ABSTRACT
Objective:
The objective of this work was to study the lactobacilli and bifidobacteria population in human milk of healthy women, and to investigate the influence that several factors (including antibioteraphy during pregnancy and lactation, country and date of birth, delivery mode, or infant age) may exert on such population.
Methods:
A total of 160 women living in Germany or Austria provided the breast milk samples. Initially, 66 samples were randomly selected and cultured on MRS-Cys agar plates. Then, the presence of DNA from the genera Lactobacillus and Bifidobacterium, and from most of the Lactobacillus and Bifidobacterium species that were isolated, was assessed by qualitative polymerase chain reaction (PCR) using genus- and species-specific primers.
Results:
Lactobacilli and bifidobacteria could be isolated from the milk of 27 (40.91%) and 7 (10.61%), respectively, of the 66 cultured samples. On the contrary, Lactobacillus and Bifidobacterium sequences were detected by PCR in 108 (67.50%) and 41 (25.62%), respectively, of the 160 samples analyzed. The Lactobacillus species most frequently isolated and detected was L salivarius (35.00%), followed by L fermentum (25.00%) and L gasseri (21.88%), whereas B breve (13.75%) was the bifidobacterial species most commonly recovered and whose DNA was most regularly found. The number of lactobacilli- or bifidobacteria-positive samples was significantly lower in women who had received antibiotherapy during pregnancy or lactation.
Conclusions:
Our results suggest that either the presence of lactobacilli and/or bifidobacteria or their DNA may constitute good markers of a healthy human milk microbiota that has not been altered by the use of antibiotics.
Keywords: antibiotics, Bifidobacterium, human milk, Lactobacillus
Bacterial gut colonization in early life is a process that exerts a short-, medium-, and long-term influence on the health status of a host, and that involves bacteria arising from different sources (1); among them, culture-dependent studies have revealed that human milk is a source of live staphylococci, streptococci, lactic acid bacteria, bifidobacteria, propionibacteria, corynebacteria, and closely related Gram-positive bacteria to the infant gut (2). Several studies have shown that there is a mother-to-infant transfer of bacterial strains belonging, at least, to the genera Lactobacillus, Staphylococcus, Enterococcus, and Bifidobacterium through breast-feeding (3–7). In fact, human milk constitutes 1 of the main sources of bacteria to the breast-fed infant gut since a baby consuming approximately 800 mL/day of milk would ingest between 1 × 105 and 1 × 107 bacteria daily (8). It has been suggested that exposure of the breast-fed infant to such a wealth of bacterial phylotypes through breast-feeding may exert beneficial effects against several diseases (9). Breast-feeding has been shown to improve infant health outcomes lowering the risk of respiratory and gastrointestinal tract infections, necrotizing enterocolitis, otitis media, and allergic disease and to prevent later health problems such as inflammatory bowel disease, obesity, and type 2 diabetes mellitus (10).
The application of culture-independent molecular techniques, and particularly those based on 16S rRNA genes, allowed a complementary biodiversity assessment of the human milk microbiome. The use of such techniques confirmed the dominance of staphylococci and streptococci, the relatively frequent presence of lactic acid bacteria and bifidobacteria, and the existence of DNA belonging to other bacterial groups, such as some Gram-negative bacteria (5,11–13). Recently, the first microbiome study focused on human milk was published and the results indicated that milk bacterial communities were generally complex (9). Among the hundreds of operational taxonomic units detected in the milk of every woman, only 9 (Streptococcus, Staphylococcus, Serratia, Pseudomonas, Corynebacterium, Ralstonia, Propionibacterium, Sphingomonas, and Bradyrhizobiaceae) were present in every sample from every woman. On the contrary, milk bacterial community was generally stable over time within an individual (9).
In contrast to staphylococci, streptococci, corynebacteria, or propionibacteria, which seem to be widespread in human milk, the presence of lactobacilli and bifidobacteria seems to be more variable among women (9,11,14,15). Such variability may be the consequence of isolation difficulties, owing to fastidious growth and incubation requirements, or may be the result of the technical bias associated to molecular studies. It could also be because of host peculiarities; it has been suggested that the human milk microbiome is influenced by several factors that significantly skew its composition (16). In this context, the objective of this study was to assess whether demographic or clinical factors, such as country and date of birth, infant age, delivery mode, or antibiotherapy during pregnancy and lactation, may exert an influence on the bifidobacterial and lactobacillic population present in the breast milk of healthy women.
METHODS
Subjects and Sampling
A total of 160 healthy women participated in the study and provided a sample of breast milk. Women were recruited to cover a moderately wide area of central Europe from randomly chosen regions in Germany and Austria to represent southern and eastern (Germany) and western and eastern (Austria) parts of both countries, which included both rural and urban settings. Recruitment was carried out by midwives, who were contacted initially via the HiPP Scientific sales force (Pfaffenhofen, Germany). All of the volunteers gave written informed consent to the protocol, which was approved by the ethical committee of Hospital Clínico (Madrid, Spain). The milk samples were collected in a sterile tube by manual expression using sterile gloves. Previously, nipples and mammary areola were cleaned with soap and sterile water and soaked in chlorhexidine. The first drops (∼500 μL) were discarded. All of the samples were kept frozen until delivery to the laboratory. All of the women filled a questionnaire designed to collect information on demographic characteristics and some other factors, such as mode of delivery, anesthesia during labor, or antibiotherapy during pregnancy and lactation (Table 1).
TABLE 1.
Demographic and clinical characteristics of the participating women
| Characteristic | n | Frequency, % |
| Infant sex | ||
| Male | 80 | 50.00 |
| Female | 80 | 50.00 |
| Month of delivery | ||
| January–March | 21 | 13.13 |
| April–June | 16 | 10.00 |
| July–September | 51 | 31.88 |
| October–December | 72 | 45.00 |
| Origin of mother | ||
| Urban | 82 | 51.25 |
| Rural | 78 | 48.75 |
| Country of sampling | ||
| Austria (East) | 15 | 9.37 |
| Austria (West) | 41 | 25.62 |
| Germany (East) | 61 | 38.12 |
| Germany (South) | 43 | 26.89 |
| Nationality* | ||
| German | 101 | 63.52 |
| Austrian | 47 | 29.56 |
| Other | 11 | 6.92 |
| Delivery mode | ||
| Vaginal | 125 | 78.13 |
| Cesarean section | 35 | 21.87 |
| Time of lactation* | ||
| <1 week | 16 | 10.00 |
| 1 to 4 weeks | 118 | 73.75 |
| >4 weeks | 25 | 15.63 |
| Breast-feeding* | ||
| Exclusively | 108 | 67.92 |
| Partial feeding | 51 | 32.08 |
| Anesthesia during delivery* | ||
| No | 103 | 64.78 |
| Yes | 56 | 35.22 |
| Antibiotherapy† | ||
| No | 95 | 59.38 |
| Yes | 65 | 40.62 |
| Last dose received at | ||
| Pregnancy | 14 | 8.75 |
| Delivery | 35 | 21.85 |
| Lactation | 16 | 10.00 |
*n = 159.
†During pregnancy and/or breast-feeding.
Count and Identification of Bacteria in the Samples
Adequate dilutions of 66 randomly selected milk samples were spread onto agar plates of Man, Rogosa, and Sharpe (Oxoid, Basingstoke, UK) supplemented with l-cysteine (0.5 g/L) (MRS-Cys) for isolation of lactobacilli and bifidobacteria. The plates were incubated for 48 hours at 37°C anaerobically (85% nitrogen, 10% hydrogen, 5% carbon dioxide) in an anaerobic workstation (MINI-MACS; DW Scientific, Shipley, UK).
After incubation and counting, 10 isolates from each culture medium were randomly selected and identified at the species level by classical morphological and biochemical tests. In addition, all the Gram-positive isolates with morphology compatible with that of lactobacilli or bifidobacteria were selected and identified at the genus level by classical morphological and biochemical tests and by demonstration of fructose-6-phosphate phosphoketolase activity in cellular extracts. Identification at the species level was performed by MALDI-TOF (Vitek MS; BioMerieux, Marcy l’Etoile, France) or by polymerase chain reaction (PCR) sequencing of a 470-bp fragment of the 16S rRNA gene using primers pbl16 (5′-AGAGTTTGATCCTGGCTCAG-3′) and mlb16 (5′-GGCTGCTGGCACGTAGTTAG-3′) (17). The PCR conditions were as follows: 96°C for 30 seconds, 48°C for 30 seconds, and 72°C for 45 seconds (40 cycles) and a final extension at 72°C for 4 minutes. The amplicons were purified using the Nucleospin Extract II kit (Macherey-Nagel, Düren, Germany) and sequenced at the Genomics Unit of the Universidad Complutense de Madrid, Spain. The resulting sequences were used to search sequences deposited in the EMBL database using the BLAST algorithm, and the identity of the isolates was determined on the basis of the highest scores (≥98%).
Somatic Cell Count (SCC) in the Samples
SCC was performed with a DeLaval cell counter DCC (DeLaval International AB, Tumba, Sweden), using single-use cell counter cassettes and instructions provided by the manufacturer. The cassette that contains small amounts of a DNA-specific stain (propidium iodide) is used to collect the sample. A piston carries the milk sample toward a counting window that is exposed to an LED light source. The fluorescence signal given by the cell nuclei is recorded as a digital image that is subjected to automated image analysis.
Bacterial DNA Isolation From the Milk Samples
Initially, a fraction of the breast milk samples (1 mL) was centrifuged at 7150 g for 20 minutes. Then, total DNA was isolated from the pellets using the QIAamp DNA Stool Mini Kit (QIAgen, Hilden, Germany) following a protocol described previously (11). DNA was eluted in 20 μL of buffer AE (provided in the kit), and the purified DNA extracts were stored at −20°C.
Qualitative PCR Assays
Genus-specific detection of DNA from the genera Lactobacillus or Bifidobacterium was accomplished using the primers and PCR conditions reported by Collado et al (18). At the species level, a 2-step multiplex PCR assay was used to detect DNA from L fermentum, L gasseri, L plantarum, L reuteri, L rhamnosus, and L salivarius, using species-specific primers and PCR conditions previously reported (19). In parallel, the presence of DNA from L casei/L paracasei was assessed using the primers and PCR conditions described by Chagnaud et al (20).
PCR detection of DNA from B longum, B infantis, B dentium, and B gallicum was carried using the primers and PCR conditions described by Matsuki et al (21), whereas the presence of DNA from B adolescentis, B angulatum, B bifidum, B breve, B catenulatum, and B pseudocatenulatum was assessed using those reported by Matsuki et al (22). Finally, PCR detection of DNA from B lactis was performed using the species-specific PCR assay developed by Ventura et al (23).
Each PCR assay included DNA extracted from a reference strain of each targeted species (positive control). Amplicons were analyzed by electrophoresis (90 V, 1 hour) on 1% agarose gels. Subsequently, the gels were stained and bands were visualized in a Gel-Doc system (Bio-Rad, Hercules, CA).
Statistical Analysis
Quantitative data were expressed as the mean and 95% confidence interval (CI) of the mean or, when they were not normally distributed, as the median and interquartile range. A correlation analysis was performed to test the relation between bacterial counts in MRS-Cys and SCCs. Proportions were compared using χ2 statistics, including the Fisher exact test and the Freeman-Halton test for contingency tables greater than 2 × 2. Differences were considered significant at P < 0.050. SAS version 9.2 (SAS Institute Inc, Cary, NC) was used to carry out the analyses cited above.
RESULTS
Characteristics of the Lactating Women Participating in the Study
The 160 women enrolled in this study had a mean age of 31.82 years (95% CI 31.10–32.54 years) and their breast-fed baby had a gestational age of 39.72 weeks (ranging from 29 to 44 weeks). The median number of children was 1 (n = 104, 65.00% of participants), and only 11 women (6.88%) had 3 or more children. Moreover, 21.87% of the infants were born by cesarean section, more than one-third of the women (35.22%) received anesthesia during delivery, and 40.62% of the women had received antibiotherapy during pregnancy and/or lactation (Table 1). Most of the participating women were exclusively breast-feeding their babies whereas 32.08% did it partially. Regarding the time of sampling, most of the breast milk samples (73.75%) were collected from women during the second to fourth week of lactation, whereas 10.00% were obtained during the first week and the remaining 15.63% after the first month of lactation. Most of the women who gave samples during the first month of lactation were exclusively breast-feeding their infants (75.37%), whereas this percentage descended notably in samples obtained after the first month of lactation (28.00%). Other demographic and clinical characteristics, such as the month of delivery, nationality and origin of the mother (urban or rural), and place where the samples were obtained, are summarized in Table 1.
Bacterial and SCCs
Bacterial growth was observed in 58 of the 66 milk samples inoculated on MRS-Cys agar plates. The mean (95% CI) value of bacterial counts obtained in such medium was 1.63 (1.49–1.77) log10 colony-forming units (CFU)/mL and ranged between 1.0 and 2.7 log10 CFU/mL (Fig. 1). In those breast milk samples where bacterial growth was observed, the mean (95% CI) value of SCCs was 36.67 (35.96–41.37) cells per microliter and ranged between 23 and 68 cells per microliter, whereas in the rest of the samples (n = 8) where bacterial growth was undetectable, lower SSCs values (mean value of 27.16 cells/μL) were observed. A weak but statistically significant correlation was noted between bacterial counts in MRS-Cys plates and SSCs (r = 0.395, P = 0.002), as shown in Figure 1. Overall, the bacterial count and SCC values found in these breast milk samples indicated that none of the women were experiencing mastitis when the samples were collected.
FIGURE 1.
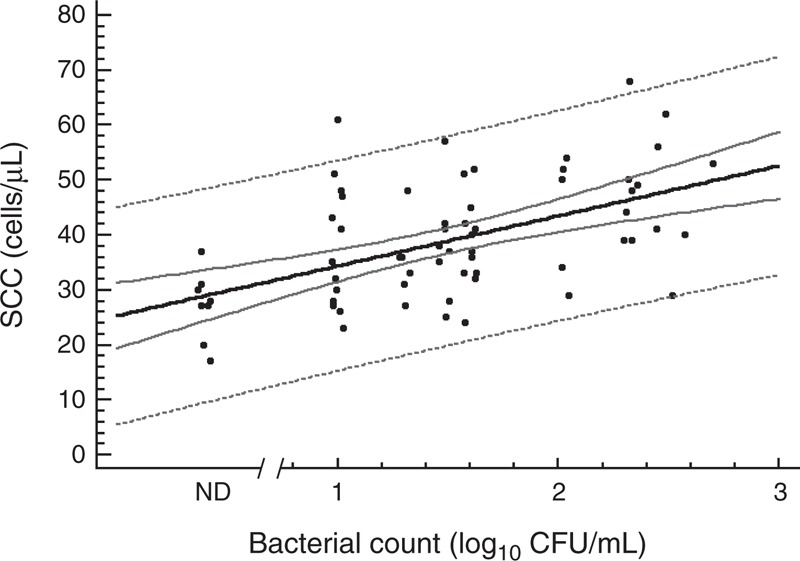
Bacterial counts in MRS-Cys and SCCs in breast milk samples (n = 66). Correlation between both parameters is shown as a solid line and follows the model: SCC (cells/μL) = 27.22 + 8.00 ∗ Bacterial counts in MRS-Cys (log10 CFU/mL), r = 0.395, P = 0.002; 95% CI for the mean value of SSC as a function of bacterial counts in MRS-Cys is shown as solid gray lines and 95% prediction intervals for new observations of bacterial counts in MRS-Cys as a function of SCC are shown as the outer dotted gray lines. Black dots = breast milk sample; ND = breast milk samples in which bacterial growth was not detected; they were not included in the correlation analysis.
Bacterial Identification
In relation to Gram-positive cocci, Staphylococcus spp were isolated from 51 samples (77.27%); Staphylococcus epidermidis was detected in all 51 samples, whereas other coagulase-negative staphylococci were isolated from <25% of the samples (data not shown). Staphylococcus aureus could not be detected in any sample. Streptococci were also isolated from 40 samples (60.61%) and most of them belonged to the species Streptococcus mitis, Streptococcus salivarius, or Streptococcus parasanguinis (data not shown).
Lactobacilli and bifidobacteria could be isolated from 27 (40.91%) and 7 (10.61%) samples of breast milk, respectively (Fig. 2). The mean (95% CI) values of bacterial counts for Lactobacillus and Bifidobacterium were 1.11 (0.99–1.23) and 0.96 (0.76–1.16) log10 CFU/mL, respectively. Identification of the isolates at the species level revealed that they belonged to the following species: B breve, B longum, L casei, L fermentum, L gasseri, L gastricus, L plantarum, L reuteri, L rhamnosus, L salivarius, and L vaginalis. The species most frequently found was L salivarius that was isolated from 9 milk samples (13.64% of the 66 cultured samples), followed by L fermentum (7 samples, 10.61%), L gasseri (6 samples, 9.09%), and B breve (5 samples, 7.58%) (Table 2). Usually, only 1 species of either lactobacillus or bifidobacteria was present in an individual breast milk sample, although 2 different species were isolated from 7 samples, and B longum, L gastricus, and L reuteri were detected simultaneously in 1 sample (Table 2).
FIGURE 2.
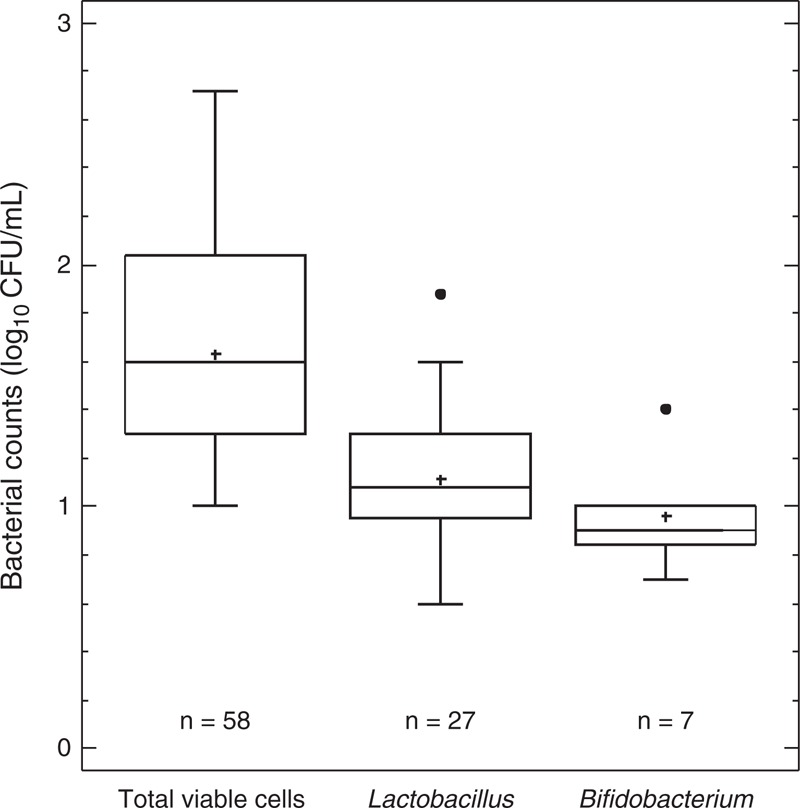
Total, lactobacilli, and bifidobacteria viable cells in breast milk samples after culturing in MRS-Cys. Bacterial growth was undetectable in 8 samples out of 66 analyzed. Mean values of bacterial counts are indicated with a “+” within each box-and-whisper plot. Median values are indicated by the line within the box plot. The box extends from the 25th to 75th percentiles, and the whiskers indicate the minimum and maximum values obtained. Outliers are represented as dots.
TABLE 2.
Viable lactobacilli and bifidobacteria diversity in the 66 breast milk samples that were cultured in this study
| Total frequency | |||
| Bacterial species | n* | N† | % |
| Bifidobacterium breve | 2 | 5 | 7.58 |
| B longum | 1 | 2 | 3.03 |
| Lactobacillus casei | 1 | 1 | 1.52 |
| L fermentum | 3 | 7 | 10.61 |
| L gasseri | 3 | 6 | 9.09 |
| L gastricus | 1 | 2 | 3.03 |
| L plantarum | 1 | 1 | 1.52 |
| L reuteri | 1 | 3 | 4.55 |
| L rhamnosus | 1 | 1 | 1.52 |
| L salivarius | 7 | 9 | 13.64 |
| L. vaginalis | 1 | 2 | 3.03 |
| B breve + L fermentum | 1 | ||
| B breve + L gasseri | 1 | ||
| B breve + L reuteri | 1 | ||
| L fermentum + L gasseri | 1 | ||
| L fermentum + L salivarius | 1 | ||
| L fermentum + L vaginalis | 1 | ||
| L gasseri + L salivarius | 1 | ||
| B longum + L gastricus + L reuteri | 1 | ||
*n = number of samples where a given combination of species was found.
†N = total number of samples where a given bacterial species was found alone or in combination with others.
PCR Assessment of Lactobacilli and Bifidobacteria Diversity in the Milk Samples
The presence of lactobacilli and bifidobacterial DNA alone or in combination was analyzed by PCR using species-specific primers in 160 breast milk samples, and it was confirmed in 113 (70.60%) samples (Fig. 3A). More specifically, Lactobacillus sequences (alone or in combination with Bifidobacterium sequences) and Bifidobacterium sequences (alone or in combination with Lactobacillus sequences) were detected in 108 and 41 samples (67.50% and 25.62% of total samples), respectively, whereas both genera were present simultaneously in 36 samples (22.50% of total samples) (Fig. 3A).
FIGURE 3.
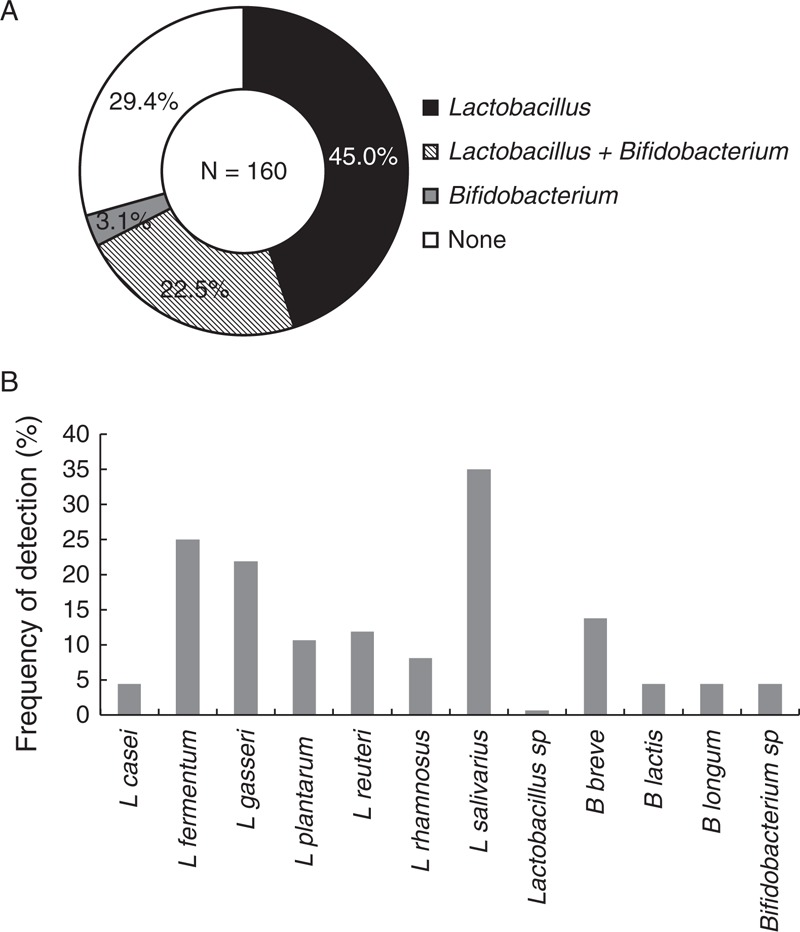
Presence of lactobacilli and/or bifidobacteria DNA in breast milk samples as determined by species-specific PCR (n = 160). Frequency of detection of DNA from genus Lactobacillus and/or Bifidobacterium (A) and lactobacilli and bifidobacterial species (B) in breast milk samples identified by species-specific PCR. The percentages are expressed over the total 160 samples analyzed. PCR = polymerase chain reaction.
The Lactobacillus species most frequently found was L salivarius (56 samples, 35.00% of total samples), followed by L fermentum (40 samples, 25.00%) and L gasseri (35 samples, 21.88%) (Fig. 3B). Other lactobacilli species detected using this approach were L reuteri (11.88%), L plantarum (10.63%), L rhamnosus (8.13%), and L casei (4.38%). Regarding bifidobacteria, B breve DNA was the most frequently found and it was present in 21 (13.75%) of the analyzed samples. B longum and B lactis were detected only in 7 samples (4.38%), each. Another 7 breast milk samples contained other species of bifidobacteria (Fig. 3B).
Globally, there was great interindividual variability regarding the lactobacilli and bifidobacterial PCR profile. In fact, up to 52 different species combinations were found among the 113 breast milk samples in which lactobacilli and/or bifidobacteria could be detected (Fig. 4). A total of 31 women displayed a profile that was not shared with any other recruited woman. The profiles of most of the samples comprised only 1 or 2 different species (38 and 43 samples, respectively) (Fig. 4). The profile comprising only L salivarius DNA was shared by 13 of the analyzed samples (representing 8% of total samples), whereas the combination of L fermentum with either L salivarius or L gasseri was present in 9 and 8 human milk samples, respectively (∼5% of total samples). In contrast, 8 breast milk samples contained DNA from 4 different Lactobacillus and Bifidobacterium species. A detailed analysis of all the combinations of lactobacilli and bifidobacterial species that were detected by PCR is presented in Figure 4.
FIGURE 4.
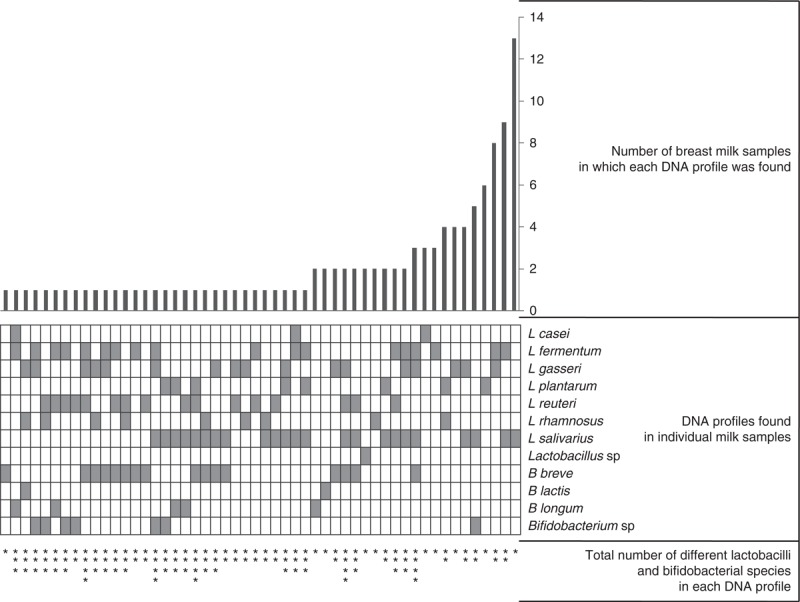
Lactobacilli and bifidobacterial diversity in breast milk samples (n = 113) as determined by PCR using species-specific primers. The 52 different combinations of DNA from lactobacilli and bifidobacterial species detected by PCR in individual breast milk samples are shown in the box grid in the middle of the figure; each file represents 1 unique combination and a gray box indicates the presence of DNA from a particular species. The bar graph at the top of the figure indicates the number of milk samples in which each specific combination of DNA from lactobacilli and bifidobacterial species was found. At the bottom of the box grid, the number of stars represents the total number of different species (lactobacilli and bifidobacteria) in each DNA profile. PCR = polymerase chain reaction.
Taken as a whole, there was a good correspondence between isolation by culture technique and PCR detection of lactobacilli or bifidobacteria, both at the genus and the species level in the 66 breast milk samples that were seeded on MRS-Cys plates (Table 3). Lactobacilli DNA was detected in 50 samples, and viable lactobacilli were isolated from approximately half of them (26 samples). The proportion of samples in which bifidobacteria were isolated by culture and in which their DNA was detected by PCR was lower (6 samples of 18 positives for bifidobacteria by PCR; Table 3). B lactis was detected only using molecular techniques (in 2 milk samples).
TABLE 3.
Comparison of the isolation of viable lactobacilli and bifidobacteria by the culture method and the detection of lactobacilli and bifidobacteria DNA by PCR using species-specific primers in breast milk samples (n = 66)
| No. positive samples | |||
| Species | Only by culture | By both culture and PCR | Only by PCR |
| Lactobacillus | 1 | 26 | 24 |
| L casei | 1 | 3 | |
| L fermentum | 7 | 12 | |
| L gasseri | 6 | 9 | |
| L gastricus | 2 | —* | —* |
| L plantarum | 1 | 6 | |
| L reuteri | 3 | 5 | |
| L rhamnosus | 1 | 6 | |
| L salivarius | 9 | 16 | |
| L vaginalis | 2 | —* | —-* |
| Bifidobacterium | 1 | 6 | 12 |
| B breve | 5 | 4 | |
| B lactis | 2 | ||
| B longum | 2 | 2 | |
| No. negative samples | |||
| Only by culture | By both culture and PCR | Only by PCR | |
| 36 | 15 | 1 | |
PCR = polymerase chain reaction.
*L gastricus and L vaginalis could not be detected by PCR because L gastricus- and L vaginalis-specific primers were not available.
Influence of Demographic and Clinical Parameters in the Detection of Lactobacilli and Bifidobacteria in Breast Milk
Associations between the presence or absence of lactobacilli or bifidobacterial DNA and relevant demographic or clinical characteristics of the women participating in the study were investigated in this study. Most of the analyzed samples (73.75%) were collected from women during a period that extended from the second to the fourth week after delivery, except 16 samples (10.00%) obtained during the first week and 25 samples (15.63%) provided by women who had been breast-feeding for more than 1 month. No differences were found regarding the distribution of bacteria belonging to either the genus Lactobacillus or Bifidobacterium or to their respective species between samples taken during the first week, from the second to the fourth week or after the first month of breast-feeding (Tables 4 and 5).
TABLE 4.
Association between demographic and clinical parameters and the presence of DNA from various Lactobacillus species in breast milk samples
| Lactobacillus | L casei | L fermentum | L gasseri | L plantarum | L reuteri | L rhamnosus | L salivarius | |||||||||
| N (%) | P* | N (%) | P | N (%) | P | N (%) | P | N (%) | P | N (%) | P | N (%) | P | N (%) | P | |
| Nationality | ||||||||||||||||
| German (n = 101) | 68 (67.33) | 0.890† | 5 (4.95) | 0.417† | 24 (23.76) | 0.300† | 21 (20.79) | 0.781† | 13 (12.87) | 0.497† | 12 (11.88) | 1.000† | 8 (7.92) | 0.639† | 36 (35.64) | 1.000† |
| Austrian (n = 47) | 33 (70.21) | 1 (2.13) | 15 (31.91) | 12 (25.53) | 4 (8.51) | 6 (12.77) | 5 (10.64) | 16 (34.04) | ||||||||
| Others (n = 11) | 7 (63.64) | 1 (9.09) | 1 (9.09) | 2 (18.18) | 0 (0.00) | 1 (9.09) | 0 (0.00) | 4 (36.36) | ||||||||
| Country of sampling | ||||||||||||||||
| Germany (East) (n = 61) | 41 (67.21) | 0.845† | 3 (4.92) | 0.856† | 14 (22.95) | 0.570 | 12 (19.67) | 0.927† | 8 (13.11) | 0.777† | 10 (16.39) | 0.302† | 6 (9.84) | 0.827† | 21 (34.43) | 0.552† |
| Germany (South) (n = 43) | 27 (62.79) | 2 (4.65) | 10 (23.26) | 10 (23.26) | 3 (6.98) | 2 (4.65) | 2 (4.65) | 15 (34.88) | ||||||||
| Austria (East) (n = 15) | 11 (73.33) | 1 (6.67) | 6 (40.00) | 3 (20.00) | 1 (6.67) | 2 (13.33) | 1 (6.67) | 3 (20.00) | ||||||||
| Austria (West) (n = 41) | 29 (70.73) | 1 (2.44) | 10 (24.39) | 10 (24.39) | 5 (12.20) | 5 (12.20) | 4 (9.76) | 17 (41.46) | ||||||||
| Origin of mother | ||||||||||||||||
| Urban (n = 82) | 55 (67.07) | 0.906 | 4 (4.88) | 1.000† | 20 (24.39) | 0.855 | 18 (21.95) | 0.981 | 7 (8.54) | 0.379 | 10 (12.20) | 0.898 | 7 (8.54) | 0.845 | 24 (29.27) | 0.119 |
| Rural (n = 78) | 53 (67.95) | 3 (3.85) | 20 (25.64) | 17 (21.79) | 10 (12.82) | 9 (11.54) | 6 (7.69) | 32 (41.03) | ||||||||
| No. children | ||||||||||||||||
| 1 (n = 104) | 72 (69.23) | 0.607 | 5 (4.81) | 0.313† | 21 (20.19) | 0.136† | 28 (26.92) | 0.119† | 11 (10.58) | 0.913† | 14 (13.46) | 0.474† | 8 (7.69) | 0.806† | 34 (32.69) | 0.468† |
| 2 (n = 45) | 30 (66.67) | 1 (2.22) | 15 (33.33) | 6 (13.33) | 5 (11.11) | 5 (11.11) | 4 (8.89) | 19 (42.22) | ||||||||
| ≥3 (n = 11) | 6 (54.55) | 1 (9.09) | 4 (36.36) | 1 (9.09) | 1 (9.09) | 0 (0.00) | 1 (9.09) | 3 (27.27) | ||||||||
| Delivery month | ||||||||||||||||
| January–March (n = 21) | 15 (71.43) | 0.121 | 1 (4.76) | 0.129† | 7 (33.33) | 0.769† | 6 (28.57) | 0.191 | 2 (9.52) | 0.190† | 4 (19.05) | 0.326† | 1 (4.76) | 0.378† | 5 (23.81) | 0.104 |
| April–June (n = 16) | 14 (87.50) | 0 (0.00) | 3 (18.75) | 6 (37.50) | 4 (25.00) | 2 (12.50) | 2 (12.50) | 8 (50.00) | ||||||||
| July–September (n = 51) | 29 (56.86) | 5 (9.80) | 13 (25.49) | 12 (23.53) | 3 (5.88) | 3 (5.88) | 2 (3.92) | 13 (25.49) | ||||||||
| October–December (n = 72) | 50 (69.44) | 1 (1.39) | 17 (23.61) | 11 (15.28) | 8 (11.11) | 10 (13.89) | 8 (11.11) | 30 (41.67) | ||||||||
| Infant sex | ||||||||||||||||
| Female (n = 80) | 52 (65.00) | 0.500 | 4 (5.00) | 1.000† | 18 (22.50) | 0.465 | 13 (16.25) | 0.085 | 8 (10.00) | 0.798 | 10 (12.50) | 0.807 | 7 (8.75) | 0.772 | 23 (28.75) | 0.097 |
| Male (n = 80) | 56 (77.00) | 3 (3.75) | 22 (27.50) | 22 (27.50) | 9 (11.25) | 9 (11.25) | 6 (7.50) | 33 (41.25) | ||||||||
| Breast-feeding | ||||||||||||||||
| Exclusively (n = 108) | 73 (67.59) | 0.896 | 4 (3.70) | 0.532† | 26 (24.07) | 0.647 | 23 (21.30) | 0.751 | 10 (9.26) | 0.395 | 13 (12.04) | 0.961 | 9 (8.33) | 1.000 | 37 (34.26) | 0.712 |
| Partial (n = 51) | 35 (68.63) | 3 (5.88) | 14 (27.45) | 12 (23.53) | 7 (13.73) | 6 (11.76) | 4 (7.84) | 19 (37.25) | ||||||||
| Time of lactation | ||||||||||||||||
| <1 week (n = 16) | 11 (68.75) | 0.577 | 1 (6.25) | 0.815† | 5 (31.25) | 0.716 | 4 (25.00) | 0.127 | 2 (12.50) | 0.570† | 3 (18.75) | 0.594† | 0 (0.00) | 0.483† | 4 (25.00) | 0.619 |
| 1–4 weeks (n = 118) | 77 (65.25) | 5 (4.24) | 29 (24.58) | 22 (18.64) | 11 (9.32) | 13 (11.02) | 10 (8.47) | 41 (34.75) | ||||||||
| >4 weeks (n = 25) | 19 (76.00) | 1 (4.00) | 5 (20.00) | 9 (36.00) | 4 (16.00) | 3 (12.00) | 3 (12.00) | 10 (40.00) | ||||||||
| Delivery mode | ||||||||||||||||
| Vaginal (n = 125) | 89 (71.20) | 0.059 | 6 (4.80) | 1.000† | 35 (28.00) | 0.098 | 29 (23.20) | 0.444 | 14 (11.20) | 0.767† | 14 (11.20) | 0.618 | 9 (7.20) | 0.483† | 48 (38.40) | 0.088 |
| Cesarean section (n = 35) | 19 (54.29) | 1 (2.86) | 5 (14.29) | 6 (17.14) | 3 (8.57) | 5 (14.29) | 4 (11.43) | 8 (22.86) | ||||||||
| Anesthesia during delivery | ||||||||||||||||
| No (n = 103) | 74 (71.84) | 0.097 | 2 (1.94) | 0.053† | 30 (29.13) | 0.068 | 22 (21.36) | 0.787 | 12 (11.65) | 0.596 | 10 (9.71) | 0.384 | 8 (7.77) | 0.799 | 44 (42.72) | 0.007 |
| Yes (n = 56) | 33 (58.93) | 5 (8.93) | 9 (16.07) | 13 (23.21) | 5 (8.93) | 8 (14.29) | 5 (8.93) | 12 (21.43) | ||||||||
| Antibiotherapy‡ | ||||||||||||||||
| No (n = 95) | 80 (84.21) | 0.000 | 4 (4.21) | 1.000† | 31 (32.63) | 0.007 | 24 (25.26) | 0.210 | 14 (14.74) | 0.065† | 12 (12.63) | 0.721 | 7 (7.37) | 0.672 | 45 (47.37) | <0.001 |
| Yes (n = 65) | 28 (43.08) | 3 (4.62) | 9 (13.85) | 11 (16.92) | 3 (4.62) | 7 (10.77) | 6 (9.23) | 11 (16.92) | ||||||||
*χ2 test.
†Fisher-Freeman-Halton exact test.
‡During pregnancy and/or breast-feeding.
TABLE 5.
Association between demographic and clinical parameters and the presence of DNA from various Bifidobacterium species in breast milk samples
| Bifidobacterium | B breve | B lactis | B longum | ||||||
| N (%) | P* | N (%) | P | N (%) | P | N (%) | P | ||
| Nationality | |||||||||
| German (n = 101) | 27 (26.73) | 0.917† | 13 (12.87) | 0.753† | 6 (5.94) | 0.659† | 6 (5.94) | 0.659† | |
| Austrian (n = 47) | 12 (25.53) | 8 (17.02) | 1 (2.13) | 1 (2.13) | |||||
| Others (n = 11) | 2 (18.18) | 1 (9.09) | 0 (0.00) | 0 (0.00) | |||||
| Country of sampling | |||||||||
| Germany (East) (n = 61) | 21 (34.43) | 0.051† | 11 (18.03) | 0.174† | 4 (6.56) | 0.815† | 3 (4.92) | 0.656† | |
| Germany (South) (n = 43) | 6 (13.95) | 2 (4.65) | 2 (4.65) | 3 (6.98) | |||||
| Austria (East) (n = 15) | 3 (20.00) | 2 (13.33) | 0 (0.00) | 0 (0.00) | |||||
| Austria (West) (n = 41) | 11 (26.83) | 7 (17.07) | 1 (2.44) | 1 (2.44) | |||||
| Origin of mother | |||||||||
| City (n = 82) | 22 (26.83) | 0.720 | 11 (13.41) | 0.900 | 3 (3.66) | 0.715† | 4 (4.88) | 1.000† | |
| Country (n = 78) | 19 (24.36) | 11 (14.10) | 4 (5.13) | 3 (3.85) | |||||
| No. children | |||||||||
| 1 (n = 104) | 28 (26.92) | 0.834† | 16 (15.38) | 0.864† | 5 (4.81) | 1.000† | 4 (3.85) | 0.518† | |
| 2 (n = 45) | 10 (22.22) | 5 (11.11) | 2 (4.44) | 2 (4.44) | |||||
| >3 (n = 11) | 3 (27.27) | 1 (9.09) | 0 (0.00) | 1 (9.09) | |||||
| Delivery month | |||||||||
| January–March (n = 21) | 7 (33.33) | 0.711 | 4 (19.05) | 0.132† | 0 (0.00) | 0.784† | 0 (0.00) | 0.114† | |
| April–June (n = 16) | 5 (31.25) | 5 (31.25) | 0 (0.00) | 0 (0.00) | |||||
| July–September (n = 51) | 11 (21.57) | 5 (9.80) | 3 (5.88) | 4 (7.84) | |||||
| October–December (n = 72) | 18 (25.00) | 8 (11.11) | 4 (5.56) | 3 (4.17) | |||||
| Infant sex | |||||||||
| Female (n = 80) | 21 (26.25) | 0.856 | 11 (13.75) | 1.000 | 4 (5.00) | 1.000† | 3 (3.75) | 1.000† | |
| Male (n = 80) | 20 (25.00) | 11 (13.75) | 3 (3.75) | 4 (5.00) | |||||
| Breast-feeding | |||||||||
| Exclusively (n = 108) | 29 (26.85) | 0.655 | 13 (12.04) | 0.339 | 5 (4.63) | 1.000† | 5 (4.63) | 1.000† | |
| Partial (n = 51) | 12 (23.53) | 9 (17.65) | 2 (3.92) | 2 (3.92) | |||||
| Time of lactation | |||||||||
| <1 week (n = 16) | 5 (31.25) | 0.817 | 2 (12.50) | 0.089† | 0 (0.00) | 1.000† | 0 (0.00) | 0.540† | |
| 1–4 weeks (n = 118) | 29 (24.58) | 13 (11.02) | 6 (5.08) | 7 (5.93) | |||||
| >4 weeks (n = 25) | 7 (28.00) | 7 (28.00) | 1 (4.00) | 0 (0.00) | |||||
| Delivery mode | |||||||||
| Vaginal (n = 125) | 34 (27.20) | 0.389 | 17 (13.60) | 0.917 | 6 (4.80) | 1.000† | 6 (4.80) | 1.000† | |
| Cesarean section (n = 35) | 7 (20.00) | 5 (14.29) | 1 (2.86) | 1 (2.86) | |||||
| Anesthesia during delivery | |||||||||
| No (n = 103) | 28 (27.18) | 0.424 | 14 (13.59) | 0.904 | 4 (3.88) | 0.698† | 5 (4.85) | 1.000† | |
| Yes (n = 56) | 12 (21.43) | 8 (14.29) | 3 (5.36) | 2 (3.57) | |||||
| Antibiotherapy‡ | |||||||||
| No (n = 95) | 30 (31.58) | 0.037 | 14 (14.74) | 0.661 | 6 (6.23) | 0.242† | 5 (5.26) | 0.702† | |
| Yes (n = 65) | 11 (16.92) | 8 (12.31) | 1 (1.54) | 2 (3.08) | |||||
*χ2 test.
†Fisher-Freeman-Halton exact test.
‡During pregnancy and/or breast-feeding.
The number of milk samples positive for lactobacilli or bifidobacteria were significantly lower in those women who had received antibiotherapy during pregnancy or lactation (χ2 test; P = 0.000 and P = 0.037 for lactobacilli and bifidobacteria, respectively) (Tables 4 and 5). The administration time of the antibiotic (pregnancy, delivery, or lactation) did not modify the frequency of samples with Lactobacillus, Bifidobacterium or their respective species, although it would be advisable to analyze a higher number of subjects to draw a conclusive evidence of this aspect. Lactobacilli were also less frequently detected in the breast milk of women who had been subjected to cesarean section and/or had received anesthesia during delivery although, in such cases, the differences were not statistically significant (χ2 test, P = 0.059 and P = 0.097, respectively) (Table 4).
To further investigate the influence of these factors on the diversity of the Lactobacillus and Bifidobacterium species detected in these human milk samples, an analysis was carried out considering the presence or absence of DNA from the different species (Tables 4 and 5). L fermentum and L salivarius DNA was detected only in 13.85% and 16.92%, respectively, of the samples obtained from women who had received antibiotherapy, whereas the percentage ascended to 32.63% and 47.37%, respectively, in the case of women who had not been treated with antibiotics (χ2 test, P = 0.007 and <0.001, respectively). Similarly, L plantarum was present in 14 milk samples (14.74%) obtained from women who did not take antibiotics but it was detected only in 3 women (4.62%) of the group that received antibiotherapy, although the difference was not statistically significant (Fisher test, P = 0.065). Detection of L fermentum and L salivarius was also higher in women who had their babies by vaginal delivery than in those that had them by cesarean section, although the differences did not reach statistical significance (P = 0.088 and P = 0.098, respectively). Receiving anesthesia during delivery had a similar impact in the presence of DNA from L fermentum and L salivarus in breast milk; the presence of these lactobacilli species was found more frequently in women who did not receive anesthesia during delivery, although the difference was statistically significant only for L salivarius (P = 0.068 and P = 0.007, respectively). Similarly, a higher frequency of B breve, B lactis, and B longum was observed in milk samples from women with a vaginal delivery but, again, the differences were not statistically significant.
Finally, the association between the presence or absence of viable lactobacilli and bifidobacterium and relevant clinical parameters in the 66 milk samples that had been cultured was investigated. There was a tendency indicating that the proportion of milk samples containing viable lactobacilli, particularly L fermentum and L salivarius, was higher in the group of women who did not receive antibiotics during pregnancy and/or breast-feeding compared with the group that received antibiotherapy, but the association did not reach significant values (Table S1). The associations between the presence of Lactobacillus and Bifidobacterium DNA and some clinical parameters that have been previously described were, however, confirmed in this smaller group of milk samples (Table S1).
DISCUSSION
In the last years, culture-dependent methods have shown that breast milk is a source of commensal, mutualistic, and probiotic bacteria with the ability to influence the initial colonization of the infant gut (2,24). In this study, a low number of viable bacteria (<103 CFU/mL) was found in most of the samples, in agreement with previous analyses (25). This fact reflects the hygienic collection and proper storage of the samples and, together with the low SCC level of milk samples, indicates that participating women did not experience mastitis. Higher counts are usually related to non-hygienic sampling, improper storage of the samples, and/or use of contaminated milk pumps for sampling collection or mastitis (26,27)
Although the main target of this study were lactobacilli and bifidobacteria, coagulase-negative staphylococci and viridans streptococci could be isolated from 51 (77.27%) and 40 (60.61%), respectively, of the 66 cultured samples. Staphylococci and streptococci constitute the dominant culturable bacteria in human milk, and related DNA sequences of both genera are also the prevailing ones in this biological fluid, albeit with substantial interindividual differences (4,8,9,11,14,16,18,28). In spite of this, staphylococci and streptococci have received a marginal attention regarding their role in the human mammary gland and in the early colonization of the infant gut, although they could be useful to reduce the acquisition of undesired pathogens, particularly in infants exposed to hospital environments (7,28–30).
With respect to lactobacilli and bifidobacteria, they could be isolated from 27 (40.91%) and 7 (10.61%) of the 66 cultured samples, respectively. Such bacterial groups constitute a subdominant culturable population in human milk (3,5–8,31), and are particularly attractive because of its use as probiotics. It is noteworthy that lactobacilli and bifidobacteria are difficult to isolate because their low concentration in human milk is close to the detection limit for culture methods (5). All of the lactobacilli and bifidobacterial species isolated in this study have already been described in human milk (3,5–8). Particularly, strains belonging to the species L fermentum, L salivarius, L gasseri, B breve, and B longum can be transferred to the infant gut through breast milk (3,6,7).
In this work, qualitative PCR analysis revealed the presence of lactobacilli and bifidobacterial DNA in 108 (67.50%) and 41 (25.62%), respectively, of the 160 samples analyzed. Globally, there was a good correspondence between isolation of viable bacteria and PCR detection of lactobacilli or bifidobacterial DNA, at both the genus and the species level. The higher number of samples from which lactobacilli or bifidobacterial DNA was amplified compared with those from which viable bacteria were isolated by culture methods was an expected result, owing to the low concentrations of lactobacilli and bifidobacteria in human milk that hinder their detection (as stated before) and to the lack of discrimination between DNA from live or dead bacteria of PCR assays. Human milk contains a wide range of free bacterial DNA signatures, which may contribute to program the neonatal immune system (25).
Previous culture-independent assessments of the bacterial diversity of human milk have shown the presence of lactobacilli and bifidobacterial DNA (5,11–14,16,18,25). In contrast, some pyrosequencing-based studies have found a low relative abundance of DNA from lactobacilli and/or bifidobacteria (9,14,32). Differences in genetic, cultural, environmental, or dietary factors (9,14), or technical bias associated with culture-independent methods (differential lysis among different bacterial strains, species, genera, and phylotypes when obtaining bacterial DNA from the biological sample and/or differential amplification rate because of the selected primers for pyrosequencing) could explain these controversial results. In fact, in a previous study, the application of universal primers or Lactobacillus-specific primers to a set of human milk samples showed different results (11,12). Therefore, standardization of procedures to be able to compare results provided by different studies focused on the same type of biological material would be advisable.
In this study, the influence of several factors on the detection of lactobacilli and bifidobacterial DNA by PCR was also assessed. The factor that exerted the strongest influence on the presence of lactobacilli or bifidobacteria was the administration of antibiotherapy to mothers during pregnancy or lactation. More specifically, detection of lactobacilli or bifidobacterial DNA in the milk samples using genus- and some species-specific (L fermentum and L salivarius) primers was significantly lower in those women who had received antibiotherapy during such periods. Lactobacilli were also less frequently detected in the breast milk of women who had been subjected to cesarean section, probably owing to the antibiotherapy associated to such surgery. It has been long known that antibiotics are responsible for dysbiosis processes in the human microbiota, leading to antibiotic-associated diarrhea and gastroenteritis, urogenital, and oral infections (33). It is becoming evident that antibiotherapy during pregnancy, intrapartum, and lactation alters the maternal microbiota, a fact that may have negative consequences for infant health (34). An increased risk of asthma exacerbation and hospitalization, requiring inhaled corticosteroids, in children if mothers used antibiotics during pregnancy, supports a role for bacterial ecology in pre- or perinatal life for the development of asthma (35).
The decrease in the lactobacilli and bifidobacterial populations of breast milk may have negative consequences for breast-fed infants because they are important members of the human gut microbiota in early life. Infants with delayed colonization or decreased numbers of these bacteria may be more susceptible to a variety of gastrointestinal or allergic conditions (36). Recently, a comprehensive analysis of the fecal microbiota in infants with colic, as compared with control infants, revealed that bifidobacteria and lactobacilli were significantly reduced in infants with colic. Moreover, the colic phenotype correlated positively with specific groups of Proteobacteria but negatively with bacteria belonging to the Firmicutes phyla, which includes some lactobacilli and canonical groups known to produce butyrate and lactate (37). Interestingly, several trials have shown that infants with infantile colic benefit from the administration of a Lactobacillus strain, claimed to be of human milk origin (38). Therefore, it is not strange that most reviews and meta-analysis have repeatedly confirmed the beneficial effects of some probiotic lactobacilli or bifidobacteria strains for the prevention or treatment of antibiotic-associated conditions (39,40).
Among the bacteria isolated from human milk, species such as L gasseri, L salivarius, L reuteri, L fermentum, or B breve are considered among those with probiotic potential and enjoy the Qualified Presumption of Safety status conceded by the European Food Safety Authority (EFSA). In contrast to other bacteria, these seem to be uniquely adapted to reside in the human digestive tract and to interact with us in symbiosis from the time we are born (24). Some studies have shown that human milk lactobacilli may play several roles in the infant gut, such as the increase in the production of functional metabolites such as butyrate, which is the main energy source for colonocytes and a relevant compound in the modulation of intestinal function (2). As a result, they improve the intestinal habit, with an increase in fecal moisture, and in stool frequency and volume (41). They can also contribute to the reduction of the incidence and severity of infections in the breast-fed infant by different mechanisms (42,43). Recently, the administration of L fermentum CECT5716, a strain isolated from human milk, in a prebiotic containing follow-on formula to infants during an intervention period of 6 months led to 46% (P = 0.032), 27% (P = 0.026), and 30% (P = 0.003) reduction in the incidence rates of gastrointestinal infections, upper respiratory tract infections, and total number of infections, respectively (44). Consistently, the same probiotic L fermentum CECT5716 added to a prebiotic containing infant formula also led to a significant reduction of incidence rates of gastrointestinal infections (−71%, P = 0.018) in infants ages 1 to 6 months (41). Breast milk bacteria may also participate in the correct maturation of the infant immune system because some strains are able to modulate both natural and acquired immune responses in animal models and humans (45–47).
Finally, the antibiotic-associated loss of lactobacilli and bifidobacteria in milk may also have negative consequences for breast health because of the overgrowth of mastitis-causing agents (48). In fact, multiresistance to antibiotics is a common feature among clinical staphylococci involved in such conditions (49). This explains why this condition used to be elusive to antibiotic therapy, and why it constitutes one of the main reasons to cease breast-feeding (50). In this context, the development of new strategies based on selected probiotic lactobacilli isolated from human milk is an efficient alternative for mastitis treatment (27). This suggests that human milk lactobacilli may play important roles in mammary homeostasis.
In conclusion, the results of this study confirm that lactobacilli and bifidobacteria are common members of the human milk microbiota of women who did not receive antibiotics during pregnancy or lactation. Therefore, the presence of such bacteria may be a marker of a healthy non–antibiotic-altered human milk microbiota, and this should be taken into account when defining a criterion standard of breast milk. As a consequence, administration of selected human milk lactobacilli or bifidobacteria to pregnant or lactating women receiving antibiotics, or to their infants, may constitute an attractive approach to restore the natural bacterial ecosystem existing in human milk.
Supplementary Material
Treatment of Jaundice in Colonial America
In 17th century colonial America was a letter written to Governor John Winthrop (1588–1649) of Massachusetts by his physician friend, Edward Stafford, recommending saffron for treating jaundice. Using John Gerard's Herball (1597) as a source, Stafford in 1643 suggested the following recipe:
For the yellow Jaundise or Jaunders - Boyle a quart
sweet milke, dissolve therein as much bay-salt, or
fine Salpeter, as shall make it brackish in taste and
putting Saffron in a fine linen clout, rubb it into
the Milke, until the Milke be very yellow; and give it the [child] to drink.
The recipe was based on the popular Doctrine of Signatures (similia similubus curantur), which subscribed to the notion that the use of a drug must resemble the disease. Thus, walnut shells were used for head injuries, bear's grease for baldness, turmeric or saffron for jaundice, powdered mummy for aging, thistle for a stitch, and rust or red wine for anemia. The Doctrine was a favorite of Paracelsus and would become the basis of Hahnemann's homeopathy.
—Contributed by Angel R. Colón, MD
Footnotes
This work was supported by a research contract funded by HiPP. Know-how derived from the FUN-C-FOOD (Consolider-Ingenio 2010) and AGL2010-15420 projects from the Ministerio de Economía y Competitividad (Spain) was applied in this study.
The authors report no conflicts of interest.
REFERENCES
- 1.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet 2012; 13:260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández L, Langa S, Martín V, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 2013; 69:1–10 [DOI] [PubMed] [Google Scholar]
- 3.Martín R, Langa S, Reviriego C, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr 2003; 143:754–758 [DOI] [PubMed] [Google Scholar]
- 4.Jiménez E, Delgado S, Maldonado A, et al. Staphylococcus epidermidis: a differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol 2008; 8:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin R, Jimenez E, Heilig H, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol 2009; 75:965–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makino H, Kushiro A, Ishikawa E, et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl Environ Microbiol 2011; 77:6788–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martín V, Maldonado A, Moles L, et al. Sharing of bacterial strains between breast milk and infant feces. J Human Lact 2012; 28:36–44 [DOI] [PubMed] [Google Scholar]
- 8.Heikkilä MP, Saris PEJ. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol 2003; 95:471–478 [DOI] [PubMed] [Google Scholar]
- 9.Hunt KM, Foster JA, Forney LJ, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011; 6:e21313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.American Academy of Paediatrics (AAP) Breastfeeding and the use of human milk. Pediatrics 2012; 129:e827–e841 [DOI] [PubMed] [Google Scholar]
- 11.Martin R, Heilig HG, Zoetendal EG, et al. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res Microbiol 2007; 158:31–37 [DOI] [PubMed] [Google Scholar]
- 12.Martín R, Heilig HG, Zoetendal EG, et al. Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol 2007; 103:2638–2644 [DOI] [PubMed] [Google Scholar]
- 13.Gueimonde M, Laitinen K, Salminen S, et al. Breast milk: a source of bifidobacteria for infant gut development and maturation? Neonatology 2007; 92:64–66 [DOI] [PubMed] [Google Scholar]
- 14.Jost T, Lacroix C, Braegger C, et al. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr 2013; 14:1–10 [DOI] [PubMed] [Google Scholar]
- 15.Ward TL, Hosid S, Ioshikhes I, et al. Human milk metagenome: a functional capacity analysis. BMC Microbiol 2013; 13:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabrera-Rubio R, Collado MC, Laitinen K, et al. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 2012; 96:544–551 [DOI] [PubMed] [Google Scholar]
- 17.Kullen MJ, Sanozky-Dawes RB, Crowell DC, et al. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J Appl Microbiol 2005; 89:511–516 [DOI] [PubMed] [Google Scholar]
- 18.Collado MC, Delgado S, Maldonado A, et al. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real time PCR. Lett Appl Microbiol 2009; 48:523–528 [DOI] [PubMed] [Google Scholar]
- 19.Song Y, Kato N, Liu C, et al. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol Lett 2000; 187:167–173 [DOI] [PubMed] [Google Scholar]
- 20.Chagnaud P, Machinis K, Coutte LA, et al. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. Microbiol Methods 2001; 44:139–148 [DOI] [PubMed] [Google Scholar]
- 21.Matsuki T, Watanabe K, Tanaka R, et al. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol 1999; 65:4506–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuki K, Watanabe R, Tanaka R, et al. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS Microbiol Lett 1998; 167:113–121 [DOI] [PubMed] [Google Scholar]
- 23.Ventura M, Reniero R, Zink R. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl Environ Microbiol 2001; 67:2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeurink PV, van Bergenhenegouwen J, Jimenez E, et al. Human milk: a source of more life than we imagine. Benef Microbes 2013; 4:17–30 [DOI] [PubMed] [Google Scholar]
- 25.Perez PF, Doré J, Leclerc M, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 2007; 119:e724–e732 [DOI] [PubMed] [Google Scholar]
- 26.Marín ML, Arroyo R, Jimenez E, et al. Cold storage of human milk: effect on its bacterial composition. J Pediatr Gastroenterol Nutr 2009; 49:343–348 [DOI] [PubMed] [Google Scholar]
- 27.Arroyo R, Martín V, Maldonado A, et al. Treatment of infectious mastitis during lactation: antibiotics versus oral administration of lactobacilli isolated from breast milk. Clin Infect Dis 2010; 50:1551–1558 [DOI] [PubMed] [Google Scholar]
- 28.Hunt KM, Preuss J, Nissan C, et al. Human milk oligosaccharides promote the growth of staphylococci. Appl Environ Microbiol 2012; 78:4763–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwase T, Uehara Y, Shinji H, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010; 465:346–349 [DOI] [PubMed] [Google Scholar]
- 30.Uehara Y, Kikuchi K, Nakamura T, et al. H2O2 produced by viridans group streptococci may contribute to inhibition of methicillin-resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin Infect Dis 2001; 32:1408–1413 [DOI] [PubMed] [Google Scholar]
- 31.Solís G, de Los Reyes-Gavilán CG, Fernández N, et al. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 2010; 16:307–310 [DOI] [PubMed] [Google Scholar]
- 32.Ward R, Ninonuevo M, Mills D, et al. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol 2006; 72:4497–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 2011; 9:233–243 [DOI] [PubMed] [Google Scholar]
- 34.Murk W, Risnes KR, Bracken MB. Prenatal or early-life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics 2011; 127:1125–1138 [DOI] [PubMed] [Google Scholar]
- 35.Stensballe LG, Simonsen J, Jensen SM, et al. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. J Pediatr 2013; 162:832–838 [DOI] [PubMed] [Google Scholar]
- 36.Arvola T, Ruuska T, Keränen J, et al. Rectal bleeding in infancy: clinical, allergological, and microbiological examination. Pediatrics 2006; 117:e760–e768 [DOI] [PubMed] [Google Scholar]
- 37.de Weerth C, Fuentes S, Puylaert P, et al. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics 2013; 131:e550–e558 [DOI] [PubMed] [Google Scholar]
- 38.Sung V, Hiscock H, Tang M, et al. Probiotics to improve outcomes of colic in the community: protocol for the Baby Biotics randomized controlled trial. BMC Pediatr 2012; 12:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hempel S, Newberry SJ, Maher AR, et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA 2012; 307:1959–1969 [DOI] [PubMed] [Google Scholar]
- 40.Goldenberg JZ, Ma SS, Saxton JD, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev 2013; 5:CD006095. [DOI] [PubMed] [Google Scholar]
- 41.Gil-Campos M, López MÁ, Rodriguez-Benítez MV, et al. Lactobacillus fermentum CECT 5716 is safe and well tolerated in infants of 1-6 months of age: a randomized controlled trial. Pharmacol Res 2012; 65:231–238 [DOI] [PubMed] [Google Scholar]
- 42.Martín R, Olivares M, Marín ML, et al. Probiotic potential of 3 lactobacilli strains isolated from breast milk. J Human Lact 2005; 21:8–17 [DOI] [PubMed] [Google Scholar]
- 43.Olivares M, Díaz-Ropero MP, Martín R, et al. Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J Appl Microbiol 2006; 101:72–79 [DOI] [PubMed] [Google Scholar]
- 44.Maldonado J, Cañabate F, Sempere L, et al. Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J Pediatr Gastroenterol Nutr 2012; 54:55–61 [DOI] [PubMed] [Google Scholar]
- 45.Díaz-Ropero MP, Martín R, Sierra S, et al. Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J Appl Microbiol 2006; 102:337–343 [DOI] [PubMed] [Google Scholar]
- 46.Olivares M, Díaz-Ropero MP, Sierra S, et al. Oral intake of Lactobacillus fermentum CECT5716 enhances the effects of influenza vaccination. Nutrition 2007; 23:254–260 [DOI] [PubMed] [Google Scholar]
- 47.Pérez-Cano FJ, Dong H, Yaqoob P. In vitro immunomodulatory activity of Lactobacillus fermentum CECT5716 and Lactobacillus salivarius CECT5713: two probiotic strains isolated from human breast milk. Inmunobiology 2010; 12:996–1004 [DOI] [PubMed] [Google Scholar]
- 48.Contreras GA, Rodríguez JM. Mastitis: comparative etiology and epidemiology. J Mammary Gland Biol Neoplasia 2011; 16:339–356 [DOI] [PubMed] [Google Scholar]
- 49.Delgado S, Arroyo R, Jiménez E, et al. Staphylococcus epidermidis strains isolated from breast milk of women suffering infectious mastitis: potential virulence traits and resistance to antibiotics. BMC Microbiol 2009; 9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.World Health Organization Mastitis: Causes and Management. Geneva:WHO; 2000 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


