least 50% of the repeating probes above detection level. The array output was received in Excel spreadsheets as lists of raw data and also as “simple detectable” data, which were the average of 6 signal values for each microRNA on the array. For each set at each time point analyzed, significant differences (probability values) among groups for a given detectable microRNA signal were calculated. Those with P ≤0.05 were analyzed using gene hierarchical clustering of the log2 value of each signal. Three separate analyses for each time point using different aliquots derived from independent pools of uterine tissue were used. A list of miRNAs from combined data from the three trials which showed significant differences across time (as determined by LC Sciences; > 1.5-fold difference in expression and level of expression > 1,000 signal intensity [scale of 1 to 100,000 based upon its fluorescence which is indicative of its relative level within the tissue]) was generated. Of those, we selected a subset of miRNAs for verification by qRT-PCR.
Quantitative Real-Time RT-PCR for miRNA
Quantitative RT-PCR (qRT-PCR) was performed30 to analyze the temporal expression and treatment effects of specific mature miRNAs. Total RNA was used from the same aliquots analyzed by microarrays (described above) or total RNA was isolated from additional samples using the same methodology as described under microarray analysis. miRNA kits for mirn99b, mirn181b, mirn429 and mirn451 were purchased from Applied Biosystems (Foster City, CA) to quantitate their fold change in expression. Total RNA (250 ng in 5μl) was reverse transcribed using reverse transcription (RT) kits (Applied Biosystems) following the manufacturer’s protocol with the following modifications. Briefly, miRNAs were reverse transcribed in a single reaction using 2 μl of each miRNA specific 5X RT primers. Resulting material was then used for independent qRT-PCR for each miRNA. To normalize for starting material, a reverse snRNA U6 was included in the miRNA RT reactions and qRT-PCR of U6 was performed. qRT-PCR reactions were completed on a 7900 HT Sequence Detection System (Applied Biosystems). Samples were run in triplicate and the average value used in subsequent calculations. Each primer set included a minus RT control. The 2-delta-delta CT method was used to calculate the fold-change values among samples.31 Experiments were performed on at least 4 different independent replicates (N ≥ 4) and data are displayed as the mean ± standard deviation of the means (SD).
Statistical analysis
Unless otherwise indicated, all data was analyzed by one-way ANOVA for comparison across time points. When an F test indicated statistical significance, post-hoc analysis was made using the Tukey HSD procedure. Unpaired t-tests were used for planned comparisons between specific treatment groups within time points. Significance was set at P < 0.05 for all comparisons.
Results
Analysis of uterine miRNA expression (N=3 separate array analyses for each time point) revealed that of the 568 murine miRNAs analyzed, 445 were detectable in uterine tissue. Of these, ten miRNAs were shown to be significantly regulated by E2 (Table 1). From these, we selected seven miRNAs (mirn429, mirn451, mirn99b, mirn181b, mirn204, mirn155 and mirn720) for further analysis based upon their fold change in response to estrogen administration (> 1.5-fold from controls).
Table 1.
Uterine microRNAs (mirn) significantly regulated by estrogen based upon microRNA microarray analysis.
| Estrogen effect | Relative level of expression1 |
|---|---|
| Up-regulated | |
| mmu-mirn451 | Intermediate (9,999 – 2,000 units) |
| mmu-mirn429 | Intermediate (9,999 – 2,000 units) |
| mmu-mirn99b | Intermediate (9,999 – 2,000 units) |
| mmu-mirn155 | Low (1,999 – 500 units) |
| mmu-mirn7a | Low (1,999 – 500 units) |
| Down-regulated | |
| mmu-mirn24 | High (100,000 – 10,000 units) |
| mmu-mirn181b | Intermediate (9,999 – 2,000 units) |
| Biphasic | |
| mmu-mirn150 | Intermediate (9,999 – 2,000 units) |
| mmu-mirn720 | Low (1,999 – 500 units) |
| mmu-mirn204 | Low (1,999 – 500 units) |
Levels of expression were arbitrarily assigned based upon Cy3/Cy5 intensity images as described in “Materials and Methods” using the scale: High (100,000 to 10,000 units), Intermediate (9,999 – 2,000 units), and Low (1,999 – 500 units). We considered signal intensities below 500 units an insignificant level of expression.
Table 2 summarizes the miRNA microarray data for those miRNAs whose expression significantly changed in response to estrogen administration. Expression of mirn155, mirn429 and mirn451 was significantly increased by estrogen administration while mirn181b and mirn204 expression was significantly reduced. To confirm the pattern of expression and fold changes compared to 0h controls, we performed qRT-PCR. qRT-PCR confirmed for all analyzed miRNAs except for mirn99b and mirn720.
Table 2.
Average fold changes in select estrogen regulated murine uterine microRNAs after estrogen treatment.
| MicroRNA (mirn) | Hours post estrogen treatment | ||||||
|---|---|---|---|---|---|---|---|
| 0h | 2h | 4h | 8h | 16h | 24h | 30h | |
| mirn99b | 1.0 | 0.71 | 1.16 | 1.29 | 1.47 | 1.79 | 1.74 |
| mirn429 | 1.0 | 0.79 | 0.96 | 1.29 | 1.43 | 1.60 | 1.71 |
| mirn155 | 1.0 | 2.20 | 1.90 | 1.41 | 1.32 | 4.62 | 3.30 |
| mirn451 | 1.0 | 8.51 | 12.71 | 21.94 | 11.22 | 15.92 | 7.53 |
| mirn181b | 1.0 | 0.85 | 0.84 | 0.54 | 0.45 | 0.38 | 0.38 |
| mirn720 | 1.0 | 0.41 | 0.49 | 0.81 | 0.93 | 1.62 | 1.23 |
| mirn204 | 1.0 | 0.56 | 2.32 | 1.11 | 0.46 | 0.45 | 1.21 |
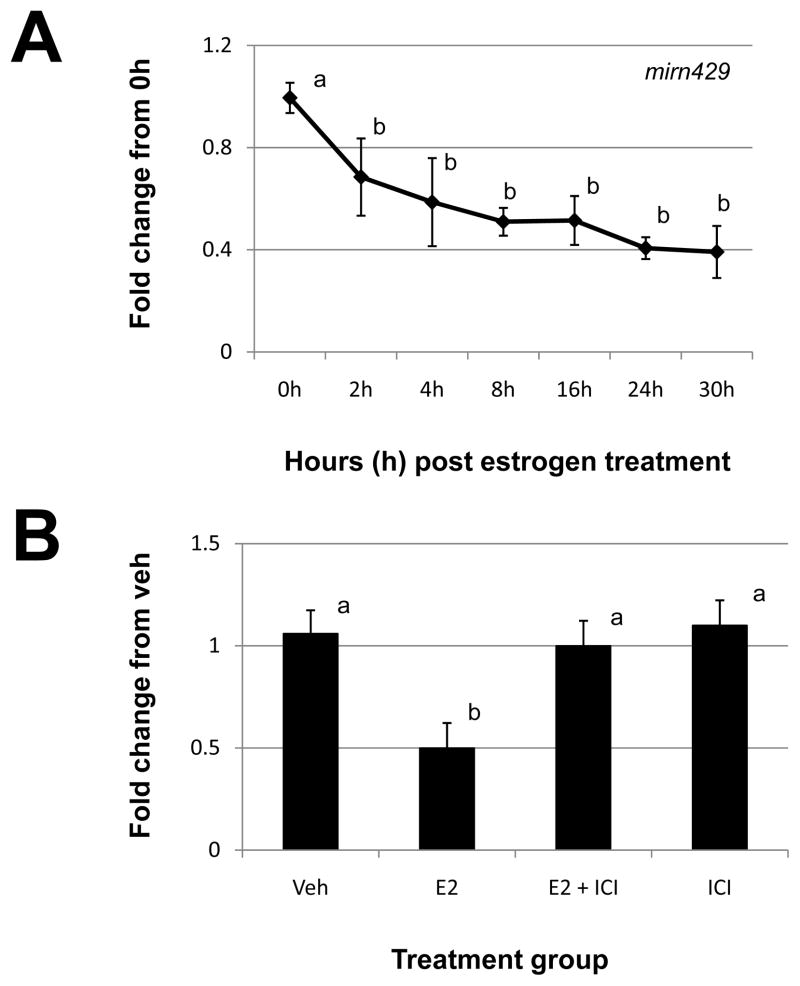
qRT-PCR analysis failed to detect significant changes across time points for either of these miRNAs (data not shown). Similarly, we were unable to confirm the miRNA microarray-derived pattern of expression for mirn429 using qRT-PCR. However, we did detect significant decreases in mirn429 expression using this approach (Fig. 1A). More specifically, mirn429 expression significantly decreased at 4h post estrogen administration and remained suppressed up to and including 30h post steroid treatment (Fig. 1A). To determine if the decrease in mirn429 expression in response to estrogen treatment was mediated via the cognate estrogen receptor pathway, we pre-treated animals with the estrogen receptor antagonist ICI 182,780 (ICI). As depicted in Figure 1B, pre-treatment with ICI blocked the estrogen decrease in mirn429 expression and ICI alone had no affect on expression of the miRNA.
Figure 1. Estrogen regulation of uterine mirn429.
(A) Uterine mirn429 expression was quantitated by qRT-PCR as described in “Materials and Methods” after estrogen administration to ovariectomized mice. (B). Specificity of estrogen modulation was confirmed using the estrogen receptor antagonist, 182,780 ICI (ICI). Data were normalized to the level of U6 expression and are expressed as the fold change from 0h (A) or vehicle (Veh; B) ± SD. Data are representative of 4 to 5 data points/time point or treatment group (N≥4). Different letters indicate statistical significance among the means as determined by one-way ANOVA. Significance was set at P<0.05 for all analysis.
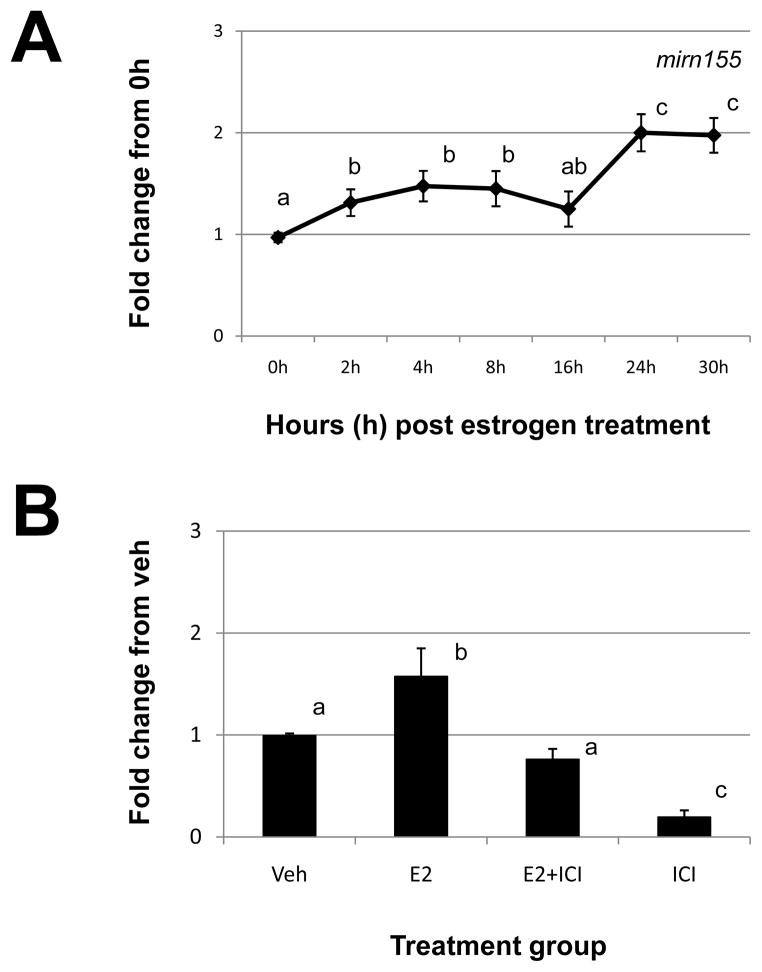
Assessment of mirn155 by qRT-PCR (Fig. 2A) revealed a similar pattern as detected by microarray analysis (Table 2). More specifically, estrogen induced a modest but significant increase between 2 and 8h post-estrogen administration and a more robust increase at 24 and 30h (Fig. 2A). This estrogen-induced increase in mirn155 expression was confirmed to occur via the estrogen receptor pathway as ICI blocked the estrogen-induced increase in the expression of this miRNA (Fig. 2B). ICI alone significantly reduced the expression of mirn155 (Fig. 2B).
Figure 2. Estrogen regulation of uterine mirn155.
A) Uterine mirn155 expression was quantitated by qRT-PCR as described in “Materials and Methods” after estrogen administration to ovariectomized mice. (B). Specificity of estrogen modulation was confirmed using the estrogen receptor antagonist, 182,780 ICI (ICI). Data were normalized to the level of U6 expression and are expressed as the fold change from 0h (A) or vehicle (Veh; B) ± SD. Data are representative of 4 to 5 data points/time point or treatment group (N≥4). Different letters indicate statistical significance among the means as determined by one-way ANOVA. Significance was set at P<0.05 for all analysis.
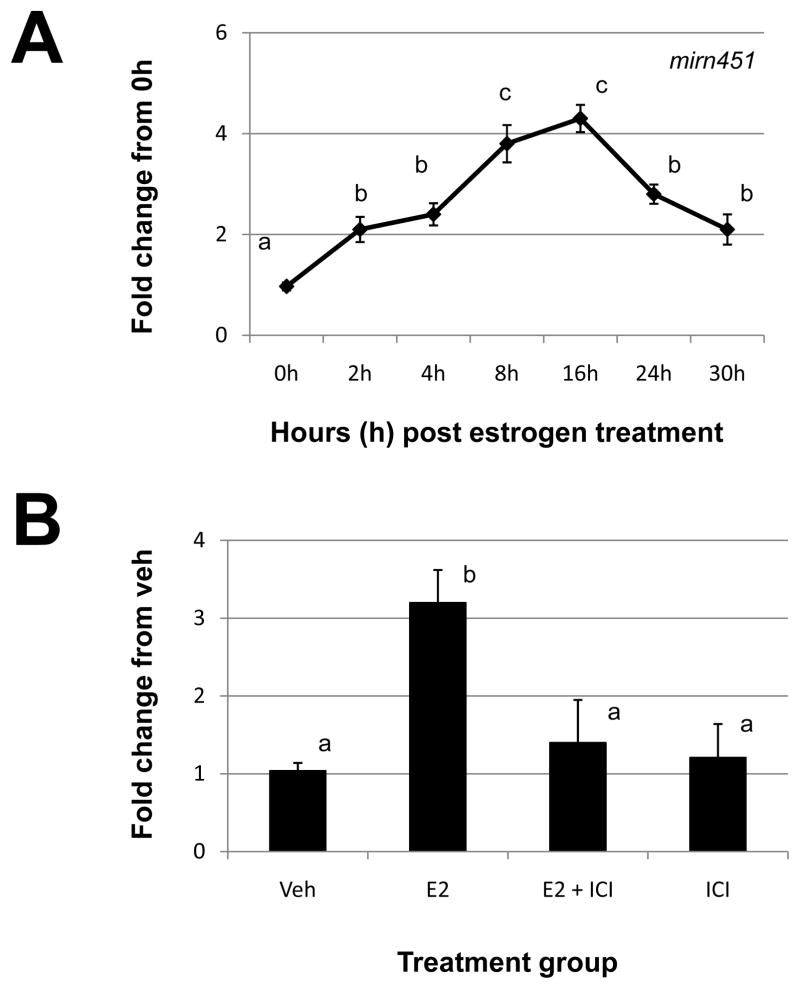
Of all uterine miRNAs studied, mirn451 exhibited the most dramatic estrogen regulation with peak expression occurring between 8 and 16h post estrogen administration (Table 2 and Fig. 3A). The increase in mirn451 expression in response to estrogen administration was confirmed to occur via the estrogen receptor pathway as ICI blocked the estrogen-induced increase in mirn451 expression (Fig. 3B). ICI alone had no significant effect on expression of this miRNA (Fig. 3B).
Figure 3. Estrogen regulation of uterine mirn451.
A) Uterine mirn451 expression was quantitated by qRT-PCR as described in “Materials and Methods” after estrogen administration to ovariectomized mice. (B). Specificity of estrogen modulation was confirmed using the estrogen receptor antagonist, 182,780 ICI (ICI). Data were normalized to the level of U6 expression and are expressed as the fold change from 0h (A) or vehicle (Veh; B) ± SD. Data are representative of 4 to 5 data points/time point or treatment group (N≥4). Different letters indicate statistical significance among the means as determined by one-way ANOVA. Significance was set at P<0.05 for all analysis.
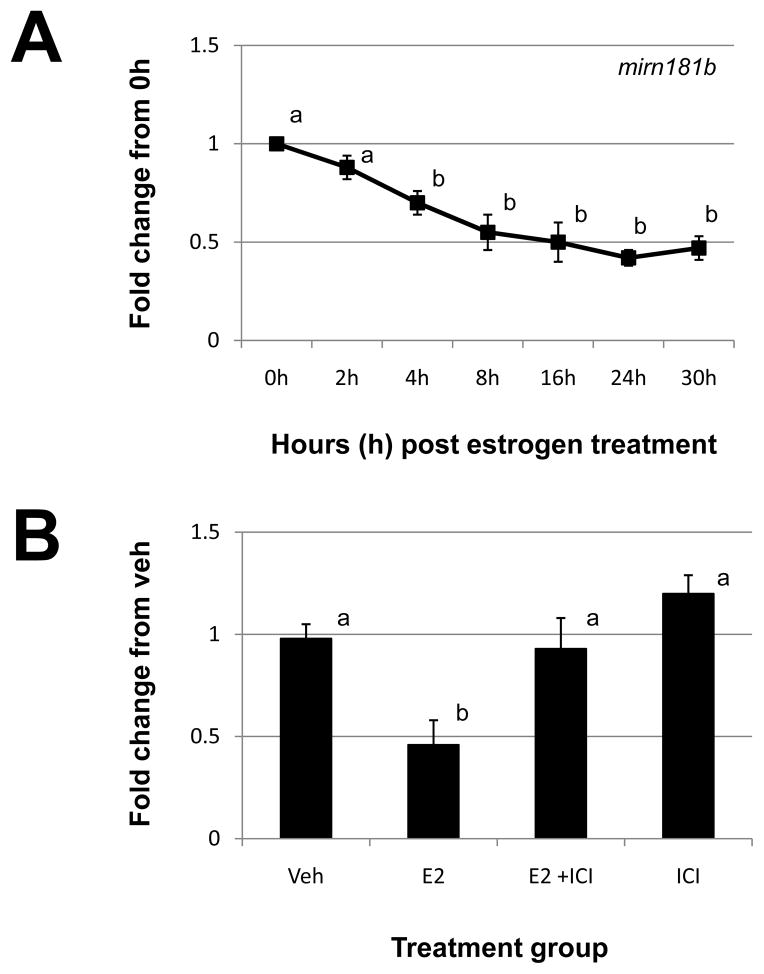
Based upon our initial miRNA microarray analysis, mirn181b was the most significantly down-regulated miRNA (Table 2). This reduction was confirmed by qRT-PCR analysis (Fig. 4A). In response to estrogen administration, mirn181b levels began to significantly decline 4h post steroid administration and remained suppressed through all time points. This decrease in mirn181b expression was confirmed to occur via the cognate estrogen receptor pathway as ICI blocked the ability of this steroid to suppress mirn181b expression (Fig. 4B). ICI alone had no affect on mirn181b expression.
Figure 4. Estrogen regulation of uterine mirn181b.
A) Uterine mirn181b expression was quantitated by qRT-PCR as described in “Materials and Methods” after estrogen administration to ovariectomized mice. (B). Specificity of estrogen modulation was confirmed using the estrogen receptor antagonist, 182,780 ICI (ICI). Data were normalized to the level of U6 expression and are expressed as the fold change from 0h (A) or vehicle (Veh; B) ± SD. Data are representative of 4 to 5 data points/time point or treatment group (N≥4). Different letters indicate statistical significance among the means as determined by one-way ANOVA. Significance was set at P<0.05 for all analysis.
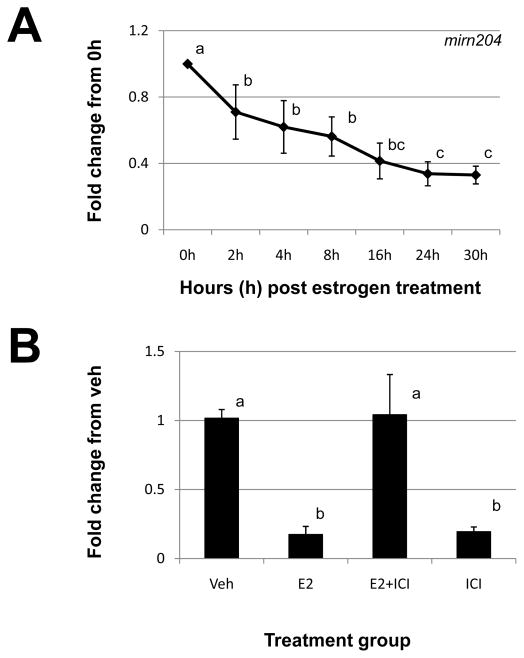
Mirn204 expression was also decreased by estrogen administration with initial microarray analysis indicating that this regulation may be biphasic (Table 2). qRT-PCR analysis confirmed that mirn204 expression was reduced in response to estrogen administration but we could not duplicate this biphasic pattern (Fig. 5A). We did find that mirn204 expression was significantly reduced at 2h post estrogen administration, continued to decline after this time point and remained significantly reduced at 30h post steroid administration (Fig. 5A). The estrogen imparted decrease in mirn204 expression was confirmed to occur via the estrogen receptor pathway as ICI blocked the estrogen decrease in mirn204 expression (Fig. 5A). However, much like that of mirn155 (Fig. 2B), ICI alone significantly reduced the expression of uterine mirn204 expression (Fig. 5B).
Figure 5. Estrogen regulation of uterine mirn204.
A) Uterine mirn204 expression was quantitated by qRT-PCR as described in “Materials and Methods” after estrogen administration to ovariectomized mice. (B). Specificity of estrogen modulation was confirmed using the estrogen receptor antagonist, 182,780 ICI (ICI). Data were normalized to the level of U6 expression and are expressed as the fold change from 0h (A) or vehicle (Veh; B) ± SD. Data are representative of 4 to 5 data points/time point or treatment group (N≥4). Different letters indicate statistical significance among the means as determined by one-way ANOVA. Significance was set at P<0.05 for all analysis.
Discussion
In the current study we demonstrate for the first time that estrogen regulates the expression of a subset of miRNAs within the murine uterus. Estrogen regulation of some of these miRNAs can be blocked by the estrogen receptor antagonist ICI 182,780 indicating modulation via the classical estrogen receptor(s)-dependent pathway. In contrast, expression of other miRNAs was directly modulated by ICI 182,780 itself which may suggest modulation via estrogen receptor-independent pathways. The major estrogen regulated uterine murine miRNAs were mirn429, mirn155, mirn451, mirn181b and mirn204. miRNAs have been proposed to play a role in modulating post-transcriptional gene regulation within the uterus.18 With respect to the impact of steroidal regulation on endometrial miRNA expression, studies have been limited to descriptive studies utilizing endometrial biopsies. One study which solely examined expression of miRNAs in endometrial biopsies from “normal” reproductive cycling patients was conducted by Kuokkanen and colleagues.20 In this study, genomic profiling of miRNAs of human endometrium epithelial cells revealed that a set of 12 miRNAs were differentially expressed between late proliferative and mid-secretory specimens. Of those 12, we could only confirm the expression and similar regulation of one miRNA, that being mirn204. In human endometrium epithelium, mirn204 (MIR204) was lower in the proliferative specimens compared to the secretory and would suggest a down-regulation by estrogen which is consistent with our findings.
In addition to the study by Kuokkanen and colleagues,20 several reports have been published which have focused on identifying miRNAs which may be expressed in uterine disease such as endometriosis19, 22–26 and endometrial cancer.27–29 One of the first studies to profile miRNA expression between endometrium from women with and without endometriosis was conducted by Pan and colleagues.26 Forty eight miRNAs were differentially expressed among eutopic and ectopic endometrium from women with versus endometrium from women without the disease. Of this group, mirn451 was one of the most significantly reduced miRNA in ectopic and eutopic endometrium from women with endometriosis. Although endometriosis is considered an estrogen-dependent disease, this pattern of expression is in contrast to that detected in our current study, where murine uterine mirn451 expression was significantly increased by estrogen.
The finding that mirn451 expression is reduced in ectopic and eutopic endometrium from women with endometriosis may be of great clinical interest. mirn451 has been proposed to regulate cell proliferation via regulation of macrophage migration inhibitory factor (MIF)32 and is also a putative regulator of MMP3 and MYC (TargetScanHuman 5.1, www.targetscan.org), both of which are over-expressed in endometriosis.33–36 Collectively, reduced expression of mirn451may allow for a post-transcriptional mis-regulation of these factors which in turn contributes to the pathophysiology of endometriosis.
Four other miRNAs were identified as being estrogen regulated in our current study, mirn429, mirn155, mirn204 and mirn181b. Our current data demonstrate that in response to estrogen, murine uterine expression of mirn429 is significantly down-regulated. mirn429 down-regulation has been associated with mesenchymal to epithelial transition37 but its role and relevant target transcripts in uterine tissue remain unknown.
Mirn155 is perhaps the most well-studied of all miRNAs. Relative to the current study, mirn155 has been proposed to function as a protective miRNA that may locally down-regulate the expression of matrix metalloproteinases.38 As such, it is tempting to speculate that mirn155 may fine tune the estrogen regulation of this system under normal physiological conditions within the endometrium39 whereas mis-expression of mirn155 may lead to abnormal expression of matrix metalloproteinases.
In the current study, both mirn204 and mirn181b expression was decreased by estrogen administration. Mirn204 was originally isolated from the eye40 and more recently its role in cancers of epithelial cell origin was reported.41 In this study by Lee and colleagues,41 mirn204 expression was reduced in 50% of the various types of cancers compared to controls. Endometrial cancer was one of the types of cancer in which mirn204 expression did not differ. While this observation would suggest that mirn204 mis-expression may not contribute to the pathology of endometrial carcinoma, it does not rule out the possibility that this miRNA may regulate important estrogen regulated targets within the endometrium such as matrix metalloproteinase-9 (MMP9). The pattern of Mmp9 expression in the mouse uterus described in our previous work42,43 coupled with the current observation that mirn204 is reduced in response to estrogen administration would suggest that this miRNA may regulate uterine Mmp9 expression.
To the best of our knowledge, our study is the first to describe the uterine expression of mirn181b. In our study, uterine expression of mirn181b was decreased by estrogen administration. Mirn181b was first described as being down-regulated in glioblastoma44 and since has been associated with leukemia and various forms of cancer.45–48 Of relevance to the current study is that mirn181b is a putative regulator of tissue inhibitor of metalloproteinase-3 (Timp3, which regulates Mmp9 activity) and that estrogen induction of uterine TIMP349 is inversely correlated with mirn181b (and mirn204) expression. These observations make it interesting to speculate that within the mouse uterus estrogen may regulate this important protease:protease inhibitor system via these miRNAs.
In addition to in vivo profiling of endometrial miRNAs, in vitro studies utilizing isolated endometrial stromal and epithelial cells have been conducted to examine steroidal regulation of select miRNAs.23,26 These studies demonstrated that endometrial stromal and/or epithelial cell miRNA expression could be modulated by estrogen and that in some cases this modulation could be blocked by the estrogen receptor antagonist 182,780 ICI (ICI). These studies also demonstrated that for some miRNAs, ICI alone acted as an estrogen agonist to either increase or decrease expression of these miRNAs. These observations are in agreement with those in the current study where in vivo administration of ICI alone resulted in significant reductions in mirn155 and mirn204. The ability of ICI to act independent of estrogen has been reported in MCF-7 breast cancer cells.50–52 One mechanism which may explain the ability of ICI to down regulate gene expression and/or miRNA expression (current study as well as those by Pan and colleagues26 and Toloubehdokhti and coworkers23) is that ICI can decrease protein levels of estrogen receptor which in turn may suppress potential ligand-independent activity of the estrogen receptor.52 Further analysis will be required to thoroughly dissect the mechanisms by which the estrogen receptor pathway regulates transcription of miRNAs.
Footnotes
Supported in part by R21 HD056387 to WBN
References
- 1.Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 2.Hewitt SC, Deroo BJ, Hansen K, et al. Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol Endocrinol. 2003;17:2070–2083. doi: 10.1210/me.2003-0146. [DOI] [PubMed] [Google Scholar]
- 3.Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 4.Das SK, Tan J, Raja S, Halder J, Paria BC, Dey SK. Estrogen targets genes involved in protein processing, calcium homeostasis and Wnt signaling in the mouse uterus independent of estrogen receptor- and -β. J Biol Chem. 2000;275:28834–28842. doi: 10.1074/jbc.M003827200. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe H, Suzuki A, Kobayashi M, et al. Analysis of temporal changes in the expression of estrogen-regulated genes in the uterus. J Mol Endocrinol. 2003;30:347–358. doi: 10.1677/jme.0.0300347. [DOI] [PubMed] [Google Scholar]
- 6.Hou X, Tan Y, Li M, Dey SK, Das SK. Canonical Wnt signaling is critical to estrogen mediated uterine growth. Mol Endocrinol. 2004;18:3035–3049. doi: 10.1210/me.2004-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 9.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rourke JR, Georges SA, Seay HR, et al. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murchison EP, Stein P, Xuan Z, et al. Critical roles for dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez G, Behringer RR. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009;76:678–688. doi: 10.1002/mrd.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagaraja AK, Andreu-Vieyra C, Franco HL, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22:2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149:6207–6212. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia HF, Jin XH, Song PP, Cui Y, Liu CM, Ma X. Temporal and spatial regulation of let-7a in the uterus during embryo implantation in the rat. J Reprod Dev. 2010;56:73–78. doi: 10.1262/jrd.09-088k. [DOI] [PubMed] [Google Scholar]
- 17.Hu SJ, Ren G, Liu JL, et al. MicroRNA expression and regulation in mouse uterus during embryo implantation. J Biol Chem. 2008;34:23473–223484. doi: 10.1074/jbc.M800406200. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Ferneaux H, Dey SK. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci U S A. 2007;104:15144–15149. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burney RO, Hamilton AE, Aghajanova LLC, et al. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;10:625–631. doi: 10.1093/molehr/gap068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuokkanen S, Chen B, Ojalvo L, Benard L, Santoro N, Pollard JW. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol Reprod. 2010:82791–801. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessey BA. Fine tuning of endometrial function by estrogen and progesterone through miRNAs. Biol Reprod. 2010;82:653–655. doi: 10.1095/biolreprod.110.083667. [DOI] [PubMed] [Google Scholar]
- 22.Filigheddu N, Gregnanin I, Porporato PE, et al. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J Biomed Biotechnol. 2010:369549. doi: 10.1155/2010/369549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toloubeydokhti T, Pan Q, Luo X, Bukulmez O, Chegini N. The expression and ovarian steroid regulation of endometrial micro-RNAs. Reprod Sci. 2008;10:993–1001. doi: 10.1177/1933719108324132. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, et al. MicroRNA-Regulated Pathways Associated with Endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Q, Chegini N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 2008;26:479–493. doi: 10.1055/s-0028-1096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;11:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 27.Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target microRNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110:206–215. doi: 10.1016/j.ygyno.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 28.Chung TK, Cheung TH, Huen NY, et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;24:1358–1365. doi: 10.1002/ijc.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu W, Lin Z, Zhuang Z, Liang X. Expression prolife of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 30.Nothnick WB, Healy CA. Steroidal regulation of uterine miRNAs is associated with modulation of the miRNA biogenesis components Exportin-5 and Dicer1. Endocrine. 2010;37:265–273. doi: 10.1007/s12020-009-9293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Bandres E, Bitarte N, Arias F, et al. MicroRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 33.Pelch KE, Schroder AL, Kimball PA, Sharpe-Timms KL, Wade-Davis J, Nagel SC. Aberrant gene expression profile in a mouse model of endometriosis mirrors that observed in women. Fertil Steril. 2009;93:1615–1627. doi: 10.1016/j.fertnstert.2009.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruner KL, Matrisian LM, Rodgers WH, Gorstein F, Osteen KG. Suppression of matrix metalloproteinases inhibits establishment of ectopic lesions by human endometrium in nude mice. J Clin Invest. 1997;99:2851–2857. doi: 10.1172/JCI119478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schenken RS, Johnson RV, Riehl RM. C-myc protooncogene polypeptide expression in endometriosis. Am J Obstet Gynecol. 1991;164:1031–1036. doi: 10.1016/0002-9378(91)90580-k. [DOI] [PubMed] [Google Scholar]
- 36.Johnson MC, Torres M, Alves A, et al. Augmented cell survival in eutopic endometrium from women with endometriosis: expression of c-myc, TGF-beta1 and bax genes. Reprod Biol Endocrinol. 2005;3:45. doi: 10.1186/1477-7827-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 38.Ceppi M, Pereira PM, Dunand-Sauthier I, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Nat Acad Sci USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Nothnick WB. The role of the metalloproteinase system within the uterus of menstruating and non-menstruating species. Front Biosci. 2005;10:353–366. doi: 10.2741/1533. [DOI] [PubMed] [Google Scholar]
- 40.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 41.Lee Y, Yang X, Huang Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Christenson LK, Nothnick WB. Regulation of matrix metalloprotein-9 (MMP-9) expression and activity in the mouse uterus by estrogen. Mol Reprod Dev. 2007;74:321–331. doi: 10.1002/mrd.20582. [DOI] [PubMed] [Google Scholar]
- 43.Nothnick WB. Regulation of uterine matrix metalloproteinase-9 and the role of microRNAs. Sem Reprod Med. 2008;26:494–499. doi: 10.1055/s-0028-1096129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 45.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakajima G, Hayashi K, Xi Y, et al. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics. 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- 47.Xi Y, Formentini A, Chien M, et al. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113–121. [PMC free article] [PubMed] [Google Scholar]
- 48.Zanette DL, Rivadavia F, Molfetta GA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 49.Nothnick WB, Zhang X, Zhou H-E. Steroidal regulation of uterine edema and TIMP-3 mRNA expression is altered in TIMP-1 deficient mice. Biol Reprod. 2004;70:500–508. doi: 10.1095/biolreprod.103.020834. [DOI] [PubMed] [Google Scholar]
- 50.Montano MM, Katzenellenbogen BS. The quinine reductase gene: A unique estrogen receptor-regulated gene that is activated by anti-estrogens. Proc Natl Acad Sci USA. 1997;94:2581–2586. doi: 10.1073/pnas.94.6.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb P, Ngyuen P, Kushner PJ. Differential SERM effects on corepressor binding dictate ERα activity in vitro. J Biol Chem. 2003;278:6912–6920. doi: 10.1074/jbc.M208501200. [DOI] [PubMed] [Google Scholar]
- 52.Frasor J, Stossi F, Kanes JM, et al. Selective estrogen receptor modulators: Discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Research. 2004:1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]