Abstract
Poly(A) tails enhance the stability and translation of most eukaryotic mRNAs, but difficulties in globally measuring poly(A)-tail lengths have impeded greater understanding of poly(A)-tail function. Here, we describe poly(A)-tail length profiling by sequencing (PAL-seq) and apply it to measure tail lengths of millions of individual RNAs isolated from yeasts, cell lines, Arabidopsis leaves, mouse liver, and zebrafish and frog embryos. Poly(A)-tail lengths were conserved between orthologous mRNAs, with mRNAs encoding ribosomal proteins and other “housekeeping” proteins tending to have shorter tails. As expected, tail lengths were coupled to translational efficiency in early zebrafish and frog embryos. However, this strong coupling diminished at gastrulation and was absent in non-embryonic samples, indicating a rapid developmental switch in the nature of translational control. This switch complements an earlier switch to zygotic transcriptional control and explains why the predominant effect of microRNA-mediated deadenylation concurrently shifts from translational repression to mRNA destabilization.
Keywords: poly(A)-tail length, translational regulation, translational efficiency, embryonic development, high-throughput sequencing, ribosome profiling
Most eukaryotic mRNAs end with poly(A) tails, which are added by a nuclear poly(A) polymerase following cleavage of the primary transcript during transcriptional termination1. These tails are then shortened by deadenylases2,3, although in some contexts, (e.g. animal oocytes and early embryos or at neuronal synapses), they can be re-extended by cytoplasmic poly(A) polymerases4,5. In the cytoplasm, the poly(A) tail promotes translation and inhibits decay2,5.
Although poly(A) tails must exceed a minimal length to promote translation, an influence of tail length beyond this minimum is largely unknown. The prevailing view is that longer tails generally lead to increased translation5,6. This idea partly stems from the known importance of cytoplasmic polyadenylation in activating certain genes in specific contexts4,5, and the increased translation observed in Xenopus oocytes and Drosophila embryos when appending synthetic tails of increasing length onto an mRNA7,8. Support for a more general coupling of tail length and translation comes from studies of yeast extracts9 and yeast cells10,11. However, the general relationship between tail length and translational efficiency has not been reported outside of yeast, primarily because transcriptome-wide measurements have been unfeasible for longer-tailed mRNAs.
Poly(A)-tail length profiling by sequencing (PAL-seq)
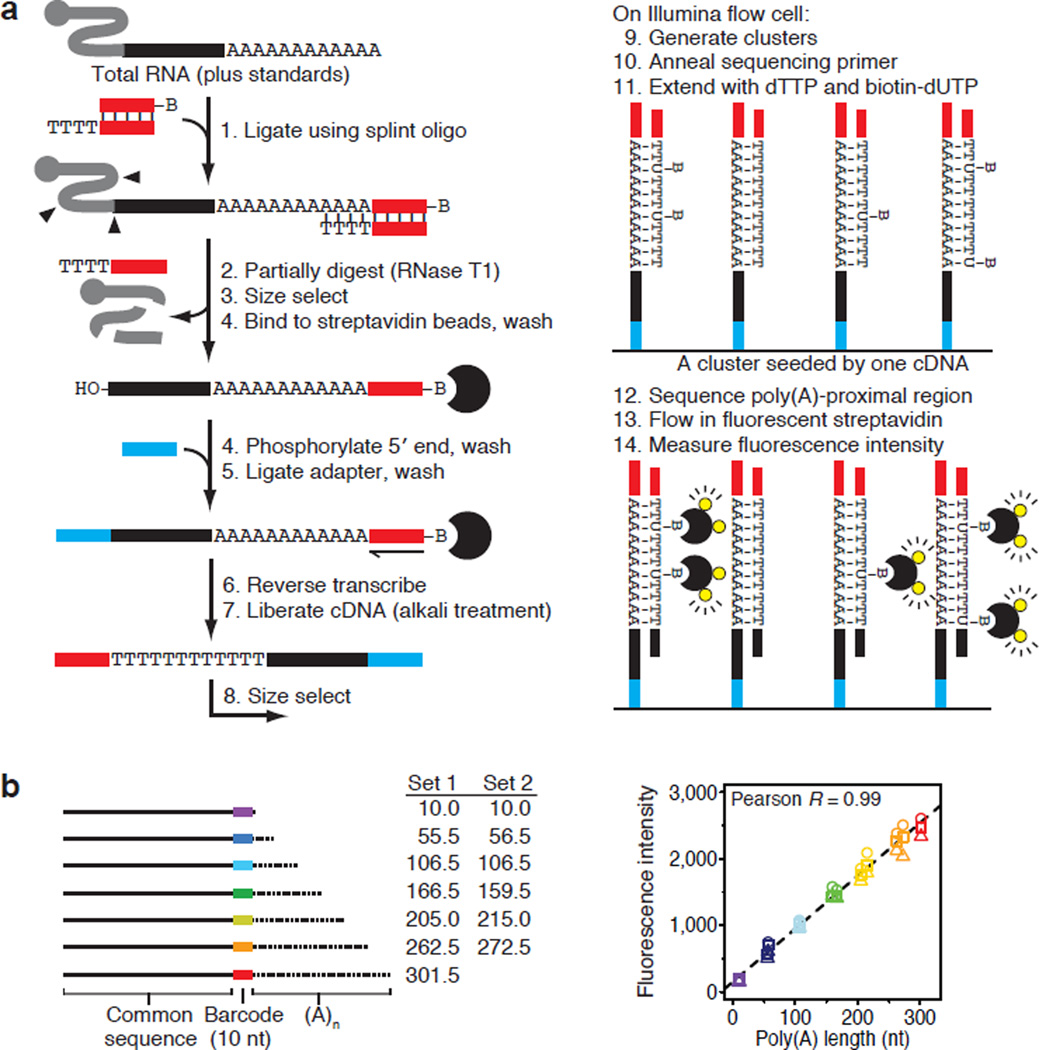
We developed a high-throughput sequencing method that accurately measures individual poly(A) tails of any physiological length (Fig. 1a). After generating sequencing clusters and before sequencing, a primer hybridized immediately 3′ of the poly(A) sequence is extended using a mixture of dTTP and biotin-conjugated dUTP as the only nucleoside triphosphates and conditions that were optimized to yield full-length extension products without terminal mismatches (Extended Data Fig. 1a). This key step quantitatively marks each cluster with biotin in proportion to the length of the poly(A) tail (Fig. 1a, step 11). After sequencing the 36 nt immediately 5′ of the poly(A) site, the flow cell is incubated with fluorophore-tagged streptavidin, which binds the biotin incorporated during primer-extension to impart fluorescence intensity proportional to the poly(A)-tract length. To account for the density of each cluster, this raw intensity is normalized to that of the fluorescent bases added during sequencing by synthesis12, thereby yielding a normalized fluorescence-intensity for the poly(A) tail of each transcript, paired with a sequencing read that identifies its poly(A) site and thus the gene of origin.
Figure 1. Global measurement of poly(A)-tail lengths.
a, Outline of PAL-seq. For each cluster, the fluorescence intensity reflects the tail length of the cDNA that seeded the cluster. Although the probability of incorporating a biotin-conjugated dU opposite each tail nucleotide is uniform, stochastic incorporation results in a variable number of biotins for each molecule within a cluster. b, Median streptavidin fluorescence intensities for two sets of mRNA-like molecules with indicated poly(A)-tail lengths, which were added to 3T3 (circle), HEK293T (triangle), and HeLa (square) samples for tail-length calibration.
Each starting sample was spiked with a cocktail of mRNA-like standards of known tail lengths (Extended Data Fig. 1b) to produce a standard curve for converting normalized fluorescence intensities to poly(A)-tail lengths (Fig. 1b). We refer to each of these tail-length measurements paired with its identifying sequence as a poly(A) tag. Although recovery of tags from the standards varied somewhat, it did not vary systematically with tail length, which indicated that length-related biases were not an issue (Extended Data Fig. 1c). Additional analyses indicated that mRNA degradation did not bias against longer poly(A) tails (Extended Data Fig. 2a).
Because alternative start sites or alternative splicing can generate different transcripts with the same poly(A) site, we considered our results with respect to unique gene models (abbreviated as ‘‘genes’’) rather than to transcripts (even though polyadenylation occurs on transcripts, not genes). Moreover, tags for alternative poly(A) sites of the same gene were pooled, unless stated otherwise. With this pipeline, analysis of RNA from NIH3T3 mouse fibroblasts (3T3 cells) yielded at least one tag from 10,094 unique protein-coding genes (including 97% of the 9,976 genes with at least one mRNA molecule per cell) and ≥100 tags from 2,873 genes, coverage typical of most samples (Supplementary Table 1).
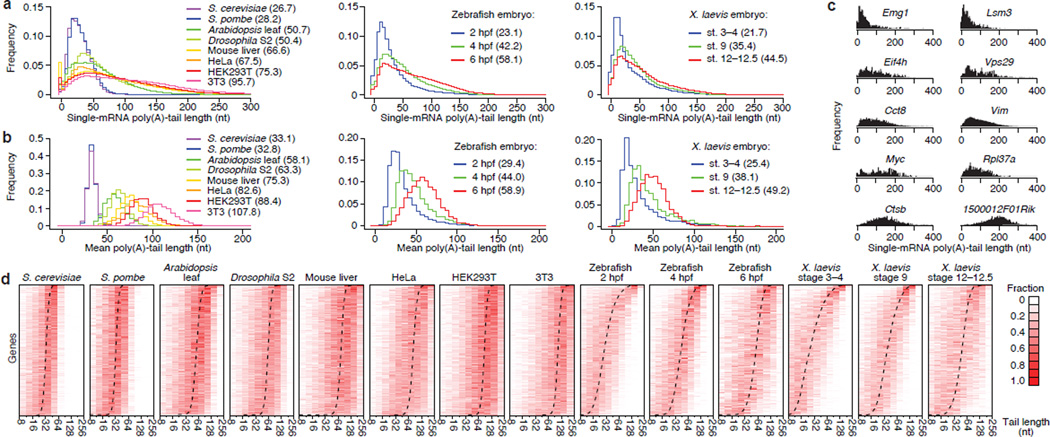
Tail-length diversity within each species
Median tail lengths in mammalian cells (range, 67–96 nt) exceeded those in Arabidopsis leaves and Drosophila S2 cells (51 and 50 nt, respectively), which exceeded those in budding and fission yeasts (27 and 28 nt, respectively) (Fig. 2a). Similar differences between mammalian, fly, plant, and yeast cells were observed when comparing tail-length averages for individual genes (Fig. 2b). For genes within each species, mean tail lengths varied, with the 10th and 90th percentiles differing by 1.4- to 1.6-fold. Variation was also observed for different mRNA transcripts from the same gene (Fig. 2c). For most genes the distributions were unimodal, with the mode approaching the mean (Fig. 2d). Poly(A)-tail lengths increased when progressing through cleavage, blastula, and gastrula stages of zebrafish embryonic development [2, 4 and 6 h post-fertilization (hpf), respectively] and analogous stages of frog development (Fig. 2a,b and d). Processed data reporting tail lengths for all genes detected in each sample are provided in the Gene Expression Omnibus (accession number GSE52809).
Figure 2. Poly(A)-tail lengths in yeast, plant, fly and vertebrate cells.
a, Global tail-length distributions. For each sample, histograms tally tail-length measurements for all poly(A) tags mapping to annotated 3′ UTRs (bin size = 5 nt). Leftmost bin includes all measurements <0 nt. Median tail lengths are in parentheses. b, Intergenic tail-length distributions. For each sample, histograms tally average tail lengths for protein-coding genes with ≥50 tags (yeasts, zebrafish and Xenopus) or ≥100 tags (other samples). Median average tail lengths are in parentheses. c, Intragenic tail-length distributions for 10 genes sampling the spectrum of average tail lengths in 3T3 cells. d, Intragenic tail-length distributions. Heatmaps show the frequency distribution of tail lengths for each gene tallied in b. The color intensity indicates the fraction of the total for the gene. Genes are ordered by average tail length (dashed line). Results from the S. cerevisiae total-RNA sample are reported in this figure.
Comparison of tail lengths for orthologous genes in human (HeLa and HEK293T) and mouse (3T3 and liver) cells revealed moderately strong correlations, indicating that tail lengths are conserved [Extended Data Table 1, Spearman R (Rs) as high as 0.46]. When analyzing gene classes that tended to have longer or shorter tails, the most striking and pervasive enrichment was for ribosomal protein and other ‘housekeeping’ genes among the short-tailed genes (Extended Data Table 2). This enrichment was strong in yeast, despite previous reports that ribosomal-protein genes tend to have long tails10,11. To address this and other discrepancies with previous yeast studies (Extended Data Fig. 3a and b), we used an independent method to measure the poly(A)-tail lengths of eight yeast genes, including four ribosomal protein genes. The results were much more consistent with our measurements than with the previous measurements (Extended Data Fig. 3 and 4). Both previous reports used the polyadenylation state microarray (PASTA) method, which fractionates RNAs by stepwise thermal elution from poly(U)-Sepharose. Although studies have successfully used poly(U)-Sepharose fractionation to detect tail-length changes for the same genes in different contexts13–15, detecting differences between different genes in the same context is more challenging. Our results suggest that PASTA, as previously implemented in yeasts10,11, is less suitable than PAL-seq for intergenic comparisons, although we cannot exclude the possibility that the discrepancies arose from different growth conditions.
The types of genes with shorter or longer tails differed between the embryonic samples and the other samples (Extended Data Table 2). Genes in the early embryo might not have the same tail lengths as their orthologs do in other contexts because prior to the maternal-to-zygotic transition (MZT), which occurs at ~3 hpf in zebrafish16 and at approximately stage 8 in X. laevis17, transcription is not yet active, and some maternal transcripts are masked for later use while others are subject to cytoplasmic polyadenylation5. At 6 hpf in zebrafish, ribosomal protein mRNAs had switched from being enriched in shorter-tail genes to being enriched in longer-tail genes (Extended Data Table 2), perhaps because these were mostly newly synthesized transcripts, which tended to have longer tails at this stage (Extended Data Fig. 5).
Because deadenylation is an important early step in eukaryotic mRNA decay2,3,18, we examined the relationship between poly(A)-tail length and published mRNA stability values (Extended Data Table 1). Tail-length and half-life were slightly negatively correlated in HeLa and 3T3 cells (Rs = −0.048 and −0.16, respectively) and variably correlated in yeast, depending on the source of the half-life measurements (Rs = ranging from −0.44 to 0.23). The weak relationships in HeLa and 3T3 cells would be expected if mRNAs with different half-lives have similar steady-state tail-length distributions with the less stable mRNAs transiting through the distributions more quickly.
No strong, easily interpretable correlations between tail length and mRNA features (3′ UTR length, ORF length, total length, splice-site number, splice-site density) or expression (steady-state accumulation and nuclear-to-cytoplasmic ratio) were observed (Extended Data Table 1). Of these, the strongest correlations were between tail length and steady-state accumulation (Rs from −0.44 to 0.25), and between tail length and mRNA length (Rs range, −0.12 to 0.36) or features related to mRNA length. Support for the latter relationship was also observed in intragenic comparisons, which revealed a weak positive relationship between tail length and the length of tandem 3′-UTR isoforms (Extended Data Fig. 6a). In early zebrafish embryos this relationship between 3′-UTR isoforms was even more pronounced when a predicted cytoplasmic polyadenylation element (CPE)4,19 was present in the unique region of the longer isoform (Extended Data Fig. 6b).
Transient coupling of tail length and translation in embryos
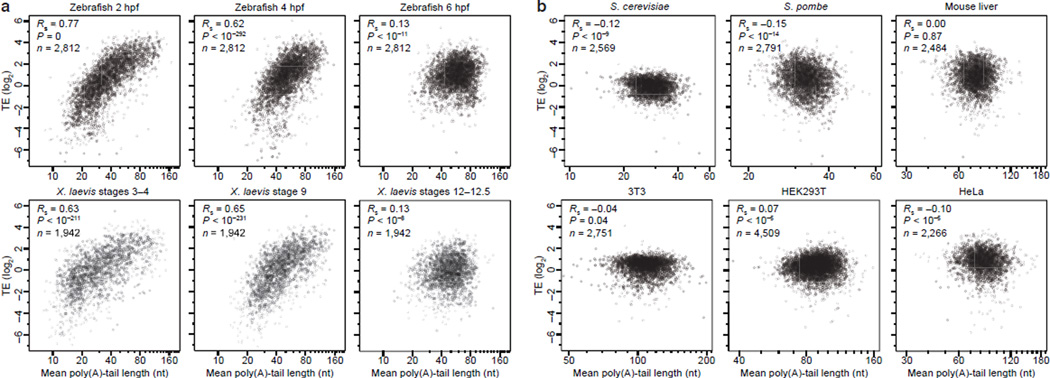
Most reports of increased translation of longer-tailed mRNAs have used oocytes and early embryos4,5. To examine whether this phenomenon reported in early embryos for a few genes applies transcriptome-wide, we performed ribosome footprint profiling and RNA-seq to measure translational efficiencies (TEs)20 from the embryonic samples used to measure tail lengths. We found that in early embryos (cleavage and blastula stages) of both fish and frog, mean poly(A)-tail length correlated strongly with TE (Fig. 3a, Rs from 0.62 to 0.77). No other mRNA feature has been reported to correlate so well with TE in any system.
Figure 3. Transient coupling between poly(A)-tail length and TE.
a, Relationship between mean tail length and TE for genes with ≥50 poly(A) tags from embryonic samples at the indicated developmental stages. For each stage, tail lengths and TEs were obtained from the same sample. MGC116473 and DDX24 fell outside the plot for X. laevis, stages 3–4, and LOC100049092 fell outside the plot for X. laevis, stages 12–12.5. b, Relationship between mean tail length and TE in the indicated cells, for genes with ≥50 (yeasts) or ≥100 (others) tags. With the exception of HeLa35, tail lengths and TEs were from the same samples. Budding yeast YBR196C, YLR355C and YDL080C, fission yeast SPCC63.04.1, mouse-liver NM_007881 and NM_145470, HEK293T NM_001007026, NM_021058, and NM_003537 and HeLa NM_001007026 fell outside their respective plots.
In these early embryonic stages a 2-fold increase in tail length corresponded to a large increase in TE—greater than 6-fold when doubling the tail from 20 to 40 nucleotides in 2 hpf zebrafish (Fig. 3a). Although longer-tailed mRNAs were more likely to contain a CPE, the relationship between tail length and TE for CPE-containing mRNAs was no different from that of other mRNAs (Extended Data Fig. 7a). In theory, this coupling might not be causal, or it might be causal but strictly due to either translational inhibition causing tail shortening or translational activity preventing tail shortening. Alternatively, all or at least some of the coupling might result from longer tail lengths causing more efficient translation in the early embryo. We favor this last possibility because it agrees with the known importance of cytoplasmic polyadenylation for activating genes in maturing oocytes8,21,22 and early embryos23,24 of Xenopus and certain other vertebrate contexts25–30. Even more importantly, it agrees with the increased translation observed in Xenopus oocytes when appending prosthetic poly(A) tails of increasing length onto an mRNA8.
The strong coupling observed in the blastula largely disappeared in gastrulating embryos (Fig. 3a, Rs = 0.13 for both fish and frog). This disappearance was not because of the more restricted tail-length range observed at gastrulation (Extended Data Fig. 7b). Moreover, we observed no positive correlation of a meaningful magnitude between mean poly(A)-tail length and TE in HeLa cells, HEK293T cells, 3T3 cells, mouse liver, budding yeast, or fission yeast (Rs = −0.10, 0.07, −0.04, 0.00, −0.12, and −0.15, respectively) (Fig. 3b). Our results in yeasts differed from those reported earlier10,11, which we again attribute to the limitations described above. In 3T3 cells, metabolic-labeling studies have been used to infer protein-synthesis rates31, which correlated with our TEs (Rs = 0.44, P <10−158) and did not correlate positively with tail length (Rs = −0.20, P <10−16). Taken together, our results suggest that beginning at gastrulation, translational control undergoes a mechanistic change that uncouples TE from poly(A)-tail length.
Intragenic comparison of tail length and translation
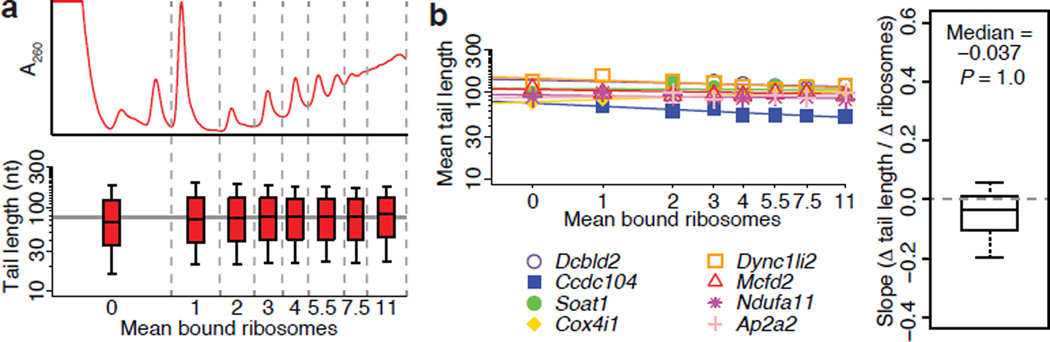
The simplest interpretation of the weak or negative correlations we observed between tail length and TE in yeast and mammalian cells is that increasing average tail length over the physiological range does not enhance translation in these contexts. However, our comparisons of average tail length and average TE between genes (Fig. 3b) might have missed a relationship that would be observed when looking at differentially translated mRNAs from the same gene. To address this possibility, we fractionated 3T3 cell lysates to isolate mRNAs associated with different numbers of ribosomes and measured the tail lengths in each fraction (Fig. 4a). To learn how poly(A)-tail length related to ribosome density for individual genes, we plotted mean tail-length values as a function of the number of bound ribosomes and fit the data for each gene with a straight line (Fig. 4b). The slopes of these lines were generally small, and most were slightly negative (Fig. 4b); positive slopes would have been expected if longer tails enhanced translation. Thus, the increase in median length observed between the lightest and heaviest fractions when considering bulk tail lengths (Fig. 4a; 66 and 82 nt, respectively) did not indicate a relationship between longer tails and enhanced translation but instead might have reflected the positive correlation between ORF length and tail length observed in 3T3 cells (Extended Data Table 1; Rs = 0.36). The trend of mostly negative slopes prevailed even when excluding data from mRNA not associated with any ribosomes (Extended Data Fig. 7c), or when examining subsets of genes with higher or lower translation efficiency, or with longer or shorter mean tail lengths (Extended Data Fig. 7d). This global intragenic analysis (Fig. 4b) supports the conclusion drawn from intergenic analyses (Fig. 3), that in all yeast and mammalian contexts examined (and presumably in most other cellular contexts), mRNAs with longer poly(A)-tails are not more efficiently translated.
Figure 4. No detectable intragenic coupling between poly(A)-tail length and TE.
a, Global analysis of tail lengths across the polysome profile for 3T3 cells. UV absorbance indicates mean number of ribosomes bound per mRNA for each fraction from the sucrose gradient (top, fractions demarcated with vertical dashed lines). Boxplots show distributions of tail lengths in each fraction for all tags mapping to annotated 3′ UTRs (bottom). Boxplot percentiles are line, median; box, 25th and 75th percentiles; whiskers, 10th and 90th percentiles. The horizontal line indicates the overall median of the median tail lengths. b, Relationship between tail lengths and ribosomes bound per mRNA for mRNAs from the same gene. For each gene, the data from a were used to plot the mean tail length as a function of bound ribosomes. Log-log plots for 8 randomly selected genes with ≥50 poly(A) tags in ≥6 fractions are shown (left), with lines indicating linear least-squared fits to the data (adding a pseudocount of 0.5 ribosomes to the fraction with 0 ribosomes). The boxplot shows the distribution of slopes for all genes with ≥50 poly(A) tags in ≥4 fractions (right; n = 4,079; one-sided, one-sample Wilcoxon test; boxplot percentiles as in a).
Explaining the shift in the ultimate effects of microRNAs
MicroRNAs (miRNAs) are 22-nt RNAs that pair to sites in mRNAs to target these messages for posttranscriptional repression32. Global measurements indicate that miRNA targeting causes mostly mRNA destabilization, with translational repression comprising a detectable but minor component of the overall repression33–36. The only known exception is the transient translational repression observed in early zebrafish embryos36. At 4 hpf miR-430 targeting causes mostly translational repression with very little mRNA destabilization, whereas by 6 hpf the outcome shifts to mostly mRNA destabilization36. Because miR-430 is induced only ~1.5 h before the 4-hpf stage, these results are interpreted as revealing the dynamics of miRNA action, in which an early phase of translational repression gives way to a later phase in which destabilization dominates36. When considering that miRNA targeting promotes poly(A)-tail shortening through the recruitment of deadenylase complexes37, our results suggests an alternative mechanism for the shift in miRNA regulatory outcomes. In this mechanism, miRNAs mediate tail shortening at both 4 and 6 hpf, but because of the switch in the nature of translational control (as well as destabilization of short-tailed mRNAs at later stages), tail shortening has very different consequences in the two stages: At 4 hpf, tail shortening predominantly decreases TE, whereas at 6 hpf, it predominantly decreases mRNA stability.
To integrate miRNA-mediated repression with effects on tail length, we injected one-cell zebrafish embryos with miRNAs that are normally not present in the early embryo and examined the influence of these injected miRNAs on ribosome-protected fragments (RPFs), mRNA levels and poly(A)-tail lengths at 2, 4 and 6 hpf. Injecting miR-155 caused RPFs from many of its predicted targets to decrease relative to RPFs from no-site control mRNAs (Fig. 5a). Despite the decrease in RPFs, target mRNA levels did not change relative to the controls at 2 and 4 hpf, indicating that at these stages miR-155 targeting caused mostly translational repression. In contrast, decreases in RPFs were accompanied by nearly commensurate mRNA reductions at 6 hpf, indicating that by this stage the outcome of repression had shifted to mostly mRNA destabilization (Fig. 5a). Thus, the shift in miRNA regulatory outcome that occurs between 4 and 6 hpf is not specific to miR-430 or its targets. With respect to mechanism, the observation of this shift between 4 and 6 hpf, even though the injected miR-155 was present and active much earlier than was miR-430, indicated that the shift reflected a transition from the unusual regulatory regime operating in pre-gastrulation embryos (in which TE is sensitive to tail length) more than it reflected the dynamics of miRNA action.
Figure 5. The influence of miR-155 on ribosomes, mRNA and tails in the early zebrafish embryo.
a, Relationship between changes in ribosome protected fragments (RPFs) and changes in mRNA levels after injecting miR-155. Changes observed between miRNA- and mock-injected embryos are plotted at the indicated stages for predicted miR-155 target genes (red, genes with ≥1 miR-155 site in their 3′ UTR) and control genes (gray, genes that have no miR-155 site yet resemble the predicted targets with respect to 3′ UTR length). To ensure that differences observed between 4 and 6 hpf were not the result of examining different genes, only site-containing genes and no-site control genes detected at both 4 and 6 hpf are shown for these stages. Lines indicate mean changes for the respective gene sets, with statistically significant differences between the sets indicated (*, P ≤0.05; **, P <10−4, one-tailed Kolmogorov–Smirnov test). Because injected miRNAs partially inhibited miR-430–mediated repression, genes with miR-430 sites were not considered. Data were normalized to the median changes observed for the controls. b, Relationship between RPF changes and mean tail-length changes after injecting miR-155. Tail-lengths were determined using PAL-seq, otherwise as in a. c, A developmental switch in the dominant mode of miRNA–mediated repression. The schematic (left) depicts the components of the bar graphs, showing how the RPF changes comprise both mRNA and TE changes. The compound bar graphs show the fraction of repression attributed to mRNA degradation (blue) and TE (green) for the indicated stage, depicting the overall impact of miR-155 (center; plotting results from a and b for genes with sites) and miR-132 (right, plotting results from Extended Data Fig. 8b for genes with sites). Slight, statistically insignificant, increases in mRNA for predicted targets resulted in blue bars extending above the axis. For samples from stages in which tail length and TE are coupled, a bracket adjacent to the compound bar indicates the fraction of repression attributable to shortened tails. Significant changes for each component are indicated with asterisks of the corresponding color (*, P ≤0.05; **, P <10−4, one-tailed Kolmogorov–Smirnov test).
The tail-length results further supported a mechanism involving shifting consequences of tail-length shortening. Predicted miR-155 targets had shortened tails at 2 and 4 hpf (Fig. 5b), which explained most of the miRNA-induced translational repression observed at these stages (Fig. 5c). By 6 hpf, the tail-length decreases observed at 4 hpf had mostly abated for predicted miR-155 targets (Fig. 5b), and these mRNAs were instead less abundant (Fig. 5a), in concordance with their extent of deadenylation at 4 hpf (Extended Data Fig. 8a). These observations agreed with the idea that tail shortening at later developmental stages destabilizes mRNAs, and suggested that the miRNA-mediated deadenylation occurring during the earlier developmental stages promotes decay later. With shorter tails no longer associated with reduced translation (Fig. 3) and instead associated with reduced mRNA levels, the ultimate consequence of miRNA-mediated repression shifted from translational repression to mRNA destabilization (Fig. 5c). Analogous results were obtained after injecting a different miRNA, miR-132 (Fig. 5c, Extended Data Fig. 8).
Because tail lengths were no longer strongly coupled to TE (Fig. 3), tail-length changes did not explain the decrease in mean TE observed at 6 hpf for miR-132 predicted targets (Fig. 5c). We conclude that when poly(A)-tail length is uncoupled from TE, the translational repression often detected as a minor component of the overall repression33–35 arises from a mechanism different from the one that dominates pre-gastrulation.
Our results provide a compelling explanation for miRNA-mediated translational repression in the pre-gastrulation zebrafish embryo: miRNAs induce poly(A) shortening, which decreases TE at this developmental period. They also explain why the pre-gastrulation zebrafish embryo is the only known context for which translational repression is the dominant outcome of miRNA-mediated regulation; in all other contexts examined, tail-length shortening causes mRNA destabilization with little or no effect on TE.
Two gene-regulatory regimes
Our results from yeast, cultured mammalian cells, and mouse liver refute the prevailing view that poly(A)-tail length broadly influences TE. In doing so, they add to the known differences between the regulatory regime operating in these cells and that in early metazoan embryos.
This absence or presence of coupling between poly(A)-tail length and TE can be rationalized in light of the potential interplay among regulatory options available in the two regulatory regimes. Yeast, mammalian, and mid-gastrulation cells were all transcriptionally active, which offers ample opportunities for nuclear control of gene expression. Moreover, active transcription enables unstable mRNAs to be replaced if required, thereby expanding the contexts in which differential mRNA stability can be exploited for gene control. Thus, an additional layer of control in which TE depends on poly(A)-tail length is dispensable. More importantly, because this type of coupling would lower output from older mRNA molecules that, in the absence of cytoplasmic polyadenylation, would often have shorter poly(A) tails, the utility of gene regulation through mRNA stability would be compromised. In this conventional regulatory regime, long-lived mRNAs would have less value if they were translated less efficiently because of their shorter tails.
For fish and frog embryos at the cleavage stage, the regulatory regime is very different. These embryos are transcriptionally inactive, which not only precludes the use of transcriptional and other nuclear processes to alter gene expression programs but also limits the use of differential mRNA stability, because degraded mRNAs cannot be replaced until zygotic transcription begins. Perhaps as a consequence, many mRNAs with short tails were observed (Fig. 2a), consistent with the known stability of short-tailed mRNAs in early embryos19,38. In these circumstances, early embryonic cells apparently harness differential tail length for global gene control. This result expands the known behavior of individual genes in Xenopus embryos23,24 and the observation that early embryonic cells have robust cytoplasmic polyadenylation,4 which increases the utility of a tail-length regulatory mechanism. Compared to metazoan cells (e.g. 6-hpf zebrafish embryos and the mammalian cells examined) subject to the standard regulatory process, cleavage-stage embryos had more uniform intragenic tail lengths and more variable intergenic lengths (Fig. 2d), as required for efficient harnessing of the tail-length regulatory regime. With their tail-length distribution also shifted towards shorter tails (Fig. 2b), cleavage-stage embryos can most efficiently exploit the tail-length differences with the greatest impact (Fig. 3a).
The transition between these two very different gene-regulatory regimes was rapid but not immediate. Despite their zygotic transcription, late-blastula embryos still coupled tail length with translation. Indeed, to the extent that newly transcribed zygotic mRNA tended to have longer tails than did the maternally inherited mRNAs (Extended Data Fig. 5), the continued coupling observed in this hybrid state would act to increase the relative output from these newly minted mRNAs, thereby sharpening the MZT.
We suspect that the tail-length regulatory regime observed in early embryos operates in other systems in which transcription is repressed (or distant) and cytoplasmic polyadenylation is active, such as early embryos of other metazoan species, maturing oocytes, and neuronal synapses5. The ability to measure poly(A)-tail lengths at single-mRNA resolution should provide important insights in these systems.
METHODS
PAL-seq
Total RNA or RNA from cytoplasmically enriched lysate (~1–50 µg) was supplemented with two mixes of tail-length standards and trace marker RNA containing an internal 32P-label (*) (ugagguaguagguuguauagu*caauccuaaucauuccaauccuaaucauucaaaaaaaaaa, IDT), which was used to monitor subsequent ligation, partial-digestion and capture steps. Polyadenylated ends in the mixture of cellular RNA and standards were ligated to a 3′-biotinylated adapter DNA oligonucleotide (p-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGACACATAC-biotin, IDT) in the presence of a splint oligonucleotide (TTCCGATCTTTTTTTTT, IDT) using T4 Rnl2 (NEB) in an overnight reaction at 18°C. The RNA was partially digested with RNase T1 (Ambion) as described39, extracted with phenol-chloroform, ethanol precipitated and then purified on a denaturing polyacrylamide gel (selecting 104–750 nt fragments), which removed residual unreacted 3′ adapter. Splinted-ligation products were captured on streptavidin M-280 Dynabeads (Invitrogen) and, while still bound to the beads, 5′ phosphorylated with 3′-phosphatase–deficient T4 polynucleotide kinase (NEB) and ligated to a 5′ adapter oligonucleotide (C3.spacer-CAAGCAGAAGACGGCATACGAGTTCAGAGTTCTAcaguccgacgauc, IDT; uppercase, DNA; lowercase, RNA) using T4 Rnl1 (NEB) in an overnight reaction at 22°C. Following reverse transcription using SuperScript II (Invitrogen) and a primer oligonucleotide (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACG, IDT), complementary DNA (cDNA) was liberated from the beads by base hydrolysis and purified on a denaturing polyacrylamide gel (selecting 166–790 nt DNA). Purified cDNA was denatured at room temperature in 5–100 mM NaOH, neutralized with addition of HT1 hybridization buffer (Illumina) and applied to an Illumina flow cell (at a typical concentration of 1.0–1.2 pM). Standard cluster generation, linearization, 3′-end blocking and primer hybridization were performed on a cBot cluster generation system (Illumina).
After transferring the flow cell to a Cluster Station designed for an Illumina Genome Analyzer, the sequencing primer was extended using a reaction mix containing 100 units/ml Klenow polymerase (NEB), 200 nM dTTP and either 10 nM (yeast samples) or 4 nM (other samples) biotin-16-dUTP (Roche). Extension was for 30 min at 37°C, flowing a fresh aliquot (≥50 µl) of reaction mix every 2 min to replenish dNTPs. Following primer extension, the flow cell was placed on a Genome Analyzer II sequencer (Illumina) for 36 cycles of standard sequencing-by-synthesis. After three additional cycles of cleavage to remove any residual sequencing fluorophores, the flow cell was washed with buffer [40.25 mM phosphate buffered saline (PBS), pH 7.4, 0.1% Tween], blocked with streptavidin-binding buffer [300 µg/ml bovine serum albumin (NEB), 40.25 mM PBS, pH 7.4, 0.1% Tween], washed with buffer again, and then imaged, as done previously12. This cycle of wash, block, wash, image was then repeated with a binding step inserted after the blocking step, in which the flow cell was incubated with 30 nM Alexa Fluor 532 Streptavidin (Invitrogen) in streptavidin-binding buffer for 10 min at 20°C. This expanded cycle was then repeated two more times, but with 100 nM streptavidin included at the binding step. Fluorescence was captured in the T and G channels because the wavelength of the excitation laser for these channels (532 nm) was identical to the fluorophore excitation wavelength. The sequential imaging confirmed that the second 100 nM streptavidin incubation did not increase mean cluster intensity (monitored in real time as part of the “first base report”), which indicated saturation of available biotin.
Calculation of poly(A)-tail lengths
Raw images taken during sequencing-by-synthesis and after binding of fluorescent streptavidin were processed with Firecrest image-analysis and Bustard base-calling software (Illumina, version 1.9.0, using default parameters) to generate a read (FASTQ) file and another file containing the position of each cluster, the read sequence, the quality score for each base, and the base intensities in all four channels for every cycle of sequencing and streptavidin binding. Reads (from all tiles) were aligned to a reference genome (hg18 for human, mm9 for mouse, dm3 for fly, danRer7 for fish, tair10 for Arabidopsis, Spombe1 for fission yeast, and sacCer3 for budding yeast) or a reference transcriptome (curated from Unigene mRNA sequences for X. laevis) using the Bowtie program for short-read mapping and the parameters ‘–l 25 –n 2 –m 1 –3 z’, where z was the number of streptavidin-binding cycles plus one. Reads containing ambiguous base calls (as indicated by characters “N” or “.”) at any position in the first 36 nt were discarded, as were reads mapping to multiple genomic loci. Reads that did not map to the genome were aligned to Bowtie indexes corresponding to the tail-length standards. For the remaining reads, mapping to the genome and standards was repeated, accounting for the possibility that the read failed to map because the sequence extended past the poly(A)-proximal fragment of the transcript and into the 5′ adaptor. This mapping was reiterated for 16 rounds (to capture tags of ≥20 nt), with each round considering previously unmapped reads in which the 5′ adaptor sequence started a nucleotide closer to the beginning of the read (requiring a perfect match to only the final 6 nt of the adaptor after the fifth round). Before each round of mapping, the adapter sequence was stripped by adjusting the Bowtie “−3” parameter. For the A. thaliana sample, the first sequenced base was of low quality, and raw images from the first cycle of sequencing were excluded for image analysis and base calling. Consequently, sequence reads were 35 nucleotides long, and only 15 rounds of iterative adapter trimming/sequence mapping were performed.
For each read carried forward as mapping to a single locus (of either the genome or the length standards), the cluster fluorescence intensity in the T channel after the first 100 nM streptavidin flow-in was recorded as the raw streptavidin fluorescence intensity. From this raw intensity, the intensity after the 0 nM flow-in was subtracted as background, and the resulting background-subtracted intensity was divided by the relative cluster intensity observed during sequencing-by-synthesis, which normalized for the density of molecules within the cluster. The relative cluster intensity was calculated by first dividing the fluorescence intensity of every sequenced base in the read by the median intensity for that base among all clusters with the same base at the same position, and then taking the average of the resulting values over the length of the read. Normalized streptavidin intensities were transformed to poly(A)-tail lengths using linear regression parameters derived from the median intensities of the standards and their mode poly(A)-tail lengths. For yeast, Arabidopsis, Drosophila, Xenopus and zebrafish samples, only the standards with tails of 10, 50 and 100 nt were used in the linear regression. For the other samples, all of the standards were used except for one with a 324-nucleotide tail (barcode sequence = CUCACUAUAC), which was typically not sufficiently abundant for accurate measurement of its tail length. Each tail length was then paired with the genomic (or standard) coordinates to yield a poly(A) tag.
Assigning poly(A) tags to genes
Reference transcript annotations were downloaded (in refFlat format) from the UCSC Genome browser or another database (Ensembl for zebrafish, Unigene for X. laevis, TAIR for Arabidopsis and PomBase for S. pombe). For human, mouse, zebrafish and fly, transcript 3′ ends were re-annotated using poly(A) sites identified by 3P-seq39 and the workflow described previously40. For each gene, a representative transcript model was chosen as the one that had the longest ORF and the longest 3′ UTR corresponding to that ORF. These reference transcript databases and a file with the sequences of the internal standards are available for anonymous download at http://web.wi.mit.edu/bartel/pub/publication.html. S. cerevisiae representative transcript models were from GSE53268. Poly(A) tags that overlapped the 3′ UTR of the representative transcript model by at least one nucleotide were assigned to that gene. Tags with tail-length measurements <−50 and >1000 nt (which included <0.0009% of the tags in any sample) were excluded from all analyses. Mean tail-length measurements <1 nt (which included measurements from 11 analyzed genes) were replaced with a value of 1.0 nt in the intragenic analysis across the polysome gradient (Fig. 4b). When considering the depth of a representative PAL-seq dataset from 3T3 cells, we considered 1.0 RPKM as the RNA-seq level indicating an average of one mRNA molecule per cell. This estimate was conservative, in that a comparison to published mRNA abundances in 3T3 cells31 indicated that 1.0 RPKM from our experiment corresponded to about 0.2 mRNA molecules per 3T3 cell.
RNA preparation for PAL-seq
For libraries made from fission yeast, HEK293T, 3T3, mouse-liver, X. laevis, and mock-, miR-132– and miR-155–injected zebrafish samples, as well as the S. cerevisiae sample analyzing cytoplasmically enriched RNA, RNA was extracted from a portion of the lysates that were also used for ribosome profiling and RNA-seq. These cleared lysates were enriched in cytoplasm. For libraries made from HeLa and polysome-gradient samples, RNA was extracted from similar cytoplasmically enriched lysates. For the polysome gradient fractionation (Fig. 4a), lysate preparation and centrifugation were performed as for ribosome profiling, but without nuclease digestion prior to fractionation. For other libraries, total RNA was used. The correlation observed when comparing PAL-seq results from HeLa cytoplasmically enriched RNA and (Extended Data Fig. 2b, Rs = 0.84) resembled that observed between biological replicates (Extended Data Fig. 2b, Rs = 0.83). The measured lengths in both types of RNA preparation were similar, despite the possibility that total RNA might have included more long-tailed mRNAs due to a population of nascent mRNAs that had full-length tails and were awaiting export. However, not all nuclear mRNAs are expected to have full-length tails (as some would still be in the process of being polyadenylated at the time of harvesting), and the nuclear population of mRNAs awaiting export presumably comprised a small fraction of the cellular mRNAs.
Tail-length standards
The common 5′ region of each standard and the unique 3′ region, consisting of the standard-specific barcode and poly(A)-tail (Fig. 1b), were synthesized separately and then ligated together to make full-length standards. To generate each 3′ region, a 5′-phosphate–bearing RNA oligonucleotide (IDT) consisting of the barcode segment followed by a 10 nt poly(A) segment was extended with E. coli poly(A)-polymerase (NEB), with ATP concentration and reaction time adjusted to yield of tails of the desired length. To narrow the tail-length distribution, extension products were sequentially purified on two denaturing polyacrylamide gels, excising products with tails of the desired length range and reducing the variability of tailed RNA to be mostly within ~5–25 nt, depending on the length of tail added (Extended Data Fig. 1b). The 5′ region of the standards was synthesized by in vitro transcription of a template containing Renilla luciferase sequence followed by that of a modified HDV ribozyme41. After gel-purification of the 5′ HDV self-cleavage product, the 2′, 3′-cyclic phosphate at its 3′ end was removed with T4 polynucleotide kinase (NEB; 3000 µl reaction containing 30,000 U enzyme and 100 mM MES-NaOH, pH 5.5, 10 mM MgCl2, 10 mM β-mercaptoethanol, 300 mM NaCl, 37°C, 6 h). After another gel purification, the dephosphorylated product was joined to the poly(A)-tailed barcode oligonucleotide by splinted ligation using T4 Rnl2 (NEB) and a bridge oligonucleotide with 10 nt of complementarity to each side of the ligation junction. Ligation products were gel-purified and mixed in desired ratios before being added to RNA samples for PAL-seq.
Ribosome footprint profiling
Immediately prior to harvesting, cultured mammalian cells were incubated with media containing 100 µg/ml cycloheximide for 10 min at 37°C to stop translation elongation. Cells were washed twice with ice-cold 9.5 mM PBS, pH 7.3 containing 100 µg/ml cycloheximide, and lysed by adding lysis buffer [10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM KCl, 2 mM dithiothreitol, 100 µg/ml cycloheximide, 1% Triton X-100, 500 units/ml RNasin Plus, and protease inhibitor (1X complete, EDTA-free, Roche)] and triturating four times with a 26-gauge needle. After centrifuging the crude lysate at 1300g for 10 min at 4°C, the supernatant was removed and flash-frozen in liquid nitrogen. Cultured S. pombe cells were grown to mid-log phase and then harvested (without cycloheximide pre-treatment) by filtering off the media and flash freezing the remaining paste, which was then manually ground into a fine powder with a mortar and pestle while being bathed in liquid nitrogen. The powder was thawed on ice, resuspended in lysis buffer and processed as described for the other lysates. Zebrafish embryos were enzymatically dechorionated and then incubated in 100 µg/ml cycloheximide in E3 buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) for 5 min at room temperature. The embryos were then transferred into lysis buffer and flash-frozen. X. laevis embryos were chemically dejellied after fertilization and flash-frozen in lysis buffer without cycloheximide pre-treatment. Once thawed, these samples were clarified as above and then processed in the same manner as other lysates. Prior to dissecting liver, a 6-week-old, male C57BL/6 mouse was sacrificed by cervical dislocation. The liver was excised, flash-frozen, and manually ground and processed as described for S. pombe. Ribosome profiling and RNA-seq were performed on cleared lysates essentially as described35, using RiboMinus-treated RNA for the S. pombe RNA-seq sample, and poly(A)-selected RNA for all others, with a detailed protocol available at http://bartellab.wi.mit.edu/protocols.html. S. cerevisiae RPF and RNA-seq data were from GSE53268 and were derived from the same sample as the S. cerevisiae PAL-seq sample analyzing cytoplasmically enriched RNA.
RPF and RNA-seq tags were mapped to the ORFs, as described previously35 (using the assemblies and transcript models used for PAL-seq), except reads overlapping the first 50 nt of each ORF were disregarded. This was done to minimize a bias from ribosomes accumulating at or shortly after the start codon, which results from translation initiation events continuing in the face of cycloheximide-inhibited elongation20. Because of this bias, genes with shorter ORFs have artifactually higher TEs if all the bound ribosomes are considered (as in conventional polysome gradient analysis). This cycloheximide effect might have distorted the TE measurements in studies that calculated ribosome densities using polysome gradient fractionation followed by microarray analysis, including those reporting a positive correlation between ribosome density and poly(A)-tail length10,11. However, this effect could not have influenced the conclusions of our polysome-gradient experiment, because our analysis focused on intragenic comparisons (Fig. 4b). TEs were considered only for genes exceeding a cutoff of 10 RPM (reads per million uniquely mapped reads) in the RNA-seq library. When calculating sequencing depth (the ‘M’ of RPM), all uniquely mapped reads that overlapped the mRNA primary or mature transcript were counted for all samples except the X. laevis samples; only the uniquely mapped reads overlapping ORFs were counted for X. laevis. For the analysis of miRNA effects, only genes exceeding a cutoff of 10 RPM in the mock-injected RNA-seq and RPF libraries, and ≥50 PAL-seq tags in the mock-injected and miRNA-injected samples were considered.
Statistics, reagents and animal models
All statistical tests were two-sided unless indicated otherwise. No power testing was done to anticipate the sample size needed for adequate statistical power. No randomization or blinding was used for miRNA injection experiments. Features of mRNAs (e.g. poly(A)-tail length, mRNA length, expression level, etc.) were not normally distributed, nor were changes in expression due to miRNA-mediated repression. Therefore, non-parametric measures or tests were used when making comparisons involving such quantities, and these tests do not make assumptions about equal variance between groups. Mammalian cell lines were obtained from ATCC, and S2 cells were the same as in ref.42 (i.e. adapted to growth in serum-free media). The BY4741 strain was used for S. cerevisiae, 972 for S. pombe, Columbia for Arabidopsis, and AB for zebrafish. All animal experiments were performed in accordance with a protocol approved by the MIT Committee on Animal Care.
Zebrafish injections
Zebrafish embryos were injected at the one-cell stage with 1 nl of 10 µM miRNA duplex (miR-155 and miR-132) or buffer alone using a PLI-100 Plus Pico-Injector. Duplexes were made by combining RNAs corresponding to either miR-132 (uaacagucuacagccauggucg) and miR-132* (accguggcauuagauuguuacu) or miR-155 (uuaaugcuaaucgugauaggggu) and miR-155* (accuaugcuguuagcauuaauc) in annealing buffer (30 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA), heating to 90°C for 1 minute, and slow cooling to room temperature over several hours. Injected embryos were incubated in E3 buffer at 28°C until time of harvesting.
Predicted miRNA targets
MicroRNA target genes were predicted using the reference transcript database used to assign zebrafish poly(A) tags. Each mRNA with a 3′ UTR that had at least one 7-nt site matching the miRNA seed region32 was predicted to be a target of that miRNA. Genes that had no 6-nt miRNA seed match anywhere within their transcript were classified as no-site genes, from which a set of no-site control genes was selected such that its 3′-UTR length distribution matched that of the predicted targets.
Calculation of the relationship between poly(A)-tail length and TE
For experiments in which zebrafish embryos were mock-injected or injected with miR-132 or miR-155, least-squares second-order polynomial regression was performed to determine the change in log2 TE for each change in log2 poly(A)-tail length. To prevent microRNA effects on TE and/or tail length from influencing any relationship, the regression analyses were performed after excluding genes for which the mRNAs contained a perfect match to either the seed (nucleotides 2–7 of the miRNA) of miR-430 (the predominant endogenous miRNA at 4 and 6 hpf) or the seed of the injected miRNA. These regression results were used to estimate the TE change attributable to tail-length change for each gene.
Poly(A)-tail measurements on RNA blots
Single-gene poly(A)-tail lengths were measured on RNA blots after directed RNase H cleavage of the interrogated mRNA. Standard methods43 were modified to enable higher resolution for shorter tails (<50 nt), such as those found on yeast mRNAs. Total RNA (3–20 µg) was heat-denatured for 5 min at 65°C in the presence or absence of 33 pmol/µg total RNA of (dT)18 (IDT), and in the presence of 25 pmol of a DNA oligonucleotide (or gapmer oligonucleotide, which had 16 DNA nucleotides flanked on each side by five 2′-O-methyl RNA nucleotides) that was complementary to a segment within the 3′-terminal region of the interrogated mRNA. After snap-cooling on ice, the RNA was treated with RNase H (Invitrogen) for 30 min at 37°C in a 20 µl reaction according to the manufacturer’s instructions. The reaction was stopped by addition of gel loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, dyes) and then analyzed on RNA blots resembling those used for small-RNA detection44 (Detailed RNA blot protocol available at http://bartellab.wi.mit.edu/protocols.html). Briefly, after separation of the RNA on a denaturing polyacrylamide gel and transfer onto a Hybond-NX membrane (GE Healthcare), the blot was treated with EDC (N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide; Sigma-Aldrich), which crosslinked the 5′ phosphate of the 3′-terminal RNase H cleavage product to the membrane45. The blot was then hybridized to a probe designed to pair to the region spanning the RNase H cleavage site and the poly(A) site. Comparison of these 3′-terminal fragments with and without poly(A) tails revealed the length of the tails.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Weinberg, V. Auyeung, I. Ulitsky, C. Jan, J.-W. Nam, A. Shkumatava, S.-J. Hong, Y. Erlich and the Whitehead Genome Technology Core (V. Dhanapal, L. Francis, S. Gupta, J. Love and T. Volkert) for helpful discussions, J.-W. Nam, I. Ulitsky and D. Weinberg for assistance with transcript annotation, C. Bresilla, X. Guo, S.-J. Hong, and A. Rothman for experimental assistance, and D. Weinberg for comments on the manuscript. Supported by NIH grant GM067031 (D.B.) and an NIH Medical Scientist Training Program fellowship T32GM007753 (A.S.). D.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions A.S. developed PAL-seq, generated tail-length measurements, and performed associated analyses. S.E. performed ribosome profiling, RNA-seq and associated analyses. G.C. performed zebrafish injections and assisted with staging. D.B. supervised with help from H.S. All authors helped design the study and write the manuscript.
Author Information Sequencing data and the processed data for each gene are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE52809.
The authors declare no competing financial interests.
REFERENCES
- 1.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nature reviews. Molecular cell biology. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 3.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley interdisciplinary reviews. RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiology and molecular biology reviews : MMBR. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nature structural & molecular biology. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 6.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley interdisciplinary reviews. RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 7.Salles FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- 8.Barkoff A, Ballantyne S, Wickens M. Meiotic maturation in Xenopus requires polyadenylation of multiple mRNAs. The EMBO journal. 1998;17:3168–3175. doi: 10.1093/emboj/17.11.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lackner DH, et al. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nutiu R, et al. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nature biotechnology. 2011;29:659–664. doi: 10.1038/nbt.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenthal ET, Tansey TR, Ruderman JV. Sequence-specific adenylations and deadenylations accompany changes in the translation of maternal messenger RNA after fertilization of Spisula oocytes. J Mol Biol. 1983;166:309–327. doi: 10.1016/s0022-2836(83)80087-4. [DOI] [PubMed] [Google Scholar]
- 14.Palatnik CM, Wilkins C, Jacobson A. Translational control during early Dictyostelium development: possible involvement of poly(A) sequences. Cell. 1984;36:1017–1025. doi: 10.1016/0092-8674(84)90051-5. [DOI] [PubMed] [Google Scholar]
- 15.Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- 16.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 17.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 18.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes & development. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 19.Aanes H, et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes & development. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- 22.Paris J, Richter JD. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol Cell Biol. 1990;10:5634–5645. doi: 10.1128/mcb.10.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- 24.Simon R, Tassan JP, Richter JD. Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes & development. 1992;6:2580–2591. doi: 10.1101/gad.6.12b.2580. [DOI] [PubMed] [Google Scholar]
- 25.Vassalli JD, et al. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes & development. 1989;3:2163–2171. doi: 10.1101/gad.3.12b.2163. [DOI] [PubMed] [Google Scholar]
- 26.Gebauer F, Xu W, Cooper GM, Richter JD. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. The EMBO journal. 1994;13:5712–5720. doi: 10.1002/j.1460-2075.1994.tb06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 28.Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795–3803. doi: 10.1242/dev.127.17.3795. [DOI] [PubMed] [Google Scholar]
- 29.Burns DM, Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes & development. 2008;22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nature cell biology. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 31.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS biology. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braun JE, Huntzinger E, Izaurralde EA. A molecular link between miRISCs and deadenylases provides new insight into the mechanism of gene silencing by microRNAs. Cold Spring Harb. Perspect. Biol. 2012;4:a012328. doi: 10.1101/cshperspect.a012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Audic Y, Omilli F, Osborne HB. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol Cell Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 39.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3'UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulitsky I, et al. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schurer H, Lang K, Schuster J, Morl M. A universal method to produce in vitro transcripts with homogeneous 3' ends. Nucleic acids research. 2002;30:e56. doi: 10.1093/nar/gnf055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salles FJ, Richards WG, Strickland S. Assaying the polyadenylation state of mRNAs. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- 44.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 45.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic acids research. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meijer HA, et al. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic acids research. 2007;35:e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, et al. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci U S A. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shalem O, et al. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol. 2008;4:223. doi: 10.1038/msb.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun M, et al. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haimovich G, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Sun M, et al. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell. 2013;52:52–62. doi: 10.1016/j.molcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Larsson E, Sander C, Marks D. mRNA turnover rate limits siRNA and microRNA efficacy. Mol Syst Biol. 2010;6:433. doi: 10.1038/msb.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.