Abstract
Salt-bridge interactions between acidic and basic amino acids contribute to the structural stability of proteins and to protein-protein interactions. A conserved salt-bridge is a canonical feature of the α-defensin antimicrobial peptide family, but the role of this common structural element has not been fully elucidated. We have investigated mouse Paneth cell α-defensin cryptdin-4 (Crp4) and peptide variants with mutations at Arg7 or Glu15 residue positions to disrupt the salt-bridge and assess the consequences on Crp4 structure, function, and stability. NMR analyses showed that both (R7G)-Crp4 and (E15G)-Crp4 adopt native-like structures, evidence of fold plasticity that allows peptides to reshuffle side chains and stabilize the structure in the absence of the salt-bridge. In contrast, introduction of a large hydrophobic side chain at position 15, as in (E15L)-Crp4 cannot be accommodated in the context of the Crp4 primary structure. Regardless of which side of the salt-bridge was mutated, salt-bridge variants retained bactericidal peptide activity with differential microbicidal effects against certain bacterial cell targets, confirmation that the salt-bridge does not determine bactericidal activity per se. The increased structural flexibility induced by salt-bridge disruption enhanced peptide sensitivity to proteolysis. Although sensitivity to proteolysis by MMP7 was unaffected by most Arg7 and Glu15 substitutions, every salt-bridge variant was degraded extensively by trypsin. Moreover, the salt-bridge facilitates adoption of the characteristic α-defensin fold as shown by the impaired in vitro refolding of (E15D)-proCrp4, the most conservative salt-bridge disrupting replacement. In Crp4, therefore, the canonical α-defensin salt-bridge facilitates adoption of the characteristic α-defensin fold, which decreases structural flexibility and confers resistance to degradation by proteinases.
Keywords: defensin, cryptdin-4, Crp4, salt-bridge, structure, folding, proteolytic stability
INTRODUCTION
Mammalian defensins are cationic and Cys-rich antimicrobial peptides (AMPs) with broad-spectrum antimicrobial activities (Ganz 2003; Selsted and Ouellette 2005). The peptide subfamilies are characterized by a central β-sheet that is constrained by three disulfide bonds whose connectivities and resultant topologies define the three subclasses α-, β-, and θ-defensins (Ganz 2003; Selsted and Ouellette 2005). α-Defensins consist of 32-40 amino acids and a characteristic CysI-CysVI, CysII-CysIV, CysIII-CysV arrangement of their disulfide connectivities (Selsted and Harwig 1989). β-defensins possess a CysI-CysV, CysII-CysIV, CysIII-CysVI array and generally exhibit conserved primary structures that enable the identification of homologues across diverse species lines (Pazgier et al. 2006). The θ-defensins are coded by truncated α-defensin genes in Old World monkeys and consist of only 18 amino acids that have a CysI-CysVI, CysII-CysV, CysIII-CysIV connectivity and a remarkable head-to-tail cyclic peptide backbone, which results from the post translational fusion of two separate hemi-precursors to form a macrocyclic peptide (Tang et al. 1999). They are one example of an increasing number of head-to-tail cyclic defense peptides that have been discovered in bacteria, plants and animals over recent years (Craik 2006).
Mammalian α-defensins are key mediators of innate immunity in phagolysosomes of neutrophils and in the small intestine where they are produced and secreted by Paneth cells (Selsted and Ouellette 2005). Paneth cell α-defensins, termed cryptdins (Crps) in mice, are secreted as components of dense core granules into the lumen of small intestinal crypts in response to cholinergic stimulation or exposure to bacteria or bacterial antigens (Satoh et al. 1986; Satoh 1988; Ayabe et al. 2000). The mode of action of these peptides involves interactions with the bacterial cell envelope leading to membrane disruption (White et al. 1995; Hadjicharalambous et al. 2008), which is dependent on peptide surface positive charge and amphipathicity (Tanabe et al. 2004), features shared by all α-defensins. The α-defensins are translated as inactive precursors that are activated post-translationally by lineage-specific proteinases (Wilson et al. 1999; Ghosh et al. 2002). Both myeloid and Paneth cell α-defensins derive from ~10 kDa prepropeptides that contain canonical signal sequences, electronegative proregions, and a 3.7-4.5 kDa mature α-defensin peptide in the C-terminal portion of the precursor. Pro-α-defensin processing and activation in mouse Paneth cells is catalyzed by matrix metalloproteinase-7 (MMP7) and takes place intracellularly and prior to secretion (Wilson et al. 1999; Ayabe et al. 2002; Shirafuji et al. 2003; Weeks et al. 2006; Figueredo et al. 2009). Disruption of the MMP7 gene abrogates intracellular proCrp activation, eliminating the accumulation of functional mature Crp peptides from the small intestine, resulting in impaired enteric innate immunity in response to oral bacterial infections (Wilson et al. 1999). In contrast to mice, trypsin activates human Paneth cell pro-α-defensins extracellularly during or after secretion into the crypt lumen (Ghosh et al. 2002).
Despite the diversity of known α-defensin primary structures, α-defensins share conserved features, including the invariant disulfide array (Selsted and Ouellette 2005), a canonical Arg-Glu salt-bridge (Wu et al. 2005), a conserved Gly residue at position CysIII+8 (Xie et al. 2005), and high Arg relative to Lys content (Llenado et al. 2009), all of which are hallmarks of the peptide family (Figure 1). Structure-function analyses by mutagenesis of these canonical features have shown consistently that bactericidal activity is independent of the conserved positions and properties, except for the high Arg content (Tanabe et al. 2004; Llenado et al. 2009). For example, disulfide-deficient Crp4 (Maemoto et al. 2004) and RMAD-4 (Kamdar et al.) mutants retain or exceed the bactericidal activity of the native molecules but are sensitive to proteolytic degradation by their activating convertases, MMP7 and azurophil serine proteinases, respectively.
Figure 1.
(a) Sequence comparison of mouse cryptdins. The conserved six cysteines, Gly, and the Arg and Glu involved in the conserved salt-bridge are highlighted in bold and the disulfide connectivities indicated by connecting lines. The sequences of the Crp4 analogues (E15G), (E15L) and (R7G) that have been characterized in this study are shown in the bottom part of the figure. (b) Structural representation of α-defensins. The structure is dominated by the central β-sheet. The side chains of the conserved disulfide array and the salt-bridge are shown in ball-and-stick. The cysteines are labelled with roman numbers I-VI in the order of appearance in the sequence.
The role of the conserved salt-bridge has been investigated in HNP-2 (Wu et al. 2005), Crp4 (Rosengren et al. 2006) and HD5 (Rajabi et al. 2008). Salt-bridge disruption did not diminish antibacterial activity or precursor folding in vitro in HNP-2 (Wu et al. 2005) or HD5, although HNP-2 and HD5 salt-bridge variants were susceptible to proteolysis by human neutrophil elastase (Wu et al. 2005) and trypsin (Rajabi et al. 2008), their respective processing enzymes. In contrast, native Crp4 and proCrp4, (E15D)-Crp4, (R7G)-Crp4, (E15G)-Crp4, (E15D)-proCrp4, (R7A)-proCrp4, and (R7G)-proCrp4 all were completely resistant to proteolysis by MMP7, the mouse Paneth cell α-defensin convertase (Rosengren et al. 2006).
Here, we have investigated the role of the canonical salt-bridge in the context of the Crp4 and proCrp4 primary structures. NMR solution structures of the Crp4 variants (R7G)-Crp4 and (E15G)-Crp4 reveal increased structural flexibility that enables both salt-bridge variants to adopt a native-like structure and fold but with marked sensitivity to trypsin proteolysis, thus complementing studies of the (E15D)-Crp4 structural variant (Rosengren et al. 2006). A comparative analysis of native proCrp4 and (E15D)-proCrp4 folding kinetics showed that removal of the salt-bridge via the conservative Asp for Glu substitution impaired peptide folding significantly. We conclude that this conserved structural feature facilitates α-defensin peptide folding and stabilizes folded molecules by limiting backbone flexibility to confer resistance to proteolytic degradation.
MATERIALS AND METHODS
Preparation of recombinant peptides
All peptide were produced recombinantly in E. coli as described previously (Rosengren et al. 2006; Figueredo et al. 2010). Peptides were purified to homogeneity by RP-HPLC and their identities were confirmed by MALDI-TOF MS and by acid-urea polyacrylamide gel electrophoresis (AU-PAGE) (Figueredo et al. 2010). Recombinant Crp4 and proCrp4 peptides were expressed in E. coli as N-terminal 6X-histidine tagged fusion proteins from the EcoRI and SalI sites of the pET28a expression vector (Novagen, Inc. Madison, WI) (Shirafuji et al. 2003; Satchell et al. 2003; Rosengren et al.). Mutations were introduced into Crp4 by PCR as described previously (Maemoto et al. 2004; Rosengren et al. ; Figueredo et al. 2010). Corresponding proCrp4 templates of these variants were prepared as described (Maemoto et al. 2004). All mutated Crp4 and proCrp4 templates were cloned in pCR-2.1 TOPO, verified by DNA sequencing, and subcloned into pET28a plasmid DNA (Novagen, Inc., Madison, WI), and transformed into E. coli BL21(DE3)-CodonPlus-RIL cells (Stratagene) for recombinant expression (Shirafuji et al. 2003; Satchell et al. 2003). His-tagged Crp4 fusion peptides were purified using nickel-nitrilotriacetic acid (Ni-NTA, Qiagen) resin affinity chromatography (Shirafuji et al. 2003). After CNBr cleavage, Crp4 peptides were purified by C18 reverse-phase high performance liquid chromatography (RP-HPLC), and molecular masses of purified peptides were determined using matrix-assisted laser desorption ionization mode mass spectrometry (Voyager-DE MALDI-TOF, PE-Biosystems, Foster City, CA) in the Mass Spectroscopy Facility, Department of Chemistry, University of California, Irvine.
Bactericidal peptide assays
Recombinant peptides were tested for microbicidal activity as before (Lehrer et al. 1988). Bacteria (~5 × 106 CFU/ml) resuspended in 10 mM PIPES (pH 7.4) supplemented with 0.01 vol of trypticase soy broth were incubated with test peptides in 50 μl for 1 h at 37°C, and surviving bacteria were counted as colony forming units per ml (CFU/ml) after overnight growth on semi-solid media (Satchell et al. 2003; Shirafuji et al. 2003).
Exposure of Glu15 Crp4 and proCrp4 variants to proteolytic enzymes
Recombinant Crp4, proCrp4, and variants with site-directed mutations at position 15 were digested with trypsin and analyzed for evidence of proteolysis by acid-urea PAGE (AU-PAGE) (Selsted et al. 1993; Shirafuji et al. 2003). Samples of Crp4 variants (5 μg) and proCrp4 variants (11 μg) were incubated with in buffer containing 10 mM N-[2-hydroxyethyl]piperizine-N'-[2-ethanesulfonic acid] (pH 7.4), 150 mM NaCl, 5 mM CaCl2 for 18-24 h at 37°C, and equimolar samples of all digests were analyzed by AU-PAGE (Shirafuji et al. 2003; Maemoto et al. 2004).
Analysis of peptide refolding kinetics
Samples (100 μg) of native proCrp4 and (E15D)-proCrp4 were reduced in 6.0 M guanidine-HCl, 0.2 M Tris base, 2 mM sodium EDTA at pH 8.2. The peptides were adjusted to 5% (vol/vol) acetic acid, and reduced forms were purified by analytical C18 RP-HPLC at ambient temperature using a 15-30% acetonitrile gradient in 0.1% TFA over 90 min at a flow rate of 1 ml/min. Samples (2-10 μg) of reduced peptides were tested for peptide quality by chromatography at ambient temperature on a Vydac C18 (3 μm particle size) column (150 mm × 2.1 mm (i.d.)) using a Waters Alliance 2690 instrument developed with a 20-40% acetonitrile gradient in 0.1%TFA for 65 min at a flow rate of 400μl/min. To compare refolding kinetics, reduced peptides were concentrated to 1 ml were diluted with 4 ml 0.1 M ammonium bicarbonate, 2 mM EDTA, 0.1 mg/ml cysteine, 0.1 mg/ml cysteine at pH 8.0. Aliquot samples (1 ml) of refolding mixtures were removed at 2 h intervals, adjusted to 5% acetic acid, concentrated to 100 μl and analyzed by C18 RP-HPLC on an Alliance 2690 as above using 20-40% acetonitrile, 0.1% TFA for 65 min at ambient temperature.
NMR Spectroscopy
Samples for structure determination contained ~1 mg of (R7G)-Crp4, ~0.3 mg of (E15G)-Crp4 or ~0.5 mg of (E15L)-Crp4 dissolved in either 90% H2O and 10% D2O (v/v) or 100% (v/v) D2O at pH ~5. All spectral data on the (E15G)-Crp4 and (E15L)-Crp4 analogues were recorded at 600 or 500 MHz on Bruker Avance NMR spectrometers, with additional data for structure determination of the (E15G)-Crp4 analogues recorded at 900 MHz on a Bruker Avance II spectrometer equipped with a cryogenic probe. Two-dimensional experiments recorded included DQF-COSY, TOCSY using an MLEV-17 spin lock sequence with a mixing time of 80 ms, and NOESY with mixing times of 100 ms, 150 ms or 200 ms. Chemical shifts were internally referenced to DSS at 0.00 ppm. Evidence for hydrogen bonds was deduced from amide temperature coefficients (ΔδHN/ΔT), which were determined by recording TOCSY spectra at 288, 293, 298, 303 and 308 K and plotting the amide shifts as a function of temperature.
Structure Determinations
The spectral data were analyzed and cross peaks were assigned and integrated in CARA (Keller 2004) and converted to distance restraints using CYANA (Guntert et al. 1997). Distance restraints for (R7G)-Crp4 were derived from a 200 ms NOESY spectra recorded at 298 K and 600 MHz, while distance restraints for (E15G)-Crp4 were deduced primarily from a NOESY spectrum recorded at 900 MHz with a mixing time of 150 ms. Backbone dihedral restraints were inferred from 3JHN-Hα coupling constants derived either from the 1D or a high digital resolution DQF-COSY. The dihedral angle φ was restrained to −120 ± 30° for 3JHN-Hα greater than 8 Hz (R7G: Tyr Cys6, Lys8, Cys11, Lys12, Glu15, Arg16, Arg18, Thr20, Cys21, Phe25, Leu26, Tyr27, Cys28, Cys29; E15G: Arg7, Arg16, Arg18, Cys21, Phe25, Leu26, Tyr27, Cys28, Cys29), to −60 ± 30° for 3JHN-Hα < 5 Hz (R7G: Lys , Ile23), and to 50 ± 40° for a coupling constant of ~7 Hz in combination with a very strong intra Hαi-HNi NOE (R7G: Arg24). Additional φ angle restraints of –100 ± 80° were included where the positive angle could be excluded based on strong sequential Hαi-1-HNi NOE compared to the intra residual Hαi-HNi NOE. Side chain χ1 angles and stereo specific assignments were determined on the basis of observed NOE and 3JHα-Hβ coupling patterns (Wagner 1990). Dihedral restraints of −60° ± 30° (R7G: Cys4, Cys11, Phe25, Cys28, Cys29; E15G: Cys4, Cys11, Phe25, Leu26, Cys28, Cys29), −180° ± 30° (R7G: Tyr27; E15G: Lys8, Tyr27) and 60 ± 30° (R7G: Tyr5, His10; E15G: Tyr5) were used for confirmed t2g3, g2t3 and g2g3 conformations, respectively. Hydrogen bonds were included into the structure calculations for all amide protons concluded to be slow exchanging, or having a ΔδHN/ΔT consistent with a hydrogen bond, only once a suitable acceptor could be identified in the preliminary structures. Three-dimensional structures were calculated using simulated annealing followed by refinement and energy minimization in a water shell using protocols within the program CNS (Brunger et al. 1998). The simulated annealing included a high temperature phase at 50000 K with 4000 steps of 0.015 ps of torsion angle dynamics, a cooling phase with 4000 steps of 0.015 ps torsion angle dynamics and finally an energy minimization phase of 5000 steps of Powell minimization. These structures were subsequently refined in a water shell in a computational process involving first heating to 500 K in steps of 100 K, each comprising 50 steps of 0.005 ps of Cartesian dynamics, second 2500 steps of Cartesian dynamics at 500 K, third lowering of the temperature in steps of 100 K, each comprising 2500 steps of 0.005 ps Cartesian dynamics, and finally 2000 steps of Powell minimization. For all calculations the force field topallhdg5.3.pro/parallhdg5.3.pro developed for the RECOORD database was used (Nederveen et al. 2005). The coordinates representing the solution structures of (R7G)-Crp4 and (E15G)-Crp4 and the experimental restraints have been submitted to the PDB and given the access codes 2ley and 2lew.
RESULTS
NMR Spectroscopy and Resonance Assignments of Crp4 Analogues
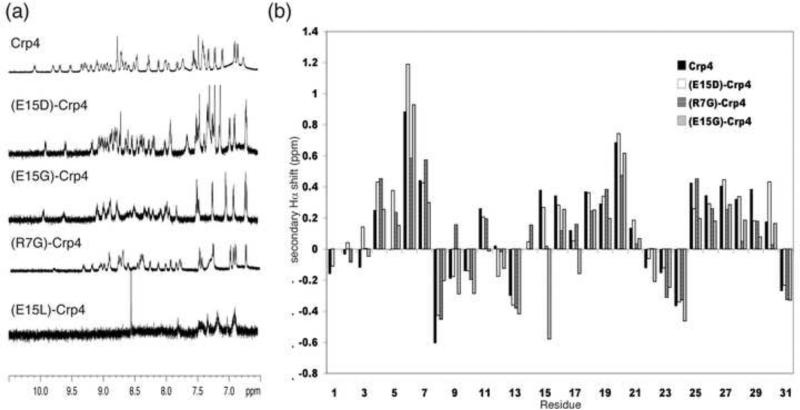
Peptides were expressed recombinantly in E. coli and purified to homogeneity by extensive RP-HPLC and their identities were confirmed by MALDI-TOF MS. To detect structural changes in the Crp4 framework as a result of mutations to the Arg7-Glu15 salt-bridge, several Crp4 analogues were studied by solution NMR spectroscopy. Homonuclear two-dimensional NMR data were recorded for (R7G)-Crp4, (E15G)-Crp4 and (E15L)-Crp4 at 500, 600 or 900 MHz. As shown in Figure 2a, the initial NMR analyses revealed that (E15G)-Crp4 and (R7G)-Crp4 showed an excellent dispersion of amide proton signals, similar to what has been reported previously for native Crp4 and (E15D)-Crp4 and consistent with a folded structure. In contrast, (E15L)-Crp4 signals were severely broadened. In fact, in the 1H 1D spectrum only the normally very sharp aromatic signals could be observed, while the amide signals were broadened beyond detection. This is consistent with a misfolded and aggregated peptide and prevented further structural analysis on (E15L)-Crp4. For both (E15G)-Crp4 and (R7G)-Crp4, peak assignments were achieved using standard sequential assignment strategies and near complete assignments of both backbone and side chain protons could be achieved. The secondary Hα proton chemical shift, i.e. the deviation of the observed chemical shifts from the chemical shifts expected for a random coil peptide (Wishart et al. 1995), are good indicators of secondary structure, with positive numbers consistent with β-sheet structure. Figure 2b shows a comparison of the secondary shifts between Crp4 and analogues, and it is clear that the overall defensin fold with three distinct strands comprising residues 4-7, 15-21 and 25-30 is retained in all analogues, and that the main differences are seen around the mutations.
Figure 2.
NMR data on Crp4 and analogues. (a) 1D 1H spectrum of Crp4, (E15D)-Crp4, (E15G)-Crp4, (R7G)-Crp4 and (E15L)-Crp4. Native Crp4 as well as the (E15D)-, (E15G)- and (R7G)- analogues show good signal dispersion. In contrast (E15L)-Crp4 is misfolded and aggregates in solution, resulting in broad lines with amide signals not visible in the 1D experiment. (b) Secondary Hα chemical shifts for Crp4 and analogues. Similar trends with stretches of positive numbers characteristic of β-sheets are seen in all peptides, indicating that the native fold is retained despite the mutations to the salt-bridge.
In native Crp4, the Hε proton of the Arg7 side chain involved in the salt-bridge is downfield shifted by > 2 ppm (to 9.61 ppm). Previously, we showed that this is not the case in (E15D)-Crp4, because the interaction between Arg7 and Glu15 is disrupted (Rosengren et al. 2006), and it is not so in (E15G)-Crp4 either. Interestingly, when all chemical shifts between the different Crp4 variants were compared, the major changes induced by different mutations are similar between the salt-bridge analogues. For example the β-protons of Cys28 were upfield shifted in (E15D)-Crp4 by 1.5 and 1.0 ppm compared to the native peptide, and in (E15G)-Crp4 and (R7G)-Crp4, these resonances are upfield shifted by 1.0 / 0.9 ppm and 1.5 / 0.6 ppm, respectively. Furthermore, the HN and Hβ protons of Cys4 are downfield shifted by 0.3-0.5 ppm in all analogues compared to native Crp4. Finally, downfield shifts of 0.3-0.5 ppm are seen for one of the Hβ protons of Tyr5 in all analogues. In contrast, different effects of mutagenesis are seen for Cys11, where one Hβ proton is up-field shifted by ~0.5 ppm in both (E15D)-Crp4 and (E15G)-Crp4 but were very similar to native Crp4 in (R7G)-Crp4.
Although the defensin fold is retained in salt-bridge variants, it has greater flexibility after salt-bridge disruption. Often, peak broadening is seen in regions of flexibility due to the averaging of the chemical shifts originating from different conformations. Although such broadening is not observed in native Crp4, the lines are generally broader in mutated analogues, as evident from the 1D spectra in Figure 2. The most affected analogue is the (E15G)-Crp4 mutant. Specifically, broadening in this analogue is seen in the loop around the mutation and involves residues 10-15, with the greatest effect at Gly14 for which the amide proton could not be detected in any of the 2D spectra recorded. Interestingly, broadening is also evident around residues 20-24 of (E15G)-Crp4, indicating that the increased flexibility may be translated through to other parts of the molecule. As expected, the degree of peak broadening is frequency dependent and significantly more severe in many cases at the high field of 900 MHz. Accordingly, a combination of spectral data from several fields were used to gain maximum assignments for (E15G)-Crp4. Conformational heterogeneity in a protein in solution is also frequently evident from the presence of additional spin systems arising from alternative conformations. However, no such additional spin systems were observed. Crp4 contains one proline residue, Pro30, which was confirmed to be in the trans conformation in all analogues based on strong sequential Hαi-1-Hδi NOEs.
The fold of the defensins are largely determined by their disulfide array. Disulfide bond connectivities can be determined from NMR data using a combination of NOEs reporting on inter-cysteine distances and χ1 dihedral angles, which define the projections of the cysteine side chains (Rosengren et al. 2003). This strategy was utilised previously to confirm the typical α-defensin disulfide connectivity of native Crp4 (Rosengren et al. 2006), and no significant differences in terms of NOE patterns or coupling constants of the cysteine residues could be observed in the data for (R7G)-Crp4 and (E15G)-Crp4, confirming that they have retained the native disulfide connectivity and thus this arrangement could be included in the structure calculations.
Structure Determination and (E15G)-Crp4 and (R7G)-Crp4 3D structures
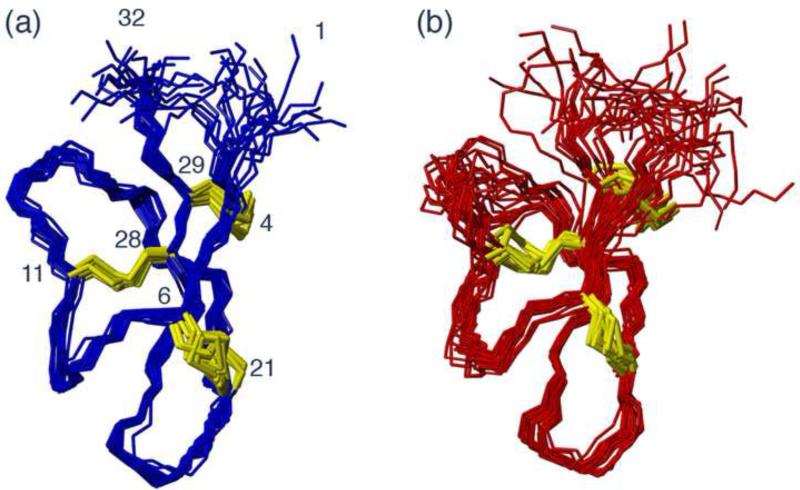
A set of structural restraints that included inter-proton distances, dihedral angles and hydrogen bond restraints were derived based on the NMR data for (E15G)-Crp4 and (R7G)-Crp4. These restraints were used to calculate a family of 50 structures of which the 20 lowest energy structures were chosen to represent the solution structure for each peptide. The preliminary structures were used to guide assignments of NOE cross peaks in cases where assignments based solely on chemical shifts were ambiguous and for the identification of hydrogen bond acceptors for amide protons identified as donors by amide exchange or temperature variation experiments. A summary of the restraints used and the structural statistics for the final round of structure calculations are presented in Table 1. Superpositions of the final 20 structures for (R7G)-Crp4 and (E15G)-Crp4 are shown in Figure 3. The characteristic native α-defensin fold, with a triple stranded β-sheet stabilized by the disulfide array, is retained in both peptides. However, differences exist in the precision of the structures in certain regions. The structural definition of the (E15G)-Crp4 structure is worse near residues 10-15 and 20-24. This is not unexpected given that these regions are broadened, and limited structural information could be gained for these residues as a result. Although the degree of disorder might be artificially augmented as a result of these missing data, a definite increase in structural flexibility is directly evident from the resonance line broadening.
Table 1.
Energies and refinement statistics for the families of NMR structures.
| NMR distance & dihedral constraints, Energies and statistics | (R7G)-Crp4 | (E15G)-Crp4 |
|---|---|---|
| Distances Total inter-residual NOE | 142 | 199 |
| Sequential NOE (|i-j| = 1) | 76 | 79 |
| Medium-range NOE (|i-j| ≤ 4) | 18 | 17 |
| Long-range NOE (|i-j| > 4) | 48 | 103 |
| Hydrogen bonds | 13 | 6 |
| Dihedral angles (backbone φ / side chain χ1) | 23 / 8 | 20 / 10 |
| Violations (NOE °0.2 A / dihedral ° 3°) | 0 / 0 | 2 / 0 |
| Energy total | −1085±24 | −885±23 |
| Bonds | 6.37±0.63 | 6.47±0.75 |
| Angles | 34.2±4.7 | 37.9±4.6 |
| Impropers | 5.69±0.84 | 5.65±1.3 |
| VdW | −84.3±4.8 | −86.9±7.1 |
| NOE | 4.65±1.4 | 7.29±3.18 |
| Experimental dihedrals | 0.24±0.15 | 0.13±0.13 |
| Dihedrals | 141±29 | 138±13 |
| Electrostatic | −1139±29 | −994±23 |
| Average Pairwise RMSD[a] (backbone atoms) | 0.65±0.20 | 1.16±0.26 |
| (heavy atoms) | 1.84±0.24 | 2.33±0.30 |
| Ramachandran Statistics: Most Favoured | 74.2% | 69.8% |
| Additionally allowed | 25% | 24% |
| Generously allowed | 0.2% | 3.8% |
| Disallowed | 0.6% | 2.5% |
[a] Pairwise RMSDs were calculated among 20 refined structures over the structured region, residues 4-29.
Figure 3.
Solution structures of (R7G)-Crp4 (a) and (E15G)-Crp4 (b). The families of 20 structures with the lowest energy from a calculated total of 50 structures are shown superimposed over residues 4-29. Disulfide bonds are shown in yellow.
Temperature variation studies of (E15G)-Crp4 and (R7G)-Crp4
Amide proton temperature coefficients (ΔδHN/ΔT), i.e., the dependence of the amide proton chemical shifts on temperature, are useful for characterizing hydrogen bond networks in proteins; internal hydrogen bonded amides are protected from the solvent and generally less sensitive to temperature changes (Clark et al. 2010). Thus, to investigate effects on the hydrogen bonding pattern of the salt-bridge variants, TOCSY spectra were recorded on (E15G)-Crp4 and (R7G)-Crp4 between 285-315 K, and the chemical shifts were plotted as a function of temperature. A comparison of the data for Crp4 and salt-bridge analogues is provided as supplementary information. Statistical analyses have revealed that >85% of amide protons with a ΔδHN/ΔT of > −4.6 ppb/K are involved in hydrogen bonds with the probability increasing to above 93% for a ΔδHN/ΔT between −1.0 and −4.0 ppb/K (Cierpicki and Otlewski). The analysis of Crp4 and variants generally agrees well with this, and, with the combined use of ΔδHN/ΔT and amide exchange data, a total of 14 hydrogen bonds involving backbone amide protons can be identified in the framework. The Crp4 salt-bridge mutants showed similar patterns of ΔδHN/ΔT indicating that in general the hydrogen bonding network was retained, except for Arg16 in (E15G)-Crp4, which appeared to lose the hydrogen bond to the carbonyl of Cys29 found in Crp4, (E15D)-Crp4 and (R7G)-Crp4.
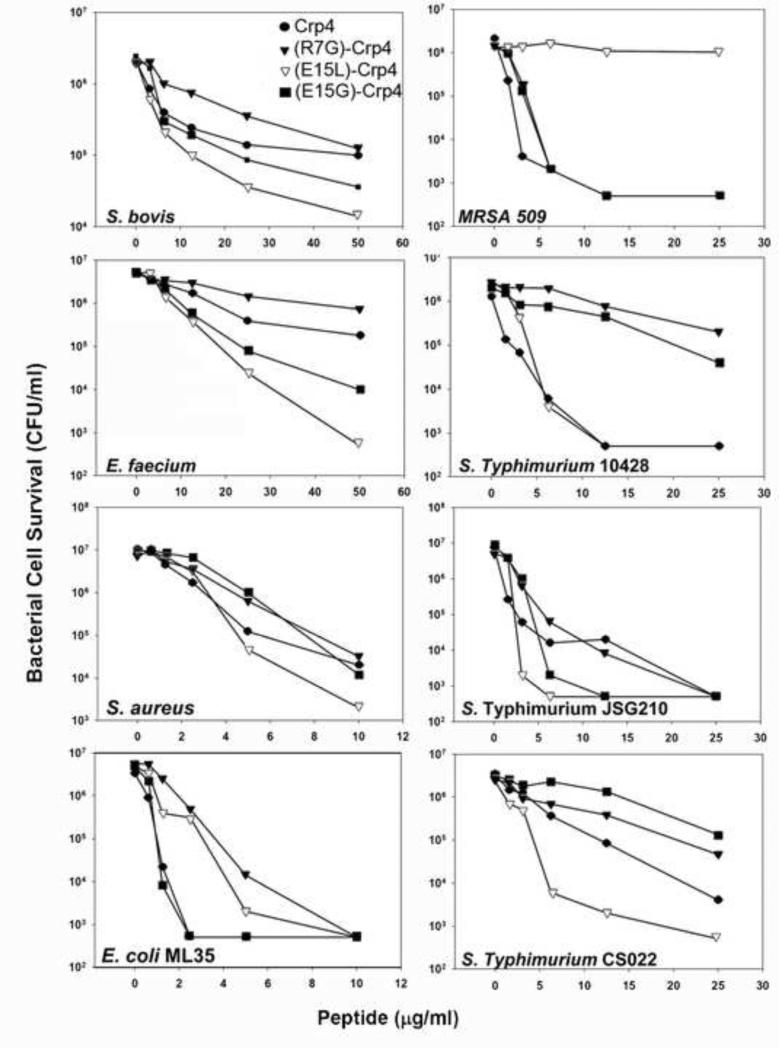
Crp4 Salt-bridge Variants Retain Bactericidal Activity
The biological effects of introducing structural flexibility by salt-bridge mutagenesis were tested by measuring bactericidal peptide activities of Crp4 and salt-bridge mutants in in vitro assays (Figure 4). For most bacterial cell targets, the effects of salt-bridge mutagenesis on bacterial cell survival were modest, with most variants retaining microbicidal activity against most bacterial species tested. In contrast, (E15L)-Crp4, a variant whose structure was most affected by salt-bridge disruption, lacks activity against MRSA509 but is the most active variant against most other species. AMPs characteristically have differential activities against varied bacterial cell targets, with potent bactericidal effects against certain species and none against others (Wilmes et al. 2011; Rosengren et al. 2002). Thus, although the structure of the (E15L)-Crp4 differs significantly from Crp4, it has somewhat augmented activity.
Figure 4.
Antimicrobial activity of Crp4 analogues. The antimicrobial activity of salt-bridge deficient analogues were tested against eight different microbes and the bactericidal effect is reported as reduction in bacterial cell survival as a function of peptide concentration.
Similarly, α-defensin disulfide variants, which are converted from the constrained α-defensin fold to highly disordered structures, have greater bactericidal peptide activity than corresponding native peptides (Maemoto et al. 2004; Hadjicharalambous et al. 2008; Kamdar et al. 2008), showing that mutations that increase peptide flexibility may not adversely affect bactericidal activity.
Salt-bridge Mutagenesis Induces Proteolytic Instability in Crp4 and ProCrp4
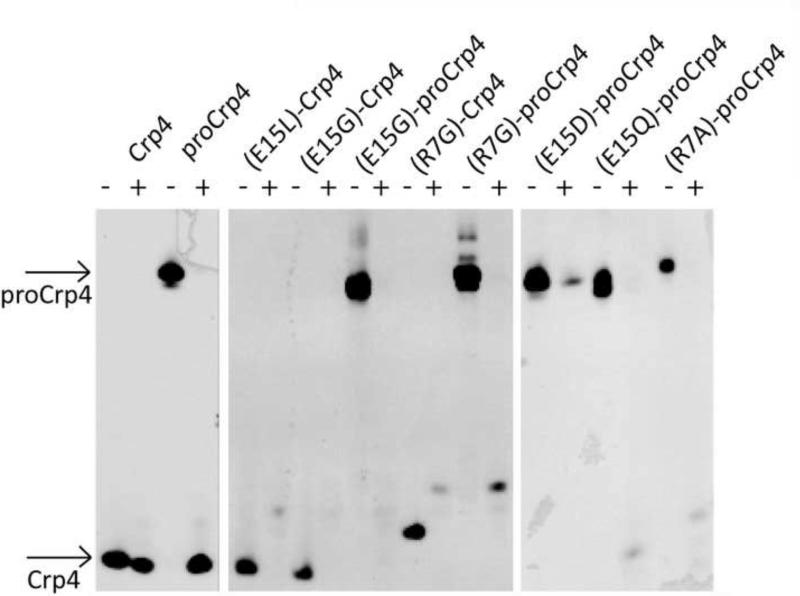
MMP7 is the mouse Paneth cell α-defensin activating convertase, and MMP7 accurately processes native and salt-bridge variants of Crp4 and proCrp4 to mature forms that, like native Crp4, resist MMP7-mediated proteolysis (Rosengren et al. 2006). Because Paneth cell trypsin activates human Paneth cell α-defensin, and native α-defensins resist degradation by trypsin and additional serine proteases (Kamdar et al. 2008; Figueredo et al. 2009; Maemoto et al. 2004), we compared the relative sensitivity of Crp4, proCrp4 and their salt-bridge variants to trypsin proteolysis. In contrast to their resistance to MMP7, every salt-bridge mutant was degraded extensively by trypsin, whether incubated with mature or precursor forms (Figure 5). Exposure of native proCrp4 to trypsin results in degradation of the precursor proregion, but the mature α-defensin domain is completely resistant. Thus, although MMP7 is the mouse Paneth cell α-defensin convertase, trypsin can also effectively process the precursor in vitro into a native like peptide that is resistant to further degradation.
Figure 5.
Proteolytic stability of Crp4, proCrp4 and various salt-bridge mutated analogues. AU-PAGE analysis of untreated peptides (−) and peptides treated with trypsin. Arrows indicate the bands corresponding to Crp4 and proCrp4. Strikingly native proCrp4 is processed into a native-like form of Crp4, which is stable to further degradation. In contrast all mutant peptides, whether subjected to trypsin as precursors or mature peptides, are fully degraded by trypsin.
Impaired Folding of the (E15D)-proCrp4 Salt-bridge Variant
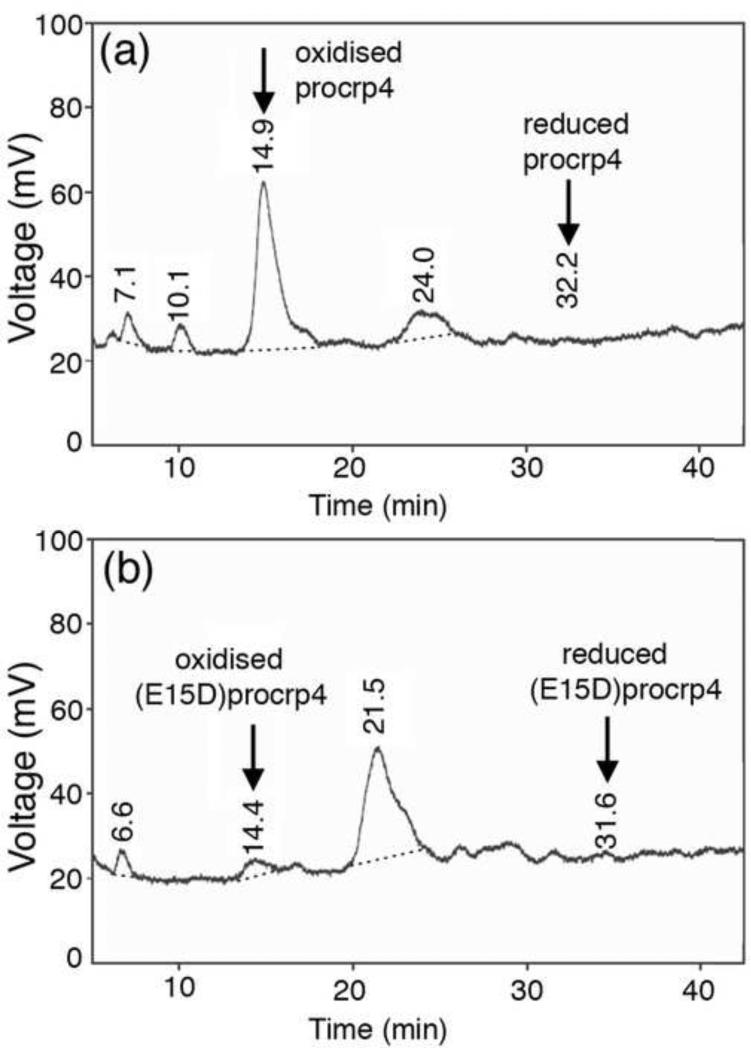
The role of the conserved salt-bridge in proCrp4 folding and formation of the disulfide array was investigated by comparing the in vitro folding behaviour of proCrp4 and the most conservative salt-bridge variant, (E15D)-proCrp4, after peptide reduction. As shown in Figure 6, the relative quantity of correctly folded precursor detected after 8 h of oxidative folding was markedly lower for (E15D)-proCrp4 compared to native proCrp4. Also, at all other time points sampled, the relative amount of folded native proCrp4 always exceeded that of the salt-bridge variant (not shown). These findings illustrate the critical role of the salt-bridge in proCrp4 folding in vitro, suggests a potential role for the salt-bridge in facilitating in situ pro-α-defensin folding in the endoplasmic reticulum.
Figure 6.
Folding of proCrp4 and (E15D)-proCrp4. HPLC traces of proCrp4 (a) and (E15G)-proCrp4 (b) after oxidative folding for 8h.
DISCUSSION
We have determined the consequences of mutational changes to the structurally conserved α-defensin salt-bridge in mouse Crp4. The canonical salt-bridge in mouse Crp4 is highly critical for efficient and correct folding of proCrp4 and for retention of a rigid polypeptide backbone structure, even though it is not essential for maintaining the α-defensin fold in the overall peptide structure once the tridisulfide array has formed. Although salt-bridge deficient analogues retain resistance towards the MMP7 activating convertase, all peptides with salt-bridge disruptions are degraded extensively by trypsin and would be unlikely to persist in the intestinal lumen as active forms. Thus, although Crp4 peptides lacking the salt-bridge have antimicrobial activity in vitro, peptide rigidity introduced by salt-bridge formation enables the molecules to confer enteric mucosal innate immunity. Previously, we demonstrated that the salt-bridge interaction between Arg7 and Glu15 was eliminated by the conservative Glu15 to Asp substitution. Although retaining electronegative charge, loss of the single side chain methylene group by mutagenesis prevented direct contact between the two side chains. Interestingly, overall Crp4 structure, bactericidal activity, or proteolytic stability to MMP7 were not compromised by the mutation. However, the structural and functional characterization of these additional salt-bridge variants of Crp4 and proCrp4, molecules, has allowed the visualization of decreased backbone rigidity due to salt-bridge disruption, which provides the structural basis for induced susceptibility to trypsin proteolysis.
The α-defensin fold is maintained after salt-bridge disruption
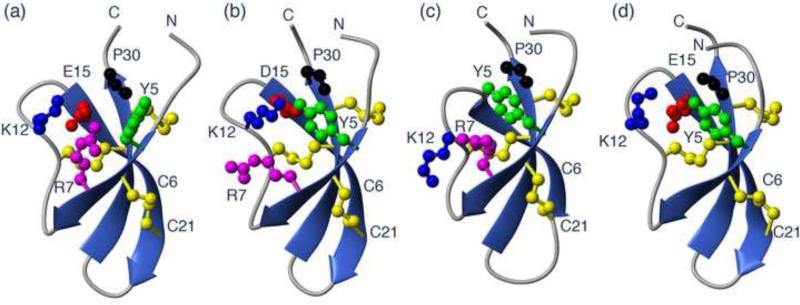
Although the conservative change of Glu15 to Asp was sufficient to remove the salt-bridge interaction between Arg7 and Glu15 (Rosengren et al. 2006), the position of the negatively charged Asp15 side chain in the core of a cluster of positive charges in Crp4 (Figure 7) is still likely to stabilize the fold. To characterize the role of such electrostatic interactions in this region further, we prepared additional mutants of both amino acids that contribute to the salt-bridge. The NMR data of (R7G)-Crp4 and (E15G)-Crp4 were of good quality, and allowed detailed structural characterization. A comparison of the native Crp4 with (E15D)-Crp4, (R7G)-Crp4 and (E15G)-Crp4 (Figure 7) shows that none of the mutations causes major conformational change of the peptide backbone. Nevertheless, side chain conformational changes are evident, and, interestingly, the changes in the mutant analogues are similar. For example, when the salt-bridge is disrupted, the Tyr5 side chain moves to fill the void left in the structure. This change is confirmed both by changes in the coupling constants for the Tyr5 residue itself and by the chemical shifts changes observed, in particular the upfield shifts of the Hβ protons of Cys28. These changes are consistent in all salt-bridge variants. The (E15L)-Crp4 molecule is severely misfolded and aggregated, and structural analysis showed that a large hydrophobic amino acid at position 15 would be highly unfavourable, given its positioning in a cluster of electropositive charge. It seems likely that introduction of hydrophobicity prevented correct disulfide pairings from forming, but the unusually low yields of oxidized (E15L)-Crp4 on refolding prevented this idea from being confirmed. Figure 7 also highlights a potential interaction between the positive charge of Lys12 with the carboxyl group of Glu15. Although Lys12 is conserved in the mouse defensins (Figure 1), this residue is highly variable in α-defensins from different species with the equivalent position accommodating residues as diverse as Asn, Ile, Arg, Ala, Tyr, Gly, Val, Leu, Pro, and Phe. Thus in this context of a general understanding of the role of the salt-bridge this position did not warrant further investigation by mutational studies.
Figure 7.
Structural effects of mutations to the conserved salt-bridge in Crp4. Comparison of the structures of Crp4 (a), (E15D)-Crp4 (b), (E15G)-Crp4 (c) and (R7G)-Crp4 (d). While the overall fold of the peptide is retained structural changes are seen in the side chains around the point of mutation in the Crp4 analogues. The structure of (E15G)-appears to be most affected and changes to the backbone seems to be translated from the residue 10-15 loop down through the β-strand to residues 20-24.
The salt-bridge variants share overall structural similarities with native Crp4, but extensive differences also were apparent and notable. For example, the spectral peak broadening observed in (E15G)-Crp4, an indicator of polypeptide chain flexibility, is evidence of increased backbone mobility, even though the fold imposed by the tridisulfide array is retained and the disulphide pairings are correct. Links between mutations to salt-bridge and backbone flexibility have been extensively characterized using molecular dynamics simulations, e.g. (Missimer et al. 2007), but experimental data are limited. Amide temperature coefficients were recorded for all salt-bridge analogues to detect changes in the backbone hydrogen bond network that could account for increased flexibility. Mostly, the temperature coefficients were consistent with slow exchange data, confirming the presence of hydrogen bonds, but, in native Crp4 Arg7 and Arg18, temperature coefficients suggest that they are not hydrogen bonded despite exchanging slowly and showing the presence of hydrogen bonds in the derived structure. These amide protons are downfield shifted significantly in the Crp4 spectrum. Such amino acid residues may appear as false negatives in temperature coefficient analyses due to a degree of thermal unfolding, which creates a shift towards random coil at higher temperatures giving an apparent higher temperature dependence than expected (Andersen et al. 1997). The only evidence for a decrease in hydrogen bonding induced by salt-bridge mutagenesis was seen for Arg16 in (E15G)-Crp4, which has a temperature coefficient that supports the existence of a hydrogen bond in both (E15D)-Crp4 and (R7G)-Crp4 but not in (E15G)-Crp4. Disruption of additional hydrogen bonds in (E15G)-Crp4 is possible for residues that were affected by resonance broadening. For example, the temperature coefficient for Cys21, which is hydrogen bonded in native Crp4, (E15D)-Crp4, and (R7G)-Crp4 could not be determined in (E15G)-Crp4 due to broadening.
Robust bactericidal peptide activity in salt-bridge variants
In in vitro antimicrobial peptide assays against varied bacterial species, Crp4 salt-bridge mutants retained or showed increased bactericidal activity against most target cells. Because completely unstructured disulfide-null α-defensin variants have potent antimicrobial activities (Kamdar et al. 2008);(Maemoto et al. 2004), these findings may have been anticipated in view of the comparatively modest structural changes induced by disrupting the salt-bridge. In contrast, charge reversal or neutralizing mutations at Arg residues leads to a stepwise decrease in activity, evidence that the mechanism of peptide action requires specific electrostatic interactions prior to bacterial cell membrane disruption, but overall peptide structure generally is not a determinant of activity. However, (E15L)-Crp4, the most structurally perturbed of the variants analyzed, was the most potent analogue tested against several species but was inactive against S. aureus, suggesting that alternative modes of action may be important in targeting certain bacterial species (Hadjicharalambous et al. 2008).
The canonical salt-bridge induces rigidity critical for proteolytic stability
Changes in thermal stability as a result of mutations to salt-bridges has been reported for several peptides and proteins, including Trp-cage peptides (Williams et al. 2011), but effects on proteolytical stability have been less studied. Native Crp4, (E15D)-Crp4 and additional salt-bridge variants are highly resistant to proteolysis by MMP7, the activating convertase (Rosengren et al.). Because MMP7 co-localizes with proCrps in Paneth cell dense core granules, resistance of the mature α-defensin domain to the activating convertase is essential for biological activity. The enhanced polypeptide flexibility induced by salt-bridge disruption prompted us to investigate proteolytic stability further to include trypsin, a robust proteinase found in small intestinal lumen into which α-defensins are secreted. All analogues, including (E15D)-Crp4, were degraded rapidly by trypsin. Thus, although Crp4 salt-bridge mutants resist MMP7-mediated proteolysis (Rosengren et al. 2006), the salt-bridge determines resistance to trypsin, and we speculate that the rigidity it establishes contributes to α-defensin persistence in the intestinal tract and thus to their biological function.
Studies indicate that slight destabilization of one part of a structure can create susceptibility to proteases not only near the mutation site but also in regions distal from the mutation as has been shown in human Paneth cell α-defensin HD5. Although native HD5 is insensitive to both trypsin and chymotrypsin, salt-bridge mutagenesis resulted in HD5 degradation by both enzymes (Rajabi et al. 2008). Although the first trypsin cleavage event occurs within the destabilized loop between Arg13 and Gln14, chymotrypsin cleaves between Tyr27 and Arg28, thirteen residues from the mutation site. Thus, eliminating the salt-bridge can destabilize the entire peptide, even after it has folded and established a correctly paired tridisulfide array. Similarly, native conotoxin MII, which comprises 16 residues with two disulfide bonds, can be degraded by endo-Gluc, which cleaves after Glu11. However, when the conotoxin MII peptide is cyclized by introduction of a 6-residue linker between the terminal Cys16 and N-terminal Gly1, the peptide becomes completely stable to endo-GluC due to added rigidity of the fold (Clark et al. 2005).
A potential way of further probing the role of the salt-bridge for peptide stability is to reverse the positions of the charged groups, i.e., making an R7E-E15R double mutant. Although such a peptide may provide further evidence for the role of the direct contact between the charged groups it was not produced here as differences in the local environment of the two residues, including the potential for clashes between the positive charge of Lys12 and the introduced positive charge at position 15, would likely influence salt-bridge formation, leading to misinterpretation of a negative result.
The conserved salt-bridge facilitates Crp4 precursor folding
The defining signature of α-defensins is the conserved disulfide array that provides the main stabilizing feature of the peptide family. Moreover, α-defensins must also fold efficiently to ensure proximity of correct Cys residue positions for formation of accurate disulfide pairings that are critical for peptide proteolytic stability and function. For example, disruption of the salt-bridge in HD5 by substitution of Glu14 to Gln14 impaired folding of the mature HD5 peptide (Rajabi et al. 2008), but introducing the same mutation into the full-length proHD5 precursor had no effect on precursor folding. This report suggested that the covalently associated proregion contributes to the precursor thereby overcoming the effects of mutagenesis on folding of the mature HD5 folding. For this reason, we investigated the effect of the most conservative E15D salt-bridge substitution on the folding of the full-length proCrp4 precursor. In contrast to the efficiency of (E14Q)-proHD5 folding, folding of (E15D)-proCrp4 was greatly impaired, evidence for a critical role for the salt-bridge in proCrp4 folding in vitro. Therefore, despite the conservation of the salt-bridge, its disruption may have differential adverse effects determined, in part, by other aspects of pro-α-defensin primary structure. Thus, one role of the canonical α-defensin salt-bridge is to facilitate Crp4 precursor folding, leading to establishment of the stabilizing disulfide array. Once folded and disulfide-stabilized, the salt-bridge provides rigidity that confers protection against robust proteinases. We speculate that the salt-bridge may contribute to efficient pro-α-defensin folding in the endoplasmic reticulum in vivo for eventual packaging into dense core granules. Considering the high levels of α-defensin gene expression in Paneth cells and promyelocytes and the extensive conservation of the salt-bridge in α-defensins, somatic mutations that disrupt the salt-bridge may induce in situ pro-α-defensin misfolding, resulting in Paneth cell endoplasmic reticulum stress, and contribute to local inflammatory responses (Kaser et al. 2011; Kaser and Blumberg 2010; McGuckin et al. 2010).
CONCLUSIONS
In summary, although the conserved salt-bridge in Crp4 is not essential for the overall peptide fold or determine bactericidal action in vitro, it has dual and critical roles in both the production and stability of the mature peptide. Compromising the salt-bridge significantly impaired proCrp4 folding in vitro, and perhaps in vivo, even when the mutational change is as subtle as E15D. Furthermore, NMR analyses of several variants revealed that disruption of the salt-bridge increased flexibility of the structure, conferring sensitivity to proteolytic degradation, an essential feature for Crp4 and α-defensins to mediate enteric immunity in protease-rich environment of the small and large intestinal lumen.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by the Linnaeus University (KJR and HSA), Australian Research Council and National Medical & Health Research Council (NHMRC) (DJC), and NIH Grants DK044632 and AI059346 (AJO). KJR is an NHMRC Biomedical CDA Fellow. NLD is a Queensland Smart State Fellow. DJC is an NHMRC Professorial Research Fellow. We gratefully acknowledge the use of the Queensland NMR Network 900 MHz spectrometer.
REFERENCES
- Andersen NH, Neidigh JW, Harris SM, Lee GM, Liu ZH, Tong H. Extracting information from the temperature gradients of polypeptide NH chemical shifts .1. The importance of conformational averaging. J Am Chem Soc. 1997;119(36):8547–8561. [Google Scholar]
- Ayabe T, Satchell DP, Pesendorfer P, Tanabe H, Wilson CL, Hagen SJ, Ouellette AJ. Activation of Paneth cell alpha-defensins in mouse small intestine. J Biol Chem. 2002;277(7):5219–5228. doi: 10.1074/jbc.M109410200. [DOI] [PubMed] [Google Scholar]
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1(2):113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Otlewski J. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J Biomol NMR. 2001;21(3):249–261. doi: 10.1023/a:1012911329730. [DOI] [PubMed] [Google Scholar]
- Clark RJ, Fischer H, Dempster L, Daly NL, Rosengren KJ, Nevin ST, Meunier FA, Adams DJ, Craik DJ. Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the alpha-conotoxin MII. Proc Nat Acad Sci USA. 2005;102(39):13767–13772. doi: 10.1073/pnas.0504613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RJ, Jensen J, Nevin ST, Callaghan BP, Adams DJ, Craik DJ. The engineering of an orally active conotoxin for the treatment of neuropathic pain. Angew Chem Int Ed Engl. 2010;49(37):6545–6548. doi: 10.1002/anie.201000620. [DOI] [PubMed] [Google Scholar]
- Craik DJ. Seamless proteins tie up their loose ends. Science. 2006;311(5767):1563–1564. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- Figueredo S, Mastroianni JR, Tai KP, Ouellette AJ. Expression and purification of recombinant alpha-defensins and alpha-defensin precursors in Escherichia coli. Methods Mol Biol. 2010;618:47–60. doi: 10.1007/978-1-60761-594-1_4. [DOI] [PubMed] [Google Scholar]
- Figueredo SM, Weeks CS, Young SK, Ouellette AJ. Anionic amino acids near the pro-alpha-defensin N terminus mediate inhibition of bactericidal activity in mouse pro-cryptdin-4. J Biol Chem. 2009;284(11):6826–6831. doi: 10.1074/jbc.M807024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3(9):710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3(6):583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Hadjicharalambous C, Sheynis T, Jelinek R, Shanahan MT, Ouellette AJ, Gizeli E. Mechanisms of alpha-defensin bactericidal action: comparative membrane disruption by Cryptdin-4 and its disulfide-null analogue. Biochemistry. 2008;47(47):12626–12634. doi: 10.1021/bi800335e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdar K, Maemoto A, Qu X, Young SK, Ouellette AJ. In vitro activation of the rhesus macaque myeloid alpha-defensin precursor proRMAD-4 by neutrophil serine proteinases. J Biol Chem. 2008;283(47):32361–32368. doi: 10.1074/jbc.M805296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Blumberg RS. Endoplasmic reticulum stress and intestinal inflammation. Mucosal Immunol. 2010;3(1):11–16. doi: 10.1038/mi.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Tomczak M, Blumberg RS. “ER stress(ed out)!”: Paneth cells and ischemia-reperfusion injury of the small intestine. Gastroenterology. 2011;140(2):393–396. doi: 10.1053/j.gastro.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RLJ. Computer aided resonance assignment tutorial. Cantina Verlag; Zurich, Switzerland: 2004. [Google Scholar]
- Lehrer RI, Ganz T, Szklarek D, Selsted ME. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target cell metabolism and divalent cations. J Clin Invest. 1988;81(6):1829–1835. doi: 10.1172/JCI113527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llenado RA, Weeks CS, Cocco MJ, Ouellette AJ. Electropositive charge in alpha-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect Immun. 2009;77(11):5035–5043. doi: 10.1128/IAI.00695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemoto A, Qu X, Rosengren KJ, Tanabe H, Henschen-Edman A, Craik DJ, Ouellette AJ. Functional analysis of the alpha-defensin disulfide array in mouse cryptdin-4. J Biol Chem. 2004;279(42):44188–44196. doi: 10.1074/jbc.M406154200. [DOI] [PubMed] [Google Scholar]
- McGuckin MA, Eri RD, Das I, Lourie R, Florin TH. ER stress and the unfolded protein response in intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G820–832. doi: 10.1152/ajpgi.00063.2010. [DOI] [PubMed] [Google Scholar]
- Missimer JH, Steinmetz MO, Baron R, Winkler FK, Kammerer RA, Daura X, van Gunsteren WF. Configurational entropy elucidates the role of salt-bridge networks in protein thermostability. Protein Sci. 2007;16(7):1349–1359. doi: 10.1110/ps.062542907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Guntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59(4):662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Human beta-defensins. Cell Mol Life Sci. 2006;63(11):1294–1313. doi: 10.1007/s00018-005-5540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabi M, de Leeuw E, Pazgier M, Li J, Lubkowski J, Lu W. The conserved salt bridge in human alpha-defensin 5 is required for its precursor processing and proteolytic stability. J Biol Chem. 2008;283(31):21509–21518. doi: 10.1074/jbc.M801851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren KJ, Daly NL, Fornander LM, Jonsson LM, Shirafuji Y, Qu X, Vogel HJ, Ouellette AJ, Craik DJ. Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4. J Biol Chem. 2006;281(38):28068–28078. doi: 10.1074/jbc.M604992200. [DOI] [PubMed] [Google Scholar]
- Rosengren KJ, Daly NL, Plan MR, Waine C, Craik DJ. Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J Biol Chem. 2003;278(10):8606–8616. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- Rosengren KJ, McManus AM, Craik DJ. The structural and functional diversity of naturally occurring antimicrobial Peptides. Curr Med Chem Anti-infective Agents. 2002;1:319–341. [Google Scholar]
- Satchell DP, Sheynis T, Kolusheva S, Cummings J, Vanderlick TK, Jelinek R, Selsted ME, Ouellette AJ. Quantitative interactions between cryptdin-4 amino terminal variants and membranes. Peptides. 2003;24(11):1795–1805. doi: 10.1016/j.peptides.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Satoh Y. Effect of live and heat-killed bacteria on the secretory activity of Paneth cells in germ-free mice. Cell Tissue Res. 1988;251(1):87–93. doi: 10.1007/BF00215451. [DOI] [PubMed] [Google Scholar]
- Satoh Y, Ishikawa K, Ono K, Vollrath L. Quantitative light microscopic observations on Paneth cells of germ-free and ex-germ-free Wistar rats. Digestion. 1986;34(2):115–121. doi: 10.1159/000199319. [DOI] [PubMed] [Google Scholar]
- Selsted ME, Harwig SS. Determination of the disulfide array in the human defensin HNP-2. A covalently cyclized peptide. J Biol Chem. 1989;264(7):4003–4007. [PubMed] [Google Scholar]
- Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6(6):551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- Selsted ME, Tang YQ, Morris WL, McGuire PA, Novotny MJ, Smith W, Henschen AH, Cullor JS. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. J Biol Chem. 1993;268(9):6641–6648. [PubMed] [Google Scholar]
- Shirafuji Y, Tanabe H, Satchell DP, Henschen-Edman A, Wilson CL, Ouellette AJ. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J Biol Chem. 2003;278(10):7910–7919. doi: 10.1074/jbc.M210600200. [DOI] [PubMed] [Google Scholar]
- Tanabe H, Qu X, Weeks CS, Cummings JE, Kolusheva S, Walsh KB, Jelinek R, Vanderlick TK, Selsted ME, Ouellette AJ. Structure-activity determinants in paneth cell alpha-defensins: loss-of-function in mouse cryptdin-4 by charge-reversal at arginine residue positions. J Biol Chem. 2004;279(12):11976–11983. doi: 10.1074/jbc.M310251200. [DOI] [PubMed] [Google Scholar]
- Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286(5439):498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- Wagner G. Nmr Investigations of Protein-Structure. Prog Nucl Mag Res Spect. 1990;22:101–139. [Google Scholar]
- Weeks CS, Tanabe H, Cummings JE, Crampton SP, Sheynis T, Jelinek R, Vanderlick TK, Cocco MJ, Ouellette AJ. Matrix metalloproteinase-7 activation of mouse paneth cell pro-alpha-defensins: SER43↓ILE44 proteolysis enables membrane-disruptive activity. J Biol Chem. 2006;281(39):28932–28942. doi: 10.1074/jbc.M602041200. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC, Selsted ME. Structure, function, and membrane integration of defensins. Curr Opin Struct Biol. 1995;5(4):521–527. doi: 10.1016/0959-440x(95)80038-7. [DOI] [PubMed] [Google Scholar]
- Williams DV, Byrne A, Stewart J, Andersen NH. Optimal salt bridge for Trp-cage stabilization. Biochemistry. 2011;50(7):1143–1152. doi: 10.1021/bi101555y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes M, Cammue BP, Sahl HG, Thevissen K. Antibiotic activities of host defense peptides: more to it than lipid bilayer perturbation. Nat Prod Rep. 2011;28(8):1350–1358. doi: 10.1039/c1np00022e. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286(5437):113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5(1):67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- Wu Z, Li X, de Leeuw E, Ericksen B, Lu W. Why is the Arg5-Glu13 salt bridge conserved in mammalian alpha-defensins? J Biol Chem. 2005;280(52):43039–43047. doi: 10.1074/jbc.M510562200. [DOI] [PubMed] [Google Scholar]
- Xie C, Prahl A, Ericksen B, Wu Z, Zeng P, Li X, Lu WY, Lubkowski J, Lu W. Reconstruction of the conserved beta-bulge in mammalian defensins using D-amino acids. J Biol Chem. 2005;280(38):32921–32929. doi: 10.1074/jbc.M503084200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.