Abstract
Recent advances in DNA targeting allow unprecedented control over gene function and expression. Targeting based on TAL effectors is arguably the most promising for systems biology and metabolic engineering. Multiple, orthogonal TAL-effector reagents of different types can be used in the same cell. Furthermore, variation in base preferences of the individual structural repeats that make up the TAL effector DNA recognition domain makes targeting stringency tunable. Realized applications range from genome editing to epigenome modification to targeted gene regulation to chromatin labeling and capture. The principles that govern TAL effector DNA recognition make TAL effectors well suited for applications relevant to plant physiology and metabolism. TAL effector targeting has merits that are distinct from those of the RNA-based DNA targeting CRISPR/Cas9 system.
Introduction
The collection of molecular tools for dissecting and manipulating plant physiology and metabolism is growing rapidly. Recent advances in DNA targeting especially have given researchers an unprecedented degree of control over gene function and expression. Among these advances, targeting based on TAL effectors, heterogeneous transcription factors produced and injected into plant cells by pathogenic bacteria, has been among the most revolutionary, and is arguably the most promising for systems biology and metabolic engineering in plants. The DNA sequence specificity of TAL effectors is virtually fully customizable, and the DNA recognition domain of TAL effectors has proven amenable to molecular fusions to direct a variety of enzymatic and other activities to desired locations in the genomes of living cells. Fusions to a nuclease for targeted DNA cleavage essential for genome editing, fusions to alternative transcriptional activation domains and to repressor domains for direct modification of the transcriptome, fusions to chromatin modifying enzymes for modification of the epigenome (and indirect modification of the transcriptome), and fusions to molecular probes for in situ labeling of DNA or to tags for capture of regional DNA protein complexes have all been developed and applied within the past five years, many within the past year. This article reviews the principles of TAL effector-based DNA targeting and highlights the applications that, integrated with emerging technologies and approaches for making molecular switches and circuits, and knowledge of plant biochemistry, hold most promise for manipulating plant physiology and metabolism. Relative merits of TAL effector-based DNA targeting and the recently developed RNA-guided DNA targeting mechanism of the CRISPR/Cas9 system are also touched upon.
Native roles of TAL effectors
TAL effectors are one of many types of proteins delivered into plant cells through the specialized type III secretion system of Gram-negative pathogenic bacteria during infection. They are unusual in two respects: first, though not the only direct host transcriptional regulators produced by plant pathogenic bacteria [1], TAL effectors are so far unique in their collective ability to transcriptionally activate diverse host genes specifically; and second, their distribution is largely limited to the genus Xanthomonas, with only closely related but less diverse functional homologs found in Ralstonia solanacearum [2], also a plant pathogen, and a more distantly related and uncharacterized homolog encoded in the genome of Burkholderia rhizoxinica, a bacterial endosymbiont of the fungal plant pathogen Rhizopus microsporus [3].
Host targets of TAL effectors whose activation is required for full expression of disease are called susceptibility (S) genes. S gene alleles with polymorphism at the effector binding site that prevents activation confer disease resistance [4]. Another type of resistance is conferred by genes that display a binding site, yet whose activation results in a rapid localized host cell death and cessation of bacterial spread [5]. Thus, TAL effectors are subject to opposing selective forces — for targeting flexibility to accommodate S gene polymorphism that might be encountered across different host genotypes, and for stringent specificity to target S genes uniquely while avoiding resistance gene activation traps [6].
TAL effector — DNA recognition
The solution nature found is a modular DNA recognition and binding mechanism that pairs individual, contiguous structural repeats in the protein with contiguous nucleotides in the DNA target [7,8]. This allows for recombinational evolution of new specificities, and at the same time variation in the overall stringency of specificity because individual modules differ both qualitatively and quantitatively in their preference for different nucleotides [7–10]. Each repeat is usually 34 amino acids, displaying variation primarily at positions 12 and 13, the ‘repeat-variable diresidue’ (RVD). Repeat specificity is encoded in the RVD. Common RVDs (using the single letter amino acid code) include HD, specifying cytosine, NG, specifying thymine, NN, specifying guanine or adenine, and NI, specifying ade-nine. Next most common are NS, and N* (in which the asterisk denotes a missing 13th residue), which have generally lax specificity. The rare RVD NH specifies G uniquely [11,12].
Repeats each constitute a two-helix bundle in which residue 12 and 13 reside on an inter-helical loop. Repeats assemble into a left-handed superhelix that wraps in a right-handed fashion around the DNA, tracking the major groove. The helical bundles pack with the inter-helical loops projected inward (and inter-bundle loops outward). Residue 12, typically N or H, stabilizes the RVD loop by binding to the carbonyl oxygen at position 8. Nucleotide preferences and binding are determined apparently exclusively by interactions of residue 13 with individual nucleotides on the plus strand, ranging from hydrogen bonding (e.g., D to cytosine) to Van der Waals contacts (e.g., G to thymine) to steric specification (e.g., I to adenine). Thus, less common RVDs such as ND or SN can generally be assumed to have the specificity of their more common counterpart that shares the same residue at position 13. The role of non-RVD residues has not been investigated in depth, but a study of the Ralstonia TAL effector homologs suggested that variation outside the RVD can quantitatively affect nucleotide preference [2].
Across the published structures however (none of which represents any Ralstonia protein), overall there is negligible structural variation outside the RVD loop from repeat to repeat. This striking modularity is at the heart of the utility of TAL effectors as DNA targeting tools. Repeats with different RVDs can be strung together in any order to create recognition domains for DNA sequences of choice, and targets can be synthesized to match particular TAL effector repeat arrays.
Design and assembly of custom TAL effector DNA recognition domains
Several web-based computational tools are available to aid in design of custom TAL effector arrays to uniquely target a given genomic site. The more sophisticated of these allow design around potential off-targets by scoring alignments of the RVD sequence to the genome using a position weight matrix that takes RVD-nucleotide association frequencies observed in naturally occurring TAL effector target pairs as probabilities. Some tools allow users to take into account experimental evidence for different relative affinity contributions of different RVDs with their preferred base [13] to limit output to arrays estimated to have good overall affinity, and one tool weighs mismatches at the N-terminal end more heavily than those at the C-terminal end in excluding possible off targets; this is based on an observed polarity of mismatch tolerance that is thought to reflect the process of target acquisition, which likely involves non-specific interaction of N-terminal portions of the protein with the DNA backbone, followed by sequential RVD-nucleotide pairings proceeding from that end [10]. But no design software yet is able to accurately estimate relative affinities of a TAL effector for different nucleotide sequences because affinity contributions of all possible RVD-nucleotide pairings individually, and position effects on those contributions, have yet to be quantified.
Most software designs require thymine before the RVD-specified sequence, in agreement with nearly all identified naturally occurring TAL effector binding sites and with experimental data showing the importance of this thymine for optimal binding and activity [7,8]. A recent report identified N-terminal mutations that can alter this specificity [14], and the Ralstonia proteins prefer G at this position [2,15], so it seems likely that further engineering will relax the requirement for T.
Several research groups have published reagents for moderately high-throughput and relatively simple assembly of custom TAL effector constructs using golden gate cloning (e.g. [16]). Very high-throughput, automatable methods have also been established [17–19]. Custom TAL effector constructs also are available commercially. Each of the construction platforms uses a small set of well-characterized RVDs, and this has proven effective for diverse applications. In the future, characterization of more RVDs and better understanding of the effects of non-RVD residues on RVD behavior, as referred to above, promises a degree of fine tuning of target specificity that will be particularly advantageous for applications such as systems biology and metabolic engineering; these fields variously may require stringent specificity for orthogonal targeting, or precisely modulated specificity to target a set of closely related sequences, not unlike the targeting requirements faced by TAL effectors in nature.
TAL effectors in genome editing
The prevailing application for TAL effectors has been genome editing, as paired fusions to the catalytic domain of the dimeric endonuclease FokI. Targeted double strand breaks in DNA made using these TAL effector nucleases (TALENs) invoke cellular DNA repair pathways that can be exploited to generate small insertions or deletions for gene knockouts, or integration of a user-supplied DNA repair template for targeted gene replacement. TALENs have been used extensively in a large variety of organisms and cell types (reviewed in Ref. [20]), including several plant species [21• ,22–24,25••].
However, TALENs have recently been or will soon be largely supplanted by the RNA-based DNA targeting system CRISPR/Cas9. The CRISPR/Cas9 system, derived from a bacterial phage defense mechanism, guides Cas9, an endonuclease, to a specific DNA sequence via an RNA molecule that folds on one end to interact with the protein and hybridizes on the other end to the target DNA via Watson–Crick base pairing. Custom targeting is achieved simply by dictating the sequence of the hybridizing region, called the spacer. Several spacers can be stacked into a single guide RNA for multiplexed targeting. Spacer length is limited to 20 bases, and Cas9 functions as a monomer, so specificity is typically not as good as that of the dimeric TALENs, and targeting range is mildly constrained by a requirement for a particular 2-3 base sequence at one end called the protospacer associated motif (PAM). But for many users the ease of generating the RNA expression constructs for targeting and the ability to multiplex outweighs these disadvantages. Further, it was recently shown that the specificity disadvantage can be ameliorated by modifications to Cas9 that convert it to a nickase, requiring simultaneous targeting of paired sites on opposing strands to achieve the double strand break necessary for editing [26,27].
TAL effector-based control of gene expression
TAL effectors naturally function as transactivators of plant genes, and designer TAL effectors (‘dTALEs’) with user assembled repeat arrays in an otherwise native protein context function well as custom transcription factors in plants [13,28,29]. In other systems, the native C-terminal acidic activation domain is often replaced with an alternative activation domain (e.g., VP16 of herpes simplex virus in human cells [9]). Replacing the activation domain with a repressor domain, such as SRDX for applications in plants, has also proven robust for targeted gene repression [12,30,31]. Simply removing the activation domain has been shown to create effective transcriptional repressors in yeast [32••] and bacteria [33].
Synthetic gene promoters bound and activated specifically by different TAL effectors have also been demonstrated to work in plants [34,35•]. Multiple distinct binding sites could be stacked into such promoters. Relative activity generally correlated with distance of a site from the transcriptional start [35•].
Recently, fusions of TAL effector DNA recognition domains to the catalytic domain of the ten-eleven trans-location protein 1 (TET1) hydroxylase were shown to be effective for targeted demethylation of specific CpGs in human gene promoters, leading to increases in associated transcript levels [36•]. Targeted histone demethylation was achieved using TAL effector fusions to lysinespecific demethylase 1 (LSD1), allowing, for example, inactivation of enhancer elements to identify genes under their control [37•]. Though less well characterized, conditional fusions of TAL effector domains to a variety of other histone effectors, including deacetylases, methylases, acetylase inhibitors, and others have also been successful [38••]. The binding itself of TAL effectors to modified chromatin has not been studied in depth, but there may be limitations when the target sequence is heterchromatic [39]. RVDs N* and NG were observed to accommodate MeC, however, enabling targeting to some methylated sequences [40].
Systems biology and metabolic engineering
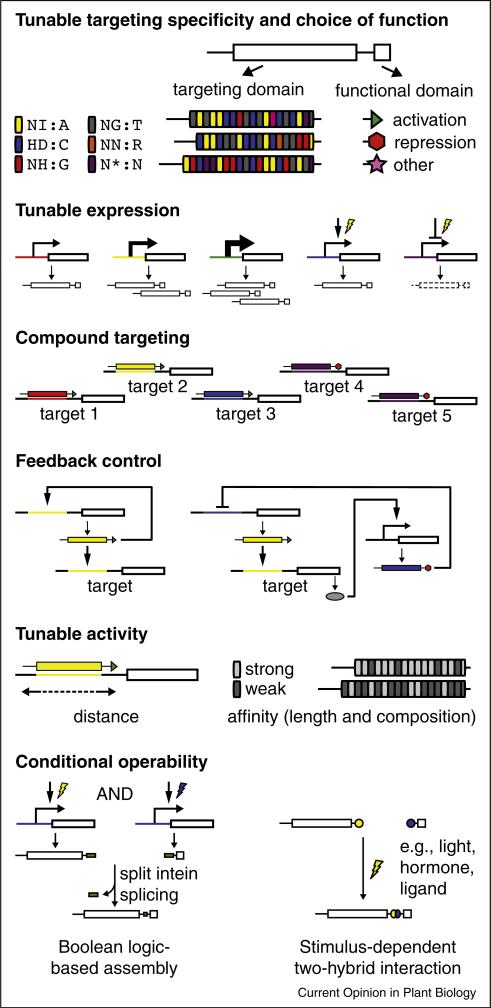
dTALEs and TAL effector-based repressors (TALERs [30], a.k.a, TALORs, i.e., TAL effector orthogonal repressors [32••]) in particular are enabling tools for systems biology (Figure 1), the engineering of gene expression circuits and switches to modify and control cellular processes. The modular targeting mechanism allows design of multiple orthogonal reagents, for exclusive targeting of distinct gene promoters, endogenous or synthetic. Activity of dTALEs or TALERs can be modulated by expressing the proteins from different strength promoters, or by calibrating their function at the target by selecting the binding site and RVD composition based on relative estimated affinity and distance of the site from the transcriptional start. Synergy among multiple TAL effector constructs targeting the same promoter can also be exploited [41• ,42•].
Figure 1.
Systems biology and metabolic engineering components feasible with TAL effector-based approaches. See text for details.
Conditional expression of the TAL effector construct can be achieved using inducible or repressible promoters. Including a dTALE binding site in the promoter of the dTALE gene itself creates a positive feedback loop. A site for a conditionally expressed TALER allows for feedback inhibition. Conditional control of activity can also be implemented at the post-transcriptional level using microRNA-based silencing of the TAL effector construct [43••]. Post-translational switches are possible as well. Leinert and colleagues [44••] recently demonstrated the feasibility of intein splicing of independently expressed intein-fused TAL effector DNA recognition and acidic activation domains to reconstitute a functional dTALE, allowing a Boolean logic based ‘AND’ switch that requires two or even three inputs for activation. And fusing light-activated, hormone-activated, or synthetic ligand-activated protein partners to different parts of the TAL effector to bring them together only in the presence of such stimuli also works [38•• ,43••].
Conditional expression might be used not only for control of target expression but in some cases for conditional alternative transcription of the target. Though the essential parameters are not completely understood, TAL effectors have been shown to initiate transcription from an alternative start site depending on the location of the TAL effector binding site [34,35• ,45,46].
Metabolic engineering
Because dTALEs and TALERs can be made orthogonal and can be expressed independently and simultaneously in the same cell, they are uniquely well-suited to metabolic engineering. Expression of genes for distinct enzymes in a complex metabolic network can be selectively upregulated or repressed in a scalable way to maximize the efficiency of some pathways, shut down others, and obtain the desired biochemical output. Not to mention, substrate specificity and activity of individual enzymes can be modified by genome editing.
Outlook
As noted earlier in this article, for genome editing, TAL effector-based reagents are likely to be replaced by CRISPR/Cas9. But what about the other types of TAL effector-based DNA targeting tools? Targeted gene activation using a modified CRISPR/Cas9 system in which Cas9 was made catalytically inactive and fused to an activation domain was recently demonstrated [27]. Though less active than dTALEs, multiple guide RNAs targeted to the same promoter were synergistic and yielded good activity. In light of this success, CRISPR/ Cas9 clearly has the potential to be developed for many of the same purposes that TAL effectors have. As a programmable complex of the three major biopolymers RNA, protein, and DNA, CRISPR/Cas9 likely will find additional, unique applications [47]. However, because guide RNA expression relies on the use of RNA polymerase III promoters, which we currently cannot manipulate for conditional expression in the same way we can manipulate RNA polymerase II promoters, and because a particular Cas9 protein will not distinguish among cog-nate guide RNAs that target different genomic loci, scalability and orthogonality depend on manipulation of the Cas9 component. A recent report identified a set of four Cas9 orthologs that recognize distinct guide RNA sequences, allowing them to be targeted independently and orthogonally [48]. Further screening of orthologs and perhaps directed evolution may yield a broader array of specificities [47]. One study reported genome editing in wheat using a guide RNA construct under the control of the 35S promoter (RNA polymerase II type) [49], but this should be independently confirmed because RNA polymerase II transcripts are rapidly shunted to the cytoplasm and would not be expected to be able to interact with the genome. For the time being, TAL effector-based DNA targeting appears to offer a distinct advantage in scalability and fine tuning of expression for systems biology and metabolic engineering.
Fine tuning of target specificity also appears to be more readily achieved with TAL effectors, though better quantification of affinity contributions of different RVD-nucleotide combinations is still necessary to take full advantage of the natural variation in targeting stringency inherent to TAL effectors. Whether CRISPR/Cas9 can be made conditionally active using inteins or other fusions and post-translational modifications remains to be seen, as does the efficacy of CRISPR/Cas9 constructs designed to modify chromatin.
Overall, TAL effectors have proven to be a broadly applicable targeting platform, useful not only in gene editing, gene regulation, and chromatin modification, but also recently for chromatin labeling [50] and immuno-precipitation of genomic regions of interest [51,52]. In summary, though CRISPR/Cas9 holds great promise of similar adaptability, the array of TAL effector based tools currently in hand is opening new doors for understanding and manipulating plant physiology and metabolism through systems biology and metabolic engineering.
Acknowledgements
Research in my laboratory on TAL effectors is funded by grants from the National Science Foundation (IOS 1238189) and the National Institutes of Health (R01 GM098861).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Nissan G, Manulis-Sasson S, Chalupowicz L, Teper D, Yeheskel A, Pasmanik-Chor M, Sessa G, Barash I. The type III effector HsvG of the gall-forming Pantoea agglomerans mediates expression of the host gene HSVGT. Mol Plant-Microbe Interact. 2012;25:231–240. doi: 10.1094/MPMI-06-11-0173. [DOI] [PubMed] [Google Scholar]
- 2.de Lange O, Schreiber T, Schandry N, Radeck J, Braun K-H, Koszinowski J, Heuer H, Strauß A, Lahaye T. Breaking the DNA binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 2013;199:773–786. doi: 10.1111/nph.12324. [DOI] [PubMed] [Google Scholar]
- 3.Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics. 2011;12:210. doi: 10.1186/1471-2164-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci U S A. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 8.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 9.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 10.Meckler JF, Bhakta MS, Kim MS, Ovadia R, Habrian CH, Zykovich A, Yu A, Lockwood SH, Morbitzer R, Elsaesser J, et al. Quantitative analysis of TALE–DNA interactions suggests polarity effects. Nucleic Acids Res. 2013;41:4118–4128. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 12.Cong L, Zhou R, Kuo Y-c, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013;200:808–819. doi: 10.1111/nph.12411. [DOI] [PubMed] [Google Scholar]
- 14.Lamb BM, Mercer AC, Barbas CF., III Directed evolution of the TALE N-terminal domain for recognition of all 50 bases. Nucleic Acids Res. 2013;41:9779–9785. doi: 10.1093/nar/gkt754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle EL, Hummel AW, Demorest ZL, Starker CG, Voytas DF, Bradley P, Bogdanove AJ. TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat — 1. PLoS ONE. 2013;8:e82120. doi: 10.1371/journal.pone.0082120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Li J, Huang H, Wang GC, Jiang MJ, Yin SY, Sun CH, Zhang HS, Zhuang FF, Xi JJ. An integrated chip for the high-throughput synthesis of transcription activator-like effectors. Angew Chem. 2012;51:8505–8508. doi: 10.1002/anie.201203597. [DOI] [PubMed] [Google Scholar]
- 20.Doyle EL, Stoddard BL, Voytas DF, Bogdanove AJ. TAL effectors: highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 2013;23:390–398. doi: 10.1016/j.tcb.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013;161:20–27. doi: 10.1104/pp.112.205179. [Using tobacco protoplasts, this study demonstrated the ability to capture targeted mutations, allelic replacements, and insertions in plants without selection, using TAL effector nucleases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendt T, Holm PB, Starker CG, Christian M, Voytas DF, Brinch-Pedersen H, Holme IB. TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol Biol. 2013;83:279–285. doi: 10.1007/s11103-013-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z, Li N, Huang G, Xu J, Pan Y, Wang Z, Tang Q, Song M, Wang X. Site-specific gene targetin using transcription activator-like effector (TALE)-based nuclease in Brassica oleracea. J Integr Plant Biol. 2013;55:1092–1103. doi: 10.1111/jipb.12091. [DOI] [PubMed] [Google Scholar]
- 24.Christian M, Qi Y, Zhang Y, Voytas DF. Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3. 2013;3:1697–1705. doi: 10.1534/g3.113.007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [The first report of genome edited whole plants, this study used TAL effector nucleases derived in part from a native TAL effector to mutate the native binding site for the TAL effector, prevent subsequent target activation during attempted infection, and confer disease resistance.] [DOI] [PubMed] [Google Scholar]
- 26.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Huang S, Zhou J, Yang B. Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv. oryzae in rice. Mol Plant. 2013;6:781–789. doi: 10.1093/mp/sst034. [DOI] [PubMed] [Google Scholar]
- 30.Crocker J, Stern DL. TALE-mediated modulation of transcriptional enhancers in vivo. Nat Methods. 2013;10:762–767. doi: 10.1038/nmeth.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, Bangarusamy DK, Zhu JK. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol. 2012;78:311–321. doi: 10.1007/s11103-011-9866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Blount BA, Weenink T, Vasylechko S, Ellis T. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS ONE. 2012;7:e33279. doi: 10.1371/journal.pone.0033279. [In this pioneering proof of concept study of TAL effectors for systems biology, carried out in yeast, a diverse set of gene promoters were synthesized and paired with corresponding, custom TAL effector-based repressors to obtain a set of orthogonal regulators of differing strength.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Politz MC, Copeland MF, Pfleger BF. Artificial repressors for controlling gene expression in bacteria. Chem Commun. 2013;49:4325–4327. doi: 10.1039/c2cc37107c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Römer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci U S A. 2009;106:20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Hummel AW, Doyle EL, Bogdanove AJ. Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 2012;195:883–893. doi: 10.1111/j.1469-8137.2012.04216.x. [Using rice, this study showed that multiple TAL effector binding sites could be stacked into a single plant gene promoter in a stably transformed plant, and that distance of a site from the transcriptional start site correlated with its relative activity (strength of gene activation).] [DOI] [PubMed] [Google Scholar]
- 36•.Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013 doi: 10.1038/nbt.2726. [One of a handful of reports establishing the utility of TAL effector targeting for site-specific chromatin modification, this study demonstrated site-specific demethylation of CpG islands in selected human gene promoters using TAL effector fusions to the TET1 hydroxylase, resulting in increases in expression of those genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013 doi: 10.1038/nbt.2701. [Another report establishing the utility of TAL effector targeting for site-specific chromatin modification, this study demonstrated site-specific histone demethylation at select enhancer loci in human cells using TAL effectors fused to the lysine specific demethylase 1 protein, inactivating the enhancers to allow identification of genes under their influence.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [This paper is of primary interest for demonstrating that by separately fusing the TAL effector DNA recognition domain and an activation domain respectively to a light sensitive protein and its interacting partner, targeted activity could be made conditional, in this case, requiring light. Conditionality is an important feature for systems biology. Also of interest, this study demonstrated successful targeting of several histone effectors, helping to establish the utility of TAL effectors for targeted chromatin modification.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, Lahaye T, Leonhardt H. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valton J, Dupuy A, Daboussi F, Thomas S, Marechal A, Macmaster R, Melliand K, Juillerat A, Duchateau P. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem. 2012;287:38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [One of the first studies demonstrating synergy and tunability of TAL effector transactivation activity at human gene promoters, using multiple custom TAL effectors each substituted with the VP64 (4 VP16) activation domain and targeted to distinct sequences in a promoter.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Maeder ML, Linder SJ, Reyon D, Angstman JF, Fu Y, Sander JD, Joung JK. Robust, synergistic regulation of human gene expression using TALE activators. Nat Methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [Published back-to-back with Ref. [41•], this study further established the robustness of TAL effector mediated gene activation in human cells, also using a VP64-substituted architecture, and similarly showed synergy among TAL effectors at the same promoter.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Li Y, Moore R, Guinn M, Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep. 2012;2:897. doi: 10.1038/srep00897. [This study demonstrated conditional control of TAL effector activity using either a two hybrid approach in one iteration of which the DNA recognition and activation domains were separately fused to components that would dimerize and reconstitute the active TAL effector only in the presence of a particular ligand, or a microRNA-mediated silencing approach in which the expression of the TAL effector was made conditional.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Lienert F, Torella JP, Chen JH, Norsworthy M, Richardson RR, Silver PA. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 2013;41:9967–9975. doi: 10.1093/nar/gkt758. [Another landmark paper in establishing the applicability of TAL effectors for systems biology, this study demonstrated successful fusions of separate TAL effector domains to inteins and invocation of intein splicing to conditionally reconstitute TAL effectors only in the presence of up to three distinct signals.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Römer P, Strauss T, Hahn S, Scholze H, Morbitzer R, Grau J, Bonas U, Lahaye T. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 2009;150:1697–1712. doi: 10.1104/pp.109.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os11N3. Plant Cell. 2010;22:3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Upadhyay SK, Kumar J, Alok A, Tuli R. RNA guided genome editing for target gene mutations in wheat. G3. 2013 doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol. 2013;20:1321–1324. doi: 10.1038/nsmb.2680. [DOI] [PubMed] [Google Scholar]
- 51.Byrum SD, Taverna SD, Tackett AJ. Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res. 2013;41:e195. doi: 10.1093/nar/gkt822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujita T, Asano Y, Ohtsuka J, Takada Y, Saito K, Ohki R, Fujii H. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci Rep. 2013;3:3171. doi: 10.1038/srep03171. [DOI] [PMC free article] [PubMed] [Google Scholar]