Abstract
TRPM1(melastatin), which encodes the founding member of the TRPM family of transient receptor potential (TRP) ion channels, was first identified by its reduced expression in a highly metastatic mouse melanoma cell line. Clinically, TRPM1 is used as a predictor of melanoma progression in humans because of its reduced abundance in more aggressive forms of melanoma. Although TRPM1 is found primarily in melanin-producing cells and has the molecular architecture of an ion channel, its function is unknown. Here we describe an endogenous current in primary human neonatal epidermal melanocytes and mouse melanoma cells that was abrogated by expression of microRNA directed against TRPM1. Messenger RNA analysis showed that at least five human ion channel–forming isoforms of TRPM1 could be present in melanocytes, melanoma, brain, and retina. Two of these isoforms are encoded by highly conserved splice variants that are generated by previously uncharacterized exons. Expression of these two splice variants in human melanoma cells generated an ionic current similar to endogenous TRPM1 current. In melanoma cells, TRPM1 is prevalent in highly dynamic intracellular vesicular structures. Plasma membrane TRPM1 currents are small, raising the possibility that their primary function is intracellular, or restricted to specific regions of the plasma membrane. In neonatal human epidermal melanocytes, TRPM1 expression correlates with melanin content. We propose that TRPM1 is an ion channel whose function is critical to normal melanocyte pigmentation and is thus a potential target for pigmentation disorders.
INTRODUCTION
Melastatin (TRPM1) was discovered because of its reduced expression in the highly metastatic mouse melanoma cell line B16-F10 relative to that in the noninvasive B16-F1 melanoma cell line (1). The messenger RNA (mRNA) that was decreased in abundance in B16-F10 cells predicted a protein with no transmembrane domains. The TRPM1 gene is located on chromosome 15 in humans and on 7 in mice (2). It consists of 27 exons and encodes a 5.4-kb mRNA transcript (2). The melanocyte transcription factor MITF (microphthalmia-associated transcription factor), which is essential for the survival and differentiation of the melanocyte lineage in mice and humans (2, 3), regulates the transcription of TRPM1 in melanocytes and in melanoma (4). TRPM1 has multiple splice variants; short transcripts corresponding primarily to either the 5′ or the 3′ end of the 5.4-kb mRNA are present in both melanocytes and pigmented metastatic melanoma cell lines, whereas the full-length 5.4-kb mRNA is detectable only in melanocytes.
TRPM1 is found primarily in melanin-producing cells, including melanocytes (1, 3–6); however, its function is unknown. Melanocytes are crucial to pigmentation of the skin, a complex process in which melanin is synthesized within melanocytes and subsequently transferred to adjacent keratinocytes. Melanocytes synthesize and package melanin in melanosomes, lysosome-derived organelles that are trafficked to the melanocyte-keratinocyte junction and transferred to recipient keratinocytes. Despite intensive study, many molecular aspects of the pigmentation process remain unclear (7–9). The transfer of melanosomes from melanocytes to keratinocytes is required to protect skin from damaging ultraviolet (UV) radiation. Moreover, the degree of melanin production in the skin is the best predictor of melanoma risk (10, 11), with more melanin corresponding to decreased risk.
TRPM1 is the founding member of the TRPM subfamily and its sequence is similar to that of other members of the TRP family of cation channels. Although other members of the TRPM family act as functional ion channels and mediate numerous physiological processes (12), TRPM1’s ion channel properties and cellular functions are unknown. Full-length TRPM1 (1533 amino acids) localizes to the plasma membrane and mediates Ca2+ entry when expressed in human embryonic kidney (HEK) cells, whereas a cytosolic N-terminal truncation variant (500 amino acids) interacts directly with and suppresses the activity of full-length TRPM1 (13). Ionic current through the TRPM1 channel has not been reported so far.
Here, we show the presence of at least five TRPM1 mRNAs that encode isoforms containing N termini of variable length, all of which include the transmembrane domains and C terminus. Moreover, we found that the amplitude of an endogenous TRP current present in B16-F1 cells was decreased in B16-F10 cells. This nonselective, outwardly rectifying current was reduced by microRNA (miRNA) targeting TRPM1 and is blocked by lanthanum. We measured a small current with the same characteristics in human melanoma cells expressing two previously undescribed TRPM1 isoforms. The endogenous outward-rectifying current was also present in primary neonatal human epidermal melanocytes (HEMs), where its reduction correlated with decreased melanin concentration, suggesting a possible role for TRPM1 in pigmentation.
RESULTS
N-terminal splice variants of TRPM1
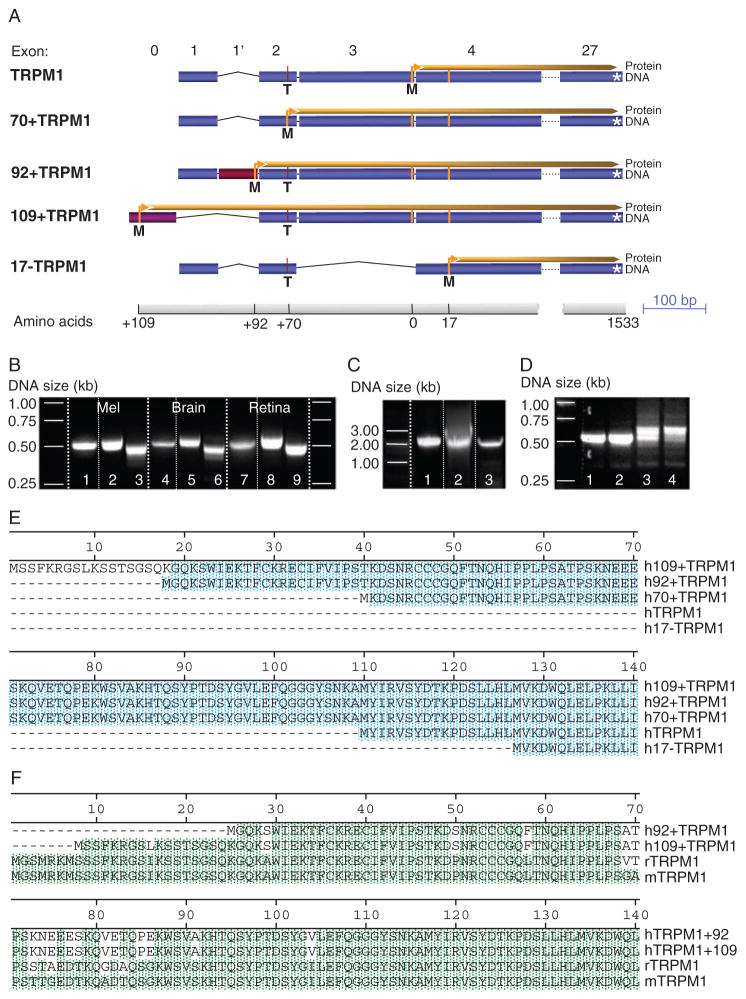
Full-length melastatin complementary DNA (cDNA) encodes a 1533–amino acid protein product with six transmembrane domains (TRPM1; NM_002420) and contains a 338–base pair (bp)–long 5′ untranslated region (UTR). Recently, the database was updated; a nucleotide change in the human TRPM1 sequence C130→T resulted in a newly identified open reading frame (ORF). An ACG (C130) within the 5′UTR became ATG, resulting in a newly identified in-frame start codon. The predicted protein contains an additional 70 amino acids at the N terminus (70+TRPM1; Fig. 1, A and E). We examined the possibility that the predicted gene product was a functional TRPM1 ion channel.
Fig. 1.
Human TRPM1 isoforms. (A) Exon composition and ORFs of TRPM1 isoforms identified in human tissue. Start sites: TRPM1, exon 3; 70+TRPM1, exon 2 (contains 70 additional N-terminal amino acids as a result of a C/T polymorphism that generates a newly identified start in-frame with the TRPM1 start site); 92+TRPM1, exon 1′ (newly identified site in-frame with the 92–amino acid N terminus of the start of TRPM1); 109+TRPM1, previously uncharacterized exon 0 (missing exons 1 and 1′, containing 109 additional N-terminal amino acids compared to TRPM1; 17−TRPM1, exon 4 (17 amino acids shorter than TRPM1, C allele at exon 2 and missing exon 3). (B) Expression of the TRPM1 N-terminal exons in human tissue. PCR from melanocyte, brain, and retina cDNA libraries using a forward primer based on exon 0 (lanes 1, 4, and 7), exon 1′ (lanes 2, 5, and 8), and exon 1 (lanes 3, 6, and 9) and a reverse primer based on exon 5. (C) Expression of human TRPM1 isoforms in melanocytes. PCR from cDNA melanocyte library or HEMs using a forward primer based on exon 0, corresponding to 109+TRPM1 (lane1), exon 1′ corresponding to 92+TRPM1 (lane 2), and exon1 corresponding to TRPM1 and 70+TRPM1 (lane 3) and a reverse primer C-terminal of the six transmembrane domains. (D) Change in the expression profile of TRPM1 in melanoma cell lines compared to primary melanocytes. PCR amplification of the N terminus of TRPM1 using a pair of oligonucleotides based on exon 1 and 5 results in a sharp band in HEM-light (lane 1) and HEM-dark (lane 2), corresponding to one dominant spliced variant. In contrast, SK-Mel19 cells (lane 3) and MALME cells (lane 4) yield a multitude of bands of slightly different molecular weight, corresponding to a multitude of splice variants. (E) Protein alignments of the N termini of the five TRPM1 splice variants shown in (A). The Met (ATG) at position 1 in 70+TRPM1 corresponds to a Thr (ACG) in most of the 92+TRPM1 sequences (95% of 20 sequences) and about 50% of the 109+TRPM1 sequences (n = 10). Blue shading corresponds to conserved amino acids. (F) The newly identified splice variants 92+TRPM1 and 109+TRPM1 are highly conserved among species. Alignment of human 92+TRPM1 and 109+TRPM1 N termini with rat and mouse TRPM1 reveals 95% sequence identity. Green shading corresponds to conserved amino acids.
We identified N-terminal variants of TRPM1 in cDNA libraries derived from human melanocytes, retina, brain, and several melanoma cell lines containing TRPM1. Using a pair of oligonucleotides based on the beginning of exon 1 and the end of exon 5, we amplified TRPM1’s N-terminal region (see exon structure in Fig. 1A) from a cDNA melanocyte library. Of 17 sequenced clones, 11 contained cytosine (C) at position 130 (TRPM1). Four of the 11 clones with C at 130 were missing exon 3, encoding a protein that lacked 17 amino acids at its N terminus (17–TRPM1; Fig. 1, A and E). Three of the sequenced clones contained 85 additional base pairs between exons 1 and 2. The last three of the 85 bp (ATG) formed a previously unrecognized start site in-frame with the start for TRPM1 and 70+TRPM1. The resulting protein has 92 additional N-terminal amino acids compared to TRPM1 (92+TRPM1). The 85 bp that generate 92+TRPM1 are flanked by corresponding splicing acceptor and donor sites and are found within intron 1, forming a new exon 1′ (Fig. 1, A and E).
To identify human sequences corresponding to mouse and rat exon 1, which encodes the N-terminal 25 amino acids of TRPM1, we performed a search (tblastn) of databases containing human genomic sequences [Nucleotide collection (nr/nt) and Reference genomic sequences (refseq_genomic)]. The search yielded DNA sequence with a high degree of identity (80 to 90%) to mouse and rat, including the 5′ donor splice site, suggesting that this sequence forms a previously uncharacterized exon in humans (exon 0). Human exon 0 contains a newly identified start codon in-frame with the start for TRPM1 and 70+TRPM1, encoding a peptide sequence almost identical to mouse and rat TRPM1, albeit seven amino acids shorter (Fig. 1E). The predicted protein initiating at the exon 0 start site, and spliced to exon 2, has an additional 109 N-terminal amino acids compared to TRPM1 (109+TRPM1) (Fig. 1A). This exon is highly conserved in more than 20 species, ranging from chicken to orangutan, including bat, cat, alpaca, and elephant. It identifies a common start of the coding sequence in a wide range of species.
TRPM1 splice variant expression
To determine whether the two newly identified splice variants (92+TRPM1 and 109+TRPM1), were expressed in human melanocytes, brain, or retina, we used forward primers within exon 0, 1′, or 1 (Fig. 1B) paired with a reverse primer within exon 5. Fragments corresponding to regions encoding the N terminus of the respective TRPM1 isoforms were found in all three tissues (Fig. 1B). The TRPM1 gene has a number of splice variants, many presumably encoding short proteins containing only the N terminus. To determine whether the detected isoforms encoded a protein containing the six transmembrane domains of the TRPM1 ion channel, we used the same forward primers described in Fig. 1B and a reverse primer based on the common C terminus of TRPM1. The amplified cDNA bands from melanocytes, brain, and retina included the regions encoding the trans-membrane domains of TRPM1 (Fig. 1C), suggesting that the newly identified exons give rise to two previously unidentified ion channel isoforms of TRPM1: 92+TRPM1 and 109+TRPM1. Moreover, the additional amino acids present in these two isoforms are highly conserved in mouse and rat (Fig. 1F), chimpanzee, dog, cat, horse, and other mammals, suggesting that these splice variants may have an important function.
We investigated whether these splice variants were expressed differentially in various tissues and in primary or transformed melanocytes. We obtained cDNA by reverse transcription polymerase chain reaction (RT-PCR), using total RNA extracted from primary neonatal HEMs. The HEMs were isolated from light-pigmented and dark-pigmented skin (HEM-light and HEM-dark, respectively) and several pigmented melanoma cell lines that express TRPM1 (SK-Mel19, SK-Mel22a, and MALME). RT-PCR amplification of the N-terminal region of TRPM1 with a forward primer based on exon 1 and a reverse primer based on exon 5 yielded a single band from HEM-dark and HEM-light, but a multitude of bands of similar size from SK-Mel 19 and MALME melanoma cell lines (Fig. 1D). This suggests that transformation of primary melanocytes into melanoma is associated with a change in the expression pattern of TRPM1 isoforms and that melanoma cells express a greater diversity of N-terminal splice variants.
To further compare the relative distribution of the splice variants, we sequenced the cDNA fragments corresponding to the N terminus of TRPM1 amplified from primary melanocytes (HEM) and melanoma cells (Table 1). Isoform composition in HEM-light and HEM-dark was similar, with 80% of the amplified cDNA copies encoding the N terminus of the 70+TRPM1 protein. The remaining 20% of the HEM isoforms were evenly divided between TRPM1 and 17–TRPM1. None of the sequenced cDNA copies coded for the 92+TRPM1 isoform. However, transcripts encoding 92+TRPM1 could be detected in HEM cells by a primer specific for exon 1′, suggesting that it is a rare isoform in primary cells (less than 2.5%). Because exon 1 and 1′ are spliced out in 109+TRPM1, we could not determine the relative abundance of this transcript.
Table 1.
Relative distribution of TRPM1 isoforms (excluding 109+TRPM1) in melanocytes, brain, and retina.
| Tissue/cell line | n | Isoform
|
|||
|---|---|---|---|---|---|
| 17−TRPM1 (%) | TRPM1 (%) | 70+TRPM1 (%) | 92+TRPM1 (%) | ||
| Melanocytes | |||||
| Primary HEM | 40 | 10 | 10 | 80 | <2.5 |
| SK-Mel19 | 18 | 16.6 | 66.6 | 0 | 16.6 |
| SK-Mel22a | 21 | 43 | 24 | 0 | 33 |
| MALME | 18 | 16 | 28 | 0 | 56 |
| Brain | 21 | 0 | >95 | 0 | <5 |
| Retina | 20 | 0 | >48 | >48 | <4 |
The relative distribution of N-terminal TRPM1 isoforms in melanoma cell lines differed substantially from that in HEMs; none of the sequences coded for the 70+TRPM1 isoform. However, the transcript encoding the 92+TRPM1 isoform was abundant (Table 1), suggesting that 92+TRPM1 is in part responsible for the cDNA bands of slightly higher molecular weight (Fig. 1D). cDNA from commercial libraries derived from human brain and retina was amplified using the same primers as above (Table 1). Similar to HEM cells, 92+TRPM1 was absent from these tissues.
We sought to identify mouse TRPM1 isoforms by homology to the human isoforms. We found evidence in the mouse genomic and transcripts sequences of N-terminal exons 0, 1, 2, and 3 (NM_001039104, BC_167220, NM_018752, respectively) (fig. S1). Exon 3 contains the start site for the mouse homolog of TRPM1 (mTRPM1), which is 86% identical to human TRPM1. mTRPM1 contains exons 1, 2, and part of exon 3 in its 5′UTR. No evidence for the C→T polymorphism that generates the start site for human 70+TRPM1 exists in mouse databases, suggesting that this isoform is not present in mice. We found a sequence similar (>95% identity) to exon 1′, which contains the start site for the human 92+TRPM1, in gorilla, orangutan, chimpanzee, and rhesus monkey. We found no sequence similar to human exon 1′ in mouse databases, suggesting lack of mouse homologs of 92+TRPM1. We found a highly conserved exon 0 in mouse, which contains an in-frame start site 116 amino acids upstream of the start site for mTRPM1 (NM_001039104). The cDNA coding for the long mouse isoform, 116+TRPM1, contains exon 0 followed by exon 2. Based on these results, mouse tissue likely expresses two isoforms of the TRPM1 ion channel, TRPM1 and 116+TRPM1. It is possible that mouse also expresses a number of splice variants, many of which lack the transmembrane domains.
TRPM1 currents
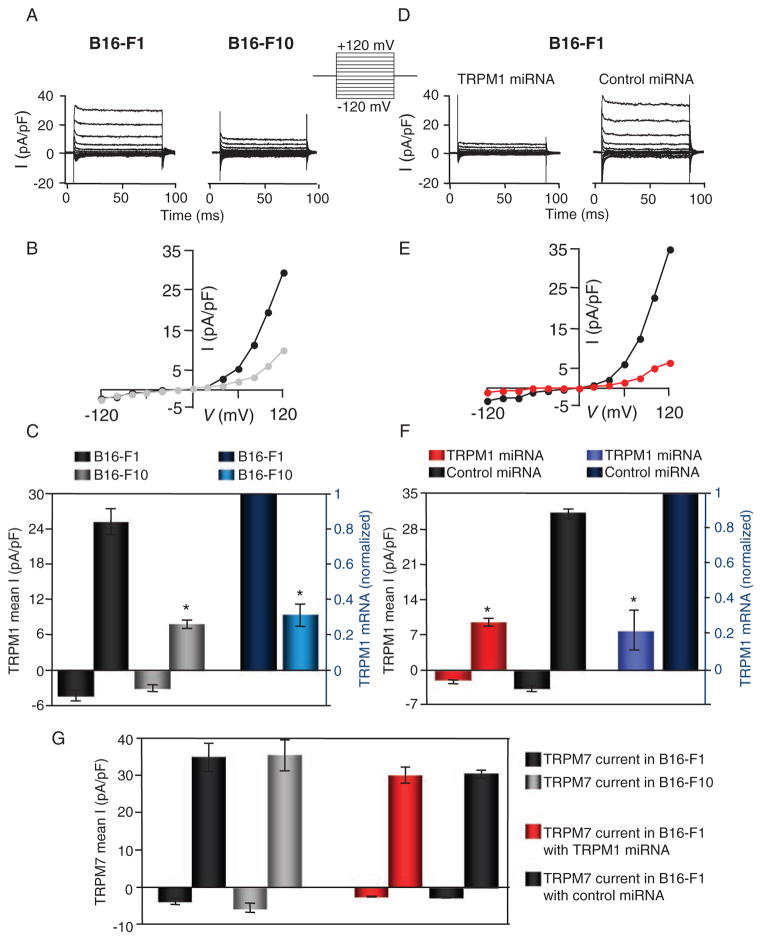
We performed whole-cell voltage clamp experiments on mouse melanoma B16-F1 (noninvasive) and B16-F10 (invasive) cell lines to compare their endogenous currents. The endogenous TRP (nonselective cationic) current measured in B16-F1 cells was significantly larger than that in B16-F10 cells (Fig. 2, A and B). In B16-F1 cells, the average outward current amplitude at 120 mV was about three times greater than that in B16-F10 cells, correlating with the difference in TRPM1 mRNA abundance in these cells (Fig. 2C). TRPM7 is a ubiquitously expressed TRP channel that frequently contaminates recordings (14), but can be blocked with intracellular Mg2+. Both TRPM6 and TRPM7 currents are denoted Mg2+-inhibitable currents (MIC) (15). To suppress endogenous TRPM7 current, internal [Mg2+] was set to 8 mM [well above the IC50 (median inhibitory concentration) of ~1 mM] (16). In separate studies of TRPM7 currents at permissive internal [Mg2+] ([Mg2+]i < 1 mM), the average amplitude of MIC currents was not significantly different in B16-F1 and B16-F10 cells (Fig. 2G and fig. S2), suggesting similar mRNA abundance of channels encoding MIC in these two cell lines.
Fig. 2.
Endogenous current density in B16-F1 and B16-F10 mouse melanoma cells correlates with TRPM1 mRNA abundance. (A) Endogenous whole-cell current recorded from B16-F1 and B16-F10 in response to the voltage step protocol shown. (B) Current-voltage (I-V) curve of the current recorded in B16-F1 (black) and B16-F10 (gray) cells. (C) Mean outward current at 120 mV (left axis) in B16-F1 (dark gray, n = 10) and B16-F10 (light gray, n = 12) correlates with TRPM1 mRNA abundance (right axis) in B16-F1 (dark blue) and B16-F10 cells (light blue, n = 4 independent experiments), respectively. B16-F10 cells had ~30% less TRPM1 mRNA than B16-F1 cells as determined by quantitative PCR (right axis) and the average outward current amplitude at 120 mV was ~30% smaller: (7.8 ± 0.7 pA/pF in B16-F10 compared to 25.0 ± 2.3 pA/pF in B16-F1 cells; left axis). *P < 0.001; t test. Error bars, ± SEM. (D) Endogenous whole-cell current is significantly reduced by TRPM1-targeted miRNA compared to control(scrambled)miRNA in response to the voltage step protocol shown at left. (E) I-V curves from B16-F1 cells expressing control (scrambled) miRNA (black) and TRPM1 miRNA (red). (F) Cells expressing TRPM1 miRNA and control (scrambled) miRNA elicited an ~75% reduction in the mRNA abundance of TRPM1 in B16-F1 cells treated with TRPM1 miRNA (right axis, light blue) and control (scrambled) miRNA (right axis, dark blue, n = 4 independent experiments). Averaged current amplitude at −120 and +120 mV for cells expressing TRPM1 miRNA and control (scrambled) miRNA was reduced ~70%: (31.4 ± 2.1 pA/pF in control cells was reduced to 9.5 ± 1.2 pA/pF in TRPM1 miRNA–expressing cells, left axis). *P < 0.001; t test. Error bars, ± SEM. (G) Averaged endogenous TRPM7 (MIC) current amplitude at −120 and +120 mV is similar in B16-F1 [(35.0 ± 7.8 pA/pF; black, n = 6) and B16-F10 (35.4 ± 8.4 pA/pF; gray, n = 5]. The amplitude of the endogenous TRPM7 current is similar in B16-F1 cells treated with TRPM1 miRNA (red, n = 9) or control (scrambled) miRNA (black, n = 9). Error bars, ± SEM.
RNA interference (RNAi) targeting of TRPM1 reduced the amplitude of the TRP current. We used a vector-based miRNA approach [green fluorescent protein (GFP) marker and two chained TRPM1-specific pre-miRNAs targeting the C terminus] to ensure targeting of all TRPM1 isoforms (see Materials and Methods). TRP current in miRNA-treated B16-F1 cells was significantly smaller when compared to that in control cells, whereas the overall shape of the I–V curve remained the same (Fig. 2, D and E). Correlating with an ~75% reduction in the mRNA abundance of TRPM1, averaged current amplitudes were reduced by ~70% in cells expressing TRPM1 miRNA compared to those expressing control miRNA (Fig. 2F). In contrast, TRPM7 current and mRNA abundance were not significantly different in TRPM1 and control miRNA–expressing cells (Fig. 2G). Lanthanum (La3+), a nonselective blocker of many TRP channels (17), reduced the current ~90% in B16-F1 and ~75% in B16-F10 cells (fig. S2). The amplitude of the outwardly rectifying current in B16-F1 and B16-F10 cells was not significantly different in the presence of 2 mM extracellular Ca2+.
TRPM1 isoform expression and cellular distribution
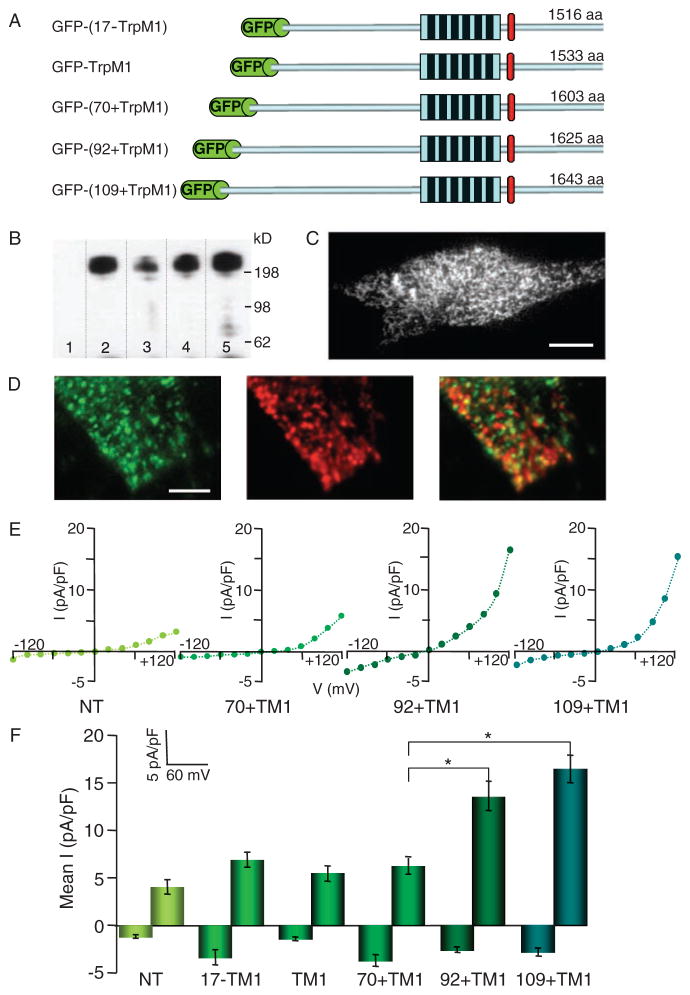
To investigate the intracellular localization of TRPM1, we attached an N-terminal GFP tag to each of the TRPM1 isoforms (Fig. 3A) and expressed these GFP-TRPM1 isoforms in HEK cells. Isoforms were immunoprecipitated with an antibody to GFP (anti-GFP), separated by SDS–polyacrylamide gel electrophoresis, and probed with a monoclonal anti-GFP (Fig. 3B). The appearance of single bands suggested that the TRPM1 splice variants are expressed as full-length proteins in these cells. However, whole-cell recordings and calcium imaging experiments using cells expressing any of the isoforms did not yield measurable currents or calcium signals. Fluorescence imaging of HEK cells expressing the GFP-TRPM1 splice variants suggests that the GFP-tagged isoforms did not reach the plasma membrane (fig. S4).
Fig. 3.
Expression of GFP-tagged TRPM1 in SK-Mel19 human melanoma. (A) Schematic representation of GFP-tagged TRPM1 ion channel isoforms. GFP is fused to the N termini of TRPM1 isoforms. All isoforms share the identical six transmembrane domains and residues thereafter to the end of the C termini. The TRPM1 isoforms have varying numbers of amino acids because of different lengths of their N termini. (B) Western blot of GFP-TRPM1 isoforms immunoprecipitated from HEK293 cells using a polyclonal anti-GFP and probed with a monoclonal anti-GFP. Immunoblotting of GFP-(17−TRPM1) (lane 2), GFP-TRPM1 (lane 3), GFP-(70+TRPM1) (lane 4), and GFP-(92+TRPM1) (lane 5) results in a single band corresponding to the full-length protein that is absent in nontransfected cells (lane 1). (C) TIRF image of SK-Mel22a melanoma cells expressing GFP-TRPM1. GFP-TRPM1 is primarily localized to intra-cellular vesicular structures (Movie S1). Calibration bar, 5 μm. (D) TIRF image of a region of an SK-Mel19 cell expressing GFP-(92+TRPM1) and immunostained with anti-GFP (left) and anti-melanosomal marker (Pmel17) (middle). Overlay of the two images (right) shows no significant colocalization of GFP-TRPM1 and melanosomes. Calibration bar, 1.2 μm. (E) I-V curves from individual SK-Mel19 cells nontransfected (NT), expressing GFP-(70+TRPM1), GFP-(92+TRPM1), or GFP-(109+TRPM1). (F) Mean inward (−120 mV) and outward (+120 mV) current amplitudes recorded from nontransfected SK-Mel19 cells (NT, n = 5) and SK-Mel19 cells expressing GFP-(17−TRPM1) (n = 4), GFP-TRPM1 (n = 10), GFP-(70+TRPM1) (n = 8), GFP-(92+TRPM1) (n = 10), and GFP-(109+TRPM1) (n = 12). *P < 0.001; t test. Error bars, ± SEM.
Prior mRNA localization studies indicate that TRPM1 is present primarily in melanin-producing cells (1, 3, 4, 6). We reasoned that the ability of TRPM1 isoforms to act as functional plasma membrane channels might depend on melanocyte-specific trafficking. When viewed with confocal microscopy, GFP-TRPM1 isoforms expressed in SK-Mel19 pigmented melanocytes (18) showed a distribution similar to that in HEK cells. To further resolve the intracellular structures containing TRPM1, we used objective-based total internal reflection fluorescent (TIRF) microscopy to selectively illuminate fluorophores within ~250 nm of the plasma membrane–glass coverslip interface. Because GFP-TRPM1 fluorescence emission was insufficient for live-cell TIRF imaging in SK-Mel19 cells, we used SK-Mel22a melanoma cells (18), which expressed larger amounts of GFP-TRPM1. To determine the mobility of the TRPM1-containing structures, we performed live TIRF imaging in SK-Mel22a melanoma cells expressing GFP-TRPM1 and found that GFP-TRPM1 was present in highly mobile tubulovesicular structures (Fig. 3C and Movie S1).
We immunostained SK-Mel19 cells expressing GFP-TRPM1 with anti-GFP. TIRF imaging of the immunostained cells revealed GFP-TRPM1 channels invesicular and tubular structures near the plasma membrane, similar to the distribution of GFP-TRPM7 channels (19). Notably, we did not see differential localization of any of the five TRPM1 isoforms. The vesicular structures containing GFP-TRPM1 resembled the size and distribution of melanosomes in SK-Mel19 pigmented melanoma cells. To determine whether GFP-TRPM1 was present in melanosomes, we co-immunostained SK-Mel19 cellswith anti-GFP and an antibody directed against the melanosome-specific protein, gp100 [also known as Pmel 17, the human homolog of the silver locus (20)]. TIRF imaging of the double-stained samples revealed vesicular distributions of both antibodies, but no significant overlap was detected between populations of vesicles labeled with antibodies directed against any of the GFP-TRPM1 isoforms and those positive for gp100 (Fig. 3D).
We then examined whether any of the five TRPM1 isoforms produced currents in SK-Mel 19 cells. Whole-cell currents elicited in response to a step voltage protocol in cells transfected with GFP fused to TRPM1, 70+TRPM1, or 17−TRPM1 were similar to those in nontransfected cells. The same result was obtained in cells expressing nontagged versions of the TRPM1 isoforms, suggesting that the lack of current could not be attributed to the GFP tag. However, transfection of SK-Mel19 cells with GFP-(92+TrpM1) or GFP-(109+TRPM1) elicited a small current. The average amplitude of the outward current at +120 mV was 13.6 ± 1.6 pA/pF for SK-Mel19 cells transfected with GFP-(92+TRPM1) and 16.4 ± 1.5 pA/pF for SK-Mel19 cells transfected with GFP-(109+TRPM1), compared to <6 pA/pF for non-transfected and GFP-TRPM1, GFP-(70+TRPM1), and GFP-(17−TRPM1) transfected cells (Fig. 3, E and F).
To characterize the biophysical properties of the ionic current elicited by GFP-(92+TRPM1) and GFP-(109+TRPM1) in SK-Mel 19 cells, we determined the selectivity of the currents for Na+ and Ca2+, the time constant of voltage activation, and the effect of La3+ on current amplitude (fig. S3). The reversal potential (Vrev) for these currents was ~0 mV, indicating that, like most TRP channels, 92+TRPM1 and 109+TRPM1 encode nonselective ion channels. In whole-cell recordings, replacement of monovalent cations by Ca2+ led to a small decrease in the current amplitude, suggesting that, like TRPM2 (21) and TRPM8 (22), the TRPM1 channel is slightly more permeable to Na+ than to Ca2+ (PNa/PCa < 10). The TRPM1 current showed fast activation kinetics, with the time constant for current activation, τ = 6.7 ± 0.6 ms, independent of voltage. La3+ (100 μM), which blocks the endogenous TRPM1-like current in B16-F1 and B16-F10 cells, reduced 92 + TRPM1 and 109 + TRPM1 currents by more than 70%, from 20.0 ± 0.9 pA/pF to 3.20 ± 0.82 pA/pF, consistent with La3+ block.
Coexpression of TRPM1 with the TRPM subfamily members TRPM3α2 and TRPM7 did not yield perceptibly different currents, suggesting that TRPM1 does not form heteromeric channels with these subunits under our recording conditions. We also tested whether TRPM1 currents could be activated or potentiated by various agonists known to elicit responses from receptors found in melanocytes. None of the melanocyte receptor agonists, α-melanocyte stimulating hormone (α-MSH, which activates the MC1 receptor to induce pigmentation), endothelin (which activates the endothelin B receptor in melanocytes), and melatonin [which might activate postulated melatonin receptors in skin (23)], affected TRPM1 currents. Similarly, the protein kinase A activator forskolin [mimics MC1 receptor activation (24)] did not elicit significantly different currents. Changes in extracellular pH (5.5 to 7.4) or osmolarity (260 to 340 mosM) also failed to significantly alter TRPM1 currents.
TRPM1 and melanin content
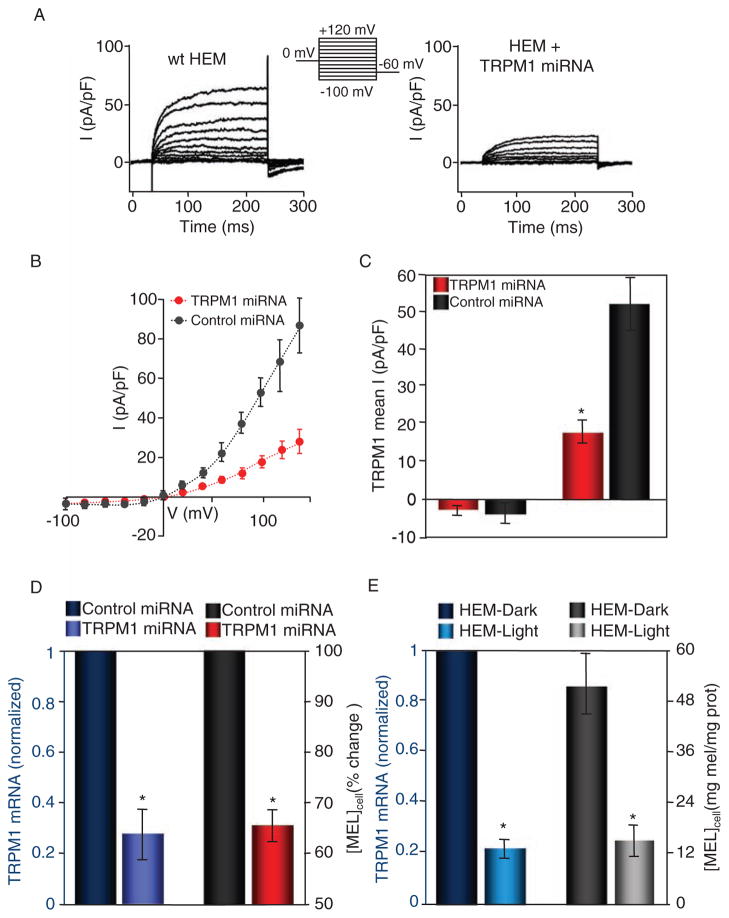
TRPM1 is found in great abundance in HEMs. In human melanomas, decreased abundance of TRPM1 correlates with increasingly aggressive melanoma (1, 5, 25). In whole-cell patch clamp measurements of HEMs isolated from neonatal foreskin, we recorded an endogenous current similar to the endogenous TRPM1 current measured in B16-F1 and B16-F10 cells, and that in SK-Mel19 cells transfected with 92+TRPM1 and 109+TRPM1 (Fig. 4A), but with slower activation kinetics. Consistent with higher TRPM1 expression in primary melanocytes (4), the amplitude of the current in HEMs was higher than that in any of the cell lines (64.6 ± 12.9 pA/pF compared to 25.0 ± 2.3 pA/pF recorded in B16-F1 cells under the same conditions). To further examine the current, we recorded from HEMs expressing either control or TRPM1 miRNA. We measured a significantly smaller current in HEMs expressing TRPM1 miRNA compared to those expressing control miRNA (Fig. 4B), suggesting that TRPM1 is responsible for the current measured in control and nontransfected cells. The average amplitude of the current at +100 mV in HEM cells expressing TRPM1-specific miRNA was reduced by ~66% (Fig. 4C).
Fig. 4.
Reduced abundance of TRPM1 mRNA correlates with decreased TRPM1 current amplitude and reduced pigmentation in primary HEM cells. (A) Representative endogenous whole-cell currents recorded from wild-type (wt, left) or TRPM1-targeting miRNA-transfected HEMs in response to a voltage step protocol (upper middle). (B) I-V relations of HEMs transfected with scrambled miRNA (control, black circles, n = 6) or anti-TRPM1 miRNA (TRPM1 miRNA, red circles, n = 6). (C) Mean inward (−100 mV) and outward (+140 mV) current recorded in response to a step protocol from HEMs expressing control (scrambled) miRNA and TRPM1 miRNA. The TRPM1 outward current was ~66% smaller in the TRPM1 miRNA–expressing cells (17.9 ± 3.1 pA/pF, n = 6, compared to 52.0 ± 7.1 pA/pF, n = 6 for control miRNA–expressing cells). *P < 0.05; t test. Error bars, ± SEM. (D) HEM-dark cells treated with TRPM1 miRNA (purple) have decreased TRPM1 mRNA abundance when compared to those treated with scrambled miRNA (dark blue) (n = 4 independent experiments). Decreased TRPM1 mRNA abundance correlates with decreased cellular melanin concentration (right axis, n = 4 independent experiments). *P < 0.05; t test. Error bars, ± SEM. (E) In HEM-dark and HEM-light cells, the endogenous abundance of TRPM1 mRNA (left axis) closely correlates with the melanin concentration (right axis, n = 3 independent experiments). An ~80% lower abundance of TRPM1 mRNA in HEM-light compared to HEM-dark cells corresponds to an ~75% reduction in the cellular melanin concentration (n = 3 independent experiments). *P < 0.05; t test. Error bars, ± SEM.
Melanocytes produce and transfer melanin to keratinocytes in response to UV radiation. We found that TRPM1 mRNA abundance was correlated with the melanin concentration of HEMs (Fig. 4D). This effect is likely to be more pronounced in miRNA-expressing cells, because the miRNA transfer efficiency in HEMs was only ~50%, whereas melanin concentration was determined for the entire population of cells. To further investigate the relationship between TRPM1 and cellular melanin concentration, we used HEMs isolated from light skin or dark skin and determined the melanin concentration and TRPM1 mRNA abundance in each cell type. We found an ~80% reduction in the abundance of TRPM1 mRNA in HEM-light compared to HEM-dark, as well as an ~72% reduction in the cellular melanin concentration (Fig. 4E). A similar correlation between TRPM1 mRNA abundance and cellular melanin concentration was found in B16-F1 and B16-F10 cells: ~60% change in TRPM1 mRNA corresponds to an ~70% decrease in melanin concentration in B16-F10 compared to that in B16-F1 (Fig. 2C and fig. S2C). Together with TRPM1’s specificity for melanocytes, the correlation between TRPM1 and melanin concentrations suggests that TRPM1 may play a role in pigmentation.
Although reduced TRPM1 abundance is associated with aggressive melanoma (5, 25), the link between TRPM1 protein function and melanoma is unknown. We wondered whether reduced amounts of the newly identified TRPM1 isoforms might correlate with increased melanocyte proliferation rates. Further in vivo studies will be required to determine whether TRPM1 has a function in melanocyte transformation, melanoma proliferation, invasion, metastasis, or apoptosis of transformed melanocytes.
DISCUSSION
In summary, we report two previously unidentified splice variants of TRPM1 that are highly conserved across species. We show that the TRPM1 splice variants are differentially expressed in primary and transformed melanocytes, suggesting that the particular splice variants expressed in melanoma cells may be biologically relevant. We also describe the basic biophysical properties of native and expressed TRPM1 ionic currents. Neither TRPM1 (as defined in the original cloning of this protein) nor 70+TRPM1 (currently listed as TRPM1 in the updated database) produced measurable currents in heterologous systems. However, the 92+TRPM1 and 109+TRPM1 isoforms resulted in a measurable current when expressed in SK-Mel19 melanoma cells. This conductance was cation nonselective and outwardly rectifying, typical for many TRP channels. We tested a number of potential regulators of TRPM1 function. These included several different G protein–coupled receptors found in melanocytes, as well as changes in the extracellular environment known to modulate other TRP ion channels (pH and osmolarity). None of the tested regulators affected TRPM1 current amplitude, voltage dependence, or activation kinetics.
Currents mediated by both native TRPM1 and heterologously expressed TRPM1 (92+TRPM1 and 109+TRPM1) were smaller than TRPM7 currents by a factor of up to 5. Cellular protein abundance is low (in Fig. 2B expressed GFP-TRPM1 isoforms could only be detected by immunoprecipitation; expression of GFP-tagged isoforms in melanoma cells resulted in low fluorescence), representing few ion channels. Given TRPM1’s low surface expression, we speculate that the primary function of these channels might be intracellular, so that only a small fraction is localized to the plasma membrane (as was found for the heterologously expressed proteins). Alternatively, the channels may be targeted to a specific region of the plasma membrane, such as melanocyte dendritic tips, so that their local density is high despite an apparently low whole-cell (plasma membrane) conductance. Another possibility is that TRPM1 channels expressed in the plasma membrane have low basal activity in the absence of specific (unknown) activators. Endogenous TRPM1 expression correlated with pigmentation in HEMs, and decreasing TRPM1 expression with RNAi decreased melanocyte melanin concentration, suggesting that TRPM1 may affect the pigmentation process and may therefore be a target for treatment of pigmentation disorders.
TRPM1 polymorphism and splice variants
The T/C (or A/G on the reverse strand) polymorphism at position 130 generates TRPM1 and 70+TRPM1 proteins and is deposited in the National Center for Biotechnology Information (NCBI) single-nucleotide polymorphism (SNP) database with the reference number rs4779816. The SNP report also contains the results of sequencing studies ascertaining the genotype diversity for different groups of the population. Remarkably, the homozygous T/T genotype present in 5% of the European or European-origin population has not been found in samples analyzed from African-American or sub-Saharan African and Asian populations. This raises the question of whether thymine at this position correlates with melanoma risk, and if so, why. Studies analyzing the allelic composition of normal and malignant melanoma tissue from the same heterozygous individuals could be used to investigate this possibility. The potential correlation between the C/T polymorphism and genetic susceptibility to melanoma could then be used as a diagnostic tool for melanoma-prone individuals.
We identified two TRPM1 N-terminal exons that generate previously unidentified splice variants: 109+TRPM1 and 92+TRPM1. The sequences coding for exon 0, which generates 109+TRPM1, and exon 1′, which generates 92+TRPM1, are conserved in many mammalian species. Because 109+TRPM1 does not include exon 1, we were not able to determine its relative abundance in different tissues or melanoma cell lines. 92+TRPM1 mRNA appears to be relatively infrequent in primary melanocytes (<2.5% of total TRPM1), brain (<5% of total TRPM1), and retina (<4% of total TRPM1), but is increased in melanoma cell lines (up to 56% of total TRPM1). One possible explanation for this increase is that, during the process of malignant transformation, melanocytes gain the ability to transcribe various TRPM1 splice variants. Notably, when melanomas become more aggressive and metastasize, the abundance of all TRPM1 isoforms decreases substantially (5).
TRPM1 localization
Our results show that TRPM1 is predominantly intracellularly localized when heterologously expressed in HEK or melanoma cell lines. The lack of specific antibodies prevented us from determining the distribution of the endogenous TRPM1 in melanocytes. However, many TRP ion channels [including TRPML1-3 (mucolipins), TRPP1-3 (polycystins), TRPV2, TRPV5, and TRPM7] have predominantly intracellular distributions, and the distribution of these channels when expressed in heterologous systems mimics that of the endogenous channels (19, 26). Even in cases where the channels appear to have a primarily intracellular function, a small portion of these channels reach the plasma membrane to generate a functional current.
We found that all TRPM1 isoforms failed to generate functional current when expressed in HEK cells. In SK-Mel19 melanoma cells, we measured a functional current only for the two previously unidentified TRPM1 isoforms, 92+TRPM1 and 109+TRPM1. The amplitude of the current generated by overexpressed 92+TRPM1 or 109+TRPM1 is small compared to that of the native TRPM1 current measured in B16-F1 and HEMs. Because TRPM1 is primarily expressed in melanin-producing cells, TRPM1 localization to the plasma membrane may require specialized protein complexes unique to these cells. The abundance of many melanocytic proteins is altered in melanoma cells, which might therefore be unable to confer proper localization of TRPM1 isoforms. Alternatively, like the mucolipins (TRPML), TRPM1 channels may be predominantly intracellular.
TRPM1 cellular function
Our results link TRPM1 to the pigmentation process. We showed that reduced expression of TRPM1 in melanocytes correlates with decreased melanin content. Recently, a mutation in TRPM1 resulting in variation in horse coat color was described, with the dark spots expressing wild-type TRPM1 and light spots expressing mutant (and presumably nonfunctional) TRPM1 (27). How might TRPM1 affect the accumulation of melanin in cells? Pigmentation requires the production and storage of melanin in melanosomes, melanosomal transport to the tip of the dendrites, and melanosomal transfer from melanocytes to the neighboring keratinocytes. The production of melanin in melanosomes is primarily controlled by the tyrosinase family of proteins (8, 28). To test whether the observed correlation between TRPM1 mRNA abundance and pigmentation is due to decreased production of melanin, we used RT-PCR to determine the abundance of tyrosinase mRNA in cells treated with control miRNA or miRNA directed against TRPM1. HEM cells treated with miRNA directed against TRPM1 do not show reduced amounts of tyrosinase mRNA, suggesting that the decreased melanin content of those cells is not due to reduced production of melanin (fig. S4). This leaves open the possibility that TRPM1 regulates the storage of melanin in melanocytes. The mechanism by which melanosomes leave the melanocytes and enter the keratinocytes is poorly understood. One possible TRPM1 function, consistent with our pigmentation data, is that TRPM1 localized to the tip of the melanocytic dendrites regulates the release of melanosomes. Such localization of the TRPM1 ion channel could also explain the small inward TRPM1 current measured in melanocytes and melanoma cells. Because most melanoma cells lose the ability to transfer melanosomes to keratinocytes, it is conceivable that the structures responsible for TRPM1 localization are also lost, preventing the proper expression of TRPM1 ion channels in these cells. Moreover, the vesicular localization of TRPM1 ion channels detected in melanoma cells suggests that the number of ion channels at the plasma membrane could be increased in response to particular signals by inserting more TRPM1-containing vesicles into the plasma membrane, as shown for TRPM7 and TRPC5 ion channels (19, 26).
Many aspects of TRPM1 ion channel properties and cellular function remain to be explored. Our results have advanced the current understanding of TRPM1 by identifying two previously unidentified and highly conserved splice variants, 92+TRPM1 and 109+TRPM1, by describing an endogenous TRPM1 current present in melanocytes and melanoma cells, and by identifying TRPM1 as a possible element of melanocyte pigmentation.
MATERIALS AND METHODS
Cell culture
HEK293T cells were cultured in medium containing Dulbecco’s modified Eagle’s medium and F12 nutrient mixture (DMEM-HAM’s F12), 10% fetal bovine serum (FBS), and 1:1 glycine and HT supplement (GHT) (Gibco). B16-F1 and B16-F10 cells were obtained from American Type Culture Collection and cultured in DMEM (Gibco) and 10% FBS. SK-Mel19 and SK-Mel22a cells were obtained from the Sloan-Kettering melanoma collection. SK-Mel19 cells were cultured in Eagle’s minimum essential medium (Gibco) supplemented with nonessential amino acids (Gibco) and 10% FBS. SK-Mel22a cells were cultured in RPMI (Gibco) supplemented with 10% FBS. Neonatal HEM-light and -dark cells were purchased from Cascade Biologics (Invitrogen), cultured in Medium 254 and human melanocyte growth supplement (HMGS; Cascade Biologics, Invitrogen), and propagated for a limited number of passages accounting for 50 to 100 cell division cycles. Cells were cultured on glass coverslips for immunostaining, imaging, and electrophysiology experiments, and on plastic Petri dishes for all the other experiments.
RNA extraction, RT-PCR, and library screening
Human Melanoma Marathon-Ready cDNA and Human Brain and Retina QUICK-clone cDNA (Clontech) were used to identify TRPM1 isoforms. cDNA from HEM and melanoma cell lines was obtained by RT-PCR with SuperScript III First-Strand Synthesis System (Invitrogen) using as template the total RNA extracted from cultured cells with the RNeasy Plus kit (Qiagen).
cDNA fragments were PCR-amplified with different pairs of oligonucleotides, cDNA from the sources listed above, deoxynucleotide triphosphate (dNTP, Roche) and Pfu Turbo or PicoMax polymerases (Stratagene). For amplification, a Touchdown protocol was used: 94°C for 30 s, five cycles of 94°C for 5 s and 70°C for 2 to 3 min, five cycles of 94°C for 5 s and 68°C for 2 to 3 min, and five cycles of 94°C for 5 s and 66°C for 2 to 3 min followed in some cases by 20 to 32 regular PCR-amplification cycles: 95°C for 30 s, 60°C for 30 s, and 72°C for 0.5 to 3 min. The amplified DNA was analyzed on 1% agarose gels and the bands were excised. DNA extracted from the gel was cloned into a plasmid vector with Zero Blunt TOPO (for Pfu Turbo-amplified fragments) or TOPO TA (for PicoMax-amplified fragments), followed by use of a PCR cloning kit (Invitrogen). Single-species amplified DNAwas sequenced. The sequences were analyzed with Lasergene software (DNASTAR, Inc.).
Cloning of TRPM1 and GFP-TRPM1 isoforms
G. Krapivinsky in the Clapham laboratory provided the full-length human TRPM1 clone obtained from a human retina library. The TRPM1 cDNA was initially cloned into the enhanced GFP/C1 vector (Clontech) as an N-terminal GFP fusion protein. After identification of the cDNA sequences that generate 70+TRPM1, 92+TRPM1, and 109+TRPM1 by library screening (see Results), we PCR-amplified the N-terminal coding sequences corresponding to these two isoforms and cloned them in-frame between GFP and TRPM1 with the Sal I site that cuts both the vector and TRPM1 1421 bp downstream of the start site.
Subsequently, we generated a series of modified pcDNA4/TO vectors: two vectors containing GFP either at the 5′ end or 3′ end of the multiple cloning site and one vector containing a hemagglutinin tag at the 5′ end of the multiple cloning site. TRPM1, 70+TRPM1, and 92+TRPM1 were then cloned into the Bam HI site of the three modified vectors and in the original pcDNA4/TO vector (Invitrogen). The GFP-TRPM1-pcDNA4/TO and TRPM1-pcDNA4TO constructs were used to clone GFP-(17−TRPM1), 17−TRPM1 and GFP-(109+TRPM1) with the use of amplified cDNA from the melanocyte library. The N-terminal cDNA sequence corresponding to 109+TRPM1 was cloned in-frame with GFP and TRPM1 with the Kpn I site of pcDNA4/TO and the Bsu 36I site of TRPM1.
Electrophysiological recordings and data analysis
Modified Ringer’s solution with low [Cl−] and zero nominal [Ca2+] contained (in millimolar) 140 sodium gluconate, 4 NaCl, 2 MgCl2, 10 Hepes-Na, and 10 glucose (pH 7.4 MES; 310 mosM). CaCl2 (2 mM) was included in the external solution where noted. Internal pipette solution contained (in millimolar) 100 Cs-MES, 10 Cs4BAPTA, 8 MgCl2, 0.3 Na-ATP (adenosine 5′-triphosphate), 1 CaCl2, and 10 Hepes-Na (pH 7.2). High internal Mg2+ concentration (8 mM) was used to prevent the activation of endogenous MIC current (29). For MIC recordings, our internal solutions contained 130 mM Cs-MeSO4, 8 mM NaCl, 4.1 mM CaCl2, 1 mM MgCl2, 10 mM EGTA, 5 mM Na2ATP, 10 mM Hepes (pH 7.2, CsOH).
Electrophysiological experiments were performed with the whole-cell patch clamp technique at room temperature. Recordings were obtained with an Axopatch 200B amplifier, Digidata 1322A analog-to-digital converter, and pClamp 8.01 software (Molecular Devices, Union City, CA). Data were filtered at 2 kHz and digitized at 5 kHz. Borosilicate glass pipettes (World Precision Instruments, Sarasota, FL) were pulled to a typical pipette resistance of 3 MΩ after heat polishing. An Ag-AgCl wire was used as the reference electrode. To minimize voltage errors, 70 to 80% of the series resistance was compensated. We applied a step protocol starting from a holding potential of 0 mV in 20-mV steps from −120 mV to +120 mV. Current-voltage (I-V) relations were obtained by step protocols. Cell membrane capacitance values were used to calculate current densities.
The relative permeability of monovalent cations was calculated from the shift in reversal potential after complete substitution of extracellular Na+ by a specific cation. Permeability of the divalent Ca2+ relative to Na+ was calculated from the absolute reversal potential measured with 30 mM Ca2+ in the extracellular solution (30). The solutions used for Na+ permeability were 150 mM NaCl, 10 mM glucose, and 10 mM Hepes (pH 7.4 with NaOH). The solutions used for Ca2+ permeability were 30 mM CaCl2, 10 mM glucose, and 10 mM Hepes (pH 7.4 with CaOH).
To determine the time constants of current activation (τ), the patch clamp data were analyzed with WinASCD (G. Droogmans; ftp://ftp.cc.kuleuven.be/pub/droogmans/winascd.zip). Whenever possible, linear background components of current traces were digitally subtracted before data analysis. Time constants of current activation (τ) were obtained from fitting the current traces with a single-exponential function.
miRNA and quantitative PCR
A set of four TRPM1-specific pairs of single strand BLOCK-iT miR RNAi oligos were designed (Invitrogen). The oligos targeted 22-bp regions: bp 925 to 946 (oligo1), bp 3421 to 3442 (oligo2), bp 3453 to 3474 (oligo3), and bp 3736 to 3757 (oligo4) of NM_002420.4 (70+TRPM1). The oligos were modified to obtain a stem-loop structure, annealed, and cloned into the pcDNA6.2-GW/EmGFP-miR expression vector (Invitrogen). The miRNA expression vector contained the coding sequence of GFP followed by the pre-miRNA insertion sites in its 3′UTR. Expression of GFP enabled identification of the miRNA-expressing cells, an important feature for single-cell measurements in cells with low miRNA transfer efficiency. Oligos 1 and 2 and oligos 3 and 4 were chained in the same vector and transfected in mammalian cells. As a negative control, we used a scrambled oligo-nucleotide with a similar fraction of CG bases as the TRPM1 oligos. The abundance of TRPM1 was determined by quantitative PCR. We found that oligo 3+4 was more effective than oligo 1+2 in reducing the abundance of endogenous TRPM1 mRNA. We used miRNA based on oligo 3+4 for all of the experiments described here.
The GFP-miRNA containing plasmid was introduced into B16-F1 and B16-F10 cells with Lipofectamine 2000 transfection reagent (Invitrogen). About 70% of all cells contained GFP, and therefore miRNA. GFP-expressing cells were selectively patched 5 to 8 days after transfection. Amaxa nucleofection with the NHEM-Neo buffer (Amaxa, Inc) was used to introduce the GFP-miRNA plasmid into HEM, according to the manufacturer’s protocol. About 50% of the HEM expressed GFP and were used for patch clamp experiments 5 to 8 days after transfection. All miRNA-treated HEM cells (whether expressing GFP or not) were used for quantitative PCR and for melanin quantification.
For quantitative PCR, we used a 7300 Real-Time PCR System (Applied Biosystems). PCR reactions were prepared according to the manufacturer’s instructions with Sybr Green PCR mix. Actin was used as a control for each PCR template and all reactions were done in triplicate. To average mRNA values from different experiments comparing the amount of mRNA in two different samples (TRPM1 versus control miRNA–treated cells or HEM-light versus HEM-dark), we normalized the largest mRNA value (control miRNA or HEM-dark) from each experiment to 1 and calculated the lower value (TRPM1 miRNA or HEM-light) as a fraction.
DNA transfection
HEK293T, SK-Mel19, and SK-Mel22a cells were transiently transfected with Lipofectamine 2000 (Invitrogen). Cells were plated at least 24 hours before the transfection and used at 80 to 90% density for HEK293Tand SK-Mel22a, and at 50 to 70% for SK-Mel19 cells. Upon addition of the transfection mix, SK-Mel22a and SK-Mel19 cells were centrifuged for 3 min at 800 rpm. Cells were then incubated with the transfection mix for 3 to 10 hours and cultured on glass or plastic 4 to 48 hours before experiments.
Immunoprecipitation and Western blotting
HEK cells plated on 35-mm Petri dishes were transfected with GFP-TRPM1 isoforms. Cells were lysed using PBS with 1% Triton X-100 and protease inhibitors, and the lysates were incubated overnight at 4°C with ImmunoPure Immobilized protein A/G (Pierce) and a monoclonal GFP antibody (Molecular Probes, Invitrogen). The samples were washed three times with PBS buffer containing 0.1% Triton X-100, eluted by boiling in NuPAGE LDS sample buffer, analyzed with the NuPAGE 4 to 12% Bis-Tris gel, and Western blotted with standard protocols. Monoclonal anti-GFP (Covance) was used as a primary antibody to detect GFP-tagged proteins. An enhanced chemoluminescence kit (Pierce) was used for detection of protein bands.
Immunostaining and fluorescent imaging
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and stained with primary antibodies for 1 hour at room temperature. Antibodies used were GFP rabbit polyclonal antibody (Molecular Probes, Invitrogen) and gp100/Pmel17 mouse monoclonal antibody (HMB 45, Dako). Samples were stained with the corresponding Alexa 488 or Alexa 568 secondary antibodies (Molecular Probes, Invitrogen), mounted, and imaged.
Live imaging experiments were performed at ~25°C with cells grown on glass coverslips and placed in a custom chamber with standard extra-cellular buffer. Extracellular buffer contained (in millimolar) 135 NaCl, 5 KCl, 1.5 CaCl2, 1.5 MgCl2, and 20 Hepes (pH 7.4). For TIRF imaging, we used a custom-built objective-based, TIRF microscope (26). Cells grown on glass coverslips were placed in a chamber with standard extracellular buffer and imaged at ~25°C. All image acquisition, and some of the analysis, was performed with MetaMorph (Universal Imaging, West Chester, PA). Time series of 200 images at 1-s intervals were recorded.
Melanin quantification
Melanin was determined for a population of cells, usually from a 35-mm Petri dish at 50 to 90% confluence. After media removal, cells were rinsed twice with PBS and then lysed with PBS + 1% Triton X-100, scraped, and collected in a microcentrifuge tube. After 30-min incubation at 4°C to allow complete lysis, the cell lysate was centrifuged at 14,000g for 30 min (4°C) to separate the soluble and insoluble fractions. The soluble fraction (supernatant) was used to determine the total protein concentration by Bradford assay (Biorad). The insoluble fraction containing the melanin pellet was re-suspended in 200 μl of 1 N NaOH and incubated at 85°C for 30 min or until fully dissolved. The optical density at 405 nm was determined for the resulting melanin solution and the melanin concentration was estimated from the melanin calibration curve. The melanin curve was determined by measuring the optical density at 405 nm for a series of concentrations of synthetic melanin (Sigma) dissolved in NaOH as described above. The average cellular melanin concentration was determined as the ratio of the sample’s melanin and total protein concentration.
Supplementary Material
Movie S1. Dynamic localization of GFP-(70+TRPM1) in human melanoma cells.
Fig. S1. Mouse TRPM1 isoforms.
Fig. S2. Lanthanum blocks endogenous TRPM1 current in B16-F1 and B16-F10 mouse melanocytes.
Fig. S3. Characterization of 92+TRPM1 and 109+TRPM1 current in SK-Mel19 human melanocytes.
Fig. S4. Cellular localization of GFP-(109+TRPM1) in HEK cells and tyrosinase mRNA abundance in HEM cells treated with TRPM1 miRNA.
Acknowledgments
We thank D. Fisher (MGH and Harvard University, Boston, MA) and M. Khaleb for providing the MALME cell line and for helpful advice with experimental techniques.
We also thank G. Krapivinsky for sharing his reagents and for technical advice. We appreciate the critical advice and constructive criticism from members of the Clapham laboratory during the development of the project.
Footnotes
REFERENCES AND NOTES
- 1.Duncan LM, Deeds J, Hunter J, Shao J, Holmgren LM, Woolf EA, Tepper RI, Shyjan AW. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res. 1998;58:1515–1520. [PubMed] [Google Scholar]
- 2.Hunter JJ, Shao J, Smutko JS, Dussault BJ, Nagle DL, Woolf EA, Holmgren LM, Moore KJ, Shyjan AW. Chromosomal localization and genomic characterization of the mouse melastatin gene (Mlsn1) Genomics. 1998;54:116–123. doi: 10.1006/geno.1998.5549. [DOI] [PubMed] [Google Scholar]
- 3.Zhiqi S, Soltani MH, Bhat KM, Sangha N, Fang D, Hunter JJ, Setaluri V. Human melastatin 1 (TRPM1) is regulated by MITF and produces multiple polypeptide isoforms in melanocytes and melanoma. Melanoma Res. 2004;14:509–516. doi: 10.1097/00008390-200412000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res. 2004;64:509–516. doi: 10.1158/0008-5472.can-03-2440. [DOI] [PubMed] [Google Scholar]
- 5.Hammock L, Cohen C, Carlson G, Murray D, Ross JS, Sheehan C, Nazir TM, Carlson JA. Chromogenic in situ hybridization analysis of melastatin mRNA expression in melanomas from American Joint Committee on Cancer stage I and II patients with recurrent melanoma. J Cutan Pathol. 2006;33:599–607. doi: 10.1111/j.1600-0560.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 6.Fang D, Setaluri V. Expression and up-regulation of alternatively spliced transcripts of melastatin, a melanoma metastasis-related gene, in human melanoma cells. Biochem Biophys Res Commun. 2000;279:53–61. doi: 10.1006/bbrc.2000.3894. [DOI] [PubMed] [Google Scholar]
- 7.Van Den Bossche K, Naeyaert JM, Lambert J. The quest for the mechanism of melanin transfer. Traffic. 2006;7:769–778. doi: 10.1111/j.1600-0854.2006.00425.x. [DOI] [PubMed] [Google Scholar]
- 8.Boissy RE. Melanosome transfer to and translocation in the keratinocyte. Exp Dermatol. 2003;12(Suppl 2):5–12. doi: 10.1034/j.1600-0625.12.s2.1.x. [DOI] [PubMed] [Google Scholar]
- 9.Berens W, Van Den Bossche K, Yoon TJ, Westbroek W, Valencia JC, Out CJ, Naeyaert JM, Hearing VJ, Lambert J. Different approaches for assaying melanosome transfer. Pigment Cell Res. 2005;18:370–381. doi: 10.1111/j.1600-0749.2005.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 11.Hearing VJ. Biogenesis of pigment granules: A sensitive way to regulate melanocyte function. J Dermatol Sci. 2005;37:3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 13.Xu XZ, Moebius F, Gill DL, Montell C. Regulation of melastatin, a TRP-related protein, through interaction with a cytoplasmic isoform. Proc Natl Acad Sci USA. 2001;98:10692–10697. doi: 10.1073/pnas.191360198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, Desai BN, Navarro B, Donovan A, Andrews NC, Clapham DE. Deletion of Trpm7 disrupts embryonic development and thymopoiesis without altering Mg2+ homeostasis. Science. 2008;322:756–760. doi: 10.1126/science.1163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerschbaum HH, Kozak JA, Cahalan MD. Polyvalent cations as permeant probes of MIC and TRPM7 pores. Biophys J. 2003;84:2293–2305. doi: 10.1016/S0006-3495(03)75035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadler MJ, Hermosura MC, Inabe K, Perraud AL, Zhu Q, Stokes AJ, Kurosaki T, Kinet JP, Penner R, Scharenberg AM, Fleig A. LTRPC7 is a Mg•ATP-regulated divalent cation channel required for cell viability. Nature. 2001;411:590–595. doi: 10.1038/35079092. [DOI] [PubMed] [Google Scholar]
- 17.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 18.Carey TE, Takahashi T, Resnick LA, Oettgen HF, Old LJ. Cell surface antigens of human malignant melanoma: Mixed hemadsorption assays for humoral immunity to cultured autologous melanoma cells. Proc Natl Acad Sci USA. 1976;73:3278–3282. doi: 10.1073/pnas.73.9.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oancea E, Wolfe JT, Clapham DE. Functional TRPM7 channels accumulate at the plasma membrane in response to fluid flow. Circ Res. 2006;98:245–253. doi: 10.1161/01.RES.0000200179.29375.cc. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi T, Urabe K, Orlow SJ, Higashi K, Imokawa G, Kwon BS, Potterf B, Hearing VJ. The Pmel 17/silver locus protein. Characterization and investigation of its melanogenic function. J Biol Chem. 1994;269:29198–29205. [PubMed] [Google Scholar]
- 21.Sano Y, Inamura K, Miyake A, Mochizuki S, Yokoi H, Matsushime H, Furuichi K. Immunocyte Ca2+ influx system mediated by LTRPC2. Science. 2001;293:1327–1330. doi: 10.1126/science.1062473. [DOI] [PubMed] [Google Scholar]
- 22.Peier AM, Moqrich A, Hergarden AC, Reeve AJ, Andersson DA, Story GM, Earley TJ, Dragoni I, McIntyre P, Bevan S, Patapoutian A. A TRP channel that senses cold stimuli and menthol. Cell. 2002;108:705–715. doi: 10.1016/s0092-8674(02)00652-9. [DOI] [PubMed] [Google Scholar]
- 23.Slominski A, Fischer TW, Zmijewski MA, Wortsman J, Semak I, Zbytek B, Slominski RM, Tobin DJ. On the role of melatonin in skin physiology and pathology. Endocrine. 2005;27:137–148. doi: 10.1385/ENDO:27:2:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, Ito S, Fisher DE. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 25.Duncan LM, Deeds J, Cronin FE, Donovan M, Sober AJ, Kauffman M, McCarthy JJ. Melastatin expression and prognosis in cutaneous malignant melanoma. J Clin Oncol. 2001;19:568–576. doi: 10.1200/JCO.2001.19.2.568. [DOI] [PubMed] [Google Scholar]
- 26.Bezzerides VJ, Ramsey IS, Kotecha S, Greka A, Clapham DE. Rapid vesicular translocation and insertion of TRP channels. Nat Cell Biol. 2004;6:709–720. doi: 10.1038/ncb1150. [DOI] [PubMed] [Google Scholar]
- 27.Bellone RR, Brooks SA, Sandmeyer L, Murphy BA, Forsyth G, Archer S, Bailey E, Grahn B. Differential gene expression of TRPM1, the potential cause of congenital stationary night blindness and coat spotting patterns (LP) in the Appaloosa horse (Equus caballus) Genetics. 2008;179:1861–1870. doi: 10.1534/genetics.108.088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasmeier C, Hume AN, Bolasco G, Seabra MC. Melanosomes at a glance. J Cell Sci. 2008;121:3995–3999. doi: 10.1242/jcs.040667. [DOI] [PubMed] [Google Scholar]
- 29.Runnels LW, Yue L, Clapham DE. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science. 2001;291:1043–1047. doi: 10.1126/science.1058519. [DOI] [PubMed] [Google Scholar]
- 30.Voets T, Prenen J, Vriens J, Watanabe H, Janssens A, Wissenbach U, Bödding M, Droogmans G, Nilius B. Molecular determinants of permeation through the cation channel TRPV4. J Biol Chem. 2002;277:33704–33710. doi: 10.1074/jbc.M204828200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Dynamic localization of GFP-(70+TRPM1) in human melanoma cells.
Fig. S1. Mouse TRPM1 isoforms.
Fig. S2. Lanthanum blocks endogenous TRPM1 current in B16-F1 and B16-F10 mouse melanocytes.
Fig. S3. Characterization of 92+TRPM1 and 109+TRPM1 current in SK-Mel19 human melanocytes.
Fig. S4. Cellular localization of GFP-(109+TRPM1) in HEK cells and tyrosinase mRNA abundance in HEM cells treated with TRPM1 miRNA.