Abstract
Glucocorticoids (GCs) strongly regulate myostatin transcript levels in mammals via glucocorticoid response elements (GREs) in the myostatin promoter, and bioinformatics methods suggest that this regulatory mechanism is conserved among many vertebrates. However, the multiple myostatin genes found in some fishes may be an exception. In rainbow trout (Oncorhynchus mykiss), two genome duplication events have produced three putatively functional myostatin genes, myostatin-1a, -1b and -2a, which are ubiquitously and differentially expressed. In addition, in silico promoter analyses of the rainbow trout myostatin promoters have failed to identify putative GREs, suggesting a divergence in myostatin function. Therefore, we hypothesized that myostatin mRNA expression is not regulated by glucocorticoids in rainbow trout. In this study, both juvenile rainbow trout and primary trout myoblasts were treated with cortisol to examine the relationship between this glucocorticoid and myostatin mRNA expression. Results suggest that exogenous cortisol does not regulate myostatin-1a and -1b expression in vivo, as myostatin mRNA levels were not significantly affected by cortisol treatment in either red or white muscle tissue. In red muscle, myostatin-2a levels were significantly elevated in the cortisol treatment group relative to the control, but not the vehicle control, at both 12 h and 24 h post-injection. As such, it is unclear if cortisol was acting alone or in combination with the vehicle. Cortisol increased myostatin-1b expression in a dose-dependent manner in vitro. Further work is needed to determine if this response is the direct result of cortisol acting on the myostatin-1b promoter or through an alternative mechanism. These results suggest that regulation of myostatin by cortisol may not be as highly conserved as previously thought and support previous work that describes potential functional divergence of the multiple myostatin genes in fishes.
Keywords: rainbow trout, myostatin, cortisol, stress, myoblasts
Introduction
Myostatin, a member of the transforming growth factor-ß superfamily, is a well-characterized inhibitor of muscle growth in mammals (McPherron et al., 1997; Rodgers and Garikipati, 2008). In fishes, myostatin has been cloned and expression patterns have been characterized in a number of fishes: zebrafish (McPherron and Lee, 1997; Xu et al., 2003), brook trout (Roberts and Goetz, 2001, 2003), gilthead seabream (Maccatrozzo et al., 2001), Atlantic salmon (Ostbye et al., 2001), rainbow trout (Garikipati et al., 2007; Garikipati et al., 2006; Rescan et al., 2001), channel catfish (Kocabas et al., 2002), sea perch (Ye et al., 2007), and orange-spotted grouper (Ko et al., 2007). In addition, the myostatin sequences are well-conserved among vertebrates, with the bioactive domain ranging from 88-100% identity, and functional studies in fish demonstrate that the myogenic functions are also conserved (Lee et al., 2009; Lee et al., 2010; Medeiros et al., 2009; Rodgers and Garikipati, 2008; Sawatari et al., 2010; Xu et al., 2003).
Genome duplication events have produced multiple myostatin genes in salmonids. Three of these genes, mystatin-1a, -1b and -2a, are putatively functional, while a premature stop codon in the open reading frame of myostatin-2b prevents the production of mature transcripts in rainbow trout and Atlantic salmon (Garikipati et al., 2007; Ostbye et al., 2007b). Interestingly, these genes appear to be ubiquitously expressed and differentially regulated during development and in response to various physiological changes (Biga et al., 2004; De Santis and Jerry, 2011; Gabillard et al., 2013; Helterline et al., 2007). Such expression patterns are suggestive of a functional divergence among fishes, as the single mammalian myostatin ortholog is predominately expressed in muscle. To identify potential mechanisms responsible for changes in spatial and temporal expression patterns, numerous studies have investigated the promoter region of myostatin in mammals and fish to characterize putative transcription factor binding sites and hormone response elements (Allen and Du, 2008; Funkenstein et al., 2009b; Gabillard et al., 2013; Garikipati et al., 2007; Garikipati et al., 2006; Hu et al., 2013; Li et al., 2012a; Li et al., 2012b; Li et al., 2012c; Ma et al., 2001a; Nadjar-Boger et al., 2012; Nadjar-Boger et al., 2013; Ostbye et al., 2007a; Rodgers and Garikipati, 2008; Spiller et al., 2002; Xue et al., 2012). Putative E-box protein, myogenic regulatory factor (MRF), and myocyte enhancing factor (MEF) binding motifs and glucocorticoid response elements (GREs) appear to be highly conserved components of the myostatin promoter in vertebrates (Rodgers and Garikipati, 2008). In this study, we were specifically interested in the presence or absence of GREs as a potential mediator in the divergence of myostatin expression among vertebrates.
In mammals, myostatin expression is highly regulated by glucocorticoids (GCs), and promoter analyses suggest that the regulation by GCs may be highly conserved among vertebrates (Funkenstein et al., 2009b; Gabillard et al., 2013; Garikipati et al., 2007; Garikipati et al., 2006; Ma et al., 2001a; Ma et al., 2003; Ostbye et al., 2007a; Rodgers and Garikipati, 2008). The actions of GCs appear to be mediated by the binding of the glucocorticoid receptor (GR) to GREs in the regulatory region of the myostatin gene to upregulate gene expression (Gilson et al., 2007; Ma et al., 2001a; Ma et al., 2003; Qin et al., 2013). However, most of the research describing GC regulation of myostatin has been done in mammals, and currently little empirical data are available regarding this relationship in other vertebrates, including fishes. The current evidence suggests that the putative GREs are not present in all of the myostatin promoters within fish species. Different approaches for identifying consensus sequences have yielded somewhat contradicting results and it is not clear if glucocorticoids directly regulate myostatin expression in teleosts (De Santis and Jerry, 2011; Funkenstein et al., 2009a; Gabillard et al., 2013; Garikipati et al., 2007; 2006; Ostbye et al., 2007b; Roberts and Goetz, 2003; Rodgers and Garikipati, 2008). Interestingly, in silico promoter analyses have failed to identify putative GREs in the promoters of any myostatin paralogs in rainbow trout, and no study has tested a relationship between glucocorticoids and myostatin expression in this species (Garikipati et al., 2007; Garikipati et al., 2006).
The current study was conducted to determine the effects of cortisol on myostatin expression in rainbow trout, using both in vivo and in vitro approaches. Based on myostatin promoter analyses, we hypothesize that myostatin mRNA expression is not affected by cortisol treatments. The presumed loss of GREs in the promoters of the rainbow trout myostatin genes make this species an excellent model system for studying the divergence in glucocorticoid regulation of myostatin. Our experiments failed to identify a clear cortisol response in vivo and only detected a significant increase in myostatin-1b expression in vitro, suggesting a change in GC regulation of myostatin relative to mammals. Although our experimental design did not directly test for the presence of GREs, our results do illustrate a potential divergence in the regulation of myostatin among the vertebrates, specifically by cortisol.
Materials and Methods
Animal Care
Juvenile rainbow trout used in the experiments detailed below were obtained from the United States Fish and Wildlife Service’s Garrison National Fish Hatchery, Riverdale, North Dakota, and housed at North Dakota State University. All fish were maintained in 800-liter flow-through tanks with a 12L:12D photoperiod and were fed AquaMax Grower (PMI Nutrition International, Inc., Brentwood, MO, USA) to apparent satiation twice daily, except 24 h before experimentation. All experiments conducted with animals were approved in advance of experimentation by the Institutional Animal Care and Use Committee at North Dakota State University, Fargo.
Experiment 1: Effects of cortisol on myostatin expression in vivo
Juvenile rainbow trout (70-100 g) were randomly assigned to three experimental tanks (60 L, 12 °C, 4 h flow-through, 12 fish per tank, ~20 kg/m3 maximum stocking density) and allowed to acclimate for one week. The three treatment groups consisted of injections of cortisol (CORT), vehicle control (VC), and no injection control (Control). Prior to injections, fish were anesthetized with buffered tricaine methylsulfonate (MS-222; 100 mg/L). The fish received either an intraperitoneal injection of cortisol (Janzen et al., 2012) dissolved in safflower oil or an injection of safflower oil at a volume consistent with the CORT group (2 μL/g BW). Post-injection, fish were place in a recovery tank for five and then returned to their appropriate experimentation tank. At 12 h and 24 h post-injection, six fish per treatment group were euthanized by overdose of MS-222 (>300 mg/mL; AVMA Guidelines for the Euthanasia of Animals, 2013) and blood plasma samples were immediately collected for glucose measurements. Additionally, tissues (skeletal muscle: red and white; and liver) were flash-frozen and stored at −80°C.
Experiment 2: Effects of cortisol on myoblast myostatin expression in vitro
Following a protocol developed by Rescan and colleagues (1995), primary myoblasts were isolated from juvenile rainbow trout (1-2.5 g). Following mechanical dissociation, white muscle tissue was washed, enzymatically digested (collagenase type IV and trypsin) and cells were filtered (100 and 40 μm). Isolated cells were counted using a hemocytometer and the trypan blue exclusion method. Isolated cells were plated on poly-L-lysine-treated (Sigma), laminin-coated (BD Biosciences) plates at a density of 2x106 cells/mL. Cultures were incubated at 18 °C in complete media (10% DMEM) under normal atmospheric conditions without CO2 supplementation. Media was changed daily for the first two days of culture. On day three, cells were treated with media containing cortisol or ethanol (vehicle control). All treatments were run in triplicate on duplicate plates and consisted of increasing concentrations of cortisol: CORT 0, 10, 100, and 1000 ng/mL. After 24 hr, media was removed and cells were harvested for total RNA isolation (RNAzol; Molecular Research Center).
Quantitative real-time PCR
Total RNA was isolated from red muscle, white muscle, liver, and myoblast cell cultures using RNAzol (Molecular Research Center, Inc.) according to the manufacturer’s instructions. Total RNA concentrations were quantified using a Nanodrop 1000 Spectrophotometer (Thermo Scientific) and 1 μg of total RNA was reverse transcribed using the ImProm-II Reverse Transcription System (Promega). Quantitative PCR (qPCR) was performed using PerfeCTa SYBR Green SuperMix (Quanta Biosciences) according to the manufacturer’s recommendations using the Mx3000P system (Stratagene). All reactions contained 2 μL sample cDNA (produced from 1 μg total RNA and diluted 1:10) or 1 μL vector at desired concentrations for standard curve. All primers used were specific for each of the three putatively functional myostatin isoforms (myostatin-1a, -1b, and -2a) and used at 300 nM. Myostatin primer sequences were used as previously described (Garikipati et al., 2007; Garikipati et al., 2006). For validation of cortisol action following injections, Hsp90 mRNA expression changes were analyzed in liver tissue by qPCR using primers previously described (Ings et al., 2011; Sathiyaa and Vijayan, 2003; Vijayan et al., 2003). Standard curves were generated by serial dilution of plasmids (pGEM-T Easy Vector, Promega) containing the amplicon of interest. Briefly, 1:10 serial dilutions of stock constructs were performed, resulting in final concentrations of 1.0 × 101 copies/μL to 1.0 × 108 copies/μL. The PCR cycling parameters were as follows: 94 °C (2 min) followed by 40 cycles at 94 °C for 20 s, 60 °C for 30 s, and 68 °C for 1 min. A dissociation curve was performed for each assay to ensure primer specificity by running a single cycle as follows: 95 °C for 1 min, 55 °C for 30 s, and 95 °C for 30 s. All data were analyzed using Mx3000P system software (Stratagene). All assays utilized a comparative baseline strategy using the ΔCq method that standardized raw data to starting input cDNA quantity (Bustin et al., 2009; De Santis et al., 2001; Meyer et al. 2013).
Plasma glucose analysis
Immediately after euthanasia, blood samples were collected via caudal venipuncture. Blood was temporarily stored in heparinized vacuum tubes on ice. Samples were then centrifuged and plasma was isolated from packed cells. Glucose levels were analyzed in plasma (5 μL) using an Accu-Chek glucose meter (Roche) as previously described (Galt et al., 2013; Meyer et al., 2013).
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 5.00, GraphPad Software, San Diego, CA, USA, www.graphpad.com. For in vivo analyses, plasma glucose and HPS90 expression were evaluated using one-way ANOVA, while myostatin expression was evaluated using two-way ANOVA treating fish as experimental units. For in vitro analyses, all expression levels were analyzed using one-way ANOVA. Tukey’s Multiple Comparison Test was conducted to compare among treatment groups as needed. All results were considered significant at P<0.05. All qPCR data are reported relative to control as percent-mean ±S.E.M.
Results
Cortisol elevated plasma glucose and Hsp90 expression in vivo, but did not affect myostatin expression
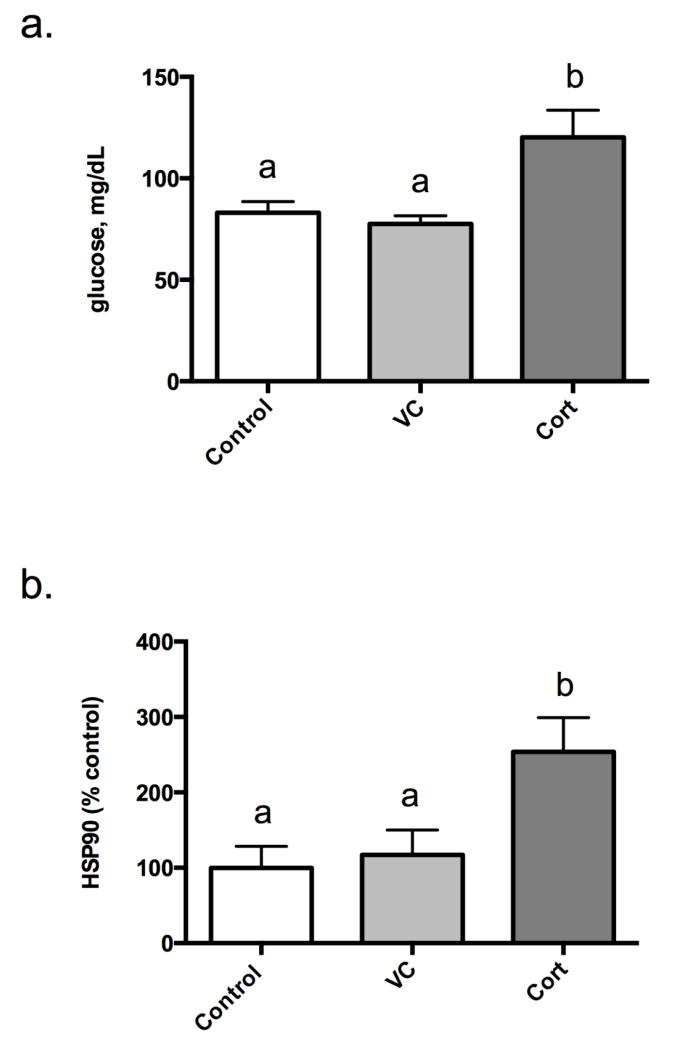
To verify that cortisol treatments initiated a physiological response, both plasma glucose levels and liver Hsp90 mRNA expression were assessed 12 h post-injection for each treatment group. Plasma glucose levels were elevated after cortisol treatment 144.8% and 154.8% relative to both the control and vehicle control, respectively (Figure 1a). In addition, cortisol treatment significantly increased Hsp90 mRNA expression 153.9% and 116.7% relative to both the control and vehicle control, respectively (Figure 1b).
Figure 1.
Circulating blood glucose levels (a) and liver HSP90 mRNA expression levels (b) 12 h post-cortisol treatment. Results are mean glucose mg/dL ±SEM (a) and percent-mean relative to control ± SEM (b) (n=6 fish/trt). Treatment groups include: no injection control (Control), vehicle control (VC; 2 μL/g BW safflower oil), and cortisol (Cort; 2 μg/g BW). Different letters are significantly (P<0.05) different.
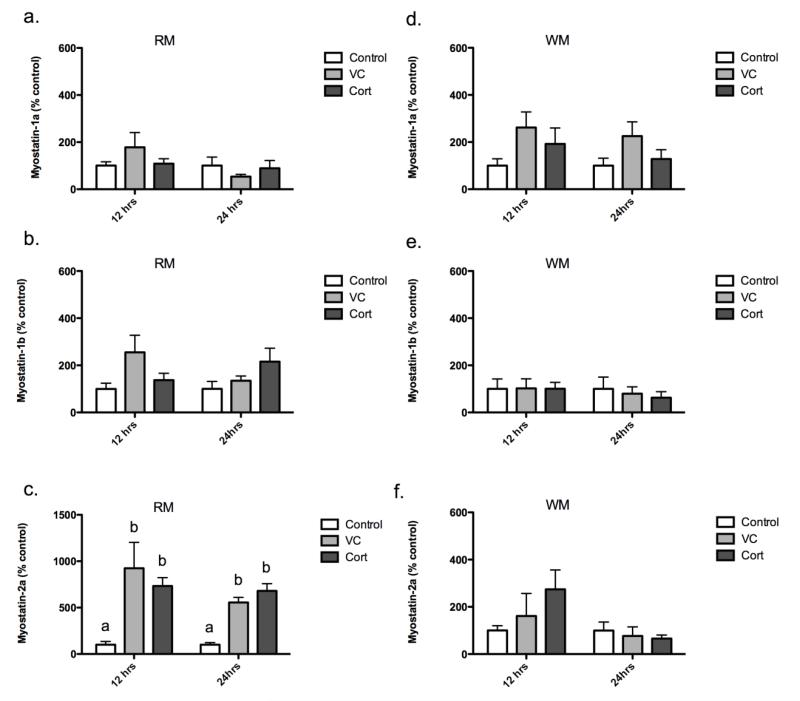
To determine if cortisol regulates myostatin expression, we quantified myostatin-1a, -1b, and -2a mRNA expression in white and red muscle 12 h and 24 h post-injection. In white muscle, cortisol treatment had no statistically significant effects on myostatin-1a, -1b, and -2a mRNA expression in vivo, both 12 h and 24 h post-injection (Figures 2d, -e, -f). In addition, no differences were detected in red muscle myostatin-1a and -1b expression at either time point (Figures 2a, -b). Myostatin-2a expression was significantly upregulated in red muscle in the vehicle and cortisol treatment groups, both 12 h (824.3% and 633.4% over control, respectively; Figure 2c) and 24 h post-injection (455.7% and 581.0% over control, respectively; Figure 2c). However, no difference was detected between these two groups.
Figure 2.
Red muscle myostatin-1a (a), -1b (b), and -2a (c) mRNA levels 12 h and 24 h post-injection. White muscle myostatin-1a (d), -1b (e), and -2a (f) mRNA levels 12 h and 24 h post-injection. Results are percent-mean relative to control ± SEM (n=6 fish/trt). Treatment groups include: no injection control (Control), vehicle control (VC; 2 μL/g BW safflower oil), and cortisol (Cort; 2 μg/g BW). Different letters are significantly (P<0.05) different.
Cortisol elevated Hsp90 and myostatin-1b expression in primary myoblasts
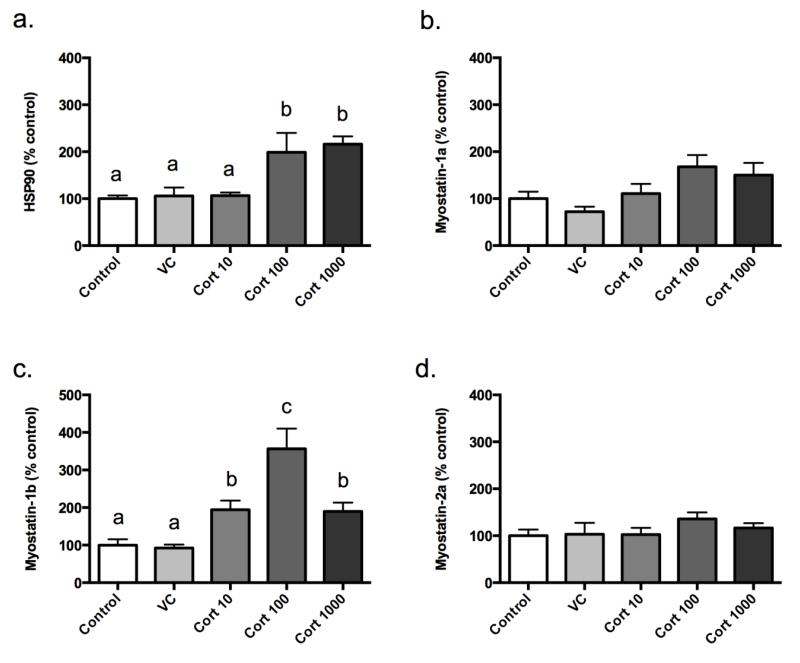
To examine whether cortisol acts at the level of the myoblast in skeletal muscle, we quantified Hsp90 mRNA expression in total myoblast lysates 24 h following treatment. Cortisol, at both 100 ng/mL and 1000 ng/mL, significantly increased Hsp90 mRNA expression relative to the no treatment control (98.8% and 116.3% over control, respectively; Figure 3a). No differences were detected between the vehicle control and cortisol treatment at 10 ng/mL relative to the no treatment control.
Figure 3.
Myoblast HSP90 mRNA (a), myostatin-1a (b), -1b (c), and -2a (d) mRNA levels expression levels 24 h post-treatment. Results are percent-mean relative to control ± SEM (n=6). Treatment groups include: Control, vehicle control (VC; ethanol), cortisol at 10 ng/mL (Cort 10), cortisol at 100 ng/mL (Cort 100), and cortisol at 1000 ng/mL (Cort 1000). Different letters are significantly (P<0.05) different.
To determine if cortisol regulates myostatin in myoblasts, we quantified myostatin-1a, -1b, and 2a mRNA expression in total myoblast lysates 24 h following treatment with increasing concentrations of cortisol. No changes were observed regarding myostatin-1a and -2a expression following our treatments (Figures 3b, -d). Myostatin-1b mRNA responded to cortisol treatment in a dose-dependent manner (Figure 3c). Specifically, expression significantly increased relative to the control at 10 ng/mL (94.5%) and at 100 ng/mL (256.6%). The mRNA abundance at 1000 ng/mL decreased to levels similar to the 10 ng/mL treatment (89.8% and 94.5%, respectively) but was still significantly higher than control levels.
Discussion
Many studies have reported a relationship between GCs and myostatin, but few have described this relationship in fishes. In this study, we conducted single exposure experiments, treating both juvenile rainbow trout and primary rainbow trout myoblasts with cortisol to determine if an interaction between cortisol and myostatin mRNA expression exists in this species. The increase in plasma glucose and hepatic Hsp90 mRNA expression demonstrate that our cortisol treatment induced a response after 12 h in vivo and 24 h in vitro. Further, the increase in myoblast Hsp90 mRNA expression was significant at both 100 ng/mL and 1000 ng/mL of cortisol, suggesting a dose-dependent response. These data corroborate previous in vivo and in vitro studies assessing Hsp90 mRNA expression patterns following glucocorticoid treatments in rainbow trout (Sathiyaa and Vijayan, 2003; Vijayan et al., 2003).
Overall, our analyses failed to detect significant expression changes in myostatin-1a or -1b mRNAs in white or red skeletal muscle following intraperitoneal injections of cortisol (relative to all controls). Because red-oxidative and white-glycolytic skeletal muscle exist as distinct tissues in teleost fish, differences in expression patterns can be analyzed easily between the two metabolically different muscle types. Several reports have demonstrated muscle fiber type-specific myostatin patterns in several teleosts (Biga et al., 2004; Ostbye et al., 2001; Patruno et al., 2008; Roberts and Goetz, 2003; Roberts et al., 2004) suggesting relevance to fiber specificity. Here we observed an increase in myostatin-2a expression specifically in red muscle, 12 h and 24 h post-injection, in both the vehicle and cortisol treatment groups. However, this response is likely attributable to the vehicle, safflower oil, which was chosen for its low viscosity relative to other lipids. The increase in myostatin-2a in response to safflower oil was unexpected considering its common use as an inert vehicle for steroids. Myostatin-2a did not respond to either treatment in white muscle and therefore suggests tissue-specific sensitivity to this or other lipid sources. Further, trends were observed in both myostatin-1a and -1b expression in response to the vehicle in both tissues, though not statistically significant. These data, along with our previous work showing changes in myostatin expression in response to a high-fat diet, provide evidence that myostatin may be responsive to lipid availability (Galt et al., 2013).
Regulation of myostatin in response to glucocorticoids occurs at many levels in mammals, including direct regulation by cortisol through a GRE located in the 5′ regulatory region of the myostatin gene (Gilson et al., 2007; Ma et al., 2001a; Ma et al., 2003; Qin et al., 2013). In this study, the lack of a clear response in vivo suggests that exogenous cortisol may not affect myostatin gene expression in rainbow trout muscle tissue. These findings are consistent and corroborate previous documentation that the rainbow trout myostatin gene promoters lack putative GREs (Garikipati et al., 2007; Garikipati et al., 2006). In fish, few studies have directly tested this relationship between glucocorticoids and myostatin expression. Immersion of tilapia larvae in cortisol resulted in decreased myostatin mRNA expression, and dexamethasone (a potent synthetic glucocorticoid) injections decreased myostatin expression in channel catfish (Rodgers et al., 2003; Weber et al., 2005). To our knowledge, no study has examined the regulatory regions of myostatin for GREs in either tilapia or channel catfish. Typically, the GR positively regulates gene expression, but in some cases negative regulation or repression of expression can occur (Schoneveld et al., 2004). In addition, myogenic regulatory factor (MRF) binding sites appear to be highly conserved in the myostatin promoters of fish and mammals. Therefore, the decrease in myostatin expression in tilapia and channel catfish may be the result of glucocorticoid-induced degradation and disruption of MRF activity thus decreasing the activation of myostatin expression (Jogo et al., 2009; Sun et al., 2008).
Studies examining myostatin expression under stress in fish have reported conflicting results. For example, chronic overcrowding of zebrafish results in elevated myostatin expression in lateral skeletal muscle (Vianello et al., 2003), while high stocking densities in zebrafish elevated both myostatin-1 and -2 in spleen tissue but not muscle (Helterline et al., 2007). The zebrafish myostatin-2 promoter contains a putative GRE, and this element may be responsible for the detected changes in myostatin expression described above (Garikipati et al., 2006). In addition to traditional physical stressors, a 30-day fasting period decreased myostatin-1 and -2 expression in rainbow trout muscle, and this decrease was ameliorated after 14 days of refeeding (Johansen and Overturf, 2006). Conversely, no change in myostatin expression was detected in adult tilapia after 30 days of fasting, but myostatin levels were consistently reduced in tilapia larvae after 9 days of fasting (Rodgers et al., 2003). In Asian sea bass, a 30-day fast differentially regulated myostatin isoform expression (De Santis and Jerry, 2011). Two studies have reported correlations between circulating cortisol levels with myostatin expression levels in fish in relation to growth hormone and probiotics (Biga et al., 2004; Carnevali et al., 2006). However, none of these studies are definitive, as only zebrafish myostatin-2 and Asian sea bass myostatin-1 have putative GREs in their promoters (De Santis and Jerry, 2011; Garikipati et al., 2006). Therefore, the differential expression patterns in response to these stressors could reflect indirect actions of the stress response, the metabolic status of the fish, or divergent adaptations among fishes.
Treatment of immortalized mammalian myoblasts (C2C12) with dexamethasone upregulates both myostatin mRNA expression and protein levels (Ma et al., 2001b). Therefore, we assessed the effects of cortisol treatment in vitro using primary rainbow trout myoblasts to further characterize the relationship between cortisol and myostatin expression. While Hsp90 mRNAs were upregulated with glucocorticoid administration, cortisol treatment had no effect on myostatin-1a or -2a expression, but myostatin-1b expression increased in a dose-dependent manner from 10 ng/mL to 100 ng/mL of cortisol. Interestingly, expression levels in response to 1000 ng/mL of cortisol were similar to levels at 10 ng/mL and significantly less than at the intermediate cortisol dose of 100 ng/mL. Previous work in primary bovine myoblasts has shown that exogenous myostatin negatively regulates myostatin mRNAs, suggests autoregulation of myostatin via a feedback loop (Forbes et al., 2006). A similar mechanism could be at play in this study, however, myostatin protein levels were not assessed. Alternatively, cortisol may negatively affect MRF activity at the high dose as previously documented (Jogo et al., 2009; Sun et al., 2008), and changes in MRF activity may indirectly affect myostatin-1b expression. While our analysis failed to detect changes in myostatin-1b expression in white muscle, it is intriguing that myoblasts isolated from this same tissue upregulated myostatin-1b when stimulated with cortisol. However, myoblasts are immature cells, and the bulk of skeletal muscle is constituted by differentiated myofibers. Additionally, the extracellular matrix plays a key role in the regulation of myostatin in vivo, and this is largely absent in cell culture-based assays (Kishioka et al., 2008; Miura et al., 2010; Sengle et al., 2011; Zeng et al., 2014). Further, the laminin used in our cell culture system has been shown to bind myostatin (Yasaka et al., 2013), and this may interfere with normal myostatin signaling.
This study provides evidence for a divergence in the GC responsiveness and tissue/cell specific regulation of myostatin in rainbow trout. Treatment of juvenile rainbow trout with intraperitoneal injections of cortisol failed to demonstrate a clear and significant effect on myostatin-1a and -1b mRNA expression. It is unclear whether the increase in red muscle myostatin-2a expression can be attributed to cortisol treatment alone, as the vehicle itself has a similar effect. Further, only myostatin-1b expression increased in primary myoblasts in response to cortisol treatment. To our knowledge, these data are the first to report that the regulation of myostatin by GCs in rainbow trout contrasts with the robust regulation that has been observed in mammals. Further studies are needed to determine if cortisol directly regulates myostatin expression in other salmonids, particularly those known to possess putative GREs in myostatin promoters.
Acknowledgements
The authors would like to thank Drs. Josep Planas and Juan Castillo for their assistance and direction with the primary myoblast culture, as well as Matthew Charging, Zachary Fowler, Brooke Franzen, Kira Marshall, and Ben Meyer for their technical assistance in dissections and isolating myogenic precursor cells from numerous fish. Funds for this work were provided, in part, to PRB by the Center for Protease Research NIH Grant # 2P20 RR015566, NIH NIAMS Grant # R03AR055350, and NDSU Advance FORWARD NSF Grant #HRD-0811239. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- GRE
glucocorticoid response element
- CORT
cortisol
- VC
vehicle control
- NO
no injection control
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen DL, Du M. Comparative functional analysis of the cow and mouse myostatin genes reveals novel regulatory elements in their upstream promoter regions. Comparative biochemistry and physiology. Comp Biochem Physiol B. 2008;150:432–439. doi: 10.1016/j.cbpb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Biga PR, Cain KD, Hardy RW, Schelling GT, Overturf K, Roberts SB, Goetz FW, Ott TL. Growth hormone differentially regulates muscle myostatin1 and -2 and increases circulating cortisol in rainbow trout (Oncorhynchus mykiss) Gen Comp Endocrinol. 2004;138:32–41. doi: 10.1016/j.ygcen.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Carnevali O, de Vivo L, Sulpizio R, Silvi GS, Cresci A. Growth improvement by probiotic in European sea bass juveniles (Dicentrarchus labrax, L.) with particular attention to IGF-1, myostatin, and cortisol gene expression. Aquaculture. 2006;258:430–438. [Google Scholar]
- De Santis C, Jerry DR. Differential tissue-regulation of myostatin genes in the teleost fish Lates calcarifer in response to fasting. Evidence for functional differentiation. Mol Cell Endocrinol. 2011;335:158–165. doi: 10.1016/j.mce.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Forbes D, Jackman M, Bishop A, Thomas M, Kambadur R, Sharma M. Myostatin auto-regulates its expression by feedback loop through Smad7 dependent mechanism. J Cell Physiol. 2006;206:264–272. doi: 10.1002/jcp.20477. [DOI] [PubMed] [Google Scholar]
- Funkenstein B, Balas V, Rebhan Y, Pliatner A. Characterization and functional analysis of the 5′ flanking region of Sparus aurata myostatin-1 gene. Comp Biochem Physiol B. 2009a;153:55–62. doi: 10.1016/j.cbpa.2008.09.031. [DOI] [PubMed] [Google Scholar]
- Funkenstein B, Balas V, Rebhan Y, Pliatner A. Characterization and functional analysis of the 5′ flanking region of Sparus aurata myostatin-1 gene. Comp Biochem Physiol A. 2009b;153:55–62. doi: 10.1016/j.cbpa.2008.09.031. [DOI] [PubMed] [Google Scholar]
- Gabillard JC, Biga PR, Rescan PY, Seiliez I. Revisiting the paradigm of myostatin in vertebrates: insights from fishes. General and comparative endocrinology. 2013;194:45–54. doi: 10.1016/j.ygcen.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Galt NJ, Froehlich JM, Meyer BM, Barrows FT, Biga PR. High-fat diet reduces local myostatin-1 paralog expression and alters skeletal muscle lipid content in rainbow trout, Oncorhynchus mykiss. Fish Physiol Biochem. 2013;XX doi: 10.1007/s10695-013-9893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garikipati DK, Gahr SA, Roalson EH, Rodgers BD. Characterization of rainbow trout myostatin-2 genes (rtMSTN-2a and -2b): genomic organization, differential expression, and pseudogenization. Endocrinology. 2007;148:2106–2115. doi: 10.1210/en.2006-1299. [DOI] [PubMed] [Google Scholar]
- Garikipati DK, Gahr SA, Rodgers BD. Identification, characterization, and quantitative expression analysis of rainbow trout myostatin-1a and myostatin-1b genes. J Endocrinol. 2006;190:879–888. doi: 10.1677/joe.1.06866. [DOI] [PubMed] [Google Scholar]
- Gilson H, Schakman O, Combaret L, Lause P, Grobet L, Attaix D, Ketelslegers JM, Thissen JP. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology. 2007;148:452–460. doi: 10.1210/en.2006-0539. [DOI] [PubMed] [Google Scholar]
- Helterline DL, Garikipati D, Stenkamp DL, Rodgers BD. Embryonic and tissue-specific regulation of myostatin-1 and -2 gene expression in zebrafish. Gen Comp Endocrinol. 2007;151:90–97. doi: 10.1016/j.ygcen.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Chen S, Zhang R, Lin Y. Single nucleotide polymorphisms in the upstream regulatory region alter the expression of myostatin. In vitro cellular & developmental biology. Animal. 2013;49:417–423. doi: 10.1007/s11626-013-9621-5. [DOI] [PubMed] [Google Scholar]
- Ings JS, Servos MR, Vijayan MM. Hepatic transcriptomics and protein expression in rainbow trout exposed to municipal wastewater effluent. Environ Sci Technol. 2011;45:2368–2376. doi: 10.1021/es103122g. [DOI] [PubMed] [Google Scholar]
- Janzen WJ, Duncan CA, Riley LG. Cortisol treatment reduces ghrelin signaling and food intake in tilapia, Oreochromis mossambicus. Domest Anim Endocrinol. 2012;43:251–259. doi: 10.1016/j.domaniend.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Jogo M, Shiraishi S, Tamura TA. Identification of MAFbx as a myogenin-engaged F-box protein in SCF ubiquitin ligase. FEBS Letters. 2009;583:2715–2719. doi: 10.1016/j.febslet.2009.07.033. [DOI] [PubMed] [Google Scholar]
- Johansen KA, Overturf K. Alterations in expression of genes associated with muscle metabolism and growth during nutritional restriction and refeeding in rainbow trout. Comp Biochem Physiol B. 2006;144:119–127. doi: 10.1016/j.cbpb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Kishioka Y, Thomas M, Wakamatsu JI, Hattori A, Sharma M, Kambadur R, Nishimura T. Decorin enhances the proliferation and differentiation of myogenic cells through suppressing myostatin activity. J Cell Physiology. 2008;215:856–867. doi: 10.1002/jcp.21371. [DOI] [PubMed] [Google Scholar]
- Ko CF, Chiou TT, Chen TT, Wu JL, Chen JC, Lu JK. Molecular cloning of myostatin gene and characterization of tissue-specific and developmental stage-specific expression of the gene in orange spotted grouper, Epinephelus coioides. Mar Biotechnol. 2007;9:20–32. doi: 10.1007/s10126-006-6059-8. [DOI] [PubMed] [Google Scholar]
- Kocabas AM, Kucuktas H, Dunham RA, Liu Z. Molecular characterization and differential expression of the myostatin gene in channel catfish (Ictalurus punctatus) Biochim Biophys Acta. 2002;1575:99–107. doi: 10.1016/s0167-4781(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Lee CY, Hu SY, Gong HY, Chen MH, Lu JK, Wu JL. Suppression of myostatin with vector-based RNA interference causes a double-muscle effect in transgenic zebrafish. Biochem Biophys Res Commun. 2009;387:766–771. doi: 10.1016/j.bbrc.2009.07.110. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim YS, Oh MY, Jeong IH, Seong KB, Jin HJ. Improving rainbow trout (Oncorhynchus mykiss) growth by treatment with a fish (Paralichthys olivaceus) myostatin prodomain expressed in soluble forms in E. coli. Aquaculture. 2010;302:270–278. [Google Scholar]
- Li H, Fan J, Liu S, Yang Q, Mu G, He C. Characterization of a myostatin gene (MSTN1) from spotted halibut (Verasper variegatus) and association between its promoter polymorphism and individual growth performance. Comp Biochem Physiol B. 2012a;161:315–322. doi: 10.1016/j.cbpb.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Li J, Deng J, Yu S, Zhang J, Cheng D, Wang H. The virtual element in proximal promoter of porcine myostatin is regulated by myocyte enhancer factor 2C. Biochem Biophys Res Commun. 2012b;419:175–181. doi: 10.1016/j.bbrc.2012.01.135. [DOI] [PubMed] [Google Scholar]
- Li J, Deng J, Zhang J, Cheng D, Wang H. [Regulation of myostatin promoter activity by myocyte enhancer factor 2] Sheng wu gong cheng xue bao = Chinese journal of biotechnology. 2012c;28:918–926. [PubMed] [Google Scholar]
- Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S. Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol. 2001a;281:E1128–1136. doi: 10.1152/ajpendo.2001.281.6.E1128. [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S. Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol. 2001b;281:E1128–1136. doi: 10.1152/ajpendo.2001.281.6.E1128. [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Bhasin S, Mahabadi V, Artaza J, Gonzalez-Cadavid N, Arias J, Salehian B. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. American journal of physiology. Endocrinol Metabol. 2003;285:E363–371. doi: 10.1152/ajpendo.00487.2002. [DOI] [PubMed] [Google Scholar]
- Maccatrozzo L, Bargelloni L, Radaelli G, Mascarello F, Patarnello T. Characterization of the myostatin gene in the gilthead seabream (Sparus aurata): sequence, genomic structure, and expression pattern. Mar Biotechnol. 2001;3:224–230. doi: 10.1007/s101260000064. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros EF, Phelps MP, Fuentes FD, Bradley TM. Overexpression of follistatin in trout stimulates increased muscling. Am J Physiol. 2009;297:R235–242. doi: 10.1152/ajpregu.91020.2008. [DOI] [PubMed] [Google Scholar]
- Meyer BM, Froehlich JM, Galt NJ, Biga PR. Inbred strains of zebrafish exhibit variation in growth performance and myostatin expression following fasting. Comp Biochem Physiol A. 2013;164:1–9. doi: 10.1016/j.cbpa.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Kishioka Y, Wakamatsu J, Hattori A, Nishimura T. Interaction between myostatin and extracellular matrix components. Anim Sci J. 2010;81:102–107. doi: 10.1111/j.1740-0929.2009.00700.x. [DOI] [PubMed] [Google Scholar]
- Nadjar-Boger E, Hinits Y, Funkenstein B. Structural and functional analysis of myostatin-2 promoter alleles from the marine fish Sparus aurata: evidence for strong muscle-specific promoter activity and post-transcriptional regulation. Mol Cell Endocrinol. 2012;361:51–68. doi: 10.1016/j.mce.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Nadjar-Boger E, Maccatrozzo L, Radaelli G, Funkenstein B. Genomic cloning and promoter functional analysis of myostatin-2 in shi drum, Umbrina cirrosa: conservation of muscle-specific promoter activity. Comp Biochem Physiol B. 2013;164:99–110. doi: 10.1016/j.cbpb.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Ostbye TK, Bardal T, Vegusdal A, Frang OT, Kjorsvik E, Andersen O. Molecular cloning of the Atlantic salmon activin receptor IIB cDNA - Localization of the receptor and myostatin in vivo and in vitro in muscle cells. Comp Biochem Physiol D. 2007a;2:101–111. doi: 10.1016/j.cbd.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Ostbye TK, Galloway TF, Nielsen C, Gabestad I, Bardal T, Andersen O. The two myostatin genes of Atlantic salmon (Salmo salar) are expressed in a variety of tissues. European journal of biochemistry / FEBS J. 2001;268:5249–5257. doi: 10.1046/j.0014-2956.2001.02456.x. [DOI] [PubMed] [Google Scholar]
- Ostbye TK, Wetten OF, Tooming-Klunderud A, Jakobsen KS, Yafe A, Etzioni S, Moen T, Andersen O. Myostatin (MSTN) gene duplications in Atlantic salmon (Salmo salar): evidence for different selective pressure on teleost MSTN-1 and -2. Gene. 2007b;403:159–169. doi: 10.1016/j.gene.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Patruno M, Sivieri S, Poltronieri C, Sacchetto R, Maccatrozzo L, Martinello T, Funkenstein B, Radaelli G. Real-time polymerase chain reaction, in situ hybridization and immunohistochemical localization of insulin-like growth factor-I and myostatin during development of Dicentrarchus labrax (Pisces: Osteichthyes) Cell Tissue Res. 2008;331:643–658. doi: 10.1007/s00441-007-0517-0. [DOI] [PubMed] [Google Scholar]
- Qin J, Du R, Yang YQ, Zhang HQ, Li Q, Liu L, Guan H, Hou J, An XR. Dexamethasone-induced skeletal muscle atrophy was associated with upregulation of myostatin promoter activity. Res Vet Sci. 2013;94:84–89. doi: 10.1016/j.rvsc.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Rescan P, Pabeouf G, Fauconneau B. Myosatellite cells of Oncorhynchus mykiss: culture and myogenesis of laminin substrates., Biology of protozoa, invertebrates, and fishes: in vitro models and applications. Coll IFREMER. 1995;18:63–68. [Google Scholar]
- Rescan PY, Jutel I, Ralliere C. Two myostatin genes are differentially expressed in myotomal muscles of the trout (Oncorhynchus mykiss) J Exp Biol. 2001;204:3523–3529. doi: 10.1242/jeb.204.20.3523. [DOI] [PubMed] [Google Scholar]
- Roberts SB, Goetz FW. Differential skeletal muscle expression of myostatin across teleost species, and the isolation of multiple myostatin isoforms. FEBS Letts. 2001;491:212–216. doi: 10.1016/s0014-5793(01)02196-2. [DOI] [PubMed] [Google Scholar]
- Roberts SB, Goetz FW. Myostatin protein and RNA transcript levels in adult and developing brook trout. Mol Cell Endocrinol. 2003;210:9–20. doi: 10.1016/j.mce.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Roberts SB, McCauley LA, Devlin RH, Goetz FW. Transgenic salmon overexpressing growth hormone exhibit decreased myostatin transcript and protein expression. J Exp Biol. 2004;207:3741–3748. doi: 10.1242/jeb.01210. [DOI] [PubMed] [Google Scholar]
- Rodgers BD, Garikipati DK. Clinical, agricultural, and evolutionary biology of myostatin: a comparative review. Endocrine Revs. 2008;29:513–534. doi: 10.1210/er.2008-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers BD, Weber GM, Kelley KM, Levine MA. Prolonged fasting and cortisol reduce myostatin mRNA levels in tilapia larvae; short-term fasting elevates. Am J Physiol. 2003;284:R1277–1286. doi: 10.1152/ajpregu.00644.2002. [DOI] [PubMed] [Google Scholar]
- Sathiyaa R, Vijayan MM. Autoregulation of glucocorticoid receptor by cortisol in rainbow trout hepatocytes. American journal of physiology. Cell Physiol. 2003;284:C1508–1515. doi: 10.1152/ajpcell.00448.2002. [DOI] [PubMed] [Google Scholar]
- Sawatari E, Seki R, Adachi T, Hashimoto H, Uji S, Wakamatsu Y, Nakata T, Kinoshita M. Overexpression of the dominant-negative form of myostatin results in doubling of muscle-fiber number in transgenic medaka (Oryzias latipes) Comp Biochem Physiol A. 2010;155:183–189. doi: 10.1016/j.cbpa.2009.10.030. [DOI] [PubMed] [Google Scholar]
- Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochim Biophys Acta. 2004;1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Sengle G, Ono RN, Sasaki T, Sakai LY. Prodomains of transforming growth factor beta (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J Biol Chem. 2011;286:5087–5099. doi: 10.1074/jbc.M110.188615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller MP, Kambadur R, Jeanplong F, Thomas M, Martyn JK, Bass JJ, Sharma M. The myostatin gene is a downstream target gene of basic helix-loop-helix transcription factor MyoD. Mol Cell Biol. 2002;22:7066–7082. doi: 10.1128/MCB.22.20.7066-7082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Trausch-Azar JS, Muglia LJ, Schwartz AL. Glucocorticoids differentially regulate degradation of MyoD and Id1 by N-terminal ubiquitination to promote muscle protein catabolism. Proc Natl Acad Sci USA. 2008;105:3339–3344. doi: 10.1073/pnas.0800165105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianello S, Brazzoduro L, Dalla Valle L, Belvedere P, Colombo L. Myostatin expression during development and chronic stress in zebrafish (Danio rerio) J Endocrinol. 2003;176:47–59. doi: 10.1677/joe.0.1760047. [DOI] [PubMed] [Google Scholar]
- Vijayan MM, Raptis S, Sathiyaa R. Cortisol treatment affects glucocorticoid receptor and glucocorticoid-responsive genes in the liver of rainbow trout. Gen Comp Endocrinol. 2003;132:256–263. doi: 10.1016/s0016-6480(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Weber TE, Small BC, Bosworth BG. Lipopolysaccharide regulates myostatin and MyoD independently of an increase in plasma cortisol in channel catfish (Ictalurus punctatus) Domest Anim Endocrinol. 2005;28:64–73. doi: 10.1016/j.domaniend.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Xu C, Wu G, Zohar Y, Du SJ. Analysis of myostatin gene structure, expression and function in zebrafish. J Exp Biol. 2003;206:4067–4079. doi: 10.1242/jeb.00635. [DOI] [PubMed] [Google Scholar]
- Xue L, Dong X, Zhang X, Diallo A. Organization and functional analysis of the 5′ flanking regions of myostatin-1 and 2 genes from Larimichthys crocea. DNA Cell Biol. 2012;31:845–855. doi: 10.1089/dna.2011.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasaka N, Suzuki K, Kishioka Y, Wakamatsu J, Nishimura T. Laminin binds to myostatin and attenuates its signaling. Animal science journal = Nihon chikusan Gakkaiho. 2013;84:663–668. doi: 10.1111/asj.12052. [DOI] [PubMed] [Google Scholar]
- Ye HQ, Chen SL, Sha ZX, Liu Y. Molecular cloning and expression analysis of the myostatin gene in sea perch (Lateolabrax japonicus) Mar Biotechnol. 2007;9:262–272. doi: 10.1007/s10126-006-6093-6. [DOI] [PubMed] [Google Scholar]
- Zeng QJ, Wang LN, Shu G, Wang SB, Zhu XT, Gao P, Xi QY, Zhang YL, Zhang ZQ, Jiang QY. Decorin-induced proliferation of avian myoblasts involves the myostatin/Smad signaling pathway. Poultry Sci. 2014;93:138–146. doi: 10.3382/ps.2013-03300. [DOI] [PubMed] [Google Scholar]