Abstract
Hypomethylation of DNA repetitive elements is a common finding in cancer, but very little is known about the DNA methylation changes of different types of DNA repetitive elements, such as interspersed repeats (LINE1 and Alu Yb8) and tandem repeats (Sat-α, NBL-2 and D4Z4). We used bisulfite-PCR Pyrosequencing to quantitatively measure the DNA methylation of 5 different DNA repetitive elements in normal tissue and cancer. In all we studied 10 different tissues from 4 individuals undergoing autopsy, 34 paired normal and tumor tissues from patients with bladder cancer, 58 patients with chronic myelogenous leukemia and 23 patients with acute promyelocytic leukemia. We found that the DNA methylation of interspersed repeats (LINE1 and Alu Yb8) was very consistent from person to person and tissue to tissue while tandem DNA repeats appeared more variable in normal tissues. In bladder cancer we found clear hypomethylation of LINE1, Alu Yb8, Sat-α, and NBL-2. Conversely, we found an increase in the DNA methylation levels of D4Z4 from normal to cancer. In contrast leukemia showed no significant changes in the DNA methylation of LINE1 and Alu Yb8, but DNA methylation increases in NBL-2 and D4Z4 tandem repeats. Our findings show that the changes in DNA methylation levels of individual DNA repetitive elements are unique for each repetitive element, which may reflect distinct epigenetic factors and may have important implications in the use of DNA methylation of repetitive elements as global DNA methylation biomarkers. Keywords: DNA methylation, DNA repetitive elements, bladder cancer, leukemia
Keywords: DNA methylation, DNA repetitive elements, bladder cancer, leukemia
Aberrant promoter hypermethylation of CpG islands is a common finding in cancer and is associated with gene silencing in human cancer 1. In contrast to CpG islands, DNA repetitive elements are normally heavily methylated in somatic tissues and account for more than one-third of the total DNA methylation in the human genome 2. However, during tumorigenesis and aging global DNA hypomethylation has been described 3–5, and is attributable a decrease of DNA methylation in DNA repetitive elements, as there is an association of DNA repetitive element methylation and total genomic 5-methylcytosine 6. The etiologies of this global decrease in DNA methylation is not known but may be induced by environmental factors and precede cancer. Hypomethylation of LINE1 and Alu repetitive elements have been associated with exposure to the carcinogen benzene 7, suggesting a link between environmental exposure and epigenetic consequence8.
The significance of this global hypomethylation of DNA repetitive elements in cancer is unclear, but it may play multiple roles in the pathogenesis of cancer. DNA hypomethylation of LINE1 promoters may lead to an increase of its transcription in vivo 9, 10. This increase of LINE1 transcription might result in increased transposition of LINE1 elements into new genomic loci causing mutational events in cancer. De novo insertion of LINE1 into the c-Myc gene and the adenomatous polyposis coli (APC) gene leads to their inactivation in somatic cells 11, 12. In addition, hypomethylation of the LINE1 antisense promoter has been shown to increase transcription of c-MET, a proto-oncogene, which leads to the generation of a truncated form of c-MET in leukemia 13. The link between repetitive element hypomethylation and cancer has also been modeled in mice carrying a hypomorphic allele and a null allele for DNA methyltransferase 1 (Dnmt1 chip/-). This mouse model shows hypomethylation-induced tumors that involve chromosome instability and retrotransposition of a intracisternal A particle into the notch1 genomic locus 14. Thus, the hypomethylation of DNA repetitive elements may promote tumorigenesis through chromosomal instability or overexpression of oncogenes 14–16.
There are many different types of DNA repetitive elements, such as interspersed repeats and tandem repeats; combined these elements comprise at least half of the human genome 17. The interspersed repeats are mostly composed of retrotransposable elements such as Long Interspersed Nuclear Elements (LINEs), Short Interspersed Nuclear Elements (SINEs), Long Terminal Repeats (LTRs), or DNA transposons 17. The ability of these elements to “jump” has lead to their abundance and their distribution throughout the genome. In contrast, tandem repeats are present as long and uninterrupted simple or complex clustered sequences, and duplicate as inter and intra chromosomal segments. Centromeric, pericentromeric, and subtelomeric regions of the human genome contain tandem repeats and reside mostly in heterochromatic regions 17. Despite the plethora of DNA repetitive elements and the large contribution of these elements to the genome, little is known about the DNA methylation of different types of DNA repetitive elements in cancer. This study systematically and comprehensively examines the changes in DNA methylation patterns of multiple DNA repetitive elements in normal tissue and cancer.
Materials and Methods
Tissue samples
Ten non-cancerous somatic tissues bladder, brain, colon, esophagus, heart, kidney, liver, lung, spleen, stomach) were collected from four individuals under going post-mortem examinations (Case 1: 52 year-old male; cirrohsis, Case 2: 35 year-old male; cirrohsis, Case 3: 58 year-old female; breast cancer, Case 4: 60 year-old female; diabetic complication). 34 bladder tumor samples along with adjacent normal bladder urothelium were obtained from 25 male and 9 female patients with bladder cancer undergoing cystectomy. 58 Chronic myelogenous leukemia (CML) and 23 acute promyelocytic leukemia (APL) samples were obtained from peripheral blood (CML) and bone marrow (APL) of patients at the time of diagnosis. See supplemental table 1 for the details about autopsy samples, bladder tumor and leukemia samples. All human tissue was collected through protocols approved by the Institutional Review Board of the University of Southern California.
DNA extraction and Sodium Bisulfite modification
DNA was extracted using standard Phenol/Chloroform methods. Bisulfite modification of genomic DNA was performed using the EZ-96 DNA Methylation-Gold Kit™ (Zymo Research, Orange, CA, USA), according to the manufacturer’s recommended protocol.
Quantification of DNA methylation by Pyrosequencing
The level of DNA methylation of LINE1, Alu Yb8 subfamily, Sat-a, D4Z4 and NBL-2 was assessed using a previously developed bisulfite-DNA Polymerase Chain Reactions (PCR) Pyrosequencing assay (See supplemental table 2 for details) 2, 18. Primers, designed to a consensus sequence for each repetitive element, amplified a global pool of repetitive elements rather than a single element or genomic locus. In brief, bisulfite treatment of DNA leads to the formation of single nucleotide polymorphism, either a C, indicating methylated cytosine, or a T, indicating unmethylated cytosine, which can be quantitated by Pyrosequencing. Biotin labeled PCR product was bound to Streptavidin Sepharose HP (Amersham Biosciences, Uppsala, Sweden). The Sepharose beads containing the immobilized PCR products were purified, washed, denatured using a 0.2 M NaOH solution, and washed again. Pyrosequencing was performed using the PSQ HS 96 Pyrosequencing System (Biotage, AB, Uppsala, Sweden) per the manufacturer’s protocol.
Statistical analysis
All values are reported as mean ± SD. Data was analyzed using the paired or unpaired Student's t test using StatView software (Abacus, Berkeley, CA, USA). The significance level was set at P<0.05.
Inter- and intra variability of DNA methylation of DNA repetitive elements in normal tissues
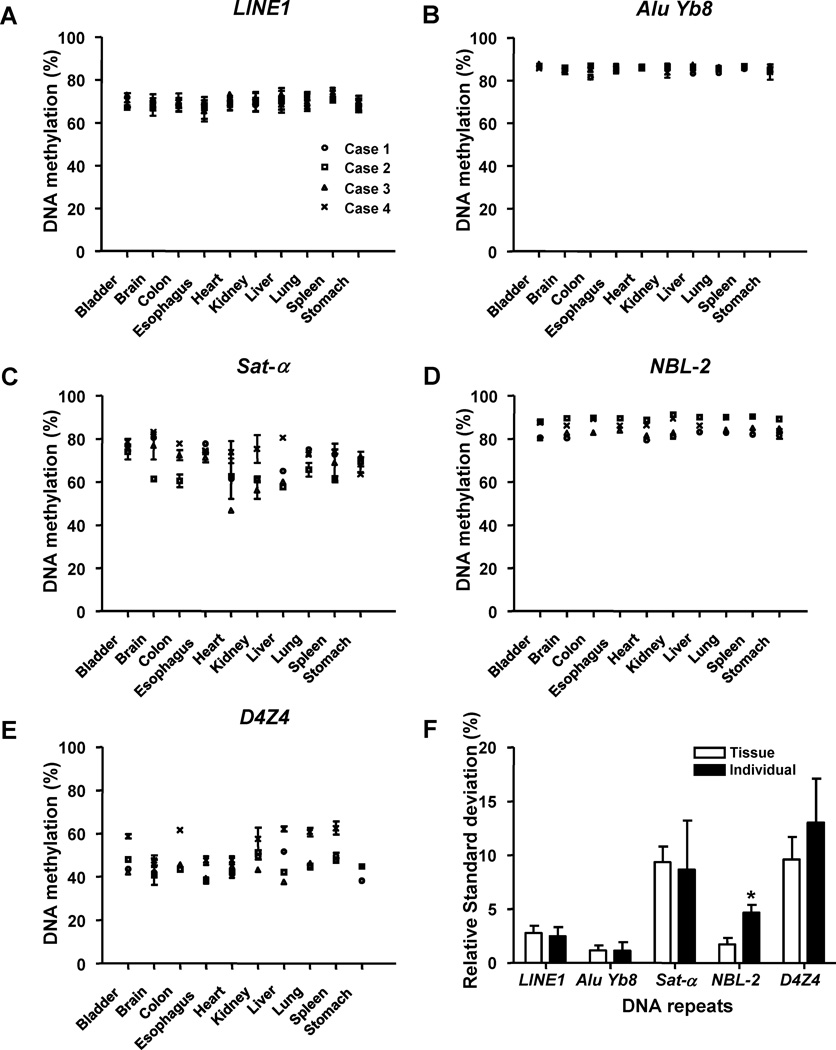
To understand the methylation of DNA repetitive elements in normal tissue, we first examined 40 unique tissues from 4 individuals without cancer undergoing autopsy (Fig. 1 and supplemental table 3). This allowed examination of the variability of DNA methylation from person to person and tissue to tissue for the first time. We employed bisulfite-PCR Pyrosequencing to quantitatively assess the level of DNA methylation of interspersed repeats (LINE1 and AluYb8) and tandem repeats (Sat-α, NBL-2 and D4Z4). We studied DNA methylation of LINE1since it is known to be hypomethylated in many types of cancer and is associated with global hypomethylation 2, 6, 19. We also have measured the DNA methylation of several Alu subfamilies including AluYb8, Alu Sx, and Alu J in normal tissues using specific polymorphisms unique to each Alu subfamily. We found the mean level of DNA methylation as measured by bisulfite PCR Pyrosequencing is 85.5%, 24% and 18.8% for Alu Yb8, Alu Sx, and Alu J respectively (data not shown). The higher DNA methylation levels of Alu Yb8 reflect the younger evolutionary age of these elements and the lower degree of C to T transition mutations compared to the older Alu Sx and Alu J elements. As previous studies have shown hypomethylation of Alu elements in cancer we chose to study the Alu Yb8 subfamily as the higher methylation levels would allow a technical advantage to study hypomethylation changes. We analyzed chromosome 1 satellite-α (Sat-α) repeats whose DNA methylation is known to be hypomethylated in cancer and associated with global DNA methylation6, 20, 21. We also studied NBL-2 and D4Z4 known to be hypomethylated in ICF (Immunodeficiency, Centromeric instability, and Facial abnormalities) syndrome patients 22. Schematic structures and estimated copy numbers of the DNA repetitive studied are shown in supplement figure 1. The bisulfite-PCR Pyrosequencing assay for the repetitive elements proved very consistent with the mean of standard deviation (SD) for LINE1, Alu Yb8, Sat-α, NBL-2 and D4Z4 DNA repeats was 2.47%, 0.93 %, 2.3 %, 0.71 % and 2.2 %, respectively.
Figure 1.
DNA methylation of different DNA repetitive elements in normal tissue. DNA methylation of 5 different DNA repetitive elements was measured in ten non-cancerous somatic tissues from four individuals under going post-mortem examinations using bisulfite-PCR Pyrosequencing. Interspersed repeats: (A) LINE1, (B) Alu Yb8; Tandem repeats: (C) Sat-α, (D) NBL-2, (E) D4Z4, (F) inter and intra variation of DNA methylation of different DNA repetitive elements. *p<0.0001. DNA methylation (%) represents the percentage of mCpG/mCpG + CpG + TpG; mC (methylated cytosine), C (unmethylated cytosine) and T (thymine that are generated from CpG by spontaneous evolutionary deamination).
The relative standard deviation (RSD) of ten different tissues (Tissue RSD, TRSD) and RSD of four individuals (Individual RSD, IRSD,) were calculated to assess inter and intra individual variability of each DNA repetitive element. The level of DNA methylation of interspersed repeats, LINE1 (TRSD=2.8%, IRSD=2.5%) and Alu Yb8 (TRSD= 1.2%, IRSD=1.1%) was more consistent among tissues and individuals than tandem repeats, Sat-α (TRSD=9.4%, IRSD=8.7%), NBL-2 (TRSD=1.7%, IRSD=4.7%) and D4Z4 (TRSD=9.6%, IRSD=13%), (Fig. 1F). This may be attributable to a sampling effect as the copy number of interspersed repeats is greater than tandem repeats, and interspersed repeats are spread more diffusely in the genome whereas tandem repeats tend to be localized to specific areas of chromosomes. DNA methylation of interspersed repeats, LINE1 and Alu Yb8, were consistent from tissue to tissue and person to person. In contrast, the DNA methylation of one of the tandem repeats, NBL-2 was more consistent from tissue to tissue within an individual (p<0.0001). It is possible that the level of DNA methylation might be determined before tissue differentiation by genetic factors such as copy number variation or an unknown genetic determinant. Thus a genetic factor or very early stem cell event would determine the level of NBL-2 methylation in all tissues within an individual. These differences between methylation of DNA repetitive elements may have important implications if DNA repetitive elements are to be used as a biomarker in population based studies.
DNA methylation of DNA repetitive elements in bladder cancer
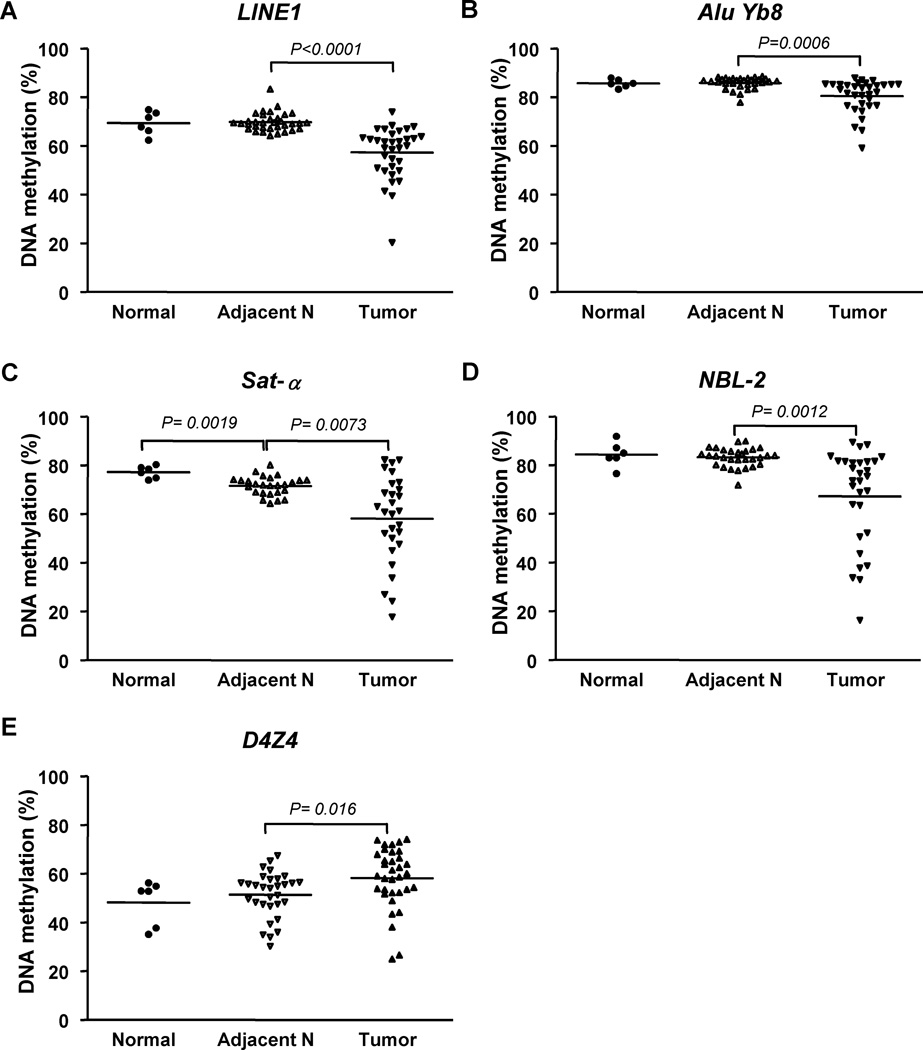
In order to determine the level of DNA methylation of DNA repetitive elements in two distinctive tumor types, we chose bladder tumor as an example of a solid tumor and leukemia as an example of a liquid tumor, both of which undergo tumor progression changes. First, we study the changes in the DNA methylation of multiple repetitive elements in cancer by studying normal bladder urothelium from people without bladder cancer with paired bladder tumors and adjacent normal tissue. We compared the level of DNA methylation using adjacent normal and tumor samples from the same individual (Fig. 2). The methylation level of interspersed repeats LINE1 and Alu Yb8 significantly decreased in bladder cancer in comparison to adjacent normal bladder urothelium. Methylation of LINE1 and Alu Yb8 decreased from 69.8 ± 3.8 % to 57.3 ± 10.6 % (p<0.0001) and from 85.9 ±2.4 to 80.5 ± 6.8 (p=0.0006), respectively, which is consistent with previous reports 19, 23 (See supplemental table 4 for details).
Figure 2.
DNA methylation changes of different DNA repetitive elements in bladder cancer. DNA methylation of 5 different repetitive elements was measured in normal bladder urothelium from people without cancer (Normal), and paired samples of normal bladder urothelium from patients with bladder cancer (Adjacent N) and bladder cancers (Tumor). Interspersed repeats: (A) LINE1, (B) Alu Yb8; Tandem repeats: (C) Sat-α, (D) NBL-2, (E) D4Z4.
In tandem repeats, the level of DNA methylation of Sat-α and NBL-2 also significantly decreased from 71.6 ± 3.9 % to 58.2 ±17.8 % (p=0.0073), and from 83.2 ± 3.9 % to 67.3% ± 19.7 % (p=0.0012), respectively. In contrast we found that DNA methylation of the tandem repeat D4Z4 increased in bladder cancer. The level of D4Z4 methylation increased from 51.3 ± 9.3% to 58.1 ± 12.4 % (p=0.016) in bladder cancer, in comparison to adjacent normal tissue. Our results are consistent with a previous report that showed D4Z4 elements are hypermethylated in some cancers24. Interestingly, we found that in adjacent normal bladder tissue from patients with cancer, DNA methylation changes could be detected in tandem repeats when compared to normal bladder tissue in people without cancer. Sat-α showed a significant decrease of 5.7 % (p=0.0019) while D4Z4 showed a mean increase of 3.0 % (p=0.45) in adjacent normal bladder from individuals with bladder cancer, compared to normal bladder. This presents the possibility that these repetitive elements can be used as an early biomarker to predict cancer. However, there is an increase in variability of DNA methylation levels of DNA repetitive elements in bladder cancer, and DNA methylation changes do not occur in every cancer, which may limit its predictive ability as early cancer marker for individual patients.
To understand the relationship among different repetitive elements within an individual, we examined the correlation between two different DNA repetitive elements in 34 pair of normal and bladder tumor tissues. Two interspersed DNA repetitive elements, LINE1 and Alu Yb8 methylation strongly correlated each other (r=0.630, p<0.0001) and also correlated with Sat-α methylation (Sat-α vs. LINE1; r=0.687, p<0.0001 and Sat-α vs. Alu Yb8; r=0.489, p=0.002), but not D4Z4 and NBL-2 (See supplemental table 5 for details). Although NBL-2 showed hypomethylation in bladder cancer, NBL-2 hypomethylation did not correlate with hypomethylation of LINE1, Alu Yb8 and Sat-α. Thus the mechanism responsible for the hypomethylation of NBL-2 is independent of hypomethylation of the other DNA repetitive elements. Again, these data support the finding that DNA methylation of tandem repeats differs from that of interspersed repeats and that different DNA repetitive elements have unique changes in methylation in bladder cancer
It is known that a germline mutation of DNMT3B causes ICF syndrome which is characterized by hypomethylation of NBL-2 and D4Z4 22, thus linking the DNA methylation of these two tandem repeats. However, the discordant DNA methylation changes of NBL-2 and D4Z4 in bladder cancer indicate that epigenetic machinery other than DNMT3B must be involved in bladder cancer. One may also consider that DNMT3B has different functions, where it is involved in increasing methylation of D4Z4 and decreasing DNA methylation of NBL-2 in bladder cancer, respectively25.
DNA methylation changes of repetitive elements in leukemia
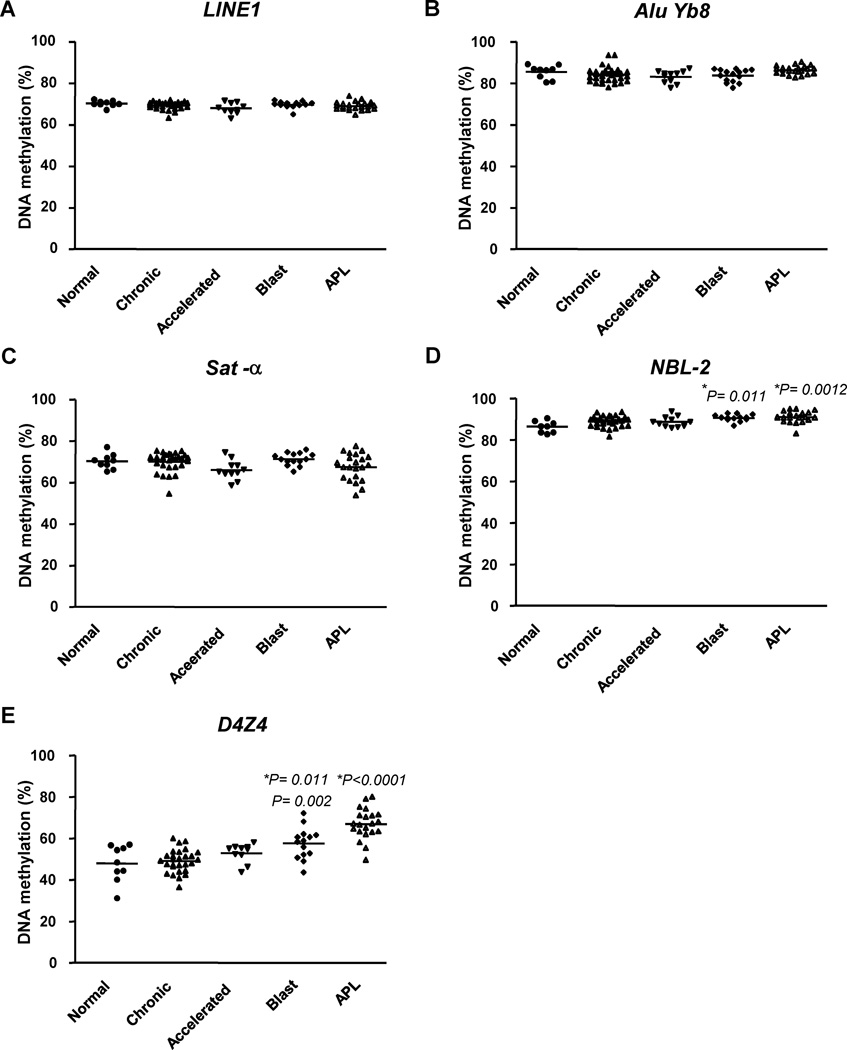
We next studied the DNA methylation changes of repetitive elements in leukemia (Fig. 3). This allowed us to compare two different cancers of the myeloid hematopoietic system and to study the changes in methylation during the progression of CML, which shows a well-defined progression from chronic to accelerated to blast phase leukemia. It has been reported that the level of LINE1 methylation decreases during CML progression 13, 27, or does not significantly change in CML patients 26. In our study, the level of DNA methylation in LINE1 and Alu Yb8 did not significantly change in CML and APL (See supplemental table 6 for details). Our finding is consistent with the previous report by Issa et al. 26, but is different from the hypomethylation of DNA repetitive elements found during CML progression 13, 27. This difference may be attributable to different techniques used to measure the level of DNA methylation: bisulfite-PCR Pyrosequencing versus methylation specific PCR (MSP).
Figure 3.
DNA methylation changes of different repetitive elements in leukemia. DNA methylation of 5 different DNA repetitive elements was measured in Normal peripheral blood (Normal), Chronic phase (Chronic) CML, Accelerated phase (Accelerated) CML, Blast crisis (Blast) CML, and APL. Interspersed repeats: (A) LINE1, (B) Alu Yb8; Tandem repeats: (C) Sat-α, (D) NBL-2, (E) D4Z4. *p vs. normal blood, †p vs. chronic phase.
As shown in bladder cancer, the tandem repeats behaved differently than the interspersed repeats in leukemia. However, in leukemia both NBL-2 and D4Z4 methylation increased in APL and during the progression of CML; the increase was slightly greater in APL. Thus, the methylation of NBL-2 decreases in bladder tumor progression, but increases during leukemogenesis. Our findings are consistent with the previous report, showing that NBL-2 repeats can be either hypomethylated in neuroblastoma and hepatocellular carcinoma or hypermethylated in ovarian epithelial carcinomas 28. Nishiyma et al. also reported that hyper- and hypomethylation at different individual CpG sites within same NBL-2 repeat in ovarian cancer 29. However, in our study we found the methylation at different individual CpG sites with NBL-2 was concordant in both bladder cancer (R2= 0.81) and in leukemia (R2= 0.50). This indicates that the behavior of DNA methylation of tandem repeats may vary from cancer to cancer and deserves further investigation.
Here we demonstrated that changes in DNA methylation of tandem repeat elements behave differently than interspersed repeats, and that the methylation changes vary between bladder cancer and leukemia. Due to the variability of DNA methylation levels in different DNA repetitive elements, methylation assessment of DNA repetitive elements may possess limited ability as a biomarker for cancer stage or disease outcome in an individual patient. However, these DNA repetitive element assays have been used as a biomarker previously in population based studies. For example, LINE1 has been used as a surrogate marker of demethylating agent treatments for MDS (myelodysplastic syndrome) and leukemia patients 30–32. LINE1 and Alu methylation assays have also been used as a biomarker of environmental exposure 7. Therefore we believe our results add to an understanding of how DNA methylation of repetitive elements change in different diseases, and can lead to a better understanding of their use as biomarkers for epigenetic drugs and population based epigenetic studies.
Supplementary Material
Acknowledgements
This work and HMB were partially funded by a grant from the Wright Foundation. ASY is the recipient of an American Society of Clinical Oncology-Association of Subspecialty Professors Career Development Award in Geriatric Oncology. SHC and this project were supported by the Southern California Environmental Health Sciences Center pilot grant funded by the National Institute of Environmental Health Sciences (Grant # 5P30ES07048). Bladder tissue samples were collected under the Program Project Grant # NCI PO1 CA86871.
References
- 1.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 2.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic acids research. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 4.Issa JP. Aging, DNA methylation and cancer. Crit Rev Oncol Hematol. 1999;32:31–43. doi: 10.1016/s1040-8428(99)00019-0. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 6.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic acids research. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 8.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Neill RJ, O'Neill MJ, Graves JA. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- 10.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 11.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 12.Morse B, Rotherg PG, South VJ, Spandorfer JM, Astrin SM. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988;333:87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- 13.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, Prosper F, Heiniger A, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 14.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2007 doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 15.Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 16.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 17.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 18.Choi SH, Byun HM, Kwan JM, Issa JP, Yang AS. Hydroxycarbamide in combination with azacitidine or decitabine is antagonistic on DNA methylation inhibition. Br J Haematol. 2007;138:616–623. doi: 10.1111/j.1365-2141.2007.06707.x. [DOI] [PubMed] [Google Scholar]
- 19.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 20.Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, Ehrlich M. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. International journal of cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 21.Qu G, Dubeau L, Narayan A, Yu MC, Ehrlich M. Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mutation research. 1999;423:91–101. doi: 10.1016/s0027-5107(98)00229-2. [DOI] [PubMed] [Google Scholar]
- 22.Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc'his D, Viegas-Pequignot E, Ehrlich M, Hanash SM. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum Mol Genet. 2000;9:597–604. doi: 10.1093/hmg/9.4.597. [DOI] [PubMed] [Google Scholar]
- 23.Jurgens B, Schmitz-Drager BJ, Schulz WA. Hypomethylation of L1 LINE sequences prevailing in human urothelial carcinoma. Cancer Res. 1996;56:5698–5703. [PubMed] [Google Scholar]
- 24.Tsumagari K, Qi L, Jackson K, Shao C, Lacey M, Sowden J, Tawil R, Vedanarayanan V, Ehrlich M. Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic acids research. 2008;36:2196–2207. doi: 10.1093/nar/gkn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 26.Issa JP, Gharibyan V, Cortes J, Jelinek J, Morris G, Verstovsek S, Talpaz M, Garcia-Manero G, Kantarjian HM. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol. 2005;23:3948–3956. doi: 10.1200/JCO.2005.11.981. [DOI] [PubMed] [Google Scholar]
- 27.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, San Jose-Eneriz E, Garate L, Cordeu L, Cervantes F, Prosper F, Heiniger A, Torres A. Repetitive DNA hypomethylation in the advanced phase of chronic myeloid leukemia. Leuk Res. 2007 doi: 10.1016/j.leukres.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Nishiyama R, Qi L, Tsumagari K, Weissbecker K, Dubeau L, Champagne M, Sikka S, Nagai H, Ehrlich M. A DNA repeat, NBL2, is hypermethylated in some cancers but hypomethylated in others. Cancer Biol Ther. 2005;4:440–448. doi: 10.4161/cbt.4.4.1622. [DOI] [PubMed] [Google Scholar]
- 29.Nishiyama R, Qi L, Lacey M, Ehrlich M. Both hypomethylation and hypermethylation in a 0.2-kb region of a DNA repeat in cancer. Mol Cancer Res. 2005;3:617–626. doi: 10.1158/1541-7786.MCR-05-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS, Cortes J, Kantarjian HM. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2'-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, Verstovsek S, Rytting M, Wierda WG, Ravandi F, Koller C, Xiao L, Faderl S, et al. Phase I/II study of the combination of 5-aza-2' - deoxycytidine with valproic acid in patients with leukemia. Blood. 2006 doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MR, Kantarjian HM, Garcia-Manero G, et al. DNA methylation changes after 5-aza-2'-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.