Abstract
Significance: Bacterial burden is believed to play a significant role in impaired wound healing of chronic wounds and the development of infection-related complications. The standard of care in the clinic relies upon cultivation-dependent methods to identify microorganisms. These assays are biased toward microorganisms that thrive in isolation under laboratory conditions.
Recent Advances: Significant advances in genomic technologies have enabled less-biased, culture-independent approaches to characterize microbial communities, or microbiomes. The aggregate sequencing and analysis of 16S ribosomal RNA genes has demonstrated that cultures under-represent true microbial diversity and load.
Critical Issues: Despite recent advances that enable culture-independent analyses of microbiomes, those organisms that are important in impaired healing remain ambiguous. Inconsistent findings across various studies highlight the need to characterize microbiomes of chronic wounds with homogenous etiology to determine differences in microbiomes that may be driven by the wound environment and that may affect wound outcomes. Rigorous analyses of wound microbiomes in light of the three dimensions of bioburden (microbial diversity, microbial load, and pathogenic organisms), clinical metadata, and wound outcomes will be a significant step forward in our quest to understand the role of microorganisms in impaired healing.
Future Directions: Longitudinal studies employing serial sampling are needed to appreciate the role of the dynamic microbial community in chronic wound healing. The value of clinical metadata needs to be examined as potential biomarkers of problematic microbiota and wound outcomes. Lastly, better characterization and understanding of wound microbiomes will open avenues for improved diagnostic and therapeutic tools for the nonhealing wound.

Elizabeth A. Grice, PhD
Scope and Significance
Microorganisms are believed to play a significant role in impaired healing of chronic wounds and the development of infection-related complications. Genomic methods of characterizing microbial communities, or microbiomes, are less biased than cultivation-based approaches and may provide a more comprehensive representation of the manner in which the microbial load, microbial diversity, and the presence of pathogens interact or converge to impact chronic wound outcomes. Here we will discuss these modern genomic methods of analyzing microbiomes and how they compare to culture-based methods for characterizing wound microbiomes. We will compare and discuss the findings of several recent studies using culture-independent methods of examining wound microbiota, focusing on those that utilize a next-generation sequencing technology, while discussing critical issues that remain to be addressed in understanding the role of the microbiome on chronic wound outcomes. Finally, we will examine the potential for improved diagnostic tools and novel therapeutic options based on the microbiome, and how they may relate to the future of wound care.
Translational Relevance
Modern genomic techniques to characterize wound bioburden, such as sequencing of bacterial 16S ribosomal RNA (16S rRNA) genes, eliminate biases associated with culture-based methods. Rapidly evolving DNA sequencing technologies, increasingly sophisticated computational approaches to analyze sequence datasets, and ever-expanding reference databases enable these methods to more precisely characterize and delineate the dynamic wound microbiome. Employing these methods has the potential to differentiate chronic wound colonization from problematic bioburden, which could ultimately guide clinical decision-making.1
Clinical Relevance
Because it is difficult to differentiate benign wound colonization from problematic bioburden using culture-based diagnostics, antibiotics are often unnecessarily given to persons with chronic wounds. This practice contributes to the growing problem of antibiotic resistance.2 However, chronic wound infections lead to complications, such as amputation, sepsis, and death.3 Culture-independent analysis of chronic wound microbiota could potentially lead to more judicial and targeted use of antibiotics than proposed in current practice guidelines, which rely on clinical signs of infection or wound cultures to drive antibiotic treatment.4 Moreover, genomic methods may lead to improved methods for diagnosing chronic wound infection and preventing infection-related complications.
Discussion of Findings and Relevant Literature
Introduction to culture-independent analysis of microbial communities
Despite 50 years of research, the role that bacterial colonization and/or infection play in wound outcomes and complications has remained elusive. Since the late 1800s, the gold standard for bacterial isolation and identification has been a culture-based methodology. Many bacterial species colonizing wounds have been identified and isolated in this manner (as reviewed by Refs.5–8). However, it is now widely accepted that culture-based techniques select for only those microorganisms that thrive under the typical nutritional and physiological conditions employed by diagnostic laboratories. Those microorganisms that thrive under these conditions are not necessarily the most abundant or influential organisms in the community. In particular, cultivation of anaerobes is problematic by routine culture-based techniques.8,9 Anaerobic organisms are postulated to be detrimental to wound repair, but require special conditions not only for growth, but also for sample transport and processing.
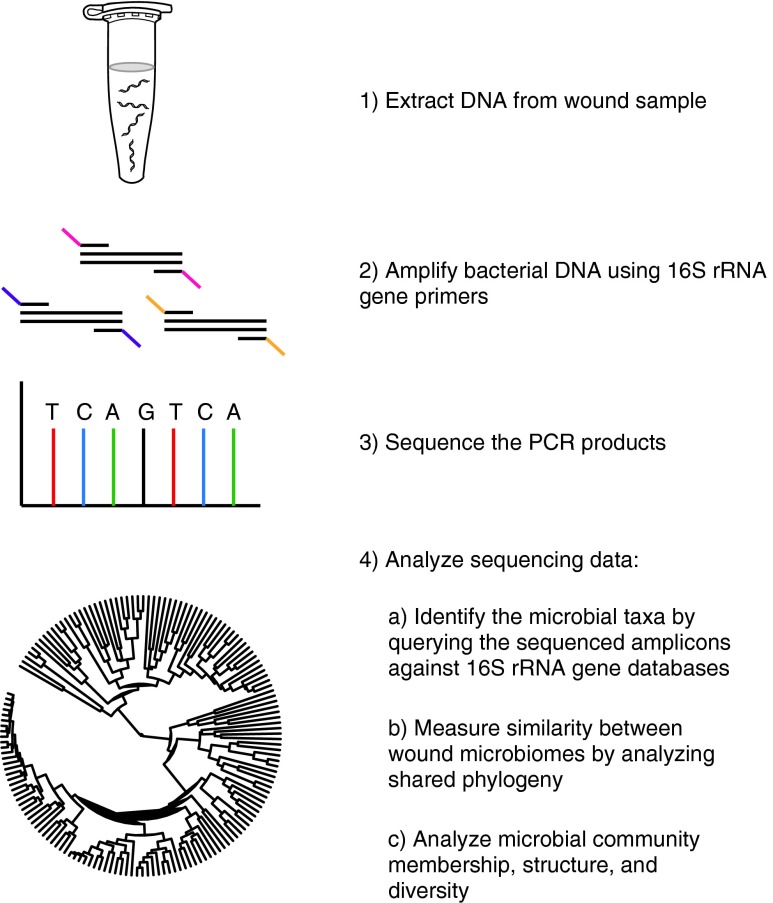
Recent advances in DNA-sequencing technology and the development of molecular techniques to identify and quantify microorganisms have revolutionized our view of the microbial world. Characterization of the bacterial microbiome takes advantage of the 16S rRNA gene, present in all prokaryotes, but not eukaryotes. The 16S rRNA gene encodes a structurally and functionally essential component of the ribosome, and contains species-specific hypervariable regions that are markers to identify bacteria, and highly conserved regions that allow broad-range amplification by polymerase chain reaction (PCR).10 Figure 1 outlines the experimental workflow of a typical bacterial microbiome-sequencing project. Following PCR amplification, 16S rRNA genes are sequenced and analyzed. Classification is enabled by databases of annotated rRNA gene sequences. For example, the Ribosomal Database Project, an online database of rRNA gene sequences, now contains >2.5 million annotated 16S rRNA sequences.11 The advent of next generation sequencing technologies and even benchtop sequencing systems (i.e., Illumina MiSeq and Life Technologies Ion Torrent platforms) has massively increased the throughput, while decreasing costs. Importantly, these techniques eliminate biases associated with cultivation-based approaches. Approaches for analyzing the fungal and viral diversity of the human microbiome are currently under development and may provide an additional insight into the role of nonbacterial microorganisms in infection-related complications and chronic wound outcomes.
Figure 1.
The workflow of a 16S ribosomal RNA (16S rRNA) gene targeted microbiome next-generation sequencing project. A heterogeneous mixture of genomic DNA is extracted from samples taken from a wound. Primers specific for the desired regions of the 16S rRNA gene are used to amplify bacterial DNA. Each sample has a unique identifying sequence on the primer known as a barcode to facilitate the multiplexing of samples on the sequencer. The resulting polymerase chain reaction (PCR) products are pooled and sequenced using platforms, such as Roche 454, Illumina MiSeq, or Life Technologies Ion Torrent. After filtering out low-quality sequences, various analyses are performed, including assignment to taxonomy, analysis of shared phylogeny, and analysis of microbial community membership, structure, and diversity. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
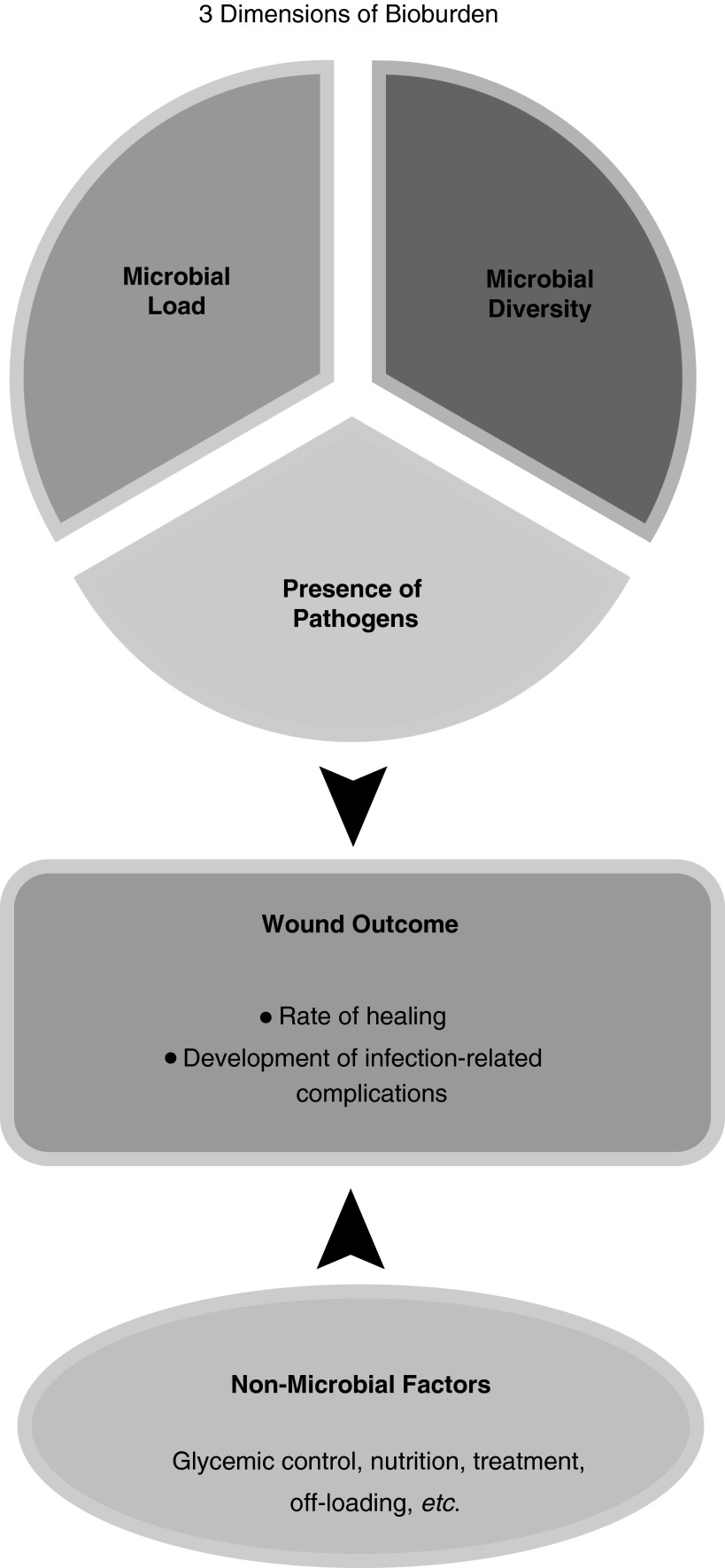
Historically, three distinct dimensions of the wound microbiota have been viewed as important in understanding the role of bioburden on wound outcomes: total microbial load, microbial diversity, and presence of pathogenic organisms (Fig. 2).6 While some studies assert that a microbial load>105 organisms per gram of tissue is related to a poor outcome,12 others have challenged this, citing interactions between different species of microbes as more important (microbe–microbe interactions or biofilms).5,9 Still others believe the presence of Staphylococcus aureus and/or anaerobes is more problematic.5 16S rRNA gene sequencing allows characterization of the microbiome based on all three dimensions: microbial load, microbial diversity, and presence of pathogens.
Figure 2.
Model of the impact of three dimensions of bioburden and nonmicrobial factors on wound outcomes. The three dimensions of bioburden are covarying components of the wound microbiome. We propose that the microbiome is one of several factors that lead to ultimate wound outcomes, such as healing or the development of infection-related complications.
Another advantage over culture-based methods is that the quantitative datasets derived from 16S rRNA gene sequencing allow each dimension, microbial load, diversity, and presence of pathogens, to be defined and measured using multiple metrics. To calculate the microbial load from real-time PCR data, a standard curve can be generated with a well-characterized isolate (i.e., Escherichia coli). The standard curve is then used to convert real-time PCR amplification data to 16S gene copy number, and from the gene copy number, the number of bacterial cells in the sample is estimated. Microbial diversity is a multidimensional measure and can encompass phylogenetic diversity, alpha diversity, and beta diversity. The phylogenetic diversity is a measure that incorporates evolutionary relationships between the organisms observed in a sample. The alpha diversity is the species diversity present in a single sample. Beta diversity is the species diversity among different samples and directly compares samples and their diversity to each other. This involves comparing the number of species common and/or unique to each sample, and their abundances. The presence of pathogens is measured by calculating the relative abundance of bacterial taxa present by comparing 16S rRNA sequences to a database of reference sequences. The relative abundance of putative pathogens in diabetic foot ulcers (DFUs; i.e., S. aureus and anaerobes) is calculated along with other bacterial taxa present, placing them in the context of the greater microbial community, while identifying additional putative pathogens among the wound microbiota.
More importantly, 16S rRNA gene sequencing allows the structure of the wound microbiome to be characterized using analyses that capture patterns that reflect varying combinations of microbial load, microbial diversity, and presence of the pathogens. To date, most studies of chronic wound bioburden have focused on one dimension of bioburden at the exclusion of the other two. Characterization of the microbiome based on all three dimensions promises to lead to a better and a more comprehensive understanding of the collective impact of the three dimensions of wound bioburden on wound outcomes than has been possible using culture-based methods.
Although 16S rRNA sequencing holds promise to move the science of chronic wound infection forward, it does have its limitations. For example, an important caveat to interpreting 16S rRNA sequence data is that it does not differentiate between viable and nonviable bacteria. It is unknown if nonviable bacteria impact wound outcomes or not. In addition, higher throughput platforms, such as Ion Torrent (Life Technologies), HiSeq and MiSeq (Illumina), and 454 (Roche), enable a much deeper sequencing of microbial communities than Sanger sequencing, but the short read lengths that are generated by these platforms are often insufficient to provide species-level discrimination. Finally, 16S gene sequence data only answers the question of “Who's there?” not “What is it doing?” nor does it provide data regarding antibiotic resistance and pathogenicity of the identified organisms.
In addition to these general limitations, there are also methodological problems to be addressed when using 16S rRNA gene sequencing to characterize the chronic wound microbiome. For example, there is little standardization among microbiome studies concerning the quality control of sequence data, usage of appropriate controls to account for contaminants and bias, and types of analyses used to assess the microbial diversity, phylogeny, and community structure. Furthermore, bias can be introduced at every step of the sample preparation, such as the DNA extraction methodology, selection of primer sequences to amplify the 16S rRNA gene, and error profiles introduced by sequencing platforms. It is therefore critical that researchers use consistent methodology if they intend to compare samples and/or studies to each other. Detailed methodology should be reported with each study published, and raw sequencing data should be freely available so that other researchers can replicate study results.
Surveying the microbiome of chronic wounds: a review of recent culture-independent studies utilizing next-generation sequencing technology
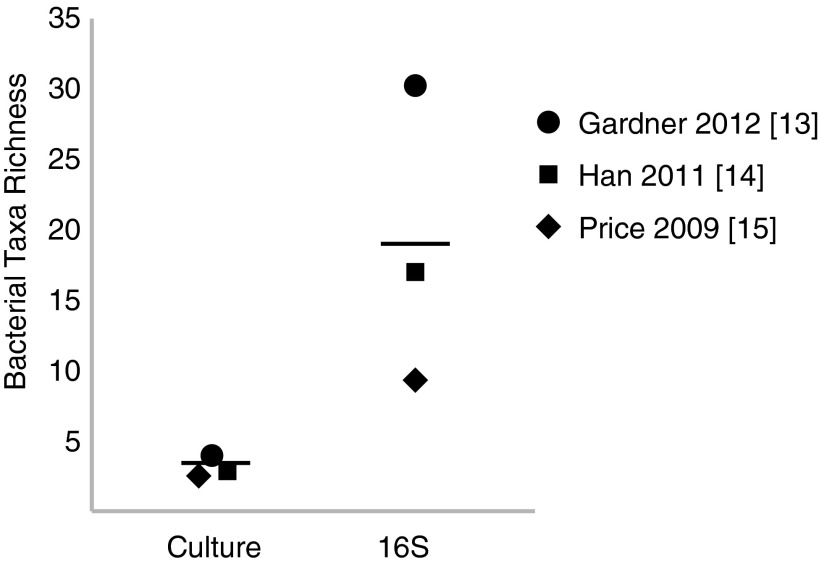
Recently, our group demonstrated that the microbiome colonizing 52 neuropathic, nonischemic DFU was associated with clinical factors.13 Not surprisingly, quantitative cultures did not fully represent DFU bioburden (microbial diversity, microbial load, and presence of pathogens) when compared to genomic techniques (Fig. 3).13–15
Figure 3.
16S rRNA gene sequencing-based methods provide a more comprehensive view of bacterial diversity compared to culture-based methods. A comparison of bacterial diversity assessed by culture-based and 16S rRNA gene sequencing results. The symbols represent the mean bacterial taxa count per sample included in each study. In Price et al.,15 the number of cultured genera was compared to the number of sequenced genera. In Han et al.,14 the number of cultured species was compared to the number of sequenced genera. In Gardner et al.,13 the number of cultured species was compared to the number of sequenced species. The horizontal line indicates the mean of the three studies surveyed. References to each study are denoted in brackets.
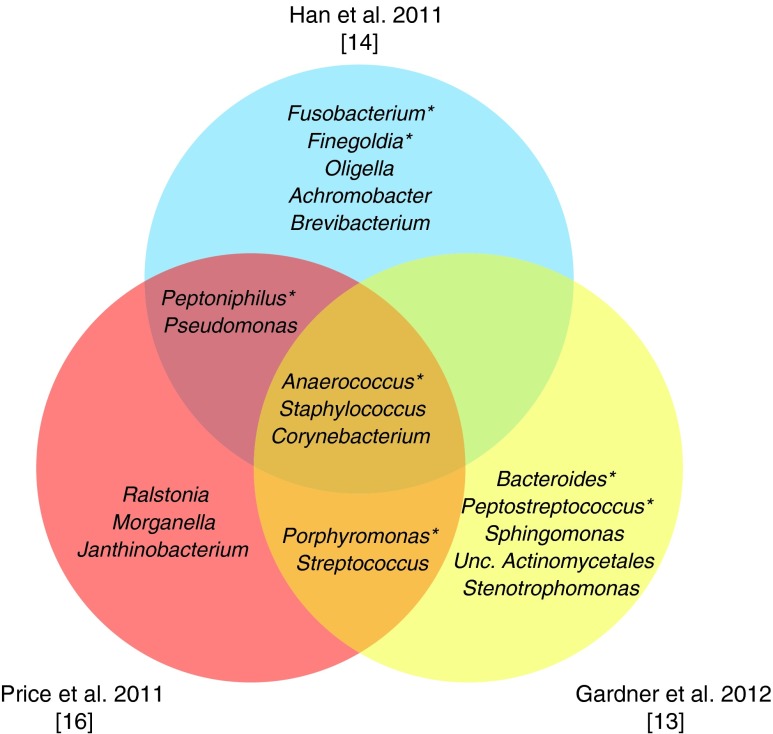
Staphylococcus, primarily of the species S. aureus, was present in 49 of 52 DFUs examined (Figs. 4 and 5).13–18 In general, relative abundance of S. aureus was negatively correlated with relative abundance of anaerobic bacteria, such as Porphyromonas, Anaerococcus, Finegoldia, Peptoniphilus, Prevotella, and Incertae Sedis XI. Deeper ulcers and those of a longer duration contained a greater microbial diversity and a higher relative abundance of anaerobic bacteria and Gram-negative Proteobacteria. Shallow ulcers and those of a shorter duration were more likely to contain a greater abundance of Staphylococcus, in most cases, the pathogenic S. aureus. Furthermore, poor control of blood glucose, as measured by hemoglobin A1c values, was associated with a higher relative abundant colonization by Staphylococcus and Streptococcus spp. Finally, the DFUs clustered into three groups based on the microbial community structure and membership. We found that these clusters were distinctive in terms of microbial load, microbial diversity, and the presence of pathogens, lending insight into the covariation of the dimensions in distinct patterns. Moreover, the clusters were associated with the glycemic control, ulcer depth, and ulcer duration identifying how dimension patterns differ along these clinical parameters.
Figure 4.
The 10 most abundant bacterial genera observed in select chronic wound microbiome publications. Each large circle represents the top 10 most abundant genera reported from the publication indicated. Only the top 10 most abundant genera are included. Staphylococcus, Corynebacterium, and Anaerococcus were common to the 10 most abundant genera in all three studies. “Unc.” denotes unclassified bacteria. *Denotes strictly anaerobic or obligately anaerobic bacteria. References to each study are denoted in brackets. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
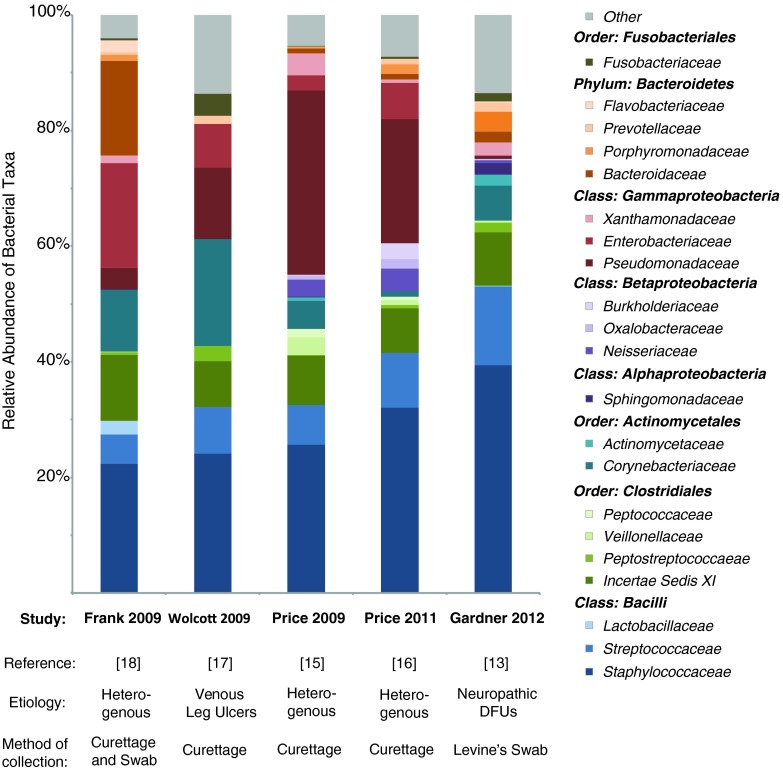
Figure 5.
Bacterial families reported to colonize chronic wounds in culture-independent studies. Twenty-one bacterial families account for the majority of microbiota colonizing chronic wounds in five studies utilizing culture-independent methods. The bacterial phyla Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria are represented. Here, we show the results of these publications as relative abundance charts, with the etiology of the wounds, and the method of collection noted. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Two other studies have examined the microbiome of DFUs using 16S rRNA gene sequencing, but did not examine the relationship between the microbiomes and other clinically relevant factors.19,20 Analyses were focused primarily on the presence of potential pathogens and did not include analyses of diversity or microbial load. In a 16S rRNA gene survey of debridement material from 40 DFUs, the most prevalent bacterial genus, Corynebacteria, was found in 75% of samples.20 Other common genera (present in at least 15 of the 40 ulcer samples) were Bacteroides, Peptoniphilus, Finegoldia, Anaerococcus, Streptococcus, and Serratia spp. This is in contrast to a study reported by the same group, published in the same year, in which Corynebacteria were absent from the top 7 reported taxa found to colonize the DFU debridement material (n=10) where the specimens from each ulcer were pooled before sequencing.19 Analyses of the community structure based on the presence of potential pathogens alone revealed 8 clusters of functionally equivalent pathogroups in 40 ulcers. That means on an average, only five ulcers were in each cluster. This is likely due to the heterogeneous nature of the ulcers in the sample as indicated by the ulcer location. For example, some ulcers were located on the ankle and dorsum of the foot indicating they may have been arterial and/or not typical DFUs.
Price et al.15 reported that Streptococcus was more prevalent in the wounds of persons with diabetes (n=12) than those free of diabetes. In this study of mixed wound types, one of the most prevalent types of bacteria, present in 25 of 32 wounds analyzed, was Clostridiales Family XI, a family of fastidious anaerobic bacteria (Fig. 5). The genera of bacteria that belong to this family, such as Anaerococcus, Peptoniphilus, Helcococcus, and Finegoldia, were not detected in parallel culture-based analyses, underscoring the utility of culture-independent examination of wound bioburden to detect those microorganisms thought to be particularly inhibitive of wound healing. In general, culture techniques underestimated an overall bacterial diversity (Fig. 3). Findings from this same study suggest that systemic antibiotics may be related to an increased relative abundance of Pseudomonadaceae (a family of Gram-negative bacteria that includes the genus Pseudomonas) in chronic wounds. This is interesting in light of the data from Gardner et al.13 that excluded subjects on antibiotics, in which Pseudomonas appears to be present in a lower relative abundance than in other studies that included subjects on antibiotics (Fig. 5). It is possible that administration of antibiotics may select for taxa such as Pseudomonas because these bacteria express abundant antibiotic resistance genes and drug efflux pumps. Antibiotic administration could also potentially select for biofilm-forming types of bacteria, such as Pseudomonas, thus structurally resisting antibiotic therapies. In another study of 13 chronic wounds of varying etiologies, Price et al. found bacterial families colonizing wounds above 1% relative abundance were (in order of decreasing abundance) Staphylococcaeae, Pseudomonadaceae, Streptococcaceae, Clostridales Incertae Sedis XI, Enterobacteriaceae, Neisseriaceae, Burkholderiaceae, Porphyromonadaceae, Oxalobacteraceae, and Corynebacteriaceae (Figs. 4 and 5).16
In a study by Han et al., microbiota colonizing 11 chronic wounds was analyzed.14 Staphylococcus was present in 10/11 wounds, but in highly variable amounts (9.6%–97% relative abundance; Figs. 4 and 5). This study also confirmed that a greater diversity of bacteria, including large numbers of anaerobic microorganisms, were detected using culture-independent approaches as compared to traditional culture-dependent methods (Fig. 3).
Critical issues to consider in wound microbiome studies
Most studies of wound microbiota have combined and analyzed heterogeneous types of diabetic ulcers (ischemic, neuropathic, and mixed type) or even heterogeneous types of chronic wounds (diabetic, pressure, and venous leg ulcers). Dissimilar wound etiology likely results in a dissimilar host/wound environment, ultimately confounding efforts to definitively associate specific microbiomes with clinical phenotypes and wound outcomes.
Another critical issue that needs to be addressed in microbiome studies of chronic wounds both cultivation-based and molecular-based is the sampling method and location. Some studies utilize debridement or curettage material to analyze microbial diversity, arguing that this is the best possible representation of wound microbiota. While some would consider punch biopsies of viable wound tissue as the best representation of wound microbiota, obtaining a punch biopsy is considered invasive, especially if employing a longitudinal study design in which the tissue needs to be removed from the wound multiple times. Furthermore, tissue specimens, whether by curettage or biopsy, contain contaminating human DNA that interferes with molecular techniques to prepare and sequence microbial DNA. Han et al. reported that 25% of their samples failed and they speculated that this might have been due to high amounts of human tissue and blood in their curettage specimens, which inhibited the PCR required to amplify the bacterial 16S rRNA gene.14 The presence of large quantities of the nonspecific template (i.e., human DNA) may outcompete bacterial DNA in the PCR reaction, in part, due to a sheer difference in the genome size. Moreover, in one study, protocols requiring the acquisition of wound tissue resulted in the loss of 28% of potential subjects because they objected to having tissue removed from their wound.21 Because longitudinal microbiome studies with larger sample sizes provide the greatest statistical power in analyzing disease-associated microbial communities, it has been recommended that researchers employ minimally invasive sample collection methods.22 Swab samples obtained by Levine's technique23 have been demonstrated to provide comparable measures of wound bioburden when compared to punch biopsies of viable wound tissue.24 Unlike other swab techniques, the Levine's technique samples wound microbiota from viable tissue by expressing tissue fluid from deep tissue layers in a one square centimeter area of wound tissue near the center of the wound.
In addition to the sampling method, there is also the question of where to sample the wound spatially. Price et al. examined a spatial variation in microbiota by obtaining curette samples in 12 subjects of a wound's leading edge, opposing leading edge, and center.16 They concluded that samples taken from different parts of the same wound are more similar than those taken from other wounds. However, they also caution that controlling for the sample site in the wound is optimal and improves the quality of the study. In comparison, a study on venous leg ulcers demonstrated that depending on the ulcer, there may be variable heterogeneity or diversity of the microbial taxa in different wound locations.17
Another challenge with existing genomic surveys of wound microbiota is associating clinical factors, such as patient and wound phenotypes, with microbiome data. If unable to access genomic techniques, clinical factors may be useful for identifying individuals at risk for problematic bioburden because the wound environment likely sustains or deters particular microbiota, which may then lead to poor wound outcomes (Fig. 2). Clinical factors that may influence wound microbiota can be extrapolated from those that have been associated with impaired wound healing. For example, poor glycemic control was found to be inversely related to the rate of healing in persons with diabetes.25 This relationship may be mediated through the influence of glycemic control on the microbiota and/or immune response within the wound environment. Another patient factor that may be related to wound microbiomes is the immune function, such as white cell count, and systemic inflammatory responses. Other potential wound factors that may alter the wound environment and influence microbial populations include wound tissue oxygen, the presence of necrotic tissue, and the location of an ulcer on the heel, the ulcer size, and ulcer duration.26–29 The wound surface area, ulcer grade, and wound duration have been associated with a failure to heal.30,31 As suggested by Christman et al.,25 it is important to study and focus on modifiable factors in wound healing that can be targeted for effective interventions. Glycemic control, smoking, antibiotic use, wound tissue oxygen, and necrotic tissue are modifiable factors that have particular potential in this regard and need to be examined more extensively in their relationship with wound microbiomes. In our study of 52 neuropathic, nonischemic DFU, we found that the glycemic control, ulcer depth, and ulcer duration were associated with the DFU microbiome.13
The potential of the microbiome as a diagnostic and therapeutic tool
The microbiome is an attractive target as a diagnostic tool, especially in the case of wounds, in that it is readily accessible, information-rich, and highly reactive to its environment/host. For example, one can imagine that the microbiome would react differently to a highly inflamed wound environment as compared to one that is devoid of inflammation or that the microbiome may drive the inflammatory state of the wound. This reaction could be measured in the types of organisms colonizing the wound, as certain organisms likely thrive in distinct environments and would therefore be expected to be more or less abundant depending upon their metabolic requirements. This reaction could also be measured in terms of the transcriptional or translational output of the microbiome, the metatranscriptome, and the metaproteome. Changes in the wound environment likely trigger responses in the microorganisms to shift the genes they express, and thus the proteins they express, such that they are able to adapt to the dynamic wound environment. We therefore expect that future studies of the chronic wound will focus on the dynamic metatranscriptome and metaproteome, to better understand the state of the microbiome and how it may relate to the wound. These studies will be critical to developing diagnostic tools that relate the microbiome to the wound outcomes and/or other clinical factors.
Therapies based on modulating the microbiome may provide an alternative to antibiotic treatments, which select for and perpetuate those strains of bacteria that are resistant to antibiotics, while causing profound nonspecific changes in the human-associated microbiome. This type of therapeutic could come in the form of a prebiotic, which would elicit changes in the wound environment thus encouraging colonization by beneficial microbes or those microbes that could prevent colonization by more pathogenic microbes. The use of probiotics for altering gut microbiota is already widespread, but the same concept could apply to wounds and other disorders of the skin. This type of therapy would directly supply specific microbiota to the environment, likely in the form of a topical ointment, in the hope that it will elicit changes in the microbiome composition or function and/or the host response.
TAKE-HOME MESSAGES.
• Culture-based approaches, the standard of care in the clinic, are biased toward those organisms that grow under standard laboratory conditions. Quantitative cultures do not fully represent the microbial diversity or microbial quantity.
• Molecular genomic methods of identifying microorganisms and delineating microbial load, such as sequencing of the bacteria-specific 16S rRNA gene, allow researchers to identify and quantify microorganisms in a less biased manner. These methods are becoming increasingly accessible, while decreasing in cost.
• Recent analyses of the microbiota colonizing neuropathic DFUs demonstrate that clinical factors, including the ulcer depth, ulcer duration, and glycemic control, were associated to various dimensions of bioburden (the microbial diversity, microbial load, and the presence of pathogenic microorganisms).
• Studies using longitudinal designs that take advantage of clinical metadata to appropriately stratify patient populations will ultimately have the most potential to reveal relationships between microbiome variation and chronic wound outcomes. Studies analyzing wounds of homogeneous etiology and pathophysiological mechanism will be critical in determining differences in the microbiome that are driven by the wound environment.
• The microbiome is a promising target for the development of diagnostic and therapeutic tools for chronic wounds.
Abbreviations and Acronyms
- DFU
diabetic foot ulcer
- PCR
polymerase chain reaction
- 16S rRNA
16S ribosomal RNA
Acknowledgments and Funding Sources
This work was supported by the National Institutes of Health: NIAMS R00 AR060873 (EAG), NINR R01 NR009448 (SEG), and NIAMS T32 AR007465 (AMM).
Author Disclosure and Ghostwriting
The authors have no competing interests. The article was written solely by its authors.
About the Authors
Elizabeth Grice, PhD, is an Assistant Professor of Dermatology at the University of Pennsylvania Perelman School of Medicine. She received her PhD in Human Genetics in 2006 from the Johns Hopkins University. She developed an expertise in culture-independent analysis of cutaneous microbiomes as a postdoctoral fellow at NIH in the laboratory of Dr. Julia Segre. Her current research interests focus on genomic and metagenomic analyses of cutaneous host–microbe interactions, especially those related to impaired wound healing. Sue Gardner, RN, PhD, is an Associate Professor in the University of Iowa College of Nursing. She received her doctorate in Nursing from the University of Iowa. Her research interests include chronic wounds, DFUs, wound infection, genomic analyses of microbial bioburden, and wound pain. Ana Misic, PhD, is a postdoctoral fellow in Dr. Grice's laboratory at the University of Pennsylvania. She received her doctorate in Biomolecular Chemistry in 2010 from the University of Wisconsin–Madison.
References
- 1.Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, and Bradley L: The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med 2007; 9:665. [DOI] [PubMed] [Google Scholar]
- 2.Rice LB: Mechanisms of resistance and clinical relevance of resistance to beta-lactams, glycopeptides, and fluoroquinolones. Mayo Clin Proc 2012; 87:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National diabetes fact sheet: national estimates and general information on diabetes and prediabetes in the United States, 2011. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2011 [Google Scholar]
- 4.Lipsky BA, Berendt AR, Cornia PB, Pile JC, Peters EJ, Armstrong DG, et al. : Executive summary: 2012 Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis 2012; 54:1679. [DOI] [PubMed] [Google Scholar]
- 5.Bowler PG. and Davies BJ: The microbiology of infected and noninfected leg ulcers. Int J Dermatol 1999; 38:573. [DOI] [PubMed] [Google Scholar]
- 6.Gardner SE. and Frantz RA: Wound bioburden and infection-related complications in diabetic foot ulcers. Biol Res Nurs 2008; 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin JM, Zenilman JM, and Lazarus GS: Molecular microbiology: new dimensions for cutaneous biology and wound healing. J Invest Dermatol 2010; 130:38. [DOI] [PubMed] [Google Scholar]
- 8.Davies CE, Wilson MJ, Hill KE, Stephens P, Hill CM, Harding KG, and Thomas DW: Use of molecular techniques to study microbial diversity in the skin: chronic wounds reevaluated. Wound Repair Regen 2001; 9:332. [DOI] [PubMed] [Google Scholar]
- 9.Bowler PG, Duerden BI, and Armstrong DG: Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001; 14:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugenholtz P. and Pace NR: Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol 1996; 14:190. [DOI] [PubMed] [Google Scholar]
- 11.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, and Tiedje JM: The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 2007; 35 (Database issue):D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robson MC, Mannari RJ, Smith PD, and Payne WG: Maintenance of wound bacterial balance. Am J Surg 1999; 178:399. [DOI] [PubMed] [Google Scholar]
- 13.Gardner SE, Hillis SL, Heilmann K, Segre JA, and Grice EA: The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 2013; 62:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han A, Zenilman JM, Melendez JH, Shirtliff ME, Agostinho A, James G, Stewart PS, Mongodin EF, Rao D, Rickard AH, and Lazarus GS: The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen 2011; 19:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, Bowers J, Rattray R, Ravel J, Kingsley C, Keim PS, Lazarus GS, and Zenilman JM: Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One 2009; 4:e6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price LB, Liu CM, Frankel YM, Melendez JH, Aziz M, Buchhagen J, Contente-Cuomo T, Engelthaler DM, Keim PS, Ravel J, Lazarus GS, and Zenilman JM: Macroscale spatial variation in chronic wound microbiota: a cross-sectional study. Wound Repair Regen 2011; 19:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolcott RD, Gontcharova V, Sun Y, and Dowd SE: Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol 2009; 9:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank DN, Wysocki A, Specht-Glick DD, Rooney A, Feldman RA, St. Amand AL, Pace NR, and Trent JD: Microbial diversity in chronic open wounds. Wound Repair Regen 2009; 17:163. [DOI] [PubMed] [Google Scholar]
- 19.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, and Wolcott RD: Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 2008; 8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, and Rhoads D: Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 2008; 3:e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner SE, Frantz RA, and Doebbeling BN: The validity of the clinical signs and symptoms used to identify localized chronic wound infection. Wound Repair Regen 2001; 9:178. [DOI] [PubMed] [Google Scholar]
- 22.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, and Knight R: Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 2012; 13:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine NS, Lindberg RB, Mason AD, Jr, and Pruitt BA, Jr: The quantitative swab culture and smear: a quick, simple method for determining the number of viable aerobic bacteria on open wounds. J Trauma 1976; 16:89. [PubMed] [Google Scholar]
- 24.Gardner SE, Frantz RA, Saltzman CL, Hillis SL, Park H, and Scherubel M: Diagnostic validity of three swab techniques for identifying chronic wound infection. Wound Repair Regen 2006; 14:548. [DOI] [PubMed] [Google Scholar]
- 25.Christman AL, Selvin E, Margolis DJ, Lazarus GS, and Garza LA: Hemoglobin A1c predicts healing rate in diabetic wounds. J Invest Dermatol 2011; 131:2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler AI, Boyko EJ, Ahroni JH, and Smith DG: Lower-extremity amputation in diabetes. The independent effects of peripheral vascular disease, sensory neuropathy, and foot ulcers. Diabetes Care 1999; 22:1029. [DOI] [PubMed] [Google Scholar]
- 27.Adam KM, Mahmoud SM, Mahadi SI, Widatalla AH, Shawer MA, and Ahmed ME: Extended leg infection of diabetic foot ulcers: risk factors and outcome. J Wound Care 2011; 20:440. [DOI] [PubMed] [Google Scholar]
- 28.Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Liechty KW, and Segre JA: Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci USA 2010; 107:14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnett A, Dave B, Ksander GA, and Vistnes LM: A concentration gradient of bacteria within wound tissues and scab. J Surg Res 1986; 41:326. [DOI] [PubMed] [Google Scholar]
- 30.Margolis DJ, Allen-Taylor L, Hoffstad O, and Berlin JA: Diabetic neuropathic foot ulcers: predicting which ones will not heal. Am J Med 2003; 115:627. [DOI] [PubMed] [Google Scholar]
- 31.Margolis DJ, Kantor J, Santanna J, Strom BL, and Berlin JA: Risk factors for delayed healing of neuropathic diabetic foot ulcers: a pooled analysis. Arch Dermatol 2000; 136:1531. [DOI] [PubMed] [Google Scholar]