Abstract
The β-site APP cleaving enzymes 1 and 2 (BACE1 and BACE2) were initially identified as transmembrane aspartyl proteases cleaving the amyloid precursor protein (APP). BACE1 is a major drug target for Alzheimer’s disease because BACE1-mediated cleavage of APP is the first step in the generation of the pathogenic amyloid-β peptides. BACE1, which is highly expressed in the nervous system, is also required for myelination by cleaving neuregulin 1. Several recent proteomic and in vivo studies usingBACE1-andBACE2-deficient mice demonstrate a much wider range of physiological substrates and functions for both proteases within and outside of the nervous system. For BACE1 this includes axon guidance, neurogenesis, muscle spindle formation, and neuronal network functions, whereas BACE2 was shown to be involved in pigmentation and pancreatic β-cell function. This review highlights the recent progress in understanding cell biology, substrates, and functions of BACE proteases and discusses the therapeutic options and potential mechanism-based liabilities, in particular for BACE inhibitors in Alzheimer’s disease.
Keywords: Alzheimer’s disease, BACE1, BACE2, protease, regulated intramembrane proteolysis, secretase
In October 2013 the first international meeting focusing on substrates, functions, trafficking and therapeutic potential of β-site APP cleaving enzyme (BACE) proteases took place in Germany at Kloster Seeon near Munich. The new and exciting data presented at the meeting prompted the present review. We start with a general introduction to BACE proteases followed by their cell biology and the complex intracellular trafficking of BACE1, which appears to be tightly linked to its ability to cleave the membrane-bound substrates. Subsequently, we describe our current knowledge about substrates and functions of BACE1, in particular in the peripheral and central nervous system (CNS), and then about substrates and functions of the homologous protease BACE2. Finally, we discuss the therapeutic potential of both proteases, highlight the current therapeutic development for BACE1 in Alzheimer’s disease (AD) and give an outlook on future BACE protease research. Regulation of BACE protease expression and activity has been partly reviewed elsewhere (Dislich and Lichtenthaler 2012; Rossner et al. 2006) and is not the topic of this review article.
β-amyloid and Alzheimer’s disease
AD is a devastating neurodegenerative disease characterized by the cerebral accumulation of two hallmark brain lesions: amyloid plaques and neurofibrillary tangles. Amyloid plaques are extracellular deposits of short 38 to 43 residue-long peptides called β-amyloid (Aβ), whereas neurofibrillary tangles are intracellular aggregates of aberrantly processed hyperphosphorylated tau, a microtubule-associated protein. Amyloid is a generic term referring to different proteins that mis-fold and self-aggregate into β-pleated sheet structures that deposit in various tissues thus causing disease, the so-called peripheral amyloidoses. Amyloid plaques define AD as an amyloidosis disease of the brain and suggest the amyloid cascade hypothesis of AD, which posits cerebral Aβ accumulation as a critical early step in AD pathogenesis that leads to neurofibrillary tangle formation, neuroinflammation, synaptic loss, neuron death, and ultimately dementia (Hardy and Selkoe 2002). If the amyloid hypothesis is true, then inhibition of cerebral Aβ accumulation should be efficacious for AD, if given early enough in the disease process.
Aβ is a normal metabolite made and secreted by most cell types, although neurons are the major producers of Aβ in the brain. Aβ is generated by endoproteolysis of the type I membrane protein amyloid precursor protein (APP; Fig. 1a). Two proteases called β- and γ-secretases cleave APP sequentially to liberate Aβ. APP is first cut by the β-secretase thus creating the amino (N)-terminus of Aβ and yielding a membrane bound carboxy (C)-terminal fragment called C99; a secreted APP ectodomain, sAPPβ is also generated (Vassar et al. 2009). Alternatively, a different protease called α-secretase may cut within the Aβ domain of APP, generating the soluble ectodomain sAPPα and the membrane bound C83 fragment, thus precluding Aβ formation. After β-secretase or α-secretase cleavages, the γ-secretase enzyme then cuts C99 or C83 to release Aβ or the non-toxic p3 fragment into the lumen of the endosome, respectively. The γ-secretase is a multi-subunit complex composed of four transmembrane proteins: presenilin, nicastrin, Pen2, and Aph1 (Sisodia and St George-Hyslop 2002; De Strooper et al. 2010). Aβ subsequently undergoes exocytosis and is secreted into the interstitial fluid of the brain. As both β- and γ-secretases are necessary for Aβ formation, these enzymes are prime drug targets for reducing cerebral Aβ levels for AD and therapeutic strategies to inhibit them are being intensely pursued. Conversely, activation of α-secretase should also lower Aβ levels, although approaches to accomplish this goal are less clear.
Fig. 1.
APP processing, FAD mutations, and β-site APP cleaving enzyme (BACE)1. (a) APP is a type-I membrane protein that is sequentially cleaved by two aspartic proteases to generate Aβ. First, the β-secretase enzyme (β) cuts APP (1) to create the N-terminus of Aβ. Two APP fragments are produced: membrane-bound C99 and secreted sAPPβ ectodomain (yellow). Second, C99 is cleaved by the γ-secretase enzyme (γ) to generate the C-terminus of Aβ. Aβ (orange) is then released into the lumen of the endosome and secreted into the extracellular medium. An intracellular domain, C59 (green), is also produced. (b) The membrane-bound APP polypeptide is represented by the gray string. APP residues that affect β-secretase processing of APP in humans are represented by gray circles, within which the wild-type residue is identified by the single-letter amino acid code. The K670N/M671L (Swedish) and A673V mutations cause FAD by increasing the rate of β-secretase cleavage and Aβ production, whereas the A673T mutation protects against Alzheimer’s disease (AD) by doing the opposite. All three mutations occur at or within one amino acid of the β-secretase cleavage site. Red, blue, and lavender notched ellipses represent α, β, and γ-secretases, respectively, cutting at their respective cleavage sites in APP. (c) BACE1 is a 501 amino acid type-I transmembrane aspartic protease. The various subdomains of BACE1 are indicated to the right of the structure. Numbers and letters refer to amino acid positions and single-letter code, respectively. The two signature aspartic protease active site motifs at positions 93 and 289 are shaded black. S–S denote positions of disulfide bridges within the catalytic domain; N represents positions of N-linked glycosylation sites; R indicates positions of acetylated arginine residues; C marks positions of S-palmitoylated cysteine residues; P indicates phosphorylation of serine 498; Ub denotes ubiquitination of lysine 501.
Human genetics has taught much about the underlying mechanisms of disease pathogenesis. In the 1970s, Brown and Goldstein discovered mutations in the LDL receptor that cause early on-set familial hypercholesterolemia, thus revealing the pathological role of high serum cholesterol levels in cardiovascular disease. These seminal studies provided the foundation for the development of one of the most widely prescribed drug classes, the statins that inhibit HMG-CoA reductase and thus lower serum cholesterol for the treatment of heart disease (Goldstein and Brown 2009). In a similar fashion, studies in the human genetics of AD have revealed that cerebral accumulation of Aβ plays a critical early role in AD pathogenesis (Tanzi 2012). Several lines of evidence draw this conclusion. First, over 200 autosomal dominant mutations that cause familial AD (FAD) have been found in the genes for APP and presenilin, the active subunit of γ-secretase. These mutations invariably lead to either increased Aβ42: Aβ40 ratio or over-production of total Aβ. Notably, the FAD mutations in APP are found near the β- and γ-secretase cleavage sites and make APP a more efficient substrate for endoproteolysis by the secretases. Of particular relevance are the K670N; M671L (Swedish) double mutation (Mullan et al. 1992a) and the A673V mutation (Di Fede et al. 2009) that are adjacent to the β-secretase cleavage site and cause FAD by increasing β-secretase processing and total Aβ production (Fig. 1b). Duplication of the APP gene in Down syndrome/trisomy 21 and rare APP locus duplications also cause FAD because of APP over-expression and total Aβ over-production. Moreover, the epsilon 4 allele of apolipoprotein E is the major genetic risk factor for late-onset AD (LOAD) and is associated with increased accumulation of cerebral Aβ. Finally, the major α-secretase enzyme in the brain, a disintegrin and metalloprotease 10 (ADAM10) (Kuhn et al. 2010; Jorissen et al. 2010), has recently been shown to harbor rare mutations in the prodomain that attenuate α-secretase activity, thereby causing increased β-secretase cleavage of APP, Aβ over-production, and LOAD (Suh et al. 2013). Together, the evidence demonstrating that AD is associated with mutations in at least five different genes that all lead to increased cerebral Aβ accumulation strongly suggests that Aβ plays a central role in AD pathogenesis.
The occurrence of mutations that cause AD implies that the converse might be true as well, namely that genetic variants exist that protect against AD. Indeed, a low-frequency mutation in APP, the A673T coding substitution, was recently shown to be associated with decreased risk of AD and reduced cognitive decline in the elderly (Jonsson et al. 2012). The A673T substitution occurs only two amino acids C-terminal to the β-secretase cleavage site (Fig. 1b) and is at the identical position as the A673V mutation that causes FAD (Di Fede et al. 2009). However, unlike A673V, APP harboring A673T is less efficiently cleaved by β-secretase, leading to a ~40% reduction in Aβ production in vitro. These results suggest that heterozygous carriers of the A673T mutation should have a life-long ~20% decrease in Aβ generation, thus protecting them from AD. Most importantly, the A673T mutation serves as proof-of-principle that modest inhibition of β-secretase cleavage of APP may prevent AD.
BACE1: the β-secretase enzyme
Soon after the characterization of APP processing and Aβ production in the early 1990s, intense efforts in academia and industry were under way to identify the β- and γ-secretase, as it was clear these enzymes are prime therapeutic targets for AD. Genetic, biochemical, and cellular assays were developed specifically for secretase identification. In 1999, five groups independently discovered the molecular identity of the β-secretase enzyme and named it BACE, Asp2, or memapsin 2 (Vassar et al. 1999; Yan et al. 1999; Sinha et al. 1999; Hussain et al. 1999; Lin et al. 2000). The groups used disparate experimental methodologies, including expression cloning, genomic strategies, and biochemical purification, to discover the β-secretase (henceforth referred to as BACE1). All arrived at the same polypeptide sequence from these different approaches, thus raising confidence that the authentic β-secretase enzyme had been identified.
Initial characterization of BACE1 demonstrated that the enzyme exhibits all the molecular and cellular properties expected for the β-secretase (Vassar et al. 2009). BACE1 is a 501 amino acid type I transmembrane aspartic protease that is related to the pepsin family and the retroviral aspartic proteases (Fig. 1c). The BACE1 catalytic domain contains two signature aspartic acid active site motifs (DT/SGS/T) that are spaced approximately 200 residues apart, similar to the pepsins. The active site of the enzyme is topologically orientated on the same side of the membrane as the β-secretase cleavage site of APP, as is required for β-secretase. Moreover, BACE1 is localized within acidic intracellular compartments including endosomes and trans-Golgi network (TGN) and has optimal enzyme activity at acidic pH, as predicted for β-secretase. Both BACE1 and β-secretase activities are relatively insensitive to the pan-aspartic protease inhibitor pepstatin. BACE1 is expressed at low levels in most cell types of the body, although it is more highly expressed in neurons, again as expected. Finally, BACE1 has the correct cleavage specificity predicted for β-secretase: BACE1 cleaves at Asp+1 and Glu+11 of the Aβ sequence; BACE1 over-expression and knockdown increases and decreases sAPPβ, C99, and Aβ production, respectively; BACE1 knockdown increases sAPPα and p3 production; BACE1 sequence specificity at P1 is Leu>>Met>Val. Taken together, these features of BACE1 made it an extremely strong β-secretase candidate.
BACE1: post-translational modifications
Similar to other aspartic proteases, BACE1 undergoes a number of post-translational modifications. BACE1 is synthesized as a zymogen in which its pre- and pro-peptide domains are removed in the endoplasmic reticulum (ER) and TGN by signal peptidase and pro-protein convertase, respectively (Benjannet et al. 2001; Bennett et al. 2000b). BACE1 also undergoes N-glycosylation at four asparagine residues (N153, N172, N223, N354) in the ER and Golgi apparatus (Haniu et al. 2000; Capell et al. 2000). During its transit through the ER, the BACE1 catalytic domain is folded and cross-linked with three disulfide bonds (C216–C420, C278–C443, C330–C380) (Haniu et al. 2000). This atypical disulfide structure, together with its transmembrane domain, makes BACE1 an unusual aspartic protease. The cytosolic domain of BACE1 can undergo phosphorylation at serine residue 498, an event that influences BACE1 trafficking in the endosomal–lysosomal system (Pastorino et al. 2002). In addition, lysine 501 in the cytosolic domain can be ubiquitinated, a process that affects BACE1 trafficking and degradation (Kang et al. 2012, 2010). In addition, BACE1 exhibits S-palmitoylation at four cysteine residues (C474, C478, C482, C485) at the junction of the transmembrane and cytosolic domains that determines localization to lipid rafts (Vetrivel et al. 2009). Finally, BACE1 may be acetylated at several arginine residues in the catalytic domain, modifications that appear to affect the regulation of the enzyme (Ko and Puglielli 2009).
Cell biology of BACE1
Cellular localization of β-secretase cleavage
BACE1 localizes to lipid rafts and this localization correlates with its β-cleavage activity toward APP. Interestingly in vitro, specific lipids that are constituents of lipid rafts stimulate BACE1 activity (Kalvodova et al. 2005). Although BACE1 is localized in various organelles, its activity is reported to be at a maximum in endosomes and to a lower extent also in the TGN. BACE1, being an aspartyl protease, requires low pH for its activity and this correlates with its localization in these two organelles that have a lumenal acidic pH (4.5–6.0) (Rajendran and Simons 2008; Kalvodova et al. 2005; Ehehalt et al. 2003). While the Swedish mutant of APP – which has a higher affinity to BACE1 than wild-type APP – can be cleaved by BACE1 in biosynthetic compartments (likely in the TGN)(Haass et al. 1995), most of the wild-type APP gets cleaved in the endocytic compartment. Besides the low endosomal pH several additional lines of evidence point to this compartment, in particular to the early endosomes (Kinoshita et al. 2003; Rajendran et al. 2006; Sannerud et al. 2011). First, the YENPTY motif in the APP cytoplasmic domain and the corresponding dileucine motif of the BACE1 cytoplasmic domain play active roles in their sorting into endosomes and inhibition of endocytosis reduces β-cleavage of APP. Second, both APP and BACE1 intermolecularly interact in endosomes. Third, retromer and retromer-associated proteins that regulate the sorting of APP from early/late endosomes, regulate BACE1-mediated cleavage of APP; other proteins that regulate the sorting of BACE1 from endosomes including Golgi-localized, gamma-ear containing, ADP-ribosylation factor binding 1 (GGA1) and GGA3 play critical roles in BACE1 cleavage. Fourth, synaptic activity that increases endocytosis also increases β-cleavage of APP. Sixth, targeting a transition-state BACE1 inhibitor to endosomes via membrane anchoring or using compounds raising the membrane-proximal endosomal pH effectively inhibit Aβ production in cultured cells and in animals (Rajendran et al. 2008; Mitterreiter et al. 2010). All this clearly suggests that BACE1 cleavage of APP mostly occurs in endosomes.
Intracellular trafficking of BACE1
The molecular events that underlie the initial sorting of BACE1 and APP to early endosomes are not fully understood. Whether BACE1 and APP are co-internalized from the cell surface or whether they have differential requirements for sorting into distinct clathrin-coated pits is still not clear. However, there is some evidence that supports the latter claim. While APP is internalized similar to tetanus toxin using cholesterol-dependent clustering and the adaptor protein-2 (Perez et al. 1999; Schneider et al. 2008), BACE1 internalization is remarkably similar to that of TfR. In addition, BACE1 sorting to early endosomes requires Arf6, again dependent on its cytosolic sorting motif pathway (Prabhu et al. 2012; Sannerud et al. 2011). It is plausible that APP and BACE1 are internalized through separate endocytic routes and that their convergence occurs at the level of either clathrin-coated vesicles or early endosomes. The existence of heterogeneity at early endosomes (Lakadamyali et al. 2006) supports the idea that APP and BACE1 separately internalize from the plasma membrane through distinct routes and then merge at the early endosomal level in order for BACE1 to cleave APP.
Once internalized into endosomes, what is the cellular fate of endosomal BACE1? BACE1 can be sorted to the Golgi via a retrograde pathway or to lysosomal compartments for degradation. Members of the GGA protein family interact with BACE1 via its acidic cluster-dileucine binding motif to regulate its trafficking from endosomes (He et al. 2005; Tesco et al. 2007; von Arnim et al. 2006) in a process also dependent on the phosphorylation status of BACE1. Interestingly both GGA1 and GGA3 negatively regulate BACE1 residency in endosomes and thus in Aβ generation. In AD cases, both proteins have been shown to be decreased confirming their involvement in Aβ generation (Kang et al. 2010; Walker and Tesco 2013). While the mechanisms underlying the decreased expression of GGA1 in AD are not fully understood, GGA3 is a caspase substrate that undergoes degradation upon apoptotic stimuli-induced caspase activation. As GGA3 is involved in endosome to lysosome sorting of BACE1, GGA3 deficiency leads to increased BACE1 levels including in endosomes and thus higher β-cleavage and Aβ generation (Kang et al. 2010).
Sorting of BACE1 to lysosomes leads to its degradation (Koh et al. 2005). Ubiquitin-proteasomal pathways have been shown to be important for its degradation, although how ubiquitination of membrane-bound BACE1 could act as a target for degradation via the proteasome is not clear. As ubiquitination is a signal for sorting into intraluminal vesicles of late endosomal compartments, BACE1 ubiquitination most likely plays a role in sorting of BACE1 to late endosomes and eventually lysosomes for its degradation (Kang et al. 2010).
Endosomal BACE1 is not only transported to the TGN and lysosomes, but also to recycling endosomes (Udayar et al. 2013). In a recent study, a paired Rab GTPase RNAi and Rab-GAP over-expression screen lead to the identification of novel membrane trafficking routes and thereby to a better understanding of the complexity of APP β-cleavage. Importantly, the recycling endosome protein Rab11 was identified as a robust player regulating β-cleavage of APP. Silencing Rab11 via RNAi reduced Aβ levels both in cell line models as well as primary neurons from wild-type mice. Mechanistic characterization suggested that BACE1 from early endosomes is recycled via Rab11-dependent recycling compartments to the cell surface and then re-internalized back to endosomes (Fig. 2).
Fig. 2.
Beta-site amyloid cleaving enzyme (BACE)1 trafficking requires recycling endosomes. BACE1 traffics from plasma membrane to early endosomes where it cleaves most of the cellular APP. From the early endosomes, BACE1 is routed to Rab11-GTPase positive recycling endosomes, through which it is sorted to plasma membrane to reinitiate another round of entry into early endosomes to cleave APP. In the absence of functional Rab11 GTPase, much of BACE1 accumulates in recycling compartments and fails to be recycled to early endosomes, as a result of which β-cleavage of APP and Aβ production are significantly decreased.
The recycling of APP or BACE1 is of interest to the AD community as inhibition of BACE1 recycling to route it for degradation could be a possibility for therapeutic intervention. On the other hand, APP recycling and sorting of APP from endosomes uses a complex machinery called the retromers. The hetero-pentameric retromer proteins associate to the cytosolic side of endosomes to mediate the retrograde transport of transmembrane proteins to the Golgi. Usually, retromers contain three VPS (Vacuolar protein sorting) proteins termed, VPS26, VPS29 and VPS35 and two sorting nexins, though the identity and the role of sorting nexins are less clear (Siegenthaler and Rajendran 2012). Loss of retromer function (VPS26, VPS35) and retromer-associated proteins (Sorting protein-related receptor containing LDLR class A repeats) has been correlated with risk for Alzheimer’s disease and elevated levels of Aβ peptide (Andersen et al. 2005; Rogaeva et al. 2007). This suggests that while retromers and retromer-associated proteins sort uncleaved APP from early endosomes to TGN (Andersen et al. 2005; Small and Gandy 2006; Rogaeva et al. 2007; Siegenthaler and Rajendran 2012), Rab11-dependent pathway recycles cellular BACE1 to early endosomes. Interestingly, a recent live cell imaging study in hippocampal neurons showed that upon internalization from the dendritic surface, BACE1 undergoes exclusive retrograde transport to the soma and that this polarized transport in dendrites requires Eps15 homology domain (EHD) proteins 1/3, whereas axonal BACE1 sorting requires Rab11 GTPase activity. Thus Rab11 and EHD proteins coordinate trafficking and axonal transport of endocytic BACE1 in recycling endosomes in polarized hippocampal neurons (Buggia-Prevot et al. 2013, 2014). Intriguingly, a variant of Rab11 has recently been identified to be associated with late-onset AD thus linking trafficking of BACE1, Aβ generation and AD (Udayar et al. 2013).
BACE1 in vivo validation
Soon after the discovery of BACE1, a related membrane-bound aspartic protease, BACE2, was identified that shares ~ 64% amino acid similarity to BACE1. The high homology between BACE1 and BACE2 suggested BACE2 might also serve as a functional β-secretase enzyme in the brain. However, unlike β-secretase, BACE2 is expressed at low levels in neurons compared to BACE1 (Bennett et al. 2000a; Laird et al. 2005). Furthermore, although BACE2 can generate Aβ in vitro, it prefers to cleave APP at residues Phe+19 and Phe+20 of Aβ (Farzan et al. 2000; Yan et al. 2001; Fluhrer et al. 2002; Basi et al. 2003). Thus, BACE2 behaves more like α-secretase in that it precludes Aβ formation. Taken together, the characteristics of BACE2 suggest it is unlikely to be a major β-secretase in the brain.
Nevertheless, it was critical to validate BACE1 as the major cerebral β-secretase in vivo. To do so, several groups employed gene targeting technology to generate BACE1 knockout (−/−) mice (Luo et al. 2001; Roberds et al. 2001; Cai et al. 2001; Dominguez et al. 2005). Initial reports suggested that BACE1−/− mice lacked an overt phenotype and were viable, fertile, and normal appearing in gross morphology, behavior, tissue histology, and blood cell and clinical chemistry. These results implied that BACE1 inhibitor drugs should be free of mechanism-based side-effects. Well-established lines of APP transgenic (Tg) mice have been generated that develop amyloid plaques with age. Several of these APP Tg lines were crossed with BACE1−/− mice to produce APP Tg/BACE1−/− bigenic mice that were shown to lack Aβ production, amyloid deposition, and Aβ-dependent memory deficits (Ohno et al. 2004; Luo et al. 2003; Laird et al. 2005; Ohno et al. 2007; McConlogue et al. 2007). These results demonstrate that BACE2 does not compensate for BACE1 deficiency for the generation of Aβ. In addition, they validate BACE1 as the major β-secretase enzyme in the brain and suggest that BACE1 inhibition should be efficacious for lowering cerebral Aβ levels in AD.
BACE protease substrates – overview
The function of a protease is determined by its substrates. Initially, APP was the only known BACE1 substrate and early reports suggested BACE1 knockout mice were normal. However, now it is evident that there are complex BACE1 null phenotypes that provide insights into important BACE1 physiological functions in vivo. Identifying all BACE1 substrates and understanding how BACE1 cleavage modulates their functions is essential as it will be these functions that will also be blocked by therapeutic BACE1 inhibition. Likewise, new phenotypes and substrates are being discovered using BACE2-deficient mice. We start by describing our current knowledge about substrates and functions of BACE1 in the peripheral nervous system, followed by the CNS. Other phenotypes in BACE1-deficient mice, which appear to be independent of the nervous system, such as increased lethality in the first month after birth and protection against diet-induced obesity will not be discussed in detail (Dominguez et al. 2005; Meakin et al. 2012). Likewise, substrates which are mostly expressed outside of the nervous system have been listed in a recent publication (Dislich and Lichtenthaler 2012). Afterward, substrates and functions of the homologous protease BACE2 will be described.
BACE1 substrates and functions in the peripheral nervous system
Hypomyelination in BACE1−/− animals
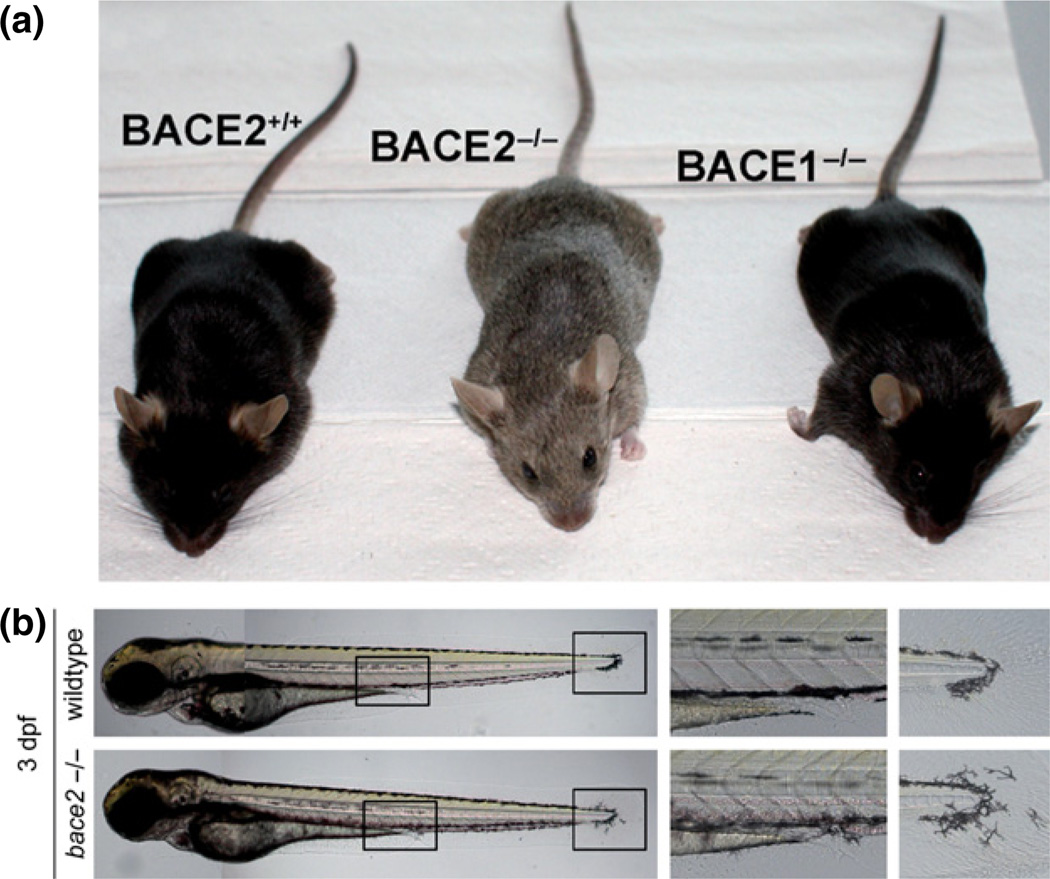
One of the best-understood functions of BACE1 is its role in the proteolytic processing and activation of neuregulin 1 (NRG1) type III (Willem et al. 2009; Fleck et al. 2012). In BACE1 knock-out mice, loss of cleavage of this substrate could be clearly functionally linked to a loss-of-function in a signaling pathway of pivotal importance during post-natal myelination (Fleck et al. 2012; Willem et al. 2009). A first hint for such a function came from a very simple experiment. A developmental western blot revealed an extremely high expression of BACE1 during the first post-natal week (Willem et al. 2006). Subsequently, BACE1 expression decreases and in adulthood only very little BACE1 could be detected. This finding along with the observations that BACE1 is preferentially expressed in neurons, myelination occurs right after birth and the major signaling pathway involved in myelination (NRG signaling) appears to require proteolytic activation (Birchmeier and Nave 2008), led to the hypothesis that BACE1 might be the protease required to regulate axonal myelination. Indeed, detailed electron microscopic analyses of sciatic nerves revealed two very striking phenotypes (Willem et al. 2006; Hu et al. 2006). First, axons in BACE1−/− animals displayed a hypomyelination phenotype (Fig. 3a; left panel); second, sorting of small-diameter axons by Schwann cell processes within Remak bundles was dramatically disturbed (Fig. 3a; right panel). Axons within Remak bundles were not separated from each other by Schwann cells, and the number of axons within Remak bundles was significantly increased (Fig. 3a; right panel). Strikingly, these myelination abnormalities within the peripheral nervous system (PNS) phenocopied a well-known knockout phenotype of one of the central players in the regulation of myelination – namely Nrg 1 type III (Birchmeier and Nave 2008; Garratt et al. 2000; Michailov et al. 2004; Taveggia et al. 2005). The neurotrophic members of the Nrg1 family signal to ErbB receptors and are involved in cardiac and neuronal development. The Nrg1 type III isoform is preferentially expressed in neurons, and axonally located Nrg1 binds to ErbB2/3 or ErbB4 receptors expressed on oligodendrocytes and Schwann cells, respectively. In line with that, reduction of Nrg1 type III in a heterozygous knockout model (Michailov et al. 2004; Taveggia et al. 2005) (the homozygous knockout is embryonically lethal) results in hypomyelination and disturbed formation of Remak bundles as described above for the BACE1 knockout. Only BACE1 but not BACE2 contributes to myelination (van Bebber et al. 2013; Willem et al. 2006; Rochin et al. 2013). Whereas the BACE1 knockout-dependent hypomyelination phenotype within the PNS could be observed in several independent BACE1−/− lines (Willem et al. 2006; Hu et al. 2006), inconsistent findings regarding BACE1 function in myelination of the CNS were obtained. Whereas one study failed to observe hypomyelination within the CNS (Willem et al. 2006), another study reported not only reduced myelination in the PNS but also within the optic nerve, hippocampus and cerebral cortex (Hu et al. 2006). To provide further insight into this issue, BACE1 knockout zebrafish have recently been generated via genome editing (van Bebber et al. 2013). Crossing of BACE1 knockout zebrafish with a reporter line, which expressed green fluorescent protein under the myelin specific claudin K promotor, allowed in vivo imaging of myelination. This model revealed a selective loss in myelination of the PNS-derived posterior lateral line nerves upon BACE1 deletion while myelination of CNS derived Mauthner axons ensheathed by oligodendrocytes was normal (Fig. 3b). Furthermore, an anti-sense gripNA-mediated knockdown of NRG1 type III in zebrafish also revealed selective hypomyelination of the PNS further supporting that BACE1 selectively affects PNS but not CNS myelination (van Bebber et al. 2013), at least in zebrafish.
Fig. 3.
Beta-site amyloid cleaving enzyme (BACE)1 controls peripheral nerve myelination and muscle spindle formation via proteolytic processing of neuregulin 1. (a) BACE1 deficiency results in hypomyelination of peripheral axons (left panel) and abnormalities in axonal-bundling (right panel) (Willem et al. 2006). (b) Delay of myelination in BACE1−/− zebrafish (van Bebber et al. 2013). At 3 days post-fertilization myelination of the posterior lateral line nerves (PNS) is severely reduced in BACE1−/−; claudin k: GFP (red arrows) whereas CNS derived oligodendrocytes ensheathing the Mauthner axons are normally myelinated (black arrows). (c) Proteolytic processing of neuregulin 1 (NRG1) type III by sheddases and intramembrane proteolysis.
Proteolytic processing of NRG1 type III by BACE1 and other sheddases
Alternative splicing generates numerous isoforms of NRG1 (Willem et al. 2009; Fleck et al. 2012). All isoforms contain an epidermal growth factor (EGF) domain, which binds and activates ErbB receptors (Falls 2003). However, signaling is only possible if the EGF domain is liberated or exposed by proteolytic cleavage. While most Nrg1 isoforms contain only one transmembrane (TM) domain, Nrg1 type III forms a hairpin-like protein with two TM domains (Falls 2003) (Fig. 3c). Nrg1 type III undergoes regulated intramembrane proteolysis (RIP) by a rather large variety of different sheddases and intramembrane cleaving proteases and may thus represent a “super-RIP” substrate (Willem et al. 2009; Fleck et al. 2012). The identification of Nrg1 type III as a physiological BACE1 substrate has already predicted that besides BACE1 other alternative sheddases cleave and activate Nrg1 type III, as the BACE1 knockout did not present with a complete amyelination phenotype but rather with hypomyelination (Willem et al. 2006; Hu et al. 2006). Indeed, studies over-expressing and knocking down/inhibiting sheddases of the ADAM (a disintegrin and metalloproteinase) family showed that the stalk region of Nrg1 type III is also cleaved by ADAM10 and ADAM17 (Fleck et al. 2013; Luo et al. 2011; La Marca et al. 2011) (Fig. 3c) in a manner very similar to the proteolytic processing of APP (Haass 2004). All three cleavages allow exposure of the EGF-like domain and consequently signaling to ErbB receptors (Fleck et al. 2013; Luo et al. 2011). The remaining C-terminal stub is then cleaved within its TM domain by γ-secretase, producing a secreted amyloid β-peptide like fragment (Nrg1 β-peptide; Fig. 3c) as well as an intracellular C-terminal cytoplasmic domain (Dejaegere et al. 2008) (C-ICD; Fig. 3c). The C-ICD translocates to the nucleus and may act as a transcriptional regulator in potential reverse signaling pathways (Bao et al. 2004, 2003; Chen et al. 2010). More recently, it was shown that the remaining membrane bound N-terminal fragment (NTF) produced by cleavage of either ADAM10, 17, or BACE1 undergoes further proteolytic cleavage to liberate a soluble (secreted) fragment containing the EGF-like domain (sEGF) (Fleck et al. 2013). Strikingly, a second cleavage site for BACE1 could be identified within the NTF of Nrg1 type III. Moreover, cleavage at the identified cleavage site appears to be very efficient that exactly matches the sequence of the Swedish mutation of APP, which is known to dramatically increase BACE1-mediated cleavage (Citron et al. 1992). BACE1 cleavage at this position of Nrg1 type III results in secretion of sEGF (Fig. 3c), which could be identified in conditioned media of primary neurons (Fleck et al. 2013). Similar to the multiple cleavages of Nrg1 type III within the stalk region by several sheddases the NTF is not only processed by BACE1 but can also be processed by ADAM17 (Fig. 3c; but not by ADAM10) (Fleck et al. 2013). The observation that dual cleavage of Nrg1 type III liberates its EGF-like domain (sEGF) raised the question whether this proteolytic fragment retains its signaling capacity. Indeed, monitoring the phosphorylation state of ErbB3 receptor and AKT in reporter cell lines and primary Schwann cells upon treatment with sEGF revealed that this fragment retains its signaling capacity in vitro. Moreover, injection of in vitro transcribed mRNAs, only encoding the sEGF-like domain, into oocytes from BACE1 knockout zebrafish (van Bebber et al. 2013) (see above) allowed rescue of the hypomyelination phenotype (Fleck et al. 2013).
A previous study suggested that the ADAM17 cleavage at the C-terminus of the EGF-like domain abolishes Schwann cell myelination (La Marca et al. 2011). However, the rescue experiments in cultured cells and in vivo using mRNAs/cDNAs encoding fragments mimicking cleavage by BACE1 or ADAM17 demonstrated that both Nrg1 derivatives are fully capable of supporting Nrg1 signaling and myelination (Fleck et al. 2013). Thus, all sheddases reported to process Nrg1 type III appear to exert redundant (activating) functions, however, BACE1 seems to be the dominant protease activating Nrg1 signaling in the PNS.
Finally, the membrane retained NTFs, which are generated either by BACE1 or ADAM17 cleavage undergo further cleavage by proteases of the signal peptide peptidase-like family (Voss, Fluhrer & Haass, unpublished observation). After intramembrane proteolysis a small peptide is secreted (C-peptide; Fig. 3c), and an N-terminal ICD (N-ICD) is released from the membrane into the cytoplasm. Whether the N-ICD has a signaling function is currently not known.
Proteolytic processing of NRG1 type I by BACE1 is required for muscle spindle formation and maintenance
Similar to NRG1 type III, NRG1 type I, which contains an Ig-like domain instead of the cysteine rich domain of NRG1 type III, is also proteolytically processed by BACE1. NRG1 type I has a large luminal N-terminal domain but lacks the second transmembrane domain, which allows NRG1 type III to form its hairpin structure (see above). Upon fusion of secreted alkaline phosphatase to NRG 1 type I and co-expression of BACE1 secreted alkaline phosphatase activity was observed in conditioned media, which could be inhibited by the addition of a BACE inhibitor (Cheret et al. 2013). As previous experiments had already shown that reduction in NRG1 in neurons or its receptor ErbB2 in muscles reduces muscles spindle formation (Andrechek et al. 2002; Hippenmeyer et al. 2002; Leu et al. 2003), these in vitro experiments strongly suggested that BACE1 might have yet another NRG1-associated function. Indeed, muscle spindle formation is gradually reduced by deletion of one or both copies of BACE1 (Cheret et al. 2013). Consequently, BACE1 deficiency in mice results in defects in coordinated movement. Moreover, pharmacological reduction in BACE1 activity in adult mice with a BACE inhibitor also reduced the number of muscle spindles (Cheret et al. 2013). Thus BACE1-mediated processing of NRG1 type 1 is not only required for the formation of muscle spindles during development but also for their maintenance during adulthood.
Therapeutic inhibition of BACE1 cleavage and NRG signaling
Finally, although BACE1 knockouts have profound and highly reproducible effects on myelination of the peripheral nervous system, this may not be a severe concern for BACE inhibition in human patients. Clearly, the BACE1 knockout phenotype on myelination is a developmental phenotype. Thus hypomyelination may not be a major concern for BACE1-directed therapies. Whether the adult function of BACE1 in spindle maintenance (Cheret et al. 2013) would be affected by BACE inhibitors in humans, must, however, be carefully monitored.
BACE1 functions in the CNS
BACE1 is concentrated in pre-synaptic terminals of different neuron types in the CNS and PNS (Kandalepas et al. 2013; Deng et al. 2013), suggesting a role for BACE1 in synaptic function. Consistent with high neuronal expression and presynaptic localization of BACE1, recent studies of BACE1−/− mice have revealed complex neurological phenotypes, many of which involve the CNS (Table 1). BACE1 knockout phenotypes are the result of deficient β-secretase processing of BACE1 substrates, and recent proteomic studies in cultured neurons have identified a large number of novel BACE1 substrates that are involved in neuronal functions (Kuhn et al. 2012; Zhou et al. 2012).
Table 1.
BACE1 null phenotypes in CNS
| Phenotype | Substrate | References |
|---|---|---|
| Astrogenesis increase, neurogenesis decrease | Jag1 | Hu et al. (2013) |
| Axon guidance defects | CHL1 | Rajapaksha et al. (2011), Cao et al. (2012), Hitt et al. (2012) |
| Hyperactivity | NRG1 | Dominguez et al. (2005), Savonenko et al. (2008) |
| Hypomyelination | NRG1 | Willem et al. (2006), Hu et al. (2006, 2008) |
| Memory deficits | – | Ohno et al. (2006, 2007, 2004), Laird et al. (2005) |
| Neurochemical deficits | – | Harrison et al. (2003) |
| Neurodegeneration w/ age | NaVβ2 | Hu et al. (2010) |
| Post-natal lethality, growth retardation | – | Dominguez et al. (2005) |
| Retinal pathology | VEGFR1 | Cai et al. (2012) |
| Schizophrenia endophenotypes | NRG1 | Savonenko et al. (2008) |
| Seizures | NaVβ2 | Kim et al. (2007), Kobayashi et al. (2008), Hu et al. (2010), Hitt et al. (2010) |
| Spine density reduction | NRG1 | Savonenko et al. (2008) |
The first phenotypes identified in BACE1−/− mice involved poor performance on spatial and temporal hippocampus-dependent memory tests (Kobayashi et al. 2008; Ohno et al. 2006, 2007, 2004; Laird et al. 2005), timid behavior (Harrison et al. 2003), and reduced serotonin and dopamine levels in hippocampus and striatum, respectively (Harrison et al. 2003). These behavioral and neurochemical phenotypes strongly suggest functions for BACE1 in specific neuronal systems of the brain, although the culpable BACE1 substrates have not been definitively identified. BACE1−/− mice also exhibit increased post-natal lethality and growth retardation (Dominguez et al. 2005), but whether these phenotypes are related to a CNS function of BACE1 is unclear.
Interestingly, lack of NRG1 processing in BACE1−/− mice, which has been described in the previous paragraphs for the peripheral nervous system, has also been associated with schizophrenia endophenotypes such as impaired pre-pulse inhibition, hypersensitivity to glutamatergic psychostimulants, spine density reduction, and hyperactivity (Savonenko et al. 2008). Overall, these NRG1 studies suggest that BACE1, in collaboration with ADAM proteases, cleaves NRG1 to release an EGF-like domain that activates ErB receptors on neighboring cells (Fleck et al. 2013; Luo et al. 2011), a signaling pathway that appears to be involved in both myelination and schizophrenia.
In addition, BACE1−/− mice exhibit spontaneous seizures and hippocampal neuron loss that increase with age, compared with wild-type littermates (Hitt et al. 2010; Hu et al. 2010; Kobayashi et al. 2008). Both generalized tonicclonic and absence seizures are observed. Also, kainate treatment induces more severe seizures and greater excitotoxic CA1 neuron death in BACE1−/− mice. BACE1 null neurons display elevated sodium current and action potential properties (Hu et al. 2010). Notably, the density of voltage-gated sodium channels (NaV) on the BACE1−/− neuron cell surface is increased, consistent with elevated sodium currents in BACE1 null neurons. Previous studies have shown that NaV β-subunits regulate the expression and cell-surface localization of sodium channels (Isom 2001; Kim et al. 2007). Moreover, β-subunits are BACE1 substrates (Gersbacher et al. 2010; Wong et al. 2005), suggesting that BACE1 processing of β-subunits controls cell-surface NaV channel density, neuronal excitability, and seizure susceptibility (Kim et al. 2011).
It is plausible that phenotypes displayed by BACE1−/− mice could be related to physiological functions of BACE1-processed APP and amyloid precursor like protein (APLP) fragments. However, studies of APP, APLP1, and APLP2 knockout mice largely suggest this is not the case. Although APP−/− mice are viable and fertile, they display a number of subtle phenotypes including reduced brain and body weight, agenesis of the corpus callosum, increased susceptibility to kainite-induced seizures, reduced locomotor activity, GABAergic abnormalities, increased L-type calcium channel levels, long-term potentiation defects, and spatial memory impairment, among others (Muller et al. 1994; Zheng et al. 1995; Li et al. 1996; Steinbach et al. 1998; Dawson et al. 1999; Magara et al. 1999; Phinney et al. 1999; Seabrook et al. 1999; Ring et al. 2007; Yang et al. 2009a). Although some APP−/− phenotypes are shared with BACE1−/− mice (e.g., reduced weight, seizures, memory deficits), most BACE1 null phenotypes are different. This is not surprising given the large set of diverse BACE1 substrates that might be functionally compromised in BACE1−/− mice.
Reminiscent of APP−/− mice, APLP1−/− and APLP2−/− mice display a subtle growth deficit and no phenotype, respectively (von Koch et al. 1997; Heber et al. 2000). In contrast, double (APLP2/APLP1 and APLP2/APP) or triple (APP/APLP1/APLP2) knockout mice die shortly after birth and show malformed neuromuscular junctions with reduced synaptic vesicle densities (von Koch et al. 1997; Heber et al. 2000; Herms et al. 2004; Wang et al. 2005; Yang et al. 2005). Like the single knockouts, APLP1/APP double knockout mice have subtle phenotypes, suggesting redundancy between APLP2 and the other family members (Heber et al. 2000). In addition to post-natal death and neuromuscular junction abnormalities, APP/APLP1/APLP2 triple knockout mice display cortical dysplasia reminiscent of type II lissencephaly in humans with partial loss of Cajal Retzius cells (Herms et al. 2004). Together, the phenotypes of the APP compound knockout mice suggest an important role for the APP family in synaptic function and maintenance.
The question of whether α- and β-secretase cleaved APP extracellular fragments have physiological functions in vivo has been recently addressed by the generation of knockin mice that solely express either the sAPPα or sAPPβ secreted ectodomain. Remarkably, the sAPPα knockin completely rescues all of the phenotypes displayed by APP−/− mice (Ring et al. 2007). In addition, the sAPPα knockin prevents the postnatal lethality shown by APLP2/APP double knockout mice, but it cannot rescue the neuromuscular junction abnormalities, spatial memory impairments, and long-term potentiation deficits of these mice (Weyer et al. 2011). Interestingly, the sAPPβ knockin is not able to rescue either the APP single or the APLP2/APP double knockout phenotypes, suggesting that sAPPα is a more physiologically relevant APP fragment than sAPPβ (Li et al. 2010). These results also suggest that both APP and APLP2 collaborate synergistically to support synaptic function in the CNS and PNS.
It has also been reported that BACE1−/− mice display retinal pathology involving lipofuscin accumulation and degeneration of retinal cell layers, phenotypes that are associated with reduced retinal microvasculature (Cai et al. 2012). In cell culture, vascular endothelial growth factor receptor 1 is processed by BACE1, suggesting that this BACE1 null retinal phenotype may result from insufficient vascular endothelial growth factor receptor 1 ectodomain shedding. However, at the recent BACE meeting several groups reported their unpublished data, which demonstrated that the retinal phenotype is not seen in all of the differently generated BACE1−/− mice. The molecular cause of these differences is presently unclear.
Defects in axon guidance and neurogenesis in the CNS of BACE1−/− mice
Another significant phenotype of BACE1−/− mice involves axon guidance defects in the hippocampus and olfactory system (Rajapaksha et al. 2011; Cao et al. 2012; Hitt et al. 2012). The mossy fiber axon pathway from dentate gyrus granule cells to CA3 pyramidal neurons is critical for learning and memory and displays one of the highest BACE1 levels in the brain. The majority of proximal mossy fiber axons pass along the CA3 pyramidal cell layer on the side with primary dendrites. However, a subset of axons called the infrapyramidal bundle (IPB) runs along the opposite (axonal) side of the CA3 cell layer for a given distance before it crosses the cell layer to join the majority of axons in the stratum lucidum. IPB length is stereotypical for a given mouse strain and correlates with memory performance: mouse strains with long IPBs obtain superior scores on hippocampus-dependent memory tests, whereas strains with short IPBs perform poorly (Crusio and Schwegler 2005). Intriguingly, BACE1−/− mice have IPBs that are ~ 30% shorter than wild-type littermates (Hitt et al. 2012), an observation consistent with BACE1 null memory deficits. In addition to short IPB length, BACE1−/− mice also display pre-mature IPB axon cross-overs of the CA3 cell layer.
In the olfactory system, olfactory sensory neurons (OSNs) project axons from the olfactory epithelium to the olfactory bulb (OB). A given OSN expresses only one of ~1000 different odorant receptor genes in a random pattern in the olfactory epithelium, yet it is able to send its axon to a topographically fixed glomerulus in the OB that represents the odor quality of the odorant molecule ligand (Sakano 2010). In wild-type mice, axon guidance of OSN axons to correct glomeruli is exquisitely precise. In contrast, BACE1 null OSN axons exhibit mis-targeting to incorrect glomeruli in a dorsal to ventral gradient in the OB (Rajapaksha et al. 2011; Cao et al. 2012), suggesting that the dependence of olfactory axon guidance on BACE1 varies according to the expressed odorant receptor. Although the significance of this finding is not yet clear, it implies that BACE1 inhibition may not affect the axon guidance of all OSNs.
The recent identification of the neural cell adhesion molecule close homolog of L1 (CHL1) as a BACE1 substrate in cultured neurons (Kuhn et al. 2012; Zhou et al. 2012) has led to important insights into the molecular basis of BACE1-dependent axon guidance. CHL1 is a type I membrane protein involved in axon outgrowth and neuronal survival (Naus et al. 2004). ADAM8 processing of CHL1 releases a soluble ectodomain fragment that may interact with neuropilin-1 and semaphorin 3A to influence axon guidance. Importantly, CHL1−/− mice have axon guidance defects in the hippocampus and OB (Heyden et al. 2008; Montag-Sallaz et al. 2002) that are highly similar to those observed in BACE1−/− mice (Hitt et al. 2012). These results suggest that release of soluble CHL1 ectodomain as a result of BACE1 processing has an important signaling role in axon guidance of hippocampal mossy fibers and OSN axons that may explain axon mis-targeting in BACE1 null mice.
Neurogenesis is a process critical for learning and memory that occurs both during development and in the adult. Interestingly, BACE1−/− mice exhibit a decrease in hippocampal neurogenesis that is accompanied by a corresponding increase in astrogenesis during post-natal development (Hu et al. 2013). This BACE1 null phenotype is associated with increased levels of Notch 1 signaling and full-length Jagged 1 protein, the Notch 1 ligand. In cell culture, BACE1 cleaves Jagged 1, proving that it is a BACE1 substrate. Thus, BACE1 may regulate post-natal neurogenesis and astrogenesis by modulating Notch 1 signaling.
Taken together, the complex neurological phenotypes of BACE1−/− mice suggest that BACE1 has diverse physiological functions in the CNS that result from deficient β-secretase processing of multiple BACE1 substrates. An important question remaining is whether these BACE1 null phenotypes derive from lack of BACE1 during development or in the adult. If the former, then BACE1 inhibitor treatment of AD adults may be relatively safe. However, processes with ongoing operation in the adult, such as axon guidance and neurogenesis, are impaired in BACE1−/− mice. This suggests that therapeutic inhibition of BACE1 may have mechanism-based side-effects. However, whether the observed BACE1−/− phenotypes might have implications for BACE1 inhibition as a future pharmacotherapy or not is a matter of ongoing discussion and will be discussed in more detail below.
Additional BACE1 substrates in the CNS
The rapidly increasing number of phenotypes in BACE1−/− mice suggests that more phenotypes are yet to be discovered. This assumption is in good agreement with the recent identification of numerous additional BACE1 substrates in proteomic screens. A first study identified over 60 candidate BACE1 substrates in tumor cell lines over-expressing BACE1 (Hemming et al. 2009). Not all of them may be physiological substrates, as over-expressed BACE1 is known to cleave in cellular compartments, such as the ER, where it normally is not localized (Vassar et al. 1999). Two recent studies identified substrates in primary neurons expressing endogenous BACE1 (Kuhn et al. 2012; Zhou et al. 2012). Zhou et al. (2012) identified 13 substrates from cultured neurons under serum- and protein-free conditions and used differential isotopic labeling of terminal amines and ε-amines for quantification. In contrast, Kuhn et al. (2012) cultured the neurons in the presence of serum proteins using B27-containing media. That study developed the novel secretome protein enrichment with click sugars (SPECS) technology which utilizes metabolic glycan labeling and click chemistry to enrich glycoproteins out of B27 containing media of primary neurons to identify 34 BACE1 substrates.
Besides APP and its homologs both studies identified the novel BACE1 substrates L1, CHL1, contactin-2, Golgi membrane protein 1 (GLG1), and peptidyl amidating monooxygenase (PAM). The CHL1-dependent phenotype of the BACE1−/− mice has already been described above. Further BACE1 and also BACE2 substrates were identified in a proteomics study in pancreatic islets and insulinoma cell lines (Stutzer et al. 2013), where both proteases are also expressed. From these studies it became clear that BACE1 does not only cleave a few neuronal membrane proteins, but has a broad range of substrates within and outside of the nervous system.
One of the proteomic studies quantified the extent to which the cleavage of the substrates depends on BACE1 (Kuhn et al. 2012). Some of the substrates, such as SEZ6, SEZ6L, and APLP1 are ‘exclusive’ BACE1 substrates, because BACE1 inhibition and BACE1-deficiency almost completely block cleavage of these substrates. For other substrates total cleavage was only partially reduced in the absence of BACE1 activity, indicating that these substrates are not only cleaved by BACE1, but also by other proteases, which are expected to be metalloproteases. Moreover, a compensatory increase in the cleavage of some substrates by another protease may occur. This is the case for APP where ADAM10 compensates for loss of BACE1 cleavage (Colombo et al. 2012; May et al. 2011). In addition, for some of the new substrates indirect effects like trans-synaptic stabilization may potentially be responsible for an apparently partial cleavage reduction upon BACE inhibition, such as has been shown in case of postsynaptic neuroligin 1 which binds to membrane bound presynaptic Neurexin-1 alpha and is increasingly cleaved upon the addition of recombinant soluble neurexin 1 alpha while treatment with a BACE inhibitor leads to reduced amounts of secreted neurexin 1 alpha (Suzuki et al. 2012; Kuhn et al. 2012; Boucard et al. 2005).
Based on the current literature, the novel BACE1 substrates can be divided into two subgroups. The first group comprises proteins that participate in synapse function, whereas the second group includes proteins that interact with the surrounding extracellular matrix of astrocytes and oligodendroglia and thereby modulate axon outgrowth and path finding as well as the formation of membrane microdomains. Finally there are substrates with other functions.
Substrates involved in synapse function
The first group of identified BACE1 substrates contains neurexin 1 alpha, the neuroligin family and the latrophilin family. Neurexin-1 alpha and latrophilins are mostly presynaptic proteins which interact with each other in cis and with post-synaptic neuroligins in trans, bridging the synaptic cleft (Boucard et al. 2012). Although it is not yet clear exactly how BACE1 cleaves these proteins and how it alters their function, it is interesting to note that some of the phenotypes of neurexin 1 alpha and neuroligin-deficient mice are recapitulated in BACE1-deficient mice. This suggests that some of the BACE1-deficiency phenotypes may result from the reduced cleavage of neurexin 1 alpha or the neuroligins. Deletion of neurexin 1 alpha reduces excitatory synaptic strength (Etherton et al. 2009). Neuroligins in concert with neurexins are needed for synapse function but not for synapse formation (Xu et al. 2012; Bang and Owczarek 2013). Triple knockout mice of neuroligin 1, 2, and 3 showed an abnormal respiration behavior leading to post-natal death owing to reduced neurotransmission in the brainstem respiratory network (Varoqueaux et al. 2006). Single knockouts of neuroligins show more subtle phenotypes. Neuroligin 1 knockout mice show reduced social interaction, impaired spatial working memory and reduced contextual- and cued-fear memory (Kim et al. 2008). Neuroligin-2 knockout mice show increased anxiety-like behaviors, reduced pain sensitivity, motor coordination, and an irregular breathing pattern, whereas neuroligin 4 mice have symptoms of autism spectrum disorders (Xu et al. 2012; Jamain et al. 2008; Blundell et al. 2009). Neurexin 1 alpha knockout mice show impaired nest building behavior and spend more time in approaching other mice which might be because of impaired interpretation of social cues or increased aggression behavior (Grayton et al. 2013). A lot of these behavioral phenotypes have as well been observed in BACE1 knockout mice and might imply that similar signaling pathways are impaired in these mice (Laird et al. 2005; Savonenko et al. 2008).
Substrates involved in axon outgrowth, axoglial interactions, and myelin microdomain structure
The second group of substrates is involved in axonal outgrowth, axoglial interactions and electric transmission. This group includes CHL1, L1, contactin-2, and the SEZ6 family. The role of BACE1 cleavage for CHL1 function has already been described above. The glycosyl phosphatidyl inositol-anchored protein contactin-2 has been confirmed as a BACE1 substrate in vitro and in vivo. Contactin-2 expressed on Schwann cells and oligodendroglia interacts with the transmembrane protein CaspR2 and Contactin-2 on the axonal membrane to maintain the voltage-gated potassium channels Kv1.1 and Kv1.2 in juxtaparanodes of myelinated axons (Poliak et al. 2003; Traka et al. 2003). Contactin-2 knockout leads to impaired learning behavior, shorter internodes, and disrupted juxtaparanodes (Savvaki et al. 2008). Thus, it appears possible that BACE1-mediated proteolytic processing of contactin-2 modulates the concentration and function of juxtaparanodal potassium channels. This possibility needs to be tested in further experiments.
Sez6 family proteins are type I membrane proteins localized to both dendritic and axonal compartments. Sez6 knockout mice display increased dendritic branching, reduced spine density on apical dendrites, and impaired excitability of layer V pyramidal neurons (Gunnersen et al. 2007). The triple knockout of the whole Sez6 family leads to motor coordination deficits reflected in electrophysiological alterations in Purkinje cell excitation in the cerebellum (Miyazaki et al. 2006). All three members of the Sez6 family are BACE1 substrates but how BACE1 modulates their functions is not yet clear as little is known about their physiological roles in vivo.
PAM is one of the candidate BACE1 substrates that has shown up in all four proteomic screens and has been shown to undergo regulated intramembrane proteolysis by an unknown sheddase and by γ-secretase in tumor cells (Rajagopal et al. 2010). This copper-dependent enzyme amidates the C-terminus of a number of neuropeptides, which is necessary for their biologic activity (Bousquet-Moore et al. 2010). PAM-deficiency leads to intrauterine death caused by malfunction of the cardiovascular system and cerebral edema that result from non-functional proaminoterminal peptide (PAM) and adrenomedullin because of their lacking amidation (Czyzyk et al. 2005). In contrast, BACE1-deficient mice may show the opposite phenotype as BACE1 negatively regulates PAM function by PAM cleavage. Given that BACE1 expression is particularly high in the brain, whereas PAM is expressed more ubiquitously, but mainly in the pituitary gland, brain, and the atrioventricular node (Braas et al. 1989), it may be possible that BACE1-deficiency mostly affects PAM function in the brain, but not necessarily in other organs.
Functional Roles of BACE2
Soon after the discovery of BACE1, a paralog termed BACE2 was identified (Bennett et al. 2000a; Farzan et al. 2000). BACE2 was shown to function like an “α-secretase” in promoting the non-amyloidogenic processing of APP (Basi et al. 2003; Farzan et al. 2000; Fluhrer et al. 2002; Yan et al. 2001). In contrast to BACE1, little attention thus has been devoted to the study of BACE2, until recently. Initial studies in BACE2 knockout mice documented no apparent phenotype, except that the early post-natal lethality observed in BACE1−/− mice is enhanced in BACE1−/−; BACE2−/− double knockout mice (Dominguez et al. 2005). Using mouse and zebrafish model systems, investigators have made significant advances recently to identify new substrates of BACE2 and disclose novel physiological roles of this aspartyl protease.
Processing of a pigment protein by BACE2 to form the amyloid matrix in melanosomes
The process of pigmentation begins with events that occur in melanosomes distributed throughout melanocytes (Hearing 2005). These endosomal organelles contain all the components required for melanin formation, including the enzyme tyrosinase (TYR) and accessory proteins including TYR-related protein 1 and TYR-related protein 2 (Hearing 2005). Melanosomes undergo a four-stage maturation process that is dependent upon the structural protein termed pigment cell-specific melanocyte protein (PMEL). Mature PMEL is synthesized as an integral membrane protein enriched in the lumen of melanosomes. Proteolysis of PMEL subsequently leads to the structural rearrangement of the melanosome from an immature, non-pigmented, round organelle to a mature, pigmented, ellipsoid melanosome (Watt et al. 2009). A stage I melanosome is characterized by its non-descript round, non-pigmented shape that resembles an endosome. Proteolytic cleavage of PMEL by a putative protease(s) within the melanosome releases an N-terminal fragment, termed Mα, competent for formation of the scaffolding, or amyloid matrix, which elongates the melanosome (stage II; elliptically shaped). Stage III commences with the deposition of eumelanin, a black/brown pigment, produced by TYR, upon the PMEL scaffold. By stage IV, the internal melanosomal structure is obscured because of the abundance of deposited eumelanin. Without the amyloid matrix, no eumelanin is able to accumulate and only pheomelanin, a red/yellow pigment, is observed in round melanosomes (Raposo and Marks 2007). Variations in the eumelanin/pheomelanin ratio account for the wide variety of coat color in all organisms. Interestingly, natural mutations in Pmel found in a variety of organisms, including mice, chicken, and horse (Theos et al. 2006; Kerje et al. 2004; Brunberg et al. 2006), shared a common phenotype: coat color dilution.
While proteolytic processing of PMEL is thought to be a critical step in the formation of the amyloid matrix, the enzyme(s) responsible remained elusive. Studies in BACE2−/− mice in which one of the BACE2 domains containing a catalytic aspartic residue is deleted of an otherwise full length protein revealed dilution of coat color suggesting that BACE2 could be a candidate (Dominguez et al. 2005) (Fig. 4a). As this pigmentation phenotype resembling mice harboring mutations in Pmel observed in Bace2−/−, but not Bace1−/− mice (Rochin et al. 2013), Rochin and coworkers addressed whether BACE2 could be a sheddase that is responsible for cleaving PMEL for proper formation of scaffold in melanosomes for pigment deposition. These investigators provided several lines of evidence in favor of this idea. In support of the notion that BACE2 resides in the same compartment as that for processing of PMEL, co-localization studies showed that BACE2 is targeted to early stages (I and II) of PMEL-containing melanosomes and associated with PMEL (Rochin et al. 2013). Secondly, both gain- and loss-of-function approaches in cultured cells revealed that BACE2 was able to cleave PMEL to release its ectodomain containing the amyloidogenic Mα-fragment in cultured cells using both gain- and loss-of-function approaches (Rochin et al. 2013). Finally, melanogenesis is impaired in Bace2−/− mice (Rochin et al. 2013). Taken together, these findings are consistent with a model in which PMEL is cleaved by BACE2 to release the luminal fragment into endosomal precursors for its formation into fibrils and finally melanosome maturation. Although BACE1 apparently does not participate in the processing of PMEL as BACE1−/−;BACE2−/− double knockout mice exhibit a similar pigmentation phenotype comparable to BACE2 null mice, a caveat is that the BACE2 null mice carry a catalytically impaired BACE2 protein (rather than the absence of BACE2 protein). Such a mutant protein could act in a dominant-negative fashion, obscuring the potential role of BACE1 in PMEL processing. BACE2 null mice lacking the complete BACE2 protein will be useful in future studies to clarify this issue.
Fig. 4.
Beta-site amyloid cleaving enzyme (BACE)2 regulates pigmentation in mice and melanophore migration in zebrafish. (a) Deletion of Bace2 in mice leads to dilution of coat color (Rochin et al. 2013). Note the black coat normally seen in the C57Bl/6 strain as observed in control and Bace1 knockout mice. (b) Deletion of Bace2 in zebrafish alters the shape and migration of melanophores (van Bebber et al. 2013). Whole fish (left panels); boxed areas are enlarged to show details near end of yolk sac extension (middle panels) and fin (right panels).
Role of BACE2 in migration of melanocytes
Recent studies in zebrafish revealed a surprising role of BACE2 in the regulation of melanocyte migration. There exists one human BACE2 ortholog in the zebrafish genome, zBACE2 and its mRNA is expressed throughout early development (van Bebber et al. 2013). Capitalizing on a collection of previously ENU (N-ethyl-N-nitrosourea)-mutagenized alleles, a C to A conversion in zBace2 was identified; this mutant led to a premature in-frame stop codon that truncated a large portion of the protein, including both catalytic domains (van Bebber et al. 2013). Thus, an N-terminal 79 amino acid peptide is predicted to be generated in such mutants (zBace2+/−). Cross-breeding of zBACE2+/− zebrafish led to the generation of homozygous mutant alleles (zBACE2−/−). Although zBACE2−/− larvae were indistinguishable from their zBACE2+/− or zBACE2+/+ littermates before the third day post-fertilization, it became clear later in development that melanophores were more dilated in larvae lacking Bace2. In addition, the migration pattern of these melanophores around the tail fin and the yolk sac extension was perturbed in zBace2−/− animals (Fig. 4b). As melanophores are neural crest derivatives, this phenotype is consistent with the expression pattern of Bace2 in neural crest cells (Thisse et al. 2004).
To assess whether compensatory mechanisms exist in regulation of migration of melanocytes between zBACE1 and zBACE2, zBACE1−/−; zBACE2−/− zebrafish were generated. Comparing defects in melanophores and melanocyte migration observed in zBACE2−/− fish, these abnormalities were not enhanced in the zBACE1−/−;zBACE2−/− animals, indicating that zBace2 serves distinct roles than those of zBace1 in term of melanocyte migration (van Bebber et al. 2013). These data support the notion that BACE1 and BACE2 serve distinct physiological roles during development and aging. Importantly, this zebrafish model is a valuable tool to assess in vivo the specificity of BACE inhibitors to identify BACE1-selective and BACE2-sparing compounds in the event that such compounds are deemed necessary to limit mechanism-based toxicity occurring in clinical trials of Alzheimer’s disease.
Regulation of pancreatic β cell mass and function by BACE2
Hyperglycemia occurring in type 2 diabetes results from the failure of the pancreas to secrete appropriate amounts of insulin to match metabolic requirements associated with insulin resistance (Kahn et al. 2006). Two major abnormalities observed in this illness are progressive degeneration of beta cell function and decrease in pancreatic beta cell mass (Kahn et al. 2006). However, the molecular mechanism underlying beta cell loss is not clearly understood. Proteolytic processing of growth factors and their receptors of β cells are thought to be important for proper autocrine and paracrine signaling. One example is the type I transmembrane protein Tmem27 (pro-proliferative plasma membrane protein), which is a target gene of the transcription factor Tcf1 (or hepatocyte nuclear factor 1a), that is linked to the most common form of maturity onset diabetes of the young (MODY3)(Shih et al. 2001). Other than the kidney (Zhang et al. 2001), expression of Tmem27 has only been documented in the pancreas (Akpinar et al. 2005). Thought to stabilize apical amino acid transporters and critical for reabsorption of amino acids (Danilczyk et al. 2006; Malakauskas et al. 2007), Tmem27 when abnormally accumulated in β cells causes an increase in pancreatic cell mass (Akpinar et al. 2005). Importantly, ectodomain shedding of Tmem27 regulates its abundance and activity in β cells as the holoprotein retains mitogenic activity (Akpinar et al. 2005). Thus, identification of the protease responsible for the shedding of Tmem27 may provide an opportunity to elevate Tmem27 levels as a strategy to modify β cell mass and function.
To identify the responsible protease, Esterhazy and coworkers employed a siRNA approach to screen all major classes of proteases in a mouse insulinoma cell line. In their initial screen, BACE2 was identified as a sheddase for Tmem27 in both mouse and human beta cells (Esterhazy et al. 2011). Importantly, when insulin-resistant mice were treated with a BACE2 inhibitor, these coworkers observed increased beta cell mass and improved control of glucose homeostasis as a result of increased insulin levels (Esterhazy et al. 2011). To complement this observation, these investigators took advantage of the availability of BACE2 knockout mice with inactive BACE2 (Dominguez et al. 2005) to examine Tmem27 processing and physiological consequences in β cells of these mutant mice. Remarkably, Esterhazy and coworkers observed that these Bace2 inactive mice have significantly lower levels of blood glucose, improved glucose tolerance and elevated β cell mass, implicating BACE2 as the sheddase that is important for the regulation of β cell maintenance (Esterhazy et al. 2011). These data thus are consistent with the notion that Bace2 controls beta cell mass and function and provide a strategy to inhibit BACE2 as a potential therapy in efforts to improve human β cell function.
Finally, Stoffel and coworkers used a proteomic approach in a pancreatic cell line to identify Seizure 6 protein family members, SEZ6L and SEZ6L2, which are islet-enriched cell surface proteins as BACE2-specific substrates (Stutzer et al. 2013). However, these substrates are cleaved by BACE1 in neurons (Kuhn et al. 2012). Why SEZ6L and SEZ6L2 are selectively cleaved by BACE2 over BACE1 in β cells is not completely clear. Likely possibilities include organ-dependent abundance of enzyme, substrate isoform or cleavage site preferences, and subcellular compartmentalization of each sheddase. These investigators also identified IGF2R and SORT1 as substrates of BACE2 in islet cells (Stutzer et al. 2013).
In summary, over the past few years, significant progress has been made in the identification of novel substrates of BACE2 and revealed that some of its physiological roles appear to be unique and distinct from those of BACE1. This will help the development of tools for evaluating BACE1 inhibitors that are selective over BACE2, thus potentially providing improved compounds that are safe and effective for the treatment of AD.
BACE inhibition as a treatment strategy for AD
Drug discovery scientists often invest years of research effort on hypotheses despite significant uncertainties owing to incomplete understanding of the fundamental biology of disease and the potential for entirely unanticipated and often insurmountable non-mechanism-based side-effects that emerge late in the development of a novel therapeutic. Therefore, it is critical that efforts are focused on hypotheses that are supported by the strongest possible data which is often derived from human genetics (Plenge et al. 2013) as is the case with the amyloid hypothesis of AD as reviewed above (Loy et al. 2013). The recently described rare protective β-secretase-cleavage-sparing variant of APP (APP-A673T) further supports the rationale for BACE1 inhibition in AD (Jonsson et al. 2012). Our understanding of the pathophysiological sequence of AD is incomplete but findings to date from ongoing natural progression studies conducted by the Alzheimer’s Disease Neuroimaging Initiative and the Dominantly Inherited Alzheimer Network (DIAN) support that Aβ plaque deposition is likely an early and potentially initiating event of AD that drives the subsequent development and/or expansion of tau pathology and inflammation that together contribute to the neurotoxic environment that causes synapse dysfunction, cell loss, and dementia (Bateman et al. 2012; Jack et al. 2012). While this interpretation of existing literature supports that early intervention with CNS Aβ lowering therapeutics will have the best chances for success, multiple studies have shown the direct neurotoxic effects of Aβ oligomers and plaques which could be operant at all stages of AD such that substantially reducing the influx of newly synthesized Aβ peptides to reduce Aβ oligomer formation and plaque growth could be beneficial even in later-stage symptomatic patients (Walsh and Selkoe 2007).
After over a decade of AD clinical research on amyloid-directed agents, has the amyloid hypothesis been adequately tested? Most clinical AD investigators will answer this with an unqualified, “No”. The primary reason given is that the amyloid-directed agents tested to date (e.g., γ-secretase inhibitors, γ-secretase modulators, plaque disruptors, and anti-Aβ immunotherapy) have failed to convincingly demonstrate an unambiguous and substantial reduction in CSF Aβ levels; CSF Aβ is the primary biomarker of soluble brain Aβ status in humans (Wan et al. 2009; Blennow 2010). For example, the strategy of γ-secretase inhibition, while strongly supported by human genetics and discovery of compounds capable of significantly reducing CNS Aβ levels in preclinical studies (Best et al. 2007) and humans such as semagacestat (Bateman et al. 2009) and avagacestat (Tong et al. 2012), ultimately failed in Phase 3 efficacy studies owing to the emergence of serious dose-limiting and thus CNS Aβ-lowering limiting mechanism-based side-effects including cancerous skin lesions, severe gastrointestinal toxicity, and ultimately cognitive worsening that were likely related to inhibition of Notch processing or other non-APP γ-secretase substrates (Doody et al. 2013; Coric et al. 2012). Indeed the dose limiting toxicities associated with γ-secretase inhibitions did not support a robust test of the amyloid hypothesis as only a transient ~25% lowering of CSF Aβ from baseline was achieved in Phase 3 efficacy trials. The confounds arising from the combination of serious mechanism-based side-effects and the likely minimal impact of semagacestat or avagecestat on CSF Aβ levels at the doses tested unfortunately limits the impact of these findings on our understanding of the validity of the amyloid hypothesis (Pomara 2013) or importantly the minimum levels of Aβ lowering that will have a benefit. Genetic studies in mice and humans suggest that moderate reductions in CNS Aβ levels can result in long-term benefits. CNS Aβ levels in BACE1 wild-type, heterozygous and knockout mice unexpectedly revealed that 50% knockdown of CNS BACE1 protein in BACE+/− mice resulted in only ~12–20% reduction in CNS Aβ level versus wild-type animals (McConlogue et al. 2007). Despite this modest effect on Aβ, aged PDAPP/ BACE1+/− mice developed about 75% less amyloid plaque burden than similarly aged PDAPP/BACE1+/+ mice (McConlogue et al. 2007). From in vitro cell studies, the rare APP-A673T protective variant was associated with a 40% reduction in Aβ production compared to APP-WT cells and an 8-fold reduced risk for AD (Jonsson et al. 2012) again suggesting that a relatively moderate reduction in Aβ production afforded by reduced BACE1 cleavage of APP may confer a long-term benefit. As PDAPP/BACE+/− mice and APP-A673T human carriers experience a lifelong reduction in steady-state CNS Aβ, it is difficult to infer whether similar levels of Aβ lowering achieved with BACE inhibition would benefit AD patients. Importantly the degree of CNS Aβ reduction required for a meaningful clinical benefit at any given point of the AD continuum remains an unknown and will require mechanisms and molecules that are capable of safely achieving the dose–response range of Aβ lowering in AD patients.
BACE inhibition – therapeutic potential of small molecules and biologics
The molecular identification of BACE1 and the subsequent reports of BACE1 knockout mice revealed an atypical member of the aspartic protease family that combined a strong therapeutic rationale for inhibitors to treat AD based on requirement of BACE1 for Aβ synthesis and the lack of overt negative phenotypes in multiple reports of BACE1 knockout mouse lines (Roberds et al. 2001; Cai et al. 2001; Luo et al. 2001). Aspartic proteases are a therapeutically important class of enzymes that includes the HIV protease and the mammalian renin enzymes that possess structural homology with the BACE1 active site (Hong et al. 2000). Renin presented major medicinal chemistry challenges and only after decades of effort was a clinically effective renin inhibitor approved for blood pressure control (Jensen et al. 2008) whereas HIV protease inhibitors are a transformational core component of HIV therapy (Ray et al. 2010). Despite these successes and significant active site structural homology among aspartic proteases, the peptidomimetic nature of classical aspartic protease inhibitor scaffolds have proven to be incompatible with the chemical and physical properties required for a clinically developable inhibitor of the CNS-localized BACE1 protease (Ghosh et al. 2012). Early examples of peptidomimetic BACE inhibitors possessed high affinity and selectivity for BACE enzymes and produced significant reductions in plasma Aβ but achieved modest reductions in brain Aβ in mice (Chang et al. 2004; Hussain et al. 2007; Sankaranarayanan et al. 2008) and non-human primates (Sankaranarayanan et al. 2009) often requiring exposure-boosting strategies. CTS-21166 is a peptidomimetic BACE inhibitor, derived from the earliest chemistry efforts on BACE inhibitors, that advanced into Phase 1 clinical trials in healthy volunteers and produced significant and sustained reductions of plasma Aβ (80% peak inhibition) following intravenous delivery however effects on CSF Aβ have not yet been reported and its development status remains unknown (Albert 2009). A novel biologics based approach to achieving highly selective BACE1 inhibition relies on a bi-functional inhibitory antibody that binds both the transferrin-receptor to improve blood-brain barrier penetration and the BACE1 enzyme domain. This antibody potently and selectively inhibits purified BACE1 and BACE1-meditated processing of APP in cells and has achieved impressive proof of concept in vivo efficacy results in transgenic APP mice (Atwal et al. 2011). The affinity optimized transferrin-receptor binding function improved antibody uptake into the brain by ~5-fold which was critical for the efficacy of this approach (Yu et al. 2011). Results to date are encouraging that this approach could serve as alternative to small molecule inhibitors of BACE1 or other CNS-localized intracellular enzymes. Importantly, this approach achieves exquisite selectivity for the BACE1 enzyme over other aspartyl proteases including BACE2. In a broader context, the blood–brain barrier enhancing strategies utilized for anti-BACE1 immunotherapy may also be harnessed to improve the performance of other CNS passive immunotherapies such as anti-Aβ and anti-tau approaches if carrier-dependent mechanism-based side-effects can be avoided (Couch et al. 2013).
Breakthrough discoveries in small molecule inhibitors of BACE came with the identification of novel acylguanidine, aminopyridine, and isothiourea pharmacophores that bound with modest affinity to the catalytic aspartic acid residues of BACE1 (Cole et al. 2006; Congreve et al. 2007; Yang et al. 2009b; Wang et al. 2010). These molecules emerged from diverse hit finding strategies using high-throughput X-ray crystallography (Congreve et al. 2007), functional screens (Cole et al. 2006), or a protein NMR-based fragment screen using an unlabeled soluble BACE1 construct (Geschwindner et al. 2007) or an 15N-labeled human BACE1 soluble enzyme domain (Wang et al. 2010). For the latter screen, the initial hit Merck D, a weak isothiourea structure that possessed a BACE1 binding Kd of ~15 µM and a functional BACE1 IC50 of ~ 200 µM (Fig. 5c), (Wang et al. 2010) launched a structure-based design effort that led to the discovery of the novel iminoheterocyclic class of aspartic protease inhibitors with more drug-like pharmacophore as exemplified by the iminohydantoin Merck E (Fig. 5c). Iminohydantoin BACE inhibitors possessed an unprecedented combination of low molecular weight and high affinity binding compared to prior peptidomimetic inhibitors in a structure that at the time of its discovery was unknown in the aspartic protease inhibitor literature, (Zhu et al. 2010). These efforts relied heavily on BACE1 X-ray crystallography and computational modeling to aid rational chemical design decisions that achieved high affinity binding coupled with robust oral pharmacokinetic and CSF Aβ lowering efficacy in rodents as exemplified by Merck F (Cumming et al. 2012). Subsequent medicinal chemistry efforts pursued ring expansion of the iminohydantoin warhead to generate the novel 6-membered C5-substituted iminopyrimidinone core, Merck J in Fig. 5c, which again maintained a low molecular weight with high affinity binding and a minimal shift in cellular potency (Stamford et al. 2012). The iminopyrimidinone SAR optimization focused on defining the appropriate combination of in vitro pharmacological properties with potent and efficacious CSF Aβ lowering in rats (Stamford et al. 2012; Stamford and Strickland 2013). The novel cyclic isothiourea core BACE inhibitor, LY2811376 Fig. 5b (May et al. 2011), that displayed modest intrinsic affinity for purified BACE1 (IC50~250 nM), more potent cellular activity and robust CNS Aβ lowering in rodents and dogs and was advanced into Phase 1 studies where it supported the first published report of BACE inhibitor-mediated lowering of CSF Aβ in healthy volunteers (May et al. 2011). Development of this compound was terminated owing to observations of retinal and neuronal and glial cell degeneration associated with intracellular accumulations of autofluorescent granules following 3 month dosing in rodent toxicology studies (May et al. 2011). These effects manifested in the eye as retinal pigment epithelium cell hypertrophy and degeneration. The histopathological findings resulting from chronic LY2811376 treatment were retained in BACE1 knockout mice implying that the effects were not likely BACE1 dependent (May et al. 2011). The histopathological findings described by May et al. are reminiscent of findings reported for cathepsin D knockout mice (Saftig et al. 1995; Shacka et al. 2007) and suggest that the moderate selectivity of LY2811376 for BACE1 over cathepsin D of ~60-fold (May et al. 2011) is not sufficient to support chronic dosing. The importance of cathepsin D selectivity is further underscored by rare cathepsin D homozygous loss of function mutations in humans that manifests as a subtype of the lysosomal storage disorder, Batten’s disease, characterized by seizures, severe developmental defects, and neurodegeneration (Siintola et al. 2006). Most genetic forms of lysosome dysfunction manifest with neurohistopathological findings of aberrant intracellular accumulation of ceroidlipofuscin deposits (Dawson and Cho 2000). Importantly, the most advanced BACE inhibitors currently in clinical trials have been described at recent scientific meetings to be virtually inactive at cathepsin D.
Fig. 5.
(a) Example of an optimized peptidomimetic β-site amyloid cleaving enzyme (BACE) inhibitor with high molecular weight and marginal in vivo Aβ lowering activity at very high doses of 250 mg/kg (Hussain et al. 2007). (b). Optimized cyclic isothiourea BACE inhibitor derived from a fragment-based screening approach (May et al. 2011). LY2811376 displays modest BACE1 potency but with robust in vivo Aβ lowering activity in animals and normal healthy volunteers in Phase 1 studies. LY2811376 development was terminated during Phase 1 owing to off target toxicities in eye and brain. (c) Merck iminoheterocyclic BACE inhibitor series evolved from a low affinity isothiourea hit (Merck D) identified in a protein NMR-based screen. Replacement off the isothiourea warhead with the more drug-like iminohydantoin core (Merck E) led to further optimization (Merck G) and ring expansion of the warhead to produce molecules like Merck J with several orders of magnitude improvements in intrinsic affinity with relatively small increase in molecular weight. MK-8931 builds upon the favorable properties described here for the iminoheterocyclic BACE inhibitor series with significant improvements in intrinsic affinity, selectivity, in vivo potency, and other key properties.
The recent proliferation of novel substrates and proposed biological functions of BACE1 and BACE2 that are reviewed herein have tempered the early expectations that chronic BACE inhibitor therapy would be devoid of any potential for mechanism-based safety concerns. Insights into the impact of reduced versus complete loss of BACE1 activity during development or adulthood can be gleaned from comparing and contrasting the impact of varying levels of chronic pharmacological inhibition of BACE1 activity in adult animals to BACE1 knockout mouse phenotypes (Willem et al. 2006; Hu et al. 2006; Savonenko et al. 2008; Laird et al. 2005; Hitt et al. 2012; Cao et al. 2012; Cai et al. 2012; Cheret et al. 2013). Phenotypes reported for BACE1 knockout mice to date reflect the impact of a lifelong 50% reduction or complete loss of BACE1 function while pharmacological inhibition of BACE activity is targeted for adults and at doses that do not achieve complete sustained inhibition. Ongoing Phase 3 clinical trials of the BACE inhibitor MK-8931 are evaluating doses that achieve a range of 50 to 75% sustained reduction in CSF Aβ. Several groups have shown that the majority of phenotypes reported for BACE1 knockout mice are not observed in BACE1 heterozygous mice suggesting that a 50% loss of BACE1 function would be tolerated. In the course of the BACE inhibitor discovery program at Merck Research Labs a number of chronic BACE inhibitor studies have been conducted (M. Kennedy et al., manuscript in preparation). These studies include: (i) chronic treatment of transgenic APP mice to examine the impact of Aβ lowering on the course of amyloid pathology and (ii) toxicology studies in multiple species at supra-therapeutic doses to assess short- and long-term safety and tolerability. These studies have evaluated structurally diverse inhibitors including MK-8931. The peripheral nerve hypomyelination phenotype reported in BACE1 knockout mice was not observed in Tg-CRND8-APPSwedish/Indiana mice following 4 months of chronic pharmacological inhibition at doses of a BACE inhibitor that produced a sustained ~ 80% brain Aβ lowering1. Other phenotypes examined in these same studies such as reduced pre-pulse inhibition were also not impacted by 4 month chronic BACE inhibitor treatment1. As described above, retinal degeneration was observed with chronic LY2811376 treatment in rodents, however, studies by this same group in BACE1 knockout mice showed this finding to be independent of BACE1 (May et al. 2011). Subsequently, a non-progressing retinal degeneration phenotype was described in young BACE1 and BACE2 knockout mice suggesting BACE activity may be important for retinal homeostasis early in life (Cai et al. 2012). However, retinal degeneration phenotypes have not been reported for all BACE1 and BACE2 knockout lines indicating that this phenotype may be strain dependent. Taken together the emerging data suggest that some functions of BACE1 may be more important during development than in adulthood which is consistent with the dramatic drop off in CNS expression of BACE1 in mice during the first few weeks of life (Willem et al. 2006) Exploring the relationship between genetic deletion of BACE1 versus pharmacological BACE inhibition in young versus adult animals remains an area of intense effort and will require collaborations that will benefit from the sharing of potent and selective BACE inhibitors. The lack of inhibitors with substantial selectivity between BACE1 and BACE2 complicates the comparison of pharmacological studies versus knockout studies. However, to date the majority of phenotypes reported for BACE1 and BACE2 knockout mice appear to be non-overlapping (Dominguez et al. 2005). Beyond the role of BACE1 in AD, recent studies using knockouts and small molecule inhibitors suggest a potential positive impact of reduced BACE activity in traumatic brain injury (Chami and Checler 2012) and peripheral nerve damage (Farah et al. 2011) (BACE1) whereas type 2 diabetes may benefit from inhibition of BACE2 (Esterhazy et al. 2011).
BACE inhibitors in clinical trials
Several BACE inhibitors have progressed into Phase 1 studies. The BACE inhibitors that are known to have advanced the furthest in clinical development are LY-2886721 (Eli Lilly), E-2069 (Eisai), and MK-8931 (Merck & Co.). Structures and detailed in vitro and in vivo profiles of these compounds have not been published in the peer-reviewed literature as this requires the release of proprietary chemical structures, however, these compounds have been extensively described at scientific meetings. All three of these molecules are high affinity and selective inhibitors of the human BACE1 enzyme and potently reduce peripheral and CNS Aβ1–40, Aβ1–42, and sAPPβ levels in rodents and large animal species such as non-human primates (E-2069 and MK-8931) or dogs (LY-2886721). While these compounds are highly selective for BACE1 over other mammalian aspartyl proteases such as cathepsins D and E and renin they are equipotent inhibitors of the human BACE2 enzyme. BACE2 does not contribute to β-secretase cleavage of APP in vivo (see above) and possesses high active site identity with BACE1 such that achieving substantial (≥ 100-fold) selectivity over BACE2 while maintaining optimal CNS drug properties has proven challenging. These pre-clinical pharmacodynamic profiles have translated well in Phase 1 studies where significant and sustained CSF Aβ lowering of ≥ 80% following single and multiple doses to healthy volunteers (LY-2886721, E2069 and MK-8931) and AD patients (MK-8931 and E-2069) has been reported. The development of LY-2886721 was recently terminated owing to observations of abnormal liver tests indicative of liver toxicity in a chronic Phase 2 study of AD patients. These findings were noted as unexpected based on the lack of any liver abnormalities in pre-clinical safety testing of LY-2886721 and were not considered related to BACE1 or BACE2 inhibition. The remaining BACE inhibitors, E-2069 and MK-8931 are progressing into long-term clinical efficacy studies in patients. Eisai has noted its commitment to explore the efficacy of E-2069 in early AD patients but has not disclosed the design or status of this trial, whereas Merck started a registration supporting Phase 2/3 safety and efficacy study of MK-8931 in mild-moderate AD patients in November of 2012. This study contains a lead-in safety cohort of 200 patients that received once daily doses of placebo, 12, 40, or 60 mg of MK8931 for 3 months. The main Phase 3 efficacy cohort will examine the impact of two dose levels of MK-8931 that will test the impact of 50% or 75% CSF Aβ lowering respectively on Alzheimer’s Disease Assesment Scale-cog and Alzheimer’s Disease Co-operative Study - Activities of Daily Living Inventory end points following 18-months treatment (for study details see NCT0173934; clinicaltrials.gov). A second parallel Phase 3 study of MK-8931 in prodromal AD patients commenced late in 2013 and will test the impact of MK-8931 on CDR-SB following 24 months of treatment at the same dose levels used in mild-moderate patients. Inclusion criteria for the prodromal AD trial require that a patient has a positive 18F-Flumetamol (Vizamyl™, GE Healthcare, Arlington Heights, IL, USA) Positron emission tomography scan or has a positive CSF tau/Aβ42 biomarker profile (for study details see NCT01953601; clincaltrials.gov).
Given the robust levels of CNS Aβ lowering achieved with BACE inhibitors such as MK-8931 it appears that a robust test of the amyloid hypothesis is underway. While a positive outcome is sorely needed for patients, even a negative outcome will provide critically needed clinical evidence that will allow AD researchers to focus more effort on alternative therapeutic mechanisms as well as novel clinical strategies aimed at studying the earliest pre-symptomatic stages of what we recognize as a decades long disease process.
Outlook
Since the identification of BACE1 and BACE2 in 1999 we have seen tremendous progress in BACE protease research, which has further intensified over the past few years as it is highlighted in this review article. Basic research demonstrates a continuously increasing spectrum of physiological functions of BACE1 and BACE2 both within and outside of the nervous system. Over the next few years we are likely to see more BACE protease functions being discovered. The recent proteomic studies have identified a large number of additional BACE protease substrates. Their validation in vitro and in vivo will help to uncover additional BACE protease functions. One challenge for the future is to link the novel substrates to the phenotypes in the BACE-deficient mice and thereby understand novel BACE protease functions at the molecular level.
It turns out that – similar to APP - many BACE substrates are not only cleaved by BACE1 or BACE2, but also by other proteases, in particular metalloproteases. Thus, another question to answer is whether the different proteases have redundant functions for an individual substrate as it seems to be the case for neuregulin. Alternatively, substrate cleavage by either BACE or a metalloprotease may lead to a different functional outcome, as it appears to happen for APP.
BACE1 has taken center stage in AD drug development programs and around a dozen companies have BACE inhibitors in clinical trials. While severe mechanism-based side-effects of BACE inhibitors have not yet been observed in clinical trials, it remains to be seen which of the BACE1 functions may be compromised upon therapeutic BACE inhibition and thereby cause side-effects. Compared to a knock-out, BACE inhibitors do not completely block BACE1 activity and may still allow residual BACE1 activity sufficient for essential signaling functions. Moreover, it is likely that many of the developmental BACE1 functions, such as myelination, are not affected in adult AD patients. However, there are developmental processes which also occur in the adult brain, such as neurogenesis and axon guidance in the hippocampus. They may be equally affected in adults upon BACE inhibition. Likewise, muscle spindle maintenance is an adult BACE1 function and is compromised in adult mice treated with a BACE inhibitor (Cheret et al. 2013). We clearly need a better understanding of adult BACE1 functions as a way to predict potential side-effects of BACE inhibitors. This will be eased by analyzing conditional BACE1-deficient mice, which are now available in different laboratories.
The growing list of BACE substrates is not only helpful for understanding BACE functions, but the substrates may have the potential to be used as companion diagnostics for individualized dosing of patients and for the potential development of BACE substrate-specific inhibitors.
Since its discovery BACE2 has been the neglected cousin of BACE1. This has changed as BACE2 substrates and functions are also being unveiled and as BACE2 is considered a potential target for diabetes. These BACE2 functions are even relevant for BACE inhibitor development programs, because most BACE inhibitors are equally or even more potent at inhibiting BACE2. In this regard, it may be necessary to generate conditional BACE1/BACE2 double knock-out mice to analyze phenotypes arising through loss of function of both proteases specifically in adulthood. Despite their similarities, BACE1 and BACE2 also have differences, for example, in substrate spectrum, expression pattern and subcellular localization. While significant work has been done on the regulation of BACE1 expression and activity, it will be interesting to see whether BACE2 is regulated in the same or a different manner and whether this may allow developing new therapeutic approaches selectively targeting BACE1. Another interesting approach would be to achieve selective inhibition of BACE1 cleavage of APP while sparing the cleavages of other physiologically relevant substrates.
Taken together, the recent discoveries demonstrate that we are in an exciting time for BACE protease research, which is no longer predominantly focusing on AD, but reaching into many other research fields. And we can expect major new insights in the next years, both for basic BACE research and for the use of BACE proteases as drug targets.
Acknowledgments
The authors thank Dr Patty Kandalepas for generating Fig. 1 and Guillaume van Niel for contribution of data for Fig. 4. This work was supported by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013)/ERC Grant Agreement No. 321366-Amyloid (to CH), the general legacy of Mrs Ammer (to the Ludwig-Maximilians University/ the chair of CH), by the BMBF (KNDD to SFL and CH), the JPND RiMod (to SFL), the DAAD (to SFL), NIH R01AG022560 (RV), NIH R01AG030142 (RV), Cure Alzheimer’s Fund (RV), BrightFocus Foundation, (RV), the Alzheimer’s Association (RV), the Swiss National Science Foundation, the Velux Foundation (both to LR).
Matthew E. Kennedy is an employee of Merck Research Labs.
Abbreviations used
- AD
Alzheimer’s disease
- APP
amyloid precursor protein
- BACE
beta-site amyloid cleaving enzyme
- CHL1
close homolog of L1
- EGF
epidermal growth factor
- ER
endoplasmic reticulum
- IPB
infrapyramidal bundle
- NRG1
neuregulin 1
- NTF
N-terminal fragment
- OSNs
olfactory sensory neurons
- TGN
trans-Golgi network
References
- Akpinar P, Kuwajima S, Krutzfeldt J, Stoffel M. Tmem27: a cleaved and shed plasma membrane protein that stimulates pancreatic beta cell proliferation. Cell Metab. 2005;2:385–397. doi: 10.1016/j.cmet.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Albert JS. Progress in the development of beta-secretase inhibitors for Alzheimer’s disease. Prog. Med. Chem. 2009;48:133–161. doi: 10.1016/s0079-6468(09)04804-8. [DOI] [PubMed] [Google Scholar]
- Andersen OM, Reiche J, Schmidt V, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl Acad. Sci. USA. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrechek ER, Hardy WR, Girgis-Gabardo AA, Perry RL, Butler R, Graham FL, Kahn RC, Rudnicki MA, Muller WJ. ErbB2 is required for muscle spindle and myoblast cell survival. Mol. Cell. Biol. 2002;22:4714–4722. doi: 10.1128/MCB.22.13.4714-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim CA, Spoelgen R, Peltan ID, et al. GGA1 acts as a spatial switch altering amyloid precursor protein trafficking and processing. J. Neurosci. 2006;26:9913–9922. doi: 10.1523/JNEUROSCI.2290-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal JK, Chen Y, Chiu C, et al. A therapeutic antibody targeting BACE1 inhibits amyloid-beta production in vivo. Sci. Transl. Med. 2011;3:84ra43. doi: 10.1126/scitranslmed.3002254. [DOI] [PubMed] [Google Scholar]
- Bang ML, Owczarek S. A matter of balance: role of neurexin and neuroligin at the synapse. Neurochem. Res. 2013;38:1174–1189. doi: 10.1007/s11064-013-1029-9. [DOI] [PubMed] [Google Scholar]
- Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. J. Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Lin H, Ouyang Y, et al. Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat. Neurosci. 2004;7:1250–1258. doi: 10.1038/nn1342. [DOI] [PubMed] [Google Scholar]
- Basi G, Frigon N, Barbour R, Doan T, Gordon G, McConlogue L, Sinha S, Zeller M. Antagonistic effects of beta-site amyloid precursor protein-cleaving enzymes 1 and 2 on beta-amyloid peptide production in cells. J. Biol. Chem. 2003;278:31512–31520. doi: 10.1074/jbc.M300169200. [DOI] [PubMed] [Google Scholar]
- Bateman RJ, Siemers ER, Mawuenyega KG, et al. A gamma-secretase inhibitor decreases amyloid-beta production in the central nervous system. Ann. Neurol. 2009;66:48–54. doi: 10.1002/ana.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RJ, Xiong C, Benzinger TL, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bebber F, Hruscha A, Willem M, Schmid B, Haass C. Loss of Bace2 in zebrafish affects melanocyte migration and is distinct from Bace1 knock out phenotypes. J. Neurochem. 2013;127:471–481. doi: 10.1111/jnc.12198. [DOI] [PubMed] [Google Scholar]
- Benjannet S, Elagoz A, Wickham L, et al. Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J. Biol. Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Babu-Khan S, Loeloff R, Louis JC, Curran E, Citron M, Vassar R. Expression analysis of BACE2 in brain and peripheral tissues. J. Biol. Chem. 2000a;275:20647–20651. doi: 10.1074/jbc.M002688200. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Denis P, Haniu M, Teplow DB, Kahn S, Louis JC, Citron M, Vassar R. A furin-like convertase mediates propeptide cleavage of BACE, the Alzheimer’s beta-secretase. J. Biol. Chem. 2000b;275:37712–37717. doi: 10.1074/jbc.M005339200. [DOI] [PubMed] [Google Scholar]
- Best JD, Smith DW, Reilly MA, et al. The novel gamma secretase inhibitor N-[cis-4-[(4-chlorophenyl)sulfonyl]-4-(2,5-difluorophenyl)cyclohexyl]-1,1,1-trifl uoromethanesulfonamide (MRK-560) reduces amyloid plaque deposition without evidence of notch-related pathology in the Tg2576 mouse. J. Pharmacol. Exp. Ther. 2007;320:552–558. doi: 10.1124/jpet.106.114330. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Nave KA. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56:1491–1497. doi: 10.1002/glia.20753. [DOI] [PubMed] [Google Scholar]
- Blennow K. Biomarkers in Alzheimer’s disease drug development. Nat. Med. 2010;16:1218–1222. doi: 10.1038/nm.2221. [DOI] [PubMed] [Google Scholar]
- Blundell J, Tabuchi K, Bolliger MF, Blaiss CA, Brose N, Liu X, Sudhof TC, Powell CM. Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 2009;8:114–126. doi: 10.1111/j.1601-183X.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Boucard AA, Ko J, Sudhof TC. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J. Biol. Chem. 2012;287:9399–9413. doi: 10.1074/jbc.M111.318659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Moore D, Mains RE, Eipper BA. Peptidylgycine alpha-amidating monooxygenase and copper: a gene-nutrient interaction critical to nervous system function. J. Neurosci. Res. 2010;88:2535–2545. doi: 10.1002/jnr.22404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braas KM, Stoffers DA, Eipper BA, May V. Tissue specific expression of rat peptidylglycine alpha-amidating monooxygenase activity and mRNA. Mol. Endocrinol. 1989;3:1387–1398. doi: 10.1210/mend-3-9-1387. [DOI] [PubMed] [Google Scholar]
- Brunberg E, Andersson L, Cothran G, Sandberg K, Mikko S, Lindgren G. A missense mutation in PMEL17 is associated with the Silver coat color in the horse. BMC Genet. 2006;7:46. doi: 10.1186/1471-2156-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggia-Prevot V, Fernandez CG, Udayar V. A function for EHD family proteins in unidirectional retrograde dendritic transport of BACE1 and Alzheimer’s disease Aβ production. Cell Rep. 2013;5:1552–1563. doi: 10.1016/j.celrep.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggia-Prevot V, Fernandez CG, Riordan S, Vetrivel KS, Roseman J, Waters J, Bindokas VP, Vassar R, Thinakaran G. Axonal BACE1 dynamics and targeting in hippocampal neurons: a role for Rab11 GTPase. Mol. Neurodegener. 2014;9:1. doi: 10.1186/1750-1326-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat. Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi X, Kociok N, et al. beta-Secretase (BACE1) inhibition causes retinal pathology by vascular dysregulation and accumulation of age pigment. EMBO Mol. Med. 2012;4:980–991. doi: 10.1002/emmm.201101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Rickenbacher GT, Rodriguez S, Moulia TW, Albers MW. The precision of axon targeting of mouse olfactory sensory neurons requires the BACE1 protease. Sci. Rep. 2012;2:231. doi: 10.1038/srep00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell A, Steiner H, Willem M, Kaiser H, Meyer C, Walter J, Lammich S, Multhaup G, Haass C. Maturation and pro-peptide cleavage of beta-secretase. J. Biol. Chem. 2000;275:30849–30854. doi: 10.1074/jbc.M003202200. [DOI] [PubMed] [Google Scholar]
- Chami L, Checler F. BACE1 is at the crossroad of a toxic vicious cycle involving cellular stress and beta-amyloid production in Alzheimer’s disease. Mol. Neurodegener. 2012;7:52. doi: 10.1186/1750-1326-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WP, Koelsch G, Wong S, et al. In vivo inhibition of Abeta production by memapsin 2 (beta-secretase) inhibitors. J. Neurochem. 2004;89:1409–1416. doi: 10.1111/j.1471-4159.2004.02452.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hancock ML, Role LW, Talmage DA. Intramembranous valine linked to schizophrenia is required for neuregulin 1 regulation of the morphological development of cortical neurons. J. Neurosci. 2010;30:9199–9208. doi: 10.1523/JNEUROSCI.0605-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheret C, Willem M, Fricker FR, et al. Bace1 and Neuregulin-1 cooperate to control formation and maintenance of muscle spindles. EMBO J. 2013;32:2015–2028. doi: 10.1038/emboj.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe DJ. Mutation of the beta-amyloid precursor protein in familial Alzheimer’s disease increases beta-protein production. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- Cole DC, Manas ES, Stock JR, et al. Acylguanidines as small-molecule beta-secretase inhibitors. J. Med. Chem. 2006;49:6158–6161. doi: 10.1021/jm0607451. [DOI] [PubMed] [Google Scholar]
- Colombo A, Wang H, Kuhn PH, Page R, Kremmer E, Dempsey PJ, Crawford HC, Lichtenthaler SF. Constitutive alpha- and beta-secretase cleavages of the amyloid precursor protein are partially coupled in neurons, but not in frequently used cell lines. Neurobiol. Dis. 2012;49C:137–147. doi: 10.1016/j.nbd.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M, Aharony D, Albert J, et al. Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase. J. Med. Chem. 2007;50:1124–1132. doi: 10.1021/jm061197u. [DOI] [PubMed] [Google Scholar]
- Coric V, van Dyck CH, Salloway S, et al. Safety and tolerability of the gamma-secretase inhibitor avagacestat in a phase 2 study of mild to moderate Alzheimer disease. Arch. Neurol. 2012;69:1430–1440. doi: 10.1001/archneurol.2012.2194. [DOI] [PubMed] [Google Scholar]
- Couch JA, Yu YJ, Zhang Y, et al. Addressing safety liabilities of TfR bispecific antibodies that cross the blood-brain barrier. Sci. Transl. Med. 2013;5:183ra157, 181–112. doi: 10.1126/scitranslmed.3005338. [DOI] [PubMed] [Google Scholar]
- Crusio WE, Schwegler H. Learning spatial orientation tasks in the radial-maze and structural variation in the hippocampus in inbred mice. Behav. Brain Funct. 2005;1:3. doi: 10.1186/1744-9081-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming JN, Smith EM, Wang L, et al. Structure based design of iminohydantoin BACE1 inhibitors: identification of an orally available, centrally active BACE1 inhibitor. Bioorg. Med. Chem. Lett. 2012;22:2444–2449. doi: 10.1016/j.bmcl.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Czyzyk TA, Ning Y, Hsu MS, Peng B, Mains RE, Eipper BA, Pintar JE. Deletion of peptide amidation enzymatic activity leads to edema and embryonic lethality in the mouse. Dev. Biol. 2005;287:301–313. doi: 10.1016/j.ydbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Danilczyk U, Sarao R, Remy C, et al. Essential role for collectrin in renal amino acid transport. Nature. 2006;444:1088–1091. doi: 10.1038/nature05475. [DOI] [PubMed] [Google Scholar]
- Dawson G, Cho S. Batten’s disease: clues to neuronal protein catabolism in lysosomes. J. Neurosci. Res. 2000;60:133–140. doi: 10.1002/(SICI)1097-4547(20000415)60:2<133::AID-JNR1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Dawson GR, Seabrook GR, Zheng H, et al. Age-related cognitive deficits, impaired long-term potentiation and reduction in synaptic marker density in mice lacking the beta-amyloid precursor protein. Neuroscience. 1999;90:1–13. doi: 10.1016/s0306-4522(98)00410-2. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Vassar R, Golde T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 2010;6:99–107. doi: 10.1038/nrneurol.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejaegere T, Serneels L, Schafer MK, et al. Deficiency of Aph1B/C-gamma-secretase disturbs Nrg1 cleavage and sensorimotor gating that can be reversed with antipsychotic treatment. Proc. Natl. Acad Sci. USA. 2008;105:9775–9780. doi: 10.1073/pnas.0800507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, He W, Tan Y, Han H, Hu X, Xia K, Zhang Z, Yan R. Increased expression of reticulon 3 in neurons leads to reduced axonal transport of beta site amyloid precursor protein-cleaving enzyme 1. J. Biol. Chem. 2013;288:30236–30245. doi: 10.1074/jbc.M113.480079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fede G, Catania M, Morbin M, et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323:1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dislich B, Lichtenthaler SF. The membrane-bound aspartyl protease BACE1: molecular and functional properties in Alzheimer’s disease and beyond. Front. Physiol. 2012;3:8. doi: 10.3389/fphys.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Tournoy J, Hartmann D, et al. Phenotypic and biochemical analyses of BACE1- and BACE2-deficient mice. J. Biol. Chem. 2005;280:30797–30806. doi: 10.1074/jbc.M505249200. [DOI] [PubMed] [Google Scholar]
- Doody RS, Raman R, Farlow M, et al. A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N. Engl. J. Med. 2013;369:341–350. doi: 10.1056/NEJMoa1210951. [DOI] [PubMed] [Google Scholar]
- Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterhazy D, Stutzer I, Wang H, et al. Bace2 is a beta cell-enriched protease that regulates pancreatic beta cell function and mass. Cell Metab. 2011;14:365–377. doi: 10.1016/j.cmet.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl Acad. Sci. USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falls DL. Neuregulins and the neuromuscular system: 10 years of answers and questions. J. Neurocytol. 2003;32:619–647. doi: 10.1023/B:NEUR.0000020614.83883.be. [DOI] [PubMed] [Google Scholar]
- Farah MH, Pan BH, Hoffman PN, et al. Reduced BACE1 activity enhances clearance of myelin debris and regeneration of axons in the injured peripheral nervous system. J. Neurosci. 2011;31:5744–5754. doi: 10.1523/JNEUROSCI.6810-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Schnitzler CE, Vasilieva N, Leung D, Choe H. BACE2, a b-secretase homolog, cleaves at the b site and within the amyloid-b region of the amyloid-b precursor protein. Proc. Natl Acad. Sci. USA. 2000;97:9712–9717. doi: 10.1073/pnas.160115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck D, Garratt AN, Haass C, Willem M. BACE1 dependent neuregulin proteolysis. Curr. Alzheimer Res. 2012;9:178–183. [PubMed] [Google Scholar]
- Fleck D, van Bebber F, Colombo A, et al. Dual cleavage of neuregulin 1 type III by BACE1 and ADAM17 liberates its EGF-like domain and allows paracrine signaling. J. Neurosci. 2013;33:7856–7869. doi: 10.1523/JNEUROSCI.3372-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhrer R, Capell A, Westmeyer G, Willem M, Hartung B, Condron MM, Teplow DB, Haass C, Walter J. A non-amyloidogenic function of BACE-2 in the secretory pathway. J. Neurochem. 2002;81:1011–1020. doi: 10.1046/j.1471-4159.2002.00908.x. [DOI] [PubMed] [Google Scholar]
- Garratt AN, Voiculescu O, Topilko P, Charnay P, Birchmeier C. A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J. Cell Biol. 2000;148:1035–1046. doi: 10.1083/jcb.148.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersbacher MT, Kim DY, Bhattacharyya R, Kovacs DM. Identification of BACE1 cleavage sites in human voltage-gated sodium channel beta 2 subunit. Mol. Neurodegener. 2010;5:61. doi: 10.1186/1750-1326-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwindner S, Olsson LL, Albert JS, Deinum J, Edwards PD, de Beer T, Folmer RH. Discovery of a novel warhead against beta-secretase through fragment-based lead generation. J. Med. Chem. 2007;50:5903–5911. doi: 10.1021/jm070825k. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Brindisi M, Tang J. Developing beta-secretase inhibitors for treatment of Alzheimer’s disease. J. Neurochem. 2012;120(Suppl 1):71–83. doi: 10.1111/j.1471-4159.2011.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayton HM, Missler M, Collier DA, Fernandes C. Altered social behaviours in neurexin 1alpha knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS ONE. 2013;8:e67114. doi: 10.1371/journal.pone.0067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnersen JM, Kim MH, Fuller SJ, et al. Sez-6 proteins affect dendritic arborization patterns and excitability of cortical pyramidal neurons. Neuron. 2007;56:621–639. doi: 10.1016/j.neuron.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Haass C. Take five–BACE and the gamma-secretase quartet conduct Alzheimer’s amyloid beta-peptide generation. EMBO J. 2004;23:483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, Lannfelt L, Selkoe DJ. The Swedish mutation causes early-onset Alzheimer’s disease by beta-secretase cleavage within the secretory pathway. Nat. Med. 1995;1:1291–1296. doi: 10.1038/nm1295-1291. [DOI] [PubMed] [Google Scholar]
- Haniu M, Denis P, Young Y, et al. Characterization of Alzheimer’s beta-secretase protein BACE. A pepsin family member with unusual properties. J. Biol. Chem. 2000;275:21099–21106. doi: 10.1074/jbc.M002095200. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harrison SM, Harper AJ, Hawkins J, et al. BACE1 (beta-secretase) transgenic and knockout mice: identification of neurochemical deficits and behavioral changes. Mol. Cell. Neurosci. 2003;24:646–655. doi: 10.1016/s1044-7431(03)00227-6. [DOI] [PubMed] [Google Scholar]
- He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J. Biol. Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- Hearing VJ. Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J. Dermatol. Sci. 2005;37:3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Heber S, Herms J, Gajic V, et al. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J. Neurosci. 2000;20:7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Identification of beta-secretase (BACE1) substrates using quantitative proteomics. PLoS ONE. 2009;4:e8477. doi: 10.1371/journal.pone.0008477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Muller U. Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. EMBO J. 2004;23:4106–4115. doi: 10.1038/sj.emboj.7600390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyden A, Angenstein F, Sallaz M, Seidenbecher C, Montag D. Abnormal axonal guidance and brain anatomy in mouse mutants for the cell recognition molecules close homolog of L1 and NgCAM-related cell adhesion molecule. Neuroscience. 2008;155:221–233. doi: 10.1016/j.neuroscience.2008.04.080. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Shneider NA, Birchmeier C, Burden SJ, Jessell TM, Arber S. A role for neuregulin1 signaling in muscle spindle differentiation. Neuron. 2002;36:1035–1049. doi: 10.1016/s0896-6273(02)01101-7. [DOI] [PubMed] [Google Scholar]
- Hitt BD, Jaramillo TC, Chetkovich DM, Vassar R. BACE1−/− mice exhibit seizure activity that does not correlate with sodium channel level or axonal localization. Mol. Neurodegener. 2010;5:31. doi: 10.1186/1750-1326-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt B, Riordan S, Kukreja L, Eimer W, Rajapaksha T, Vassar R. beta-site amyloid precursor protein (APP) cleaving enzyme 1 (BACE1) deficient mice exhibit a close homolog of L1 (CHL1) loss-of-function phenotype involving axon guidance defects. J. Biol. Chem. 2012;287:38408–38425. doi: 10.1074/jbc.M112.415505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Koelsch G, Lin X, Wu S, Terzyan S, Ghosh AK, Zhang XC, Tang J. Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor. Science. 2000;290:150–153. doi: 10.1126/science.290.5489.150. [DOI] [PubMed] [Google Scholar]
- Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- Hu X, He W, Diaconu C, Tang X, Kidd GJ, Macklin WB, Trapp BD, Yan R. Genetic deletion of BACE1 in mice affects remyelination of sciatic nerves. FASEB J. 2008;22:2970–2980. doi: 10.1096/fj.08-106666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhou X, He W, Yang J, Xiong W, Wong P, Wilson CG, Yan R. BACE1 deficiency causes altered neuronal activity and neurodegeneration. J. Neurosci. 2010;30:8819–8829. doi: 10.1523/JNEUROSCI.1334-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, He W, Luo X, Tsubota KE, Yan R. BACE1 regulates hippocampal astrogenesis via the Jagged1-Notch pathway. Cell Rep. 2013;4:40–49. doi: 10.1016/j.celrep.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I, Powell D, Howlett DR, et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol. Cell. Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- Hussain I, Hawkins J, Harrison D, et al. Oral administration of a potent and selective non-peptidic BACE-1 inhibitor decreases beta-cleavage of amyloid precursor protein and amyloid-beta production in vivo. J. Neurochem. 2007;100:802–809. doi: 10.1111/j.1471-4159.2006.04260.x. [DOI] [PubMed] [Google Scholar]
- Isom LL. Sodium channel beta subunits: anything but auxiliary. Neuroscientist. 2001;7:42–54. doi: 10.1177/107385840100700108. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Vemuri P, Wiste HJ, et al. Shapes of the trajectories of 5 major biomarkers of Alzheimer disease. Arch. Neurol. 2012;69:856–867. doi: 10.1001/archneurol.2011.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc. Natl Acad. Sci. USA. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen C, Herold P, Brunner HR. Aliskiren: the first renin inhibitor for clinical treatment. Nat. Rev. Drug Discovery. 2008;7:399–410. doi: 10.1038/nrd2550. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Atwal JK, Steinberg S, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- Jorissen E, Prox J, Bernreuther C, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- Kalvodova L, Kahya N, Schwille P, Ehehalt R, Verkade P, Drechsel D, Simons K. Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J. Biol. Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- Kandalepas PC, Sadleir KR, Eimer WA, Zhao J, Nicholson DA, Vassar R. The Alzheimer’s beta-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol. 2013;126:329–352. doi: 10.1007/s00401-013-1152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang EL, Cameron AN, Piazza F, Walker KR, Tesco G. Ubiquitin regulates GGA3-mediated degradation of BACE1. J. Biol. Chem. 2010;285:24108–24119. doi: 10.1074/jbc.M109.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang EL, Biscaro B, Piazza F, Tesco G. BACE1 protein endocytosis and trafficking are differentially regulated by ubiquitination at lysine 501 and the Di-leucine motif in the carboxyl terminus. J. Biol. Chem. 2012;287:42867–42880. doi: 10.1074/jbc.M112.407072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerje S, Sharma P, Gunnarsson U, et al. The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics. 2004;168:1507–1518. doi: 10.1534/genetics.104.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Carey BW, Wang H, et al. BACE1 regulates voltage-gated sodium channels and neuronal activity. Nat. Cell Biol. 2007;9:755–764. doi: 10.1038/ncb1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung SY, Lee YK, et al. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proc. Natl Acad. Sci. USA. 2008;105:9087–9092. doi: 10.1073/pnas.0803448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Gersbacher MT, Inquimbert P, Kovacs DM. Reduced sodium channel Na(v)1.1 levels in BACE1-null mice. J. Biol. Chem. 2011;286:8106–8116. doi: 10.1074/jbc.M110.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Fukumoto H, Shah T, Whelan CM, Irizarry MC, Hyman BT. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J. Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- Ko MH, Puglielli L. Two endoplasmic reticulum (ER)/ER Golgi intermediate compartment-based lysine acetyltransferases post-translationally regulate BACE1 levels. J. Biol. Chem. 2009;284:2482–2492. doi: 10.1074/jbc.M804901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, Zeller M, Cole T, Buttini M, McConlogue L, Sinha S, Freedman S, Morris RG, Chen KS. BACE1 gene deletion: impact on behavioral function in a model of Alzheimer’s disease. Neurobiol. Aging. 2008;29:861–873. doi: 10.1016/j.neurobiolaging.2007.01.002. [DOI] [PubMed] [Google Scholar]
- von Koch CS, Zheng H, Chen H, Trumbauer M, Thinakaran G, van der Ploeg LH, Price DL, Sisodia SS. Generation of APLP2 KO mice and early postnatal lethality in APLP2/APP double KO mice. Neurobiol. Aging. 1997;18:661–669. doi: 10.1016/s0197-4580(97)00151-6. [DOI] [PubMed] [Google Scholar]
- Koh YH, von Arnim CA, Hyman BT, Tanzi R, Tesco G. BACE is degraded via the lysosomal pathway. J. Biol. Chem. 2005;280:32499–32504. doi: 10.1074/jbc.M506199200. [DOI] [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Koroniak K, Hogl S, et al. Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J. 2012;31:3157–3168. doi: 10.1038/emboj.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca R, Cerri F, Horiuchi K, et al. TACE (ADAM17) inhibits Schwann cell myelination. Nat. Neurosci. 2011;14:857–865. doi: 10.1038/nn.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird FM, Cai H, Savonenko AV, et al. BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 2005;25:11693–11709. doi: 10.1523/JNEUROSCI.2766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu M, Bellmunt E, Schwander M, Farinas I, Brenner HR, Muller U. Erbb2 regulates neuromuscular synapse formation and is essential for muscle spindle development. Development. 2003;130:2291–2301. doi: 10.1242/dev.00447. [DOI] [PubMed] [Google Scholar]
- Li ZW, Stark G, Gotz J, Rulicke T, Gschwind M, Huber G, Muller U, Weissmann C. Generation of mice with a 200-kb amyloid precursor protein gene deletion by Cre recombinase-mediated site-specific recombination in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1996;93:6158–6162. doi: 10.1073/pnas.93.12.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang B, Wang Z, Guo Q, Tabuchi K, Hammer RE, Sudhof TC, Zheng H. Soluble amyloid precursor protein (APP) regulates transthyretin and Klotho gene expression without rescuing the essential function of APP. Proc. Natl Acad. Sci. USA. 2010;107:17362–17367. doi: 10.1073/pnas.1012568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc. Natl Acad Sci. USA. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy CT, Schofield PR, Turner AM, Kwok JB. Genetics of dementia. Lancet. 2013;383:828–840. doi: 10.1016/S0140-6736(13)60630-3. [DOI] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Kahn S, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat. Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Damore MA, Fitzpatrick D, Liu H, Zhang J, Yan Q, Vassar R, Citron M. BACE1 (beta-secretase) knockout mice do not acquire compensatory gene expression changes or develop neural lesions over time. Neurobiol. Dis. 2003;14:81–88. doi: 10.1016/s0969-9961(03)00104-9. [DOI] [PubMed] [Google Scholar]
- Luo X, Prior M, He W, et al. Cleavage of neuregulin-1 by BACE1 or ADAM10 protein produces differential effects on myelination. J. Biol. Chem. 2011;286:23967–23974. doi: 10.1074/jbc.M111.251538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magara F, Muller U, Li ZW, Lipp HP, Weissmann C, Stagljar M, Wolfer DP. Genetic background changes the pattern of forebrain commissure defects in transgenic mice underexpressing the beta-amyloid-precursor protein. Proc. Natl Acad. Sci. USA. 1999;96:4656–4661. doi: 10.1073/pnas.96.8.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakauskas SM, Quan H, Fields TA, McCall SJ, Yu MJ, Kourany WM, Frey CW, Le TH. Aminoaciduria and altered renal expression of luminal amino acid transporters in mice lacking novel gene collectrin. Am. J. Physiol. Renal Physiol. 2007;292:F533–F544. doi: 10.1152/ajprenal.00325.2006. [DOI] [PubMed] [Google Scholar]
- May PC, Dean RA, Lowe SL, et al. Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor. J. Neurosci. 2011;31:16507–16516. doi: 10.1523/JNEUROSCI.3647-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConlogue L, Buttini M, Anderson JP, et al. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J. Biol. Chem. 2007;282:26326–26334. doi: 10.1074/jbc.M611687200. [DOI] [PubMed] [Google Scholar]
- Meakin PJ, Harper AJ, Hamilton DL, et al. Reduction in BACE1 decreases body weight, protects against diet-induced obesity and enhances insulin sensitivity in mice. Biochem. J. 2012;441:285–296. doi: 10.1042/BJ20110512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailov GV, Sereda MW, Brinkmann BG, et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- Mitterreiter S, Page RM, Kamp F, et al. Bepridil and amiodarone simultaneously target the Alzheimer’s disease beta-and gamma-secretase via distinct mechanisms. J. Neurosci. 2010;30:8974–8983. doi: 10.1523/JNEUROSCI.1199-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Hashimoto K, Uda A, et al. Disturbance of cerebellar synaptic maturation in mutant mice lacking BSRPs, a novel brain-specific receptor-like protein family. FEBS Lett. 2006;580:4057–4064. doi: 10.1016/j.febslet.2006.06.043. [DOI] [PubMed] [Google Scholar]
- Montag-Sallaz M, Schachner M, Montag D. Misguided axonal projections, neural cell adhesion molecule 180 mRNA upregulation, and altered behavior in mice deficient for the close homolog of L1. Mol. Cell. Biol. 2002;22:7967–7981. doi: 10.1128/MCB.22.22.7967-7981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan M, Crawford F, Houlden H, Axelman K, Lilius L, Winblad B, Lannfelt L. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of beta-amyloid. Nat. Genet. 1992a;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- Muller U, Cristina N, Li ZW, Wolfer DP, Lipp HP, Rulicke T, Brandner S, Aguzzi A, Weissmann C. Behavioral and anatomical deficits in mice homozygous for a modified beta-amyloid precursor protein gene. Cell. 1994;79:755–765. doi: 10.1016/0092-8674(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Naus S, Richter M, Wildeboer D, Moss M, Schachner M, Bartsch JW. Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J. Biol. Chem. 2004;279:16083–16090. doi: 10.1074/jbc.M400560200. [DOI] [PubMed] [Google Scholar]
- Ohno M, Sametsky EA, Younkin LH, Oakley H, Younkin SG, Citron M, Vassar R, Disterhoft JF. BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer’s disease. Neuron. 2004;41:27–33. doi: 10.1016/s0896-6273(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Ohno M, Chang L, Tseng W, Oakley H, Citron M, Klein WL, Vassar R, Disterhoft JF. Temporal memory deficits in Alzheimer’s mouse models: rescue by genetic deletion of BACE1. Eur. J. Neurosci. 2006;23:251–260. doi: 10.1111/j.1460-9568.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- Ohno M, Cole SL, Yasvoina M, Zhao J, Citron M, Berry R, Disterhoft JF, Vassar R. BACE1 gene deletion prevents neuron loss and memory deficits in 5XFAD APP/PS1 transgenic mice. Neurobiol. Dis. 2007;26:134–145. doi: 10.1016/j.nbd.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Ikin AF, Nairn AC, Pursnani A, Buxbaum JD. The carboxyl-terminus of BACE contains a sorting signal that regulates BACE trafficking but not the formation of total A (beta) Mol. Cell. Neurosci. 2002;19:175–185. doi: 10.1006/mcne.2001.1065. [DOI] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J. Biol. Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- Phinney AL, Calhoun ME, Wolfer DP, Lipp HP, Zheng H, Jucker M. No hippocampal neuron or synaptic bouton loss in learning-impaired aged beta-amyloid precursor protein-null mice. Neuroscience. 1999;90:1207–1216. doi: 10.1016/s0306-4522(98)00645-9. [DOI] [PubMed] [Google Scholar]
- Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discovery. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- Poliak S, Salomon D, Elhanany H, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J. Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomara N. Semagacestat for treatment of Alzheimer’s disease. N. Engl. J. Med. 2013;369:1661. doi: 10.1056/NEJMc1310845. [DOI] [PubMed] [Google Scholar]
- Prabhu Y, Burgos PV, Schindler C, Farias GG, Magadan JG, Bonifacino JS. Adaptor protein 2-mediated endocytosis of the beta-secretase BACE1 is dispensable for amyloid precursor protein processing. Mol. Biol. Cell. 2012;23:2339–2351. doi: 10.1091/mbc.E11-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal C, Stone KL, Mains RE, Eipper BA. Secretion stimulates intramembrane proteolysis of a secretory granule membrane enzyme. J. Biol. Chem. 2010;285:34632–34642. doi: 10.1074/jbc.M110.145334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapaksha TW, Eimer WA, Bozza TC, Vassar R. The Alzheimer’s beta-secretase enzyme BACE1 is required for accurate axon guidance of olfactory sensory neurons and normal glomerulus formation in the olfactory bulb. Mol. Neurodegener. 2011;6:88. doi: 10.1186/1750-1326-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L, Simons K. Membrane Trafficking and Targeting in Alzheimer’s disease. Heidelberg, Berlin: Springer; 2008. Foundation Ipsen Series. [Google Scholar]
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl Acad. Sci. USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L, Schneider A, Schlechtingen G, et al. Efficient inhibition of the Alzheimer’s disease beta-secretase by membrane targeting. Science. 2008;320:520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS. Melanosomes–dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Fatima Z, Saxena A. Drugs for AIDS. Mini Rev. Med. Chem. 2010;10:147–161. doi: 10.2174/138955710791185145. [DOI] [PubMed] [Google Scholar]
- Ring S, Weyer SW, Kilian SB, et al. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J. Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberds SL, Anderson J, Basi G, et al. BACE knockout mice are healthy despite lacking the primary beta-secretase activity in brain: implications for Alzheimer’s disease therapeutics. Hum. Mol. Genet. 2001;10:1317–1324. doi: 10.1093/hmg/10.12.1317. [DOI] [PubMed] [Google Scholar]
- Rochin L, Hurbain I, Serneels L, et al. BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc. Natl Acad Sci. USA. 2013;110:10658–10663. doi: 10.1073/pnas.1220748110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression–implications for Alzheimer’s disease. Prog. Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Saftig P, Hetman M, Schmahl W, et al. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 1995;14:3599–3608. doi: 10.1002/j.1460-2075.1995.tb00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H. Neural map formation in the mouse olfactory system. Neuron. 2010;67:530–542. doi: 10.1016/j.neuron.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S, Price EA, Wu G, et al. In vivo beta-secretase 1 inhibition leads to brain Abeta lowering and increased alpha-secretase processing of amyloid precursor protein without effect on neuregulin-1. J. Pharmacol. Exp. Ther. 2008;324:957–969. doi: 10.1124/jpet.107.130039. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S, Holahan MA, Colussi D, et al. First demonstration of cerebrospinal fluid and plasma A beta lowering with oral administration of a beta-site amyloid precursor protein-cleaving enzyme 1 inhibitor in nonhuman primates. J. Pharmacol. Exp. Ther. 2009;328:131–140. doi: 10.1124/jpet.108.143628. [DOI] [PubMed] [Google Scholar]
- Sannerud R, Declerck I, Peric A, et al. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc. Natl Acad. Sci. USA. 2011;108:E559–E568. doi: 10.1073/pnas.1100745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savonenko AV, Melnikova T, Laird FM, Stewart KA, Price DL, Wong PC. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc. Natl Acad. Sci. USA. 2008;105:5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvaki M, Panagiotaropoulos T, Stamatakis A, Sargiannidou I, Karatzioula P, Watanabe K, Stylianopoulou F, Karagogeos D, Kleopa KA. Impairment of learning and memory in TAG-1 deficient mice associated with shorter CNS internodes and disrupted juxtaparanodes. Mol. Cell. Neurosci. 2008;39:478–490. doi: 10.1016/j.mcn.2008.07.025. [DOI] [PubMed] [Google Scholar]
- Schneider A, Rajendran L, Honsho M, Gralle M, Donnert G, Wouters F, Hell SW, Simons M. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J. Neurosci. 2008;28:2874–2882. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabrook GR, Smith DW, Bowery BJ, et al. Mechanisms contributing to the deficits in hippocampal synaptic plasticity in mice lacking amyloid precursor protein. Neuropharmacology. 1999;38:349–359. doi: 10.1016/s0028-3908(98)00204-4. [DOI] [PubMed] [Google Scholar]
- Shacka JJ, Klocke BJ, Young C, Shibata M, Olney JW, Uchiyama Y, Saftig P, Roth KA. Cathepsin D deficiency induces persistent neurodegeneration in the absence of Bax-dependent apoptosis. J. Neurosci. 2007;27:2081–2090. doi: 10.1523/JNEUROSCI.5577-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DQ, Screenan S, Munoz KN, Philipson L, Pontoglio M, Yaniv M, Polonsky KS, Stoffel M. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- Siegenthaler BM, Rajendran L. Retromers in Alzheimer’s disease. Neurodegener. Dis. 2012;10:116–121. doi: 10.1159/000335910. [DOI] [PubMed] [Google Scholar]
- Siintola E, Partanen S, Stromme P, Haapanen A, Haltia M, Maehlen J, Lehesjoki AE, Tyynela J. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain. 2006;129:1438–1445. doi: 10.1093/brain/awl107. [DOI] [PubMed] [Google Scholar]
- Sinha S, Anderson JP, Barbour R, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- Sisodia SS, St George-Hyslop PH. gamma-Secretase, Notch, Abeta and Alzheimer’s disease: where do the presenilins fit in? Nat. Rev. Neurosci. 2002;3:281–290. doi: 10.1038/nrn785. [DOI] [PubMed] [Google Scholar]
- Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamford A, Strickland C. Inhibitors of BACE for treating Alzheimer’s disease: a fragment-based drug discovery story. Curr. Opin. Chem. Biol. 2013;17:320–328. doi: 10.1016/j.cbpa.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Stamford AW, Scott JD, Li SW, et al. Discovery of an orally available, brain penetrant BACE1 inhibitor that affords robust CNS abeta reduction. ACS Med. Chem. Lett. 2012;3:897–902. doi: 10.1021/ml3001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach JP, Muller U, Leist M, Li ZW, Nicotera P, Aguzzi A. Hypersensitivity to seizures in beta-amyloid precursor protein deficient mice. Cell Death Differ. 1998;5:858–866. doi: 10.1038/sj.cdd.4400391. [DOI] [PubMed] [Google Scholar]
- Stutzer I, Selevsek N, Esterhazy D, Schmidt A, Aebersold R, Stoffel M. Systematic proteomic analysis identifies beta-site amyloid precursor protein cleaving enzyme 2 and 1 (BACE2 and BACE1) substrates in pancreatic beta-cells. J. Biol. Chem. 2013;288:10536–10547. doi: 10.1074/jbc.M112.444703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Choi SH, Romano DM, Gannon MA, Lesinski AN, Kim DY, Tanzi RE. ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron. 2013;80:385–401. doi: 10.1016/j.neuron.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Hayashi Y, Nakahara S, et al. Activity-dependent proteolytic cleavage of neuroligin-1. Neuron. 2012;76:410–422. doi: 10.1016/j.neuron.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Tanzi RE. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Kang EL, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos AC, Berson JF, Theos SC, et al. Dual loss of ER export and endocytic signals with altered melanosome morphology in the silver mutation of Pmel17. Mol. Biol. Cell. 2006;17:3598–3612. doi: 10.1091/mbc.E06-01-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Tong G, Wang JS, Sverdlov O, et al. Multicenter, randomized, double-blind, placebo-controlled, single-ascending dose study of the oral gamma-secretase inhibitor BMS-708163 (Avagacestat): tolerability profile, pharmacokinetic parameters, and pharmacodynamic markers. Clin. Ther. 2012;34:654–667. doi: 10.1016/j.clinthera.2012.01.022. [DOI] [PubMed] [Google Scholar]
- Traka M, Goutebroze L, Denisenko N, et al. Association of TAG-1 with Caspr2 is essential for the molecular organization of juxtaparanodal regions of myelinated fibers. J. Cell Biol. 2003;162:1161–1172. doi: 10.1083/jcb.200305078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayar V, Buggia-Prevot V, Guerreiro RL, et al. A Paired RNAi and RabGAP overexpression screen identifies Rab11 as a regulator of beta-amyloid production. Cell Rep. 2013;5:1536–1551. doi: 10.1016/j.celrep.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J. Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrivel KS, Meckler X, Chen Y, et al. Alzheimer disease Abeta production in the absence of S-palmitoylation-dependent targeting of BACE1 to lipid rafts. J. Biol. Chem. 2009;284:3793–3803. doi: 10.1074/jbc.M808920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KR, Tesco G. Molecular mechanisms of cognitive dysfunction following traumatic brain injury. Front. Aging Neurosci. 2013;5:29. doi: 10.3389/fnagi.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Selkoe DJ. A beta oligomers - a decade of discovery. J. Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- Wan HI, Jacobsen JS, Rutkowski JL, Feuerstein GZ. Translational medicine lessons from flurizan’s failure in Alzheimer’s disease (AD) trial: implication for future drug discovery and development for AD. Clin. Transl. Sci. 2009;2:242–247. doi: 10.1111/j.1752-8062.2009.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Yang G, Mosier DR, et al. Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J. Neurosci. 2005;25:1219–1225. doi: 10.1523/JNEUROSCI.4660-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Strickland C, Voigt JH, et al. Application of fragment-based NMR screening, X-ray crystallography, structure-based design, and focused chemical library design to identify novel microM leads for the development of nM BACE-1 (beta-site APP cleaving enzyme 1) inhibitors. J. Med. Chem. 2010;53:942–950. doi: 10.1021/jm901472u. [DOI] [PubMed] [Google Scholar]
- Watt B, van Niel G, Fowler DM, et al. N-terminal domains elicit formation of functional Pmel17 amyloid fibrils. J. Biol. Chem. 2009;284:35543–35555. doi: 10.1074/jbc.M109.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer SW, Klevanski M, Delekate A, et al. APP and APLP2 are essential at PNS and CNS synapses for transmission, spatial learning and LTP. EMBO J. 2011;30:2266–2280. doi: 10.1038/emboj.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem M, Garratt AN, Novak B, et al. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- Willem M, Lammich S, Haass C. Function, regulation and therapeutic properties of beta-secretase (BACE1) Semin. Cell Dev. Biol. 2009;20:175–182. doi: 10.1016/j.semcdb.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Wong HK, Sakurai T, Oyama F, et al. beta Subunits of voltage-gated sodium channels are novel substrates of beta-site amyloid precursor protein-cleaving enzyme (BACE1) and gamma-secretase. J. Biol. Chem. 2005;280:23009–23017. doi: 10.1074/jbc.M414648200. [DOI] [PubMed] [Google Scholar]
- Xu JY, Xia QQ, Xia J. A review on the current neuroligin mouse models. Sheng Li Xue Bao. 2012;64:550–562. [PubMed] [Google Scholar]
- Yan R, Bienkowski MJ, Shuck ME, et al. Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- Yan R, Munzner JB, Shuck ME, Bienkowski MJ. BACE2 functions as an alternative alpha-secretase in cells. J. Biol. Chem. 2001;276:34019–34027. doi: 10.1074/jbc.M105583200. [DOI] [PubMed] [Google Scholar]
- Yang G, Gong YD, Gong K, et al. Reduced synaptic vesicle density and active zone size in mice lacking amyloid precursor protein (APP) and APP-like protein 2. Neurosci. Lett. 2005;384:66–71. doi: 10.1016/j.neulet.2005.04.040. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang Z, Wang B, Justice NJ, Zheng H. Amyloid precursor protein regulates Cav1.2 L-type calcium channel levels and function to influence GABAergic short-term plasticity. J. Neurosci. 2009a;29:15660–15668. doi: 10.1523/JNEUROSCI.4104-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Fucini RV, Fahr BT, et al. Fragment-based discovery of nonpeptidic BACE-1 inhibitors using tethering. Biochemistry. 2009b;48:4488–4496. doi: 10.1021/bi900017q. [DOI] [PubMed] [Google Scholar]
- Yu YJ, Zhang Y, Kenrick M, et al. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 2011;3:84ra44. doi: 10.1126/scitranslmed.3002230. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wada J, Hida K, et al. Collectrin, a collecting duct-specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J. Biol. Chem. 2001;276:17132–17139. doi: 10.1074/jbc.M006723200. [DOI] [PubMed] [Google Scholar]
- Zheng H, Jiang M, Trumbauer ME, et al. beta-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell. 1995;81:525–531. doi: 10.1016/0092-8674(95)90073-x. [DOI] [PubMed] [Google Scholar]
- Zhou L, Barao S, Laga M, et al. The neural cell adhesion molecules L1 and CHL1 are cleaved by BACE1 protease in vivo. J. Biol. Chem. 2012;287:25927–25940. doi: 10.1074/jbc.M112.377465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Sun ZY, Ye Y, et al. Discovery of cyclic acylguanidines as highly potent and selective beta-site amyloid cleaving enzyme (BACE) inhibitors: part I–inhibitor design and validation. J. Med. Chem. 2010;53:951–965. doi: 10.1021/jm901408p. [DOI] [PubMed] [Google Scholar]