Abstract
The identification of activating NOTCH1 mutations in T-cell acute lymphoblastic leukemia (T-ALL) led to clinical testing of γ-secretase inhibitors (GSI) that prevent NOTCH1 activation1–3. However, responses have been transient4,5, suggesting that resistance limits clinical efficacy. Here we modeled T-ALL resistance, identifying GSI-tolerant ‘persister’ cells that expand in the absence of NOTCH signaling. Rare persisters are already present in naïve T-ALL populations, and the reversibility of the phenotype suggests an epigenetic mechanism. Relative to GSI-sensitive cells, persisters activate distinct signaling and transcriptional programs, and exhibit chromatin compaction. A knockdown screen identified chromatin regulators essential for persister viability, including BRD4. BRD4 binds enhancers near critical T-ALL genes, including MYC and BCL2. The BRD4 inhibitor JQ1 down-regulates these targets and induces growth arrest and apoptosis in persisters, at doses well tolerated by GSI-sensitive cells. Consistently, the GSI-JQ1 combination was found to be effective against primary human leukemias in vivo. Our findings establish a role for epigenetic heterogeneity in leukemia resistance that may be addressed by incorporating epigenetic modulators in combination therapy.
T-ALL is an aggressive malignancy frequently associated with activating mutations in NOTCH1, a critical oncogene in this disease1. Gamma-secretase inhibitors (GSIs) that prevent NOTCH1 cleavage and activation have been tested in clinical trials and mouse models, but responses have been modest and transient5. To understand mechanisms by which T-ALL cells overcome NOTCH1 inhibition, we modeled GSI-resistance in vitro and in vivo, and investigated functional and molecular characteristics of resistant cells. By chronically exposing NOTCH-dependent T-ALL cells to GSI (compound E) in vitro, we isolated a population of ‘persister’ cells that tolerate GSI concentrations more than 50-fold higher than naïve cells (Figure 1a,b; S1a; S2a).
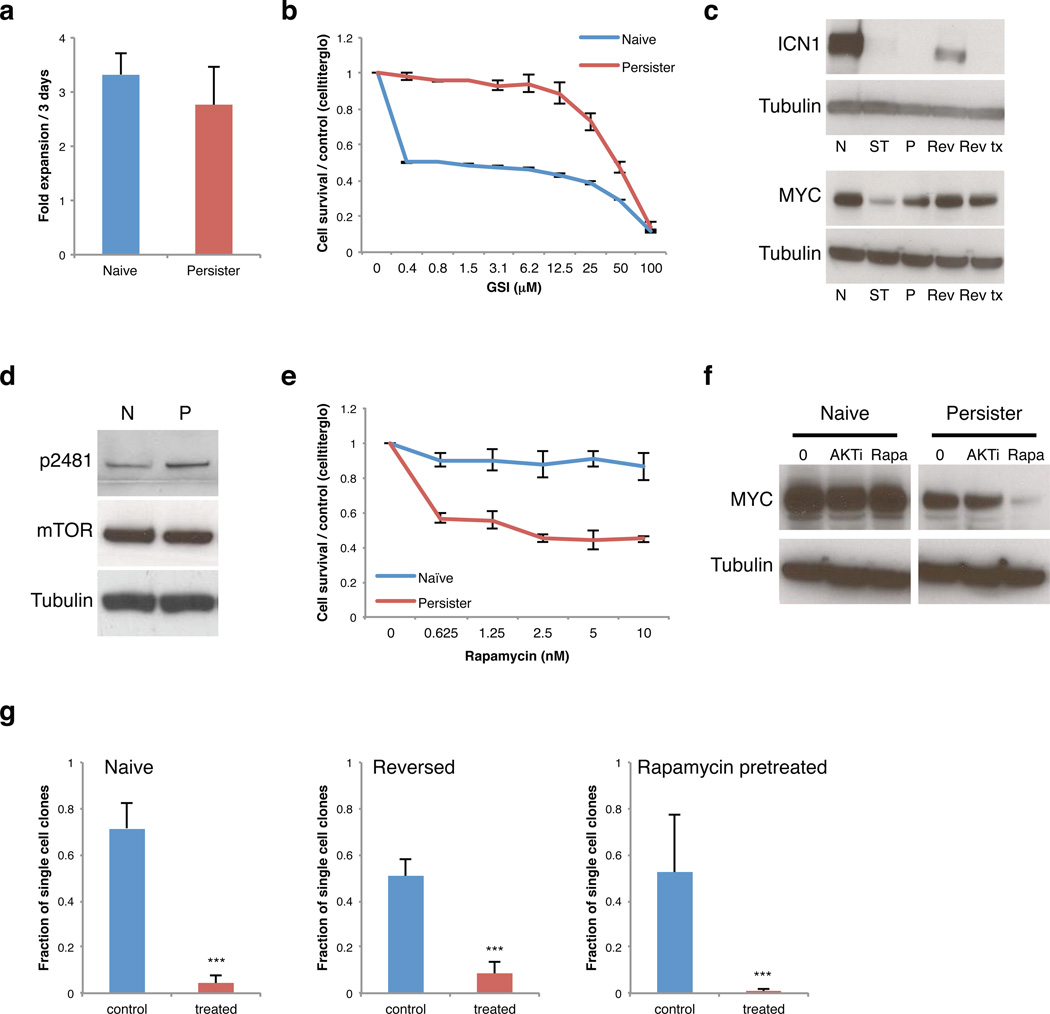
Figure 1. A sub-population of drug tolerant T-ALL cells mediates resistance to NOTCH inhibition.
a. Bar plot compares expansion rates of naïve T-ALL cells (grown in the absence of GSI) to persister cells grown continuously in 1 µM GSI (2 replicates, error bar reflect s.d.). b. Proliferation of naïve and persister cells treated with the indicated doses of NOTCH inhibitor for 6 days (2 replicates, error bars reflect s.d.). c. Western blot shows activated intracellular NOTCH1 (ICN1) and MYC levels in naïve cells (N), short-term treated cells (ST, 5 days of 1 µM GSI), persister cells in 1 µM GSI (P), ‘reversed’ persister cells removed from GSI for 2 weeks (Rev), and reversed cells re-exposed to GSI (Rev tx, 5 days). Reactivation of NOTCH signaling in reversed cells suggests that the persister phenotype is reversible. d. Western blot shows phospho-mTOR (p2481), a marker of activated mTOR signaling, total mTOR and Tubulin in naïve (N), and persister (P) cells. e. Proliferation of naïve and persister cells treated with indicated concentrations of Rapamycin for 6 days (4 replicates, error bar reflect s.d.). f. Western blots show MYC expression in naïve and persister cells after 3 days treatment with 2 µM AKT inhibitor, MK-2206 (AKTi), or 10 nM mTOR inhibitor, Rapamycin (Rapa). (Data shown for DND-41 cells; Data for KOPT-K1 in Figure S2). g. Rare drug tolerant cells pre-exist in naïve T-ALL populations. Single cells from a naïve DND-41 population, a reversed population (reversed for 1 month) or a naïve population pre-treated with 10 nM Rapamycin for 3 days were sorted into individual wells of 96 well plates and cultured in 1 µM GSI (treated) or without drug (control) for 4 – 6 weeks. Bar plot indicates the fraction of single cells that form colonies (sorted single cells per condition: naïve n = 936, reversed n = 936, Rapamycin pretreated n = 858, pooled from 2 independent experiments; *** = p value < 0.001, error bars reflect s.d.). These data suggest that GSI tolerance is already evident in a small fraction of naïve T-ALL cells and is reversible.
To characterize these drug tolerant cells, we first examined the active intracellular form of NOTCH1 (ICN1)6. ICN1 is present at high levels in naïve T-ALL cells, but is essentially undetectable in persisters (Figure 1c, top; S2b, top). NOTCH1 target genes, including DTX1 and HES4, are also down-regulated in persisters (Figure S1b,c; S3i). The phenotype is reversible, as persister cells rapidly re-express ICN1 and NOTCH1 target genes after removal of GSI (Figure 1c; S1b,d; S2b). Re-exposure of these ‘reversed’ cells to GSI leads to down-regulation of NOTCH1 target genes and growth arrest, as in the original naïve T-ALL population (Figure S1b). This reversibility suggests that the resistance phenotype is mediated epigenetically.
Induction of MYC is thought to be a major mechanism whereby constitutive NOTCH1 activation results in leukemic T-cell transformation7–9. Although MYC levels are dramatically reduced by short-term GSI treatment, they are maintained at moderate levels in the persisters despite continued GSI treatment (Figure 1c; S2b). Consistently, the strong MYC transcriptional signature of naïve T-ALL cells is reduced in persisters (Table S1). Conversely, gene signatures associated with MAPK, JNK, PI3K and MTOR signaling are enhanced (Table S1). Genetic alterations of PTEN and consequent AKT pathway activation have been associated with GSI resistance10,11. Although the T-ALL cells modeled here do not harbor PTEN mutations11, the persisters have increased levels of phosphorylated PTEN and are sensitive to AKT inhibitors (Figure S1e; S2d). mTOR activation has also been observed in T-ALL12,13. Persisters have increased levels of p2481-phosphorylated mTOR and are sensitive to the mTOR inhibitor Rapamycin (Figure 1d,e; S2c). Inhibition of mTOR markedly reduces MYC levels in persisters, but not in naïve T-ALL cells (Figure 1f). In contrast, persisters do not exhibit enhanced sensitivity to Doxorubicin or Vorinostat (Figure S1f, S2e). These data suggest that rewired signaling programs maintain MYC expression and cell proliferation in the absence of NOTCH activity.
Next, we sought to distinguish whether resistance is induced by GSI or, alternatively, whether a fraction of unexposed cells is already drug tolerant. In support of the latter, when we isolated single cells from a naïve T-ALL population, we found that ~4% expand in GSI (Figure 1g). In contrast, the remaining cells fail to grow even after extended periods. An analogous analysis of reversed cells, which were exposed to GSI but then expanded in the absence of drug for 1 month, again revealed a small fraction of GSI-tolerant single cells (~9%). We hypothesized that pre-existing GSI-tolerant cells share phenotypic characteristics with persisters, such as mTOR dependency. In support of this, pretreatment with Rapamycin reduced the frequency of GSI-tolerant single cells by ~5-fold (Figure 1g). These data suggest that GSI resistance depends on a pre-existing sub-population of T-ALL cells with distinct signaling programs that readily expand in the absence of NOTCH signaling. The reversibility of the phenotype suggests that this heterogeneity is mediated by epigenetic rather than genetic alterations.
In addition to altered signaling, persisters exhibit morphologic changes, including decreased cell and nuclear size (Figure 2a,b; S3a,b). We postulated that these changes might reflect global chromatin changes associated with NOTCH inactivity. We found that the persisters have higher global levels of repressive histone modifications and heterochromatin protein HP1 (Figure 2c,d; S3c,e). Chromatin in persisters is also relatively inaccessible to micrococcal nuclease (MNase) digestion (Figure S3g), consistent with a compact state. Chromatin compaction likely reflects the absence of NOTCH1 activity, as it is also evident in short-term GSI-treated cells and in T-ALL cells in which NOTCH signaling is inhibited by dominant-negative mastermind-like 1 (DN-MAML1) (Figure 2c; S1c; S3c,e,i).
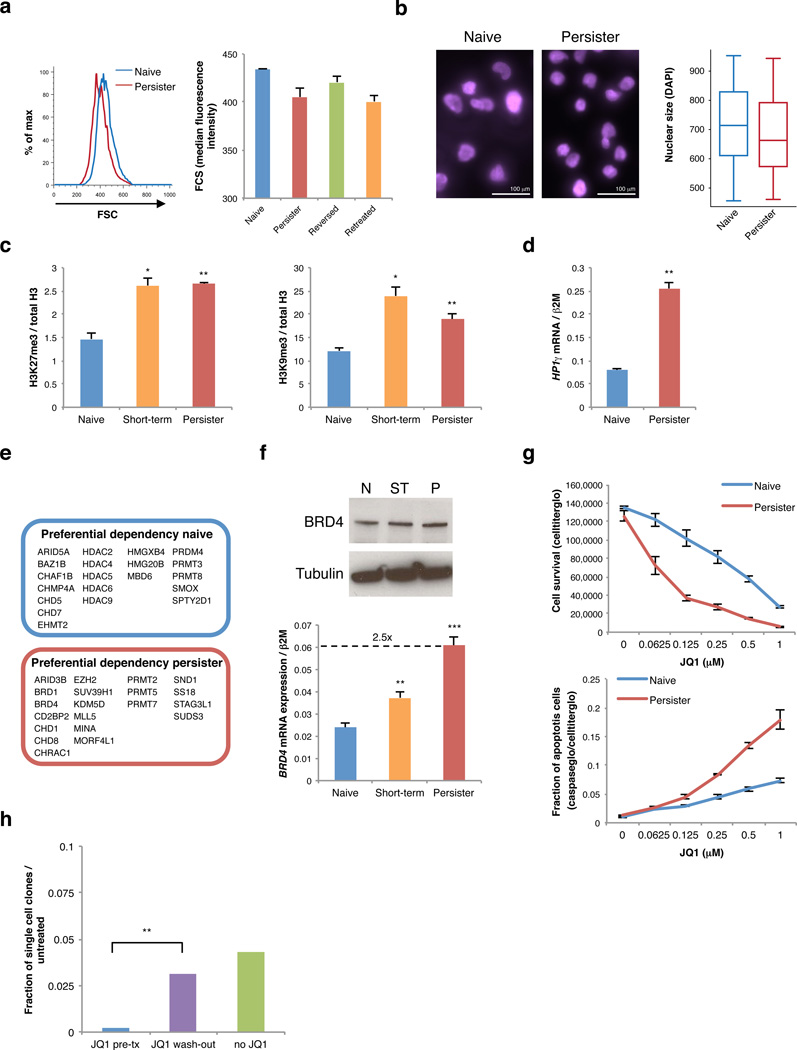
Figure 2. Drug tolerant T-ALL cells adopt an altered chromatin state and are BRD4 dependent.
a. Forward scatter analysis indicates size distributions of naïve (blue), persister (red), reversed (green) cells, and reversed cells re-exposed to GSI for 5 days (retreated, orange). b. Size of naïve (left) and persister (right) cell nuclei is shown by DAPI stain and quantified in box plot (far right; naïve n = 585, persister n = 496; p value < 10−4). c. Bar plots indicate relative levels of repressive histone modifications per ELISA of bulk histones from naïve, short-term treated (3 days) and persister cells (2 replicates, error bars reflect s.d., * = p value < 0.05, ** = p value < 0.01, *** = p value < 0.001). d. HP1γ mRNA expression shown for naïve and persister T-ALL cells (2 replicates, error bars reflect s.d.). e. shRNA screen identified chromatin regulators preferentially required for naïve or persister cell survival. Top hits for each cell state are indicated. f. BRD4 expression is shown for naïve (N), short-term treated (ST, 5 days) and persister (P) cells (top: protein expression; bottom: mRNA expression, 3 replicates, error bars reflect s.d., ** = p value < 0.01, *** = p value < 0.001). (Data shown for DND-41 cells; Data for KOPT-K1 in Figure S3). g. Proliferation (6 days treatment, 4 replicates) and rate of apoptosis (4 days treatment, 3 replicates) shown for naïve and persister cells treated with indicated doses of JQ1 (error bars reflect s.d.). h. Naïve DND-41 populations were pretreated with 0.5 µM JQ1 for 4 days (JQ1 pre-tx), pretreated with 0.5 µM JQ1 for 4 days but then cultured for 24 hours without JQ1 (JQ1 wash-out), or never exposed to JQ1 (no JQ1). Single cells from each of these populations were sorted into individual wells of 96 well plates and cultured with GSI (1 µM) or without GSI (control) for 6 weeks. Bar plot indicates the fraction of single cells that form colonies in GSI, relative to control (sorted single cells per condition: n = 936, data pooled from 2 independent experiments, error bars reflect s.d.). Pre-treatment with JQ1 eliminates pre-existing GSI tolerant cells from naïve T-ALL populations, but this effect is reversed by 24 hour JQ1 wash-out.
We postulated that the altered chromatin state in persisters might uncover new susceptibilities that could be targeted by emerging epigenetic therapies. To test this, we designed a lentiviral short-hairpin RNA (shRNA) knockdown screen that targeted ~350 chromatin regulators with an average of 5 independent hairpins per gene (see Methods). We identified ~15 genes for which knockdown compromised survival in naïve and persister cells, and were thus presumed to be generally required in T-ALL (Table S2). We also identified genes preferentially required for survival of either naïve or persister cells. For example, naïve cells are preferentially dependent on several histone deacetylases, while persisters are more dependent on certain arginine methyltransferases and histone methyltransferases (Figure 2e; Table S2).
We identified BRD4 as a top hit in the screen, with 3 shRNAs significantly reducing persister cell proliferation without affecting the naïve population (Table S2; Figure S4a,b). BRD4 is a member of the BET family of bromodomains that bind acetylated histones14,15. BRD4 has been implicated in several malignancies, including acute myeloid leukemia and lymphoma16,17. BRD4 expression increases with NOTCH1 inhibition and is higher in persisters (Figure 2f, S3f). This BRD4 dependency is of particular interest given the development of small molecule BET inhibitors, such as JQ118,19. Indeed, we found that persisters exhibit a ~5-fold enhanced sensitivity to JQ1, and undergo proliferation arrest and apoptosis in response to JQ1 concentrations well tolerated by naïve T-ALL cells (Figure 2g, top; S2e, S4c). We therefore considered whether the pre-existing GSI-tolerant cells, identified in naïve T-ALL populations by single cell analysis (Figure 1g), might also be BRD4 dependent. Indeed, when we pre-treated naïve T-ALL cells with JQ1 for 4 days and then removed the JQ1 immediately before isolating individual cells into 96 well plates, we were unable to detect any GSI-tolerant clones (Figure 2h). However, when we pre-treated naïve cells, but then removed the JQ1 24 hours prior to isolating individual cells, we were able to detect GSI-tolerant clones in a proportion similar to control naïve T-ALL. Thus, pre-existing GSI-tolerant cells in naïve T-ALL populations are highly BRD4 dependent, like persisters, and exist in dynamic equilibrium with the more prevalent GSI-sensitive cells.
To investigate its regulatory functions, we mapped BRD4 binding in naïve and persister T-ALL populations by chromatin immunoprecipitation and sequencing (ChIP-seq). We also mapped histone modifications associated with promoters (H3K4me3), transcripts (H3K36me3), enhancers (H3K4me1, H3K27ac) and Polycomb-repressed loci (H3K27me3)20,21. In both cell states, BRD4 binds promoters (~30% of sites) and putative enhancers enriched for H3K4me1 and H3K27ac (~60%) (Figure 3a,b; S5a). We collated the largest BRD4 binding sites in persister cells, as these may reflect super-enhancers with critical functions in cell type-specific gene regulation22. Proximal genes encode many known T-ALL regulators, including the transcription factor ETV6, the cell cycle regulator CDK6, the signaling molecule RPTOR, and the pro-survival protein BCL2 (Figure 3b,c,d; S5b,d; Table S3). Although most large BRD4 peaks are shared between the populations, some are specific to either naïve (e.g., those in NOTCH target loci) or persister cells (Figure S5c). Thus, BRD4 binds and may sustain the activity of regulatory elements and target genes required for T-ALL proliferation.
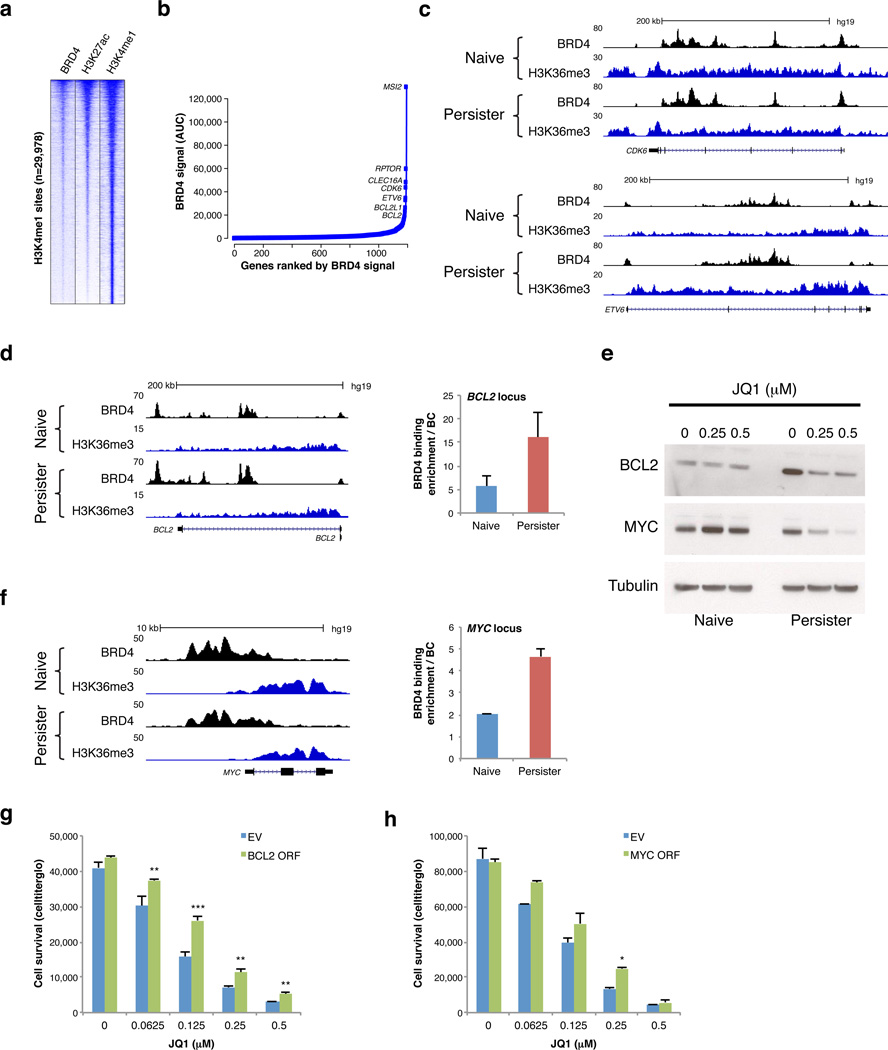
Figure 3. BRD4 binds enhancers near developmental, cell cycle and pro-survival genes in T-ALL.
a. Heatmap shows enrichment signals for BRD4, H3K27ac and H3K4me1 over 29,978 H3K4me1-marked distal sites in persister cells (rows; 10 kb regions, centered on H3K4me1 peaks, ranked by overall signal intensities of BRD4 and H3K27ac). b. BRD4 peaks were ranked by signal intensity of area under the curve (AUC) in persister cells averaging BRD4 signal intensities of DND-41 and KOPT-K1 persisters to identify candidate ‘super-enhancers’. Plot depicts top ranked sites linked to an active gene in T-ALL (see Methods). c. Tracks show BRD4 binding and H3K36me3 enrichment (marks transcribed regions) over the CDK6 and ETV6 loci, both of which contain BRD4-bound ‘super-enhancers’. d, f. Tracks show BRD4 binding and H3K36me3 enrichment across the BCL2 and MYC loci in naïve and persister cells (left). Plots (middle) show BRD4 enrichment as measured by ChIP-qPCR over putative regulatory elements in the BCL2 and MYC loci in naïve and persister cells (2 replicates, error bars reflect s.d.). e. Western blots show BCL2 and MYC expression in naïve and persister cells after 4 days treatment with indicated JQ1 doses. (Data shown for KOPT-K1 cells; Data for DND-41 in Figure S5). g. Plot shows relative proliferation of DND-41 persister cells transfected with BCL2 expression vector (BLC2 ORF) or empty vector control (EV) after 6 days treatment with indicated JQ1 doses (3 replicates, error bars reflect s.d., ** = p value < 0.01, *** = p value < 0.001). h. Plot shows relative proliferation of DND-41 persister cells transfected with MYC expression vector (MYC ORF) or empty vector control (EV) after 6 days treatment with indicated JQ1 doses (2 replicates, error bars reflect s.d., * = p value < 0.05).
We next considered the molecular basis for the preferential BRD4 dependency in the persisters. The relatively consistent BRD4 binding patterns in naïve and persister cells suggest that BRD4 re-localization is unlikely to explain the differential dependency. However, persister cells exhibit an altered chromatin environment with increased compaction, increased levels of repressive histone modifications and reduced levels of the enhancer-associated H3K27ac (Figure 2c; S3c,d,e,g). Consistently, ChIP-seq data reveal that persisters have modestly higher levels of repressive histone modifications in potential regulatory regions (Figure S3h). BRD4 is believed to play an important role as a ‘bookmark’ of active regulatory elements, maintaining their chromatin state as cells progress through mitosis15,23. Thus, we suggest that generalized chromatin repression in persisters renders enhancers particularly dependent on BRD4 for their epigenetic maintenance (Figure S6).
We next focused on individual genes that might account for the BRD4 dependency. BCL2 is an established BRD4-dependent gene in mixed lineage leukemia24. BCL2 is near a top ranked BRD4 site and expressed at higher levels in persisters (Figure 3b,d,e; S3i; S5d). JQ1 treatment markedly reduces BCL2 expression in persisters, but has little effect on naïve cells (Figure 3e). BCL2 down-regulation appears to contribute to the reduced survival of JQ1-treated persister cells, as exogenous BCL2 over-expression partially rescues these cells from JQ1 treatment (Figure 2f, 3g). The MYC oncogene is an established driver in T-ALL and a canonical BRD4 target18. A large enhancer in the MYC locus is strongly enriched for BRD4 binding (Figure 3f, S5f). JQ1 significantly reduces MYC expression in persisters at doses that do not alter MYC expression in naïve cells (Figure 3e). This effect can be partially rescued by MYC over-expression (Figure 3h). Hence, compromised survival of JQ1-treated persister cells depends on down-regulation of BCL2 and MYC.
To investigate the in vivo relevance of GSI resistance and associated epigenetic changes, we injected luciferase-expressing KOPT-K1 T-ALL cells orthotopically into NOD-scid IL-2Rgnull (NSG) mice and followed bioluminescence over time. GSI resistance developed rapidly in vivo after a short period of slowed leukemic growth (Figure S7a). ICN1 levels were drastically reduced in bone marrow of GSI-treated mice and NOTCH1 target genes were down-regulated in the leukemia cells, indicating that resistance is not due to NOTCH1 reactivation (Figure 4a,b). These 'in vivo persisters' also share other phenotypic characteristics with their in vitro counterparts, including reactivation of MYC expression, increased HP1γ, and increased BCL2 expression (Figure 4a,c).
Figure 4. Combination therapy targeting NOTCH and BRD4 in relapsed/induction failure primary T-ALL.
a. NSG mice were injected with luciferase-transfected KOPT-K1 T-ALL cells and leukemic mice were treated with NOTCH inhibitor DBZ for 5 days (ST, 3 doses), DBZ for 3 weeks (LT, every other day dosing), or vehicle (Veh); (5 mice per group). LT mice were harvested when bioluminescence had plateaued and then significantly increased (see methods; Figure S7a). Images show H&E stains and IHC for activated NOTCH1 (ICN1) and MYC of bone marrow from the respective leukemic mice. b. NOTCH1 target gene expression shown for leukemia cells sorted from spleen of vehicle (Veh) or long-term (LT) treated mice. Datapoints reflect averages of 2 – 3 mice (2 replicates, error bars reflect s.d., * = p value < 0.05, ** = p value < 0.01, *** = p value < 0.001). c, left. HP1γ expression shown for leukemia cells as in panel b. c, right. Intracellular BCL2 expression measured by flow cytometry shown for leukemia cells as in panel b. d. Proliferation of three primary T-ALLs grown in vitro for 6 days in the presence of 1 µM GSI, 0.25 µM JQ1 or both compounds (2 – 3 replicates, error bars reflect s.d.). e. Experimental design for primary T-ALL xenotransplantation trials. Primary T-ALL samples from three primary pediatric T-ALLs (T-ALL-x-9, T-ALL-x-11 and T-ALL-x-14; see Table S4) were transplanted into NSG mice. Once leukemic burden reached 25% in peripheral blood by human CD45 staining, mice were randomized into 4 treatment groups (vehicle, JQ1, DBZ or DBZ+JQ1 combination). Mice were treated for 3 consecutive weeks, and were then monitored for disease and sacrificed when they became moribund. f. Kaplan-Meier survival curves for sample T-ALL-x-9, T-ALL-x-11 and T-ALL-x-14 (p value < 0.01 as assessed by the log-rank test, for details see Table S4; 4–5 mice per group). Combination treatment significantly prolonged survival over single agents and vehicle in all three trials.
We hypothesized that combined inhibition of NOTCH1 and BRD4 might be effective treatment for T-ALL, including relapsed or induction failure disease, where conventional therapy has failed. We therefore examined the efficacy of GSI-JQ1 combination therapy in primary T-ALLs in vitro and in vivo. We examined three tumors from pediatric patients at diagnosis or upon relapse (Figure S7b, Table S4), including a relapsed early thymic progenitor (ETP)-T-ALL patient, who failed cytotoxic therapy. First, we examined the sensitivity of these leukemias to GSI, JQ1 or combined treatment in vitro. We found the combination to be significantly more effective at inhibiting growth/survival (Figure 4d). To examine the effects in vivo, we engrafted NSG mice with the primary T-ALLs and initiated treatment when a leukemic burden of ~25% in peripheral blood was reached. Randomized groups were treated with vehicle, NOTCH inhibitor (DBZ), BRD4 inhibitor (JQ1) or the DBZ+JQ1 combination for 3 weeks and effects on overall survival determined (Figure 4e,f). Combination therapy significantly prolonged survival over vehicle or single agent therapy for all three T-ALLs (Figure 4f; Table S4). Notably, primary leukemia cells treated with single agent GSI in vivo have increased H3K27me3, analogous to the in vitro treated lines (Figure S7c). These data support the in vivo relevance of our study and the potential of this combination of targeted therapy to augment current and emerging therapies for T-ALL.
Therapeutic resistance plagued early GSI trials in humans5 and is a major challenge in cancer treatment, pertinent to conventional chemotherapy and targeted therapy25. We have shown that T-ALLs can acquire GSI resistance by a fully reversible epigenetic mechanism. Persister cells rely on alternative signaling pathways to proliferate in the absence of NOTCH1 signaling, and appear to arise from rare drug-tolerant cells already present in naïve T-ALL populations. Persisters also exhibit an altered chromatin state and enhanced sensitivity to BRD4 inhibition. Although the chromatin changes are reminiscent of an established model of drug tolerant lung cancer cells26, the underlying mechanisms appear distinct, as we do not observe KDM5a up-regulation or sensitivity to histone deacetylase inhibitors (Figure S1f; S2e). The in vivo relevance of our findings is supported by the efficacy of combinatorial therapy with NOTCH and BRD4 inhibitors in patient-derived xenograft models of pediatric T-ALL. The effect of combination therapy in vivo is particularly striking given the short duration of treatment, the fact that treatment was initiated upon significant leukemic burden and the refractory nature of the relapsed samples examined. Our study provides a framework for understanding epigenetic alterations and heterogeneity in tumor pathogenesis, and suggests that drug resistance may be addressed by combination therapies that incorporate epigenetic modulators.
Online Methods
Cell culture
The human T-cell leukemia cell lines DND-41 and KOPT-K1 were a kind gift of Dr. A. T. Look. Cells were grown in RPMI 1640 containing 10% FCS at 37°C with 5% CO2 and were maintained between a density of 5 × 105 cells/ml and 2 × 106 cells/ml. Persister cells were established by treating DND-41 or KOPT-K1 cells with 1 µM GSI for at least 7 weeks, replenishing the inhibitor every 3 – 4 days (Compound E, EMD4 Biosciences). ‘Reversed’ cells were generated from persister cells by culturing without GSI for a minimum of 2 weeks. Cell numbers were determined using Nexcelom Bioscience Cellometer Auto T4. Single cells were sorted into 96 well round bottom plates and continuously treated with 1 µM Compound E or control. The number of clones that outgrew after 4 – 6 weeks of culture was determined using a Zeiss Observer.Z1 with AxioCam MRm with a 2.5× objective imaging system. At least 500 wells were analyzed per condition in 2 independent experiments. Primary T-ALL cells obtained from pediatric patients with T-ALL with IRB approval were passaged through NOD-scid IL2Rgnull (NSG) mice and maintained in MEMα containing 10% FCS, 10% heat inactivated human AB+ serum, 10 ng/mL human IL-7, 50 ng/ml human SCF, 20 ng/ml human FLT3-ligand, 20 nM insulin, 100 ng/mL IL-2 at 37°C with 5% CO2.
Cell proliferation and apoptosis assays
Viable T-ALL cell lines were plated in 2 – 4 replicates in black or white opaque flat bottom 96 well tissue culture plates. For proliferation assays, cells were titrated to allow log phase growth for a period of 4 to 9 days prior to readout (5,000 to 15,000 cells per well). Viable primary T-ALL cells were plated in 2 – 3 replicates in black opaque flat bottom 96 well tissue culture plates at 50,000 – 100,000 cells per well for 6 days. Cell proliferation was measured using the CellTiter-Glo Luminescent Cell viability assay from Promega as described by the manufacturer. Apoptosis was measured as a function of Caspase 3/7 cleavage using the Caspase-Glo 3/7 Luminescent Assay from Promega as described by the manufacturer. End-point luminescence was measured on SpectraMax M5 plate reader (Molecular Diagnostics). Statistical analyses were performed using student t-test assuming equal variance. A 2-sided p value < 0.05 was considered statistically significant. Error bars reflect standard deviation.
In vitro inhibitor assays, western blots and ELISAs
GSI (Compound E) was obtained from EMD4 Biosciences. Rapamycin and the AKT inhibitor MK-2206 were purchased from Selleck Chemicals. Stock aliquots (1 – 100 mM) were prepared in DMSO (Sigma Aldrich) and diluted in appropriate medium before use. Matching DMSO concentrations were used for control. For western blots, cells were harvested after treatment and processed as described previously27. Total protein in each sample was determined using the SDS compatible Bio-Rad DC Protein Assay. Membranes were developed using Amersham ECL Plus Western Blotting Detection Reagents (GE Healthcare, Life Sciences) and visualized using photographic film. Western blots used antibodies against α-Tubulin (ab7291), BCL2 (ab18210), BRD4 (ab75898), total H3 (ab1791), all from Abcam; cleaved NOTCH1 (ICN1; 4147), total mTOR (5536), phospho-mTOR 2481 (9964), total PTEN (9552), phospho-PTEN (9551), HP1γ (2616), all from Cell Signaling; MYC from Cell Signaling (9402) and Santa Cruz (sc-764), H3K27ac from ActiveMotif (39133), β-Actin from Sigma (A5441). Histone ELISAs for H3K27me3, H3K9me3 and total H3 were obtained from ActiveMotif. Signal for H3K27me3 and H3K9me3 for each sample was determined as a fraction of total histone H3, following the protocol described by the manufacturer. Statistical analyses were performed using student t-test assuming equal variance. A 2-sided p value < 0.05 was considered statistically significant. Error bars reflect standard deviation.
Quantitative RT-PCR analyses
Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and reverse transcribed into cDNA using Superscript III First-Strand Synthesis system for RT-PCR. Quantitative PCR was performed with FastStart Universal SYBR Green Master (Roche) on an ABI 7500 (primer sequences are listed in Table S5). Gene expression was calculated by determining the log2 (Ct) of desired transcript compared to either GAPDH or β2-microglobulin (β2M). Statistical analyses were performed using student t-test assuming equal variance. A 2-sided p value < 0.05 was considered statistically significant. Error bars reflect standard deviation.
Flow cytometry and cell sorting
Flow cytometric analysis and sorting was run on a BD FACSCalibur, BD LSRII and BD FACSAriaII, respectively, using FITC, PE, APC conjugated antibodies against CD45, BCL2 and appropriate isotype controls (all from BD Pharmingen). Intracellular staining was performed according to the manufacturer. For flow cytometric analysis of spleens, single cell solutions were generated and red cell lysis was performed prior to staining. Cell sorting for CD45 positive and mcherry positive cells was done on an AriaII. Single cells were sorted into 96 well round bottom plates using the AriaII.
DAPI staining
For DAPI staining 5 × 107 cells were fixed in 4% paraformaldehyde (PFA) in PBS and permeabilized with 1% Triton X-100. Cells were mounted on slides using vectashield mounting medium with 1.5 µg/mL DAPI (Vector Laboratories). Slides were imaged on an Olympus IX51 microscope with Q Capture ×65 camera (400×) and Q Imaging software. Nuclear size was quantified using ImageJ.
MNase digestion
Mononucleosome isolation was performed as described with modifications28. Naïve and persister cells were incubated with 0.5% Triton X-100/buffer 1 (10 mM 2-(N-morpholino)ethane-sulfonic acid, pH 6.5, 10 mM sodium butyrate, 60 mM potassium chloride (KCl), 15mM sodium chloride (NaCl), 5 mM magnesium chloride (MgCl2), 0.25 M sucrose) on ice. Nuclei were then layered onto 2.5× volume of 30% sucrose/buffer 1 and centrifuged at 4,000 rpm. Purified nuclei were resuspended into buffer 2 (20 mM (4-(2-hydro- xyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.8, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 2 mM calcium chloride, 1 mM dithiothreitol) with 200 U of micrococcal nuclease (Roche Diagnostics) at room temperature for 5min, 10min and 15min, and then quenched by addition of ethylene glycol tetraacetic acid (EGTA) to a final concentration of 1 mM. Digested DNA was phenol/chloroform extracted and run on an agarose gel.
Chromatin immunoprecipitation (ChIP) assays
We performed ChIP-qPCR and ChIP-seq analysis in naïve and persister DND-41 and KOPT-K1 cells, as described27. Chromatin from formaldehyde-fixed cells (1 – 5 × 106 per histone mark, 107 for BRD4) was fragmented to a size range of 200 – 700 bases with a Branson 250 Sonifier. Solubilized chromatin was immunoprecipitated with antibody against H3K4me3 (Millipore 07-473CA), H3K36me3 (Abcam ab9050), H3K27me3 (Millipore 07-449), H3K27ac (Active Motif ab4729), H3K4me1 (Abcam ab8895), BRD4 (Bethyl A301-985A). Each of these antibodies was validated by western blot and dot blot as described29. Antibody-chromatin complexes were pulled down with Protein A-Sepharose, washed and then eluted. After crosslink reversal and Proteinase K treatment, immunoprecipitated DNA was extracted with phenol/chloroform, precipitated with ethanol, and treated with RNase. ChIP DNA was quantified with PicoGreen.
ChIP enrichments were assessed by quantitative PCR analysis on an ABI 7500 with 0.2 ng ChIP DNA and an equal mass of unenriched input DNA (whole cell extract; WCE). Enrichment was calculated by determining the log2 (Ct) of ChIP DNA relative to WCE. Primer sequences are listed in Table S5.
For ChIP-seq, libraries were prepared according to Illumina's instructions. ChIP DNA and input controls were sequenced with the Hi-Seq Illumina Genome Analyzer. ChIP-seq reads were aligned to the reference genome (hg19) using SOAP230, allowing at most two mismatches. Positions were randomly selected for reads with multiple hits. Reads aligned to the same position and strand were only counted once. We extended aligned reads by 250 bp to approximate fragment sizes and then derived a 25-bp resolution density map by counting the number of fragments overlapping each position. We calculated the H3K27me3 density across ± 5 kb regions of distal elements marked by H3K4me1 only to analyze local dynamics of H3K27me3, normalized to the background level, as described31.
BRD4, H3K27ac and H3K4me1 enriched intervals were identified by scanning the genome for enriched 1kb windows and then merging overlapping enriched windows to define enriched intervals, as described31. Candidate distal regulatory elements were assigned from enriched intervals that do not overlap ±2 kb region of an annotated TSS. The heatmaps for BRD4, H3K27ac and H3K4me1 signals were plotted over 10kb regions surrounding all distal H3K4me1 intervals. The heatmaps are ranked by the average signal intensities of BRD4 and H3K27ac. A list of genes near large BRD4 peaks was collated by (i) calculating the area under the curve of BRD4 peaks; (ii) assigning intergenic peaks to the nearest expressed promoter within 100 kb; and (iii) summing for each expressed gene the total area under the curve of its intragenic BRD4 peaks and the BRD4 peaks assigned to its promoter.
Gene expression analyses
RNA from biological replicates of naïve and persister DND-41 and KOPT-K1 cells was isolated using TRIzol. Gene expression profiles were generated with Affymetrix Human Genome U133 Plus 2.0 Array. Data were analysed using Genepattern and Geneset Enrichment analysis (with gene sets containing at least 50 genes per set)32–34.
Lentiviral / retroviral knock-down and over-expression
shRNA target sequences against BRD4 and control hairpin sequences are listed in Table S5. Lentiviral particles were generated with the use of standard procedures35. Infection efficiency was greater than 80% after 3 days of puromycin selection. BCL2 ORF was cloned into lentiviral expression system with puromycin resistance sites using the gateway cloning system. Cells were spin infected with virus and selected with puromycin. Cell viability was monitored with CellTiter-Glo (Promega) and cells were harvested after 6 – 8 days selection in puromycin. Overexpression was confirmed by measuring BCL2 protein expression by western blot (Figure S5g).
The retroviral constructs MigR1 (EV) and MigR1-DN-MAML1 (DN-MAML) and MigR1-myc (MYC) have been described1,7. Pseudotyped retrovirus was produced by transfection of 293T cells as described36. DND-41 and KOPT-K1 cells were infected with retrovirus by spinoculation. GFP+ cells were isolated by cell sorting. Overexpression of MYC was confirmed by RT-PCR (Figure S5f). Persister cells were cultured in the presence of 1 µM GSI throughout the entire experiment.
shRNA screen
shRNA screens were performed in naïve and persister DND-41 cells in an arrayed format using Cell Titer-Glo (Promega) to assess proliferation/viability phenotypes as described37,38. Screening included three replicates per cell line (1 replicate without puromycin treatment, 2 replicates treated with 1 µg/mL puromycin). Hairpins with an infection efficiency of greater than 50% were used for analysis. Screening data was normalized using the statistical z-score as described39. Riger analysis was used to determine hairpins that selectively affected survival in either naïve only or persister cells only40.
Cell line xenograft experiments
Xenograft experiments were performed as previously described41. In brief, a total of 2 × 106 mcherry-KOPT-K1-LucNeo cells that had been engineered to stably express firefly luciferase and mcherry were injected intravenously via the lateral tail vein into male NSG mice. Mice were imaged after injection of 75 mg/kg of D-luciferin (Promega, Madison, WI) using a Xenogen IVIS Spectrum (Caliper Life Sciences). Bioluminescence was quantified using the Living Images software package (Caliper Life Sciences). Mice with established disease were divided into treatment groups (n = 5 per group, randomized; investigator was not blinded). Mice were treated every other day with either GSI ((S)-2-[2-(3,5-Difluoro-phenyl)-acetylamino]-N-((S)-5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl)-propionamide; DBZ) at 10 µMol/kg intraperitoneal or vehicle (PBS) control. Vehicle and short-term treated mice were sacrificed after 5 days (3 doses DBZ), long-term treated mice after 3 weeks (11 doses DBZ) once slowed tumor growth was no longer observed by bioluminescence. Studies were performed under the auspices of protocols approved by the DFCI IACUC.
Primary human T-ALL xenograft studies
Primary human T-ALL cells were obtained from children with T-ALL enrolled on clinical trials of the Dana-Farber Cancer Institute. Samples were collected with informed consent and with approval of the Institutional Review Board. Patient consent forms were required from all patients or their legal guardians (if minors) for all samples collected for the study. Leukemic blasts were isolated from peripheral blood or bone marrow samples by Ficoll-Hypaque centrifugation and cryopreserved in fetal bovine serum (FBS) containing 10% DMSO and stored in liquid nitrogen. NSG mice from in-house colonies were maintained on a regimen of acidified antibiotic water. Fresh or frozen leukemic blasts were expanded in NSG mice by transplanting 0.5 – 5 × 106 cells via intravenous injection. Human engraftment in the spleen and bone marrow was greater than 80% by staining with human CD45 (BD Bioscience).
For in vivo xenograft studies, NSG mice were injected with 1 × 106 leukemic blasts via intravenous injection and bled weekly to determine the percentage of circulating human CD45+ cells in the peripheral blood. Once the leukemic burden reached 25% in the periphery mice were randomized (investigator was not blinded) to receive vehicle, JQ1 (50 mg/kg daily, diluted in 10:90 DMSO:10%hydroxypropyl-beta-cyclodextrin), DBZ (10 µM/kg on a 3 days on 4 days off regimen diluted in 0.5% methocel E4M/0.1% Tween-80), or both DBZ and JQ1 for 3 weeks by intraperitoneal injection. Mice were monitored and weighed daily and sacrificed when determined to be moribund. Kaplan-Meier survival curves and statistical analyses were performed using GraphPad Prism software, Version 5.0. A 2-sided p < 0.05 was considered statistically significant. All animal procedures used in this study were approved by The University of Massachusetts Medical School Institutional Animal Care and Use Committee.
For assessment of global histone levels NSG mice engrafted with T-ALL-x-9 at 25% disease burden were treated with vehicle or DBZ for three weeks. Mice were harvested 2 hours after GSI dosing and leukemia cells were isolated from spleen (purity 86% in vehicle treated and 85% in GSI treated mice by staining for human CD45, respectively).
Immunohistochemical detection of ICN1 and MYC
Standard 4 µm paraffin embedded tissue sections were stained using the Ventana Benchmark XT platform (Ventana Medical Systems) with extended heat-induced epitope retrieval (CC1 Buffer). Slides were incubated for 1 hour at room temperature with anti-ICN1 rabbit monoclonal antibody (clone D3B8, Cell Signaling Technology; final concentration, 8.5 µg/mL) that is specific for the neoepitope that is created by gamma-secretase cleavage of NOTCH1 or anti-MYC rabbit monoclonal antibody (Epitomics)42. Signals were then amplified (Ventana Amplification Kit) and visualized (Ventana Ultraview Universal DAB detection kit) per the manufacturer’s instructions. All staining runs included two positive control samples (xenografted REC-1 tumor cells, a mantle cell lymphoma with an activating rearrangement of NOTCH1, and tonsillar mucosa)43.
Supplementary Material
Acknowledgments
We are grateful to T. Look, M. Harris, L. Silverman and S. Sallan for providing the pediatric T-ALL samples. We thank E. Rheinbay, M. Suva, R. Ryan, N. Riggi, A. Goren, O. Ram, J. Wu, L. Pan and M. Rivera for helpful discussions; V. Mootha for critical comments on the manuscript; S. Muller-Knapp and the Structural Genomics Consortium, Oxford, UK for their assistance with inhibitor experiments; L. Hamm and the Broad RNAi platform for help with the shRNA screen; R. Issner, X. Zhang, C. Epstein, N. Shoresh, T. Durham and the Broad Genome Sequencing Platform for technical assistance; A. Christie for help with animal experiments; L. Gaffney for help with illustrations, and O. Weigert for providing the BCL2-ORF. B.K. was supported by a NIH T32 Training Grant (HL007574-30) and a St. Baldrick’s fellowship. J.E.R. was supported by a NIH T32 Training Grant (CA130807). H.W. is supported by a NIH T32 training grant (HL007627). This work was supported by the National Human Genome Research Institute (ENCODE U54 HG004570 to B.E.B.), the NIH Common Fund for Epigenomics (U01 ES017155 to B.E.B.), the NIH/NCI (CA096899 to M.A.K; 5P01 CA109901-10 to J.E.B.), the Howard Hughes Medical Institute (B.E.B.), and the Starr Cancer Consortium (B.E.B.). The LLS SCOR supports B.E.B., J.C.A., J.E.B., K.S., and A.L.K.
Footnotes
Author Contributions
B.K. and J.R. designed and performed experiments and analyzed the data. M.A.K. and B.E.B. designed the experimental strategy and supervised the study and analysis. J.Z. and D.F. carried out computational analyses. B.K., J.R., M.A.K and B.E.B drafted the manuscript. K.E.W., S.M.G, M.J.C., J.G.L., M.K., H.W., F.P., S.J.S., M.J., D.P., M.J.K., C.J.O. contributed to experiments and data analysis. L.D.S., M.A.B., D.L.G., A.G., K.S., A.L.K., D.E.R., J.E.B., J.C.A. provided reagents, contributed to analysis and gave conceptual advice. All authors discussed the results and implications and reviewed the manuscript.
Accessions
Gene expression and ChIP-Seq data were deposited in GEO (accession number GSE54380).
Competing Financial Interests
D.L.G. and M.A.B. are consultants for the Jackson Laboratory. Drug-like derivatives of JQ1 have been licensed by Dana-Farber Cancer Institute to Tensha Therapeutics, for clinical development. J.E.B. and Dana-Farber Cancer Institute have received equity in Tensha associated with this license. The remaining authors declare no competing financial interests.
References
- 1.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 2.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. The New England journal of medicine. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 4.Rao SS, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer research. 2009;69:3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- 5.Palomero T, Ferrando A. Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia. Clinical lymphoma & myeloma. 2009;9(Suppl 3):S205–S210. doi: 10.3816/CLM.2009.s.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nature reviews. Genetics. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & development. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Molecular and cellular biology. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez A, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature medicine. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalaitzidis D, et al. mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell stem cell. 2012;11:429–439. doi: 10.1016/j.stem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature cell biology. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blobel GA, Kalota A, Sanchez PV, Carroll M. Short hairpin RNA screen reveals bromodomain proteins as novel targets in acute myeloid leukemia. Cancer cell. 2011;20:287–288. doi: 10.1016/j.ccr.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews. Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 21.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voigt P, Reinberg D. BRD4 jump-starts transcription after mitotic silencing. Genome biology. 2011;12:133. doi: 10.1186/gb-2011-12-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 26.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Molecular and cellular biology. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reich M, et al. GenePattern 2.0. Nature genetics. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 33.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nature methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 36.Pear WS, Scott ML, Nolan GP. Generation of high titer, helper free retroviruses by transient transfection. In: Robbins P, editor. Methods in Molecular Biology: Methods in Gene Therapy. Tonawa, NJ: Humana Press; 1997. pp. 41–58. [DOI] [PubMed] [Google Scholar]
- 37.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 38.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 39.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nature biotechnology. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 40.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi YJ, et al. The requirement for cyclin D function in tumor maintenance. Cancer cell. 2012;22:438–451. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kluk MJ, et al. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PloS one. 2012;7:e33813. doi: 10.1371/journal.pone.0033813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kluk MJ, et al. Gauging NOTCH1 Activation in Cancer Using Immunohistochemistry. PloS one. 2013;8:e67306. doi: 10.1371/journal.pone.0067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.