Abstract
The application of primary organoid cultures containing epithelial and mesenchymal elements to cancer modeling holds promise for combining the accurate multilineage differentiation and physiology of in vivo systems with the facile in vitro manipulation of transformed cell lines. Here, a single air-liquid interface culture method was used without modification to engineer oncogenic mutations into primary epithelial/mesenchymal organoids from mouse colon, stomach and pancreas. Pancreatic and gastric organoids exhibited dysplasia upon KrasG12D expression and/or p53 loss, and readily generated adenocarcinoma upon in vivo transplantation. In contrast, primary colon organoids required combinatorial Apc, p53, KrasG12D and Smad4 mutations for progressive transformation to invasive adenocarcinoma-like histology in vitro and tumorigenicity in vivo, recapitulating multi-hit models of colorectal cancer (CRC), and versus more promiscuous transformation of small intestinal organoids. Colon organoid culture functionally validated the microRNA miR-483 as a dominant driver oncogene at the Insulin-like growth factor-2 (IGF2) 11p15.5 CRC amplicon, inducing dysplasia in vitro and tumorigenicity in vivo. These studies demonstrate the general utility of a highly tractable primary organoid system for cancer modeling and driver oncogene validation in diverse gastrointestinal tissues.
INTRODUCTION
The in vitro culture of primary, non-transformed tissues as three-dimensional (3D) structures that accurately recapitulate organ structure, multilineage differentiation and physiology has diverse applications ranging from basic biology to therapy1,2. 3D cultures of glandular organs can be subdivided into those with exclusively epithelial components, versus those with both epithelial and mesenchymal components. While the term “organoid” has been used generically for 3D structures possessing multiple cell lineages and tissue architecture, a recent proposal suggests that this term be restricted to cultures containing both epithelium and mesenchyme3.
Recent studies have described methodology for 3D culture of purely epithelial cell preparations from primary gastrointestinal tissues such as pancreas, stomach and intestine, often using specific growth factor supplementation to supply paracrine/mesenchymal signals2,4–8. In contrast, we have robustly cultured organoids with both epithelial and mesenchymal components from small intestine, colon and stomach using an air-liquid interface (ALI) methodology that does not require exogenous growth factor supplementation9,10. In this system, intestinal organoids exhibit multilineage differentiation and supporting mesenchyme, sustained growth for >350 days, recapitulated intestinal stem cells and their endogenous Wnt/Notch paracrine signaling niche, and exhibited peristalsis9. Similarly, air-liquid interface gastric organoids accurately recapitulate differentiation and ultrastructure of stomach cell lineages for >30 days 10.
Despite advantages of accurate organ ultrastructure, stromal composition and ease of experimental manipulation, primary organoid culture of diverse normal tissues has been underutilized for in vitro modeling of cancer. Recently, Ghajar and Bissell advanced the holistic notion of “cancer engineering”, describing the need for complex in vitro cell culture models of cancer that incorporate heterologous interactions between epithelium and diverse stromal cell types to interrogate both epithelial and microenvironmental aspects of malignancy11. Certainly, potential applications of such highly accurate epithelial/mesenchymal models include cancer therapeutic validation and functional validation of putative oncogenic loci from an untransformed baseline tabula rasa. Indeed, the complexity and chaos of tumor genomes revealed by genome-scale sequencing efforts such as The Cancer Genome Atlas Project (TCGA) and others12 has recently highlighted the necessity for novel in vitro methods to functionally validate putative oncogenic loci and to distinguish them from passenger mutations.
In the present study, we address these needs through the demonstration that a single air-liquid interface method can robustly model diverse gastrointestinal malignancies from pancreas, stomach and colon in primary epithelial/mesenchymal organoid culture, yielding detailed in situ histologic endpoints for oncogenic transformation in vitro, and tumorigenicity upon transplantation in vivo. Further, we systematically assess the requirements for combinatorial oncogenic transformation in colon organoids, obtaining in vitro reprogramming of primary intestinal epithelium to adenocarcinoma and recapitulating multi-step colon tumorigenesis. Finally, we demonstrate proof-of-principle for the application of primary organoid culture to driver oncogene validation, through interrogation of the 11p15.5 colon cancer amplicon containing IGF2 and miR-483.
RESULTS
Growth and transformation of primary pancreatic organoids
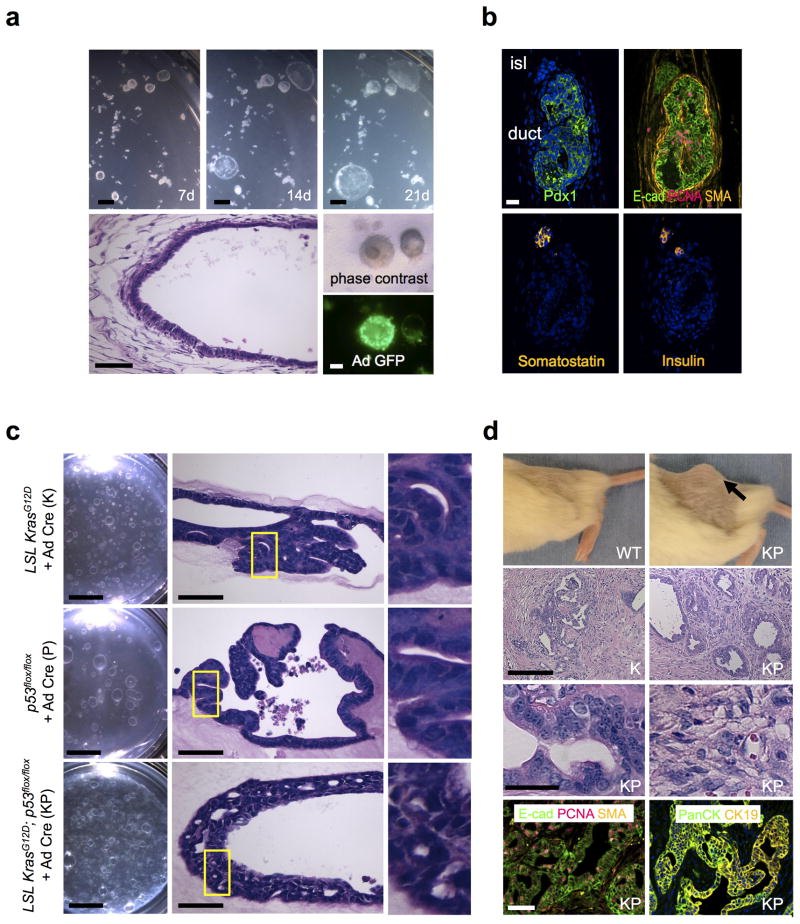
Various methods have been successfully employed for primary culture of pancreatic ductal cells without stromal components, ranging from monolayer and 3D culture of ductal populations versus single cells5–8,13,14. However, long-term pancreatic organoid culture containing epithelial and mesenchymal elements has not been previously described. Here, we cultured mouse pancreatic explants using our colon and gastric air-liquid interface culture method utilizing an inner collagen gel-containing transwell with direct air exposure9,10. Accordingly, neonatal pancreatic organoids from wild-type C57BL/6 mice were cultured in an air-liquid interface where they exhibited progressive expansion for >30 days as cystic structures containing an epithelial layer and surrounding fibroblasts, similar to our prior studies with intestine and stomach9,10 and were readily infected with adenovirus (Fig. 1a). These organoids predominantly (>90%) consisted of E-cadherin+ (E-cad+) and Pdx1+ ductal epithelium with a cystic morphology, PCNA+ proliferating cells, in association with α-smooth muscle actin+ (SMA+) stromal cells (Fig. 1a,b). Somatostatin and insulin expression were observed in rare islet-like regions that were either independent from or associated with ductal structures (Fig. 1b); sporadic immunoreactivity for glucagon and amylase were occasionally observed (data not shown).
Figure 1. In vitro oncogenic transformation of primary pancreatic organoids and in vivo tumorigenesis.
(a) In vitro culture of wild-type neonatal pancreatic organoids. Top row. Time course stereomicroscopy demonstrating progressive growth of primary pancreatic organoids at days 7, 14 and 21 of air-liquid interface culture. Scale bars, 1 mm. Bottom row. Left, day 7 H&E staining, histologic characterization demonstrates a well-differentiated cystic epithelium with surrounding fibroblastic stroma. Scale bar, 50 μm. Right middle, phase contrast. Right bottom, adeno Cre-GFP infection of pancreatic organoids. Scale bar, 1 mm. (b) Immunofluorescence demonstrates a well-differentiated cystic epithelium with surrounding fibroblastic stroma and a preponderance of epithelial E-cadherin+ and Pdx1+ ductal structures with PCNA+ proliferative cells and SMA+ stromal cells. Bottom row, somatostatin+ and insulin+ endocrine cells in a rare islet structure. Day 7 immunofluorescence is depicted. Scale bar, 25 μm. (c) Transformation of primary pancreatic organoid cultures in vitro. Pancreatic organoids from neonatal mice bearing LSL KrasG12D or/and p53flox/flox alleles (K, P, KP) were cultured with adenovirus Cre-GFP at d0 of primary plating. Left column. Stereomicroscopy of K, P and KP organoids after 20 days of primary culture and an additional 30 days of secondary passage (50 days total). Scale bars, 5 mm. Middle column. H&E staining for the indicated genotypes at day 50. As opposed to wild-type organoids, P organoids exhibited mildly stratified nuclei with moderate nuclear enlargement and K and KP organoids contained enlarged pleomorphic nuclei with stratification and cribriform growth. Scale bars, 50 μm. Right column. Enlargement of the yellow boxed fields. (d) Transformed pancreatic organoids exhibit in vivo tumorigenicity. Top row. In vivo tumor growth of the indicated genotypes, day 30 after s.c. organoid implantation into NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac mice (NOG) mice. Arrow denotes tumor. 2nd row. H&E staining of tumors. Scale bar, 100 μm. (Left) a representative region of gland-forming invasive adenocarcinoma infiltrating fibrous tissue. (Right) Region from a KP tumor exhibiting poorly differentiated invasive adenocarcinoma with areas of sarcomatoid carcinoma. 3rd row. Enlarged views of glandular (left) and poorly-differentiated regions (right) from a KP tumor. Scale bar, 25 μm. Bottom row. Glandular epithelium from a KP tumor with E-cadherin, PanCK and CK19 immunofluorescence. Immunoreactivity for PCNA-positive proliferative cells and SMA-positive stromal cells is also present. Scale bar, 50 μm.
The predominance of ductal structures within the pancreatic organoids suggested their utility for modeling pancreatic ductal adenocarcinoma (PDAC). Accordingly, we generated primary pancreatic organoids from mice with floxed alleles of KrasG12D (lox-stop-lox (LSL) KrasG12D), p53 (p53flox/flox) or both (LSL KrasG12D; p53flox/flox), by analogy to prior seminal in vivo studies15. The organoids were infected at the time of plating with adenovirus expressing Cre-GFP (Ad Cre-GFP) or a control immunoglobulin Fc fragment (Ad Fc). Appropriate epithelial expression of KrasG12D and P53 was assessed by immunofluorescence (Supplementary Fig. 1a). Throughout this manuscript, Cre-treated organoids are denoted as “K” (i.e. KrasG12D-expressing), “P” (p53-null) or “KP” (KrasG12D-expressing and p53-null)
Oncogene-transformed K, P and KP pancreatic organoids could be serially passaged (> 6 passages, longest time evaluated) in contrast to Ad Fc controls (Supplementary Fig. 1b). By 50 days of culture, K, and KP pancreatic organoids exhibited significant dysplasia with enlarged pleomorphic nuclei with stratification, cribriform growth and marked areas of invasion. P organoids did not exhibit invasion but did possess mildly stratified nuclei with moderate nuclear enlargement; these alterations encompassed ~10% of the epithelium for K and P, and > 30% for KP (Fig. 1c). The oncogene-transformed pancreatic organoids could be readily infected with retrovirus or lentivirus at the time of passage, in contrast to Ad Fc controls (Supplementary Fig. 1c and data not shown).
We re-passaged FACS-sorted EpCAM+ epithelium from these primary pancreatic organoids into 96-well air-liquid interface culture, allowing both robust secondary organoid formation and multiplexed quantitation of proliferation using the fluorescent dye Calcein, AM. The use of light-impermeable FluoroBlok membranes in the transwell allows simultaneous quantitation of vertical invasion through the filter (bottom fluorescence) versus proliferation (total fluorescence). In this assay, KP organoids exhibited significantly increased proliferation and invasion (P < 0.05 KP vs. K or P) (Supplementary Fig. 2a,b).
Importantly, dissociated K, P and KP organoids all demonstrated in vivo tumorigenicity within 30 days of subcutaneous (s.c.) transplantation into immunodeficient NOG mice, indicating full oncogenic transformation. These transplanted tumors all represented moderately or poorly differentiated invasive adenocarcinoma, containing foci of invasive glands lined by cells with enlarged pleomorphic nuclei and of focal cytoplasmic mucin, admixed with adjacent less-differentiated regions exhibiting glandular effacement in agreement with prior in vivo studies15; KP and P were more poorly differentiated than K, and KP tumor size exceeded K or P (Fig. 1d, Supplementary Fig. 2c). Areas of glandular differentiation expressed E-cadherin and the ductal markers PanCK8 and CK1915, confirming the epithelial origin of these adenocarcinomas (Fig. 1d); in non-glandular regions E-cadherin expression was lost (Supplementary Fig. 2d). These studies demonstrate not only the development of a model for sustained primary pancreatic organoid cultures, but also its successful application to PDAC modeling with detailed histologic endpoints in vitro and tumorigenicity in vivo.
Oncogenic transformation of primary gastric organoid cultures
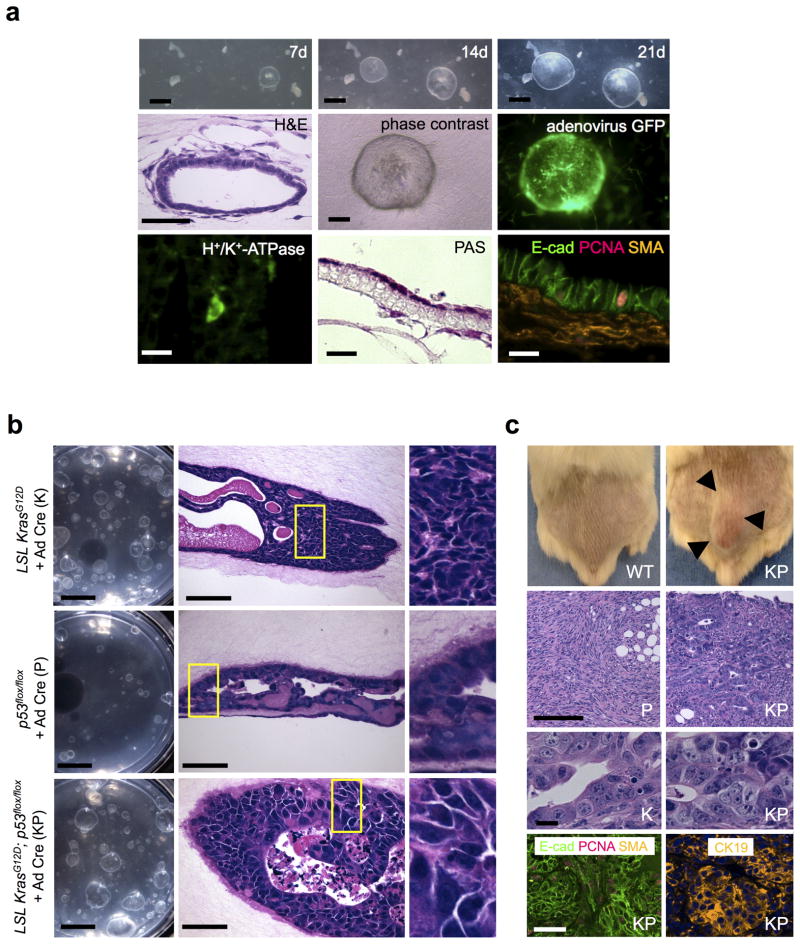
We similarly examined the oncogenic transformation of gastric organoids in air-liquid interface culture. As one of us (A. O.) has recently described10, primary gastric organoids from wild-type neonatal mice can be robustly propagated as progressively enlarging cystic structures for >30 days. Histologically, the gastric organoids exhibited a well-differentiated epithelial layer comprised of surface mucous cells and to a lesser extent mucous neck cells with peripherally-located myofibroblasts, which were confirmed by E-cadherin and SMA immunofluorescence (Fig. 2a). Regions of H+/K+-ATPase expression and PAS-positive mucus-producing cells were also observed (Fig. 2a).
Figure 2. In vitro oncogenic transformation of primary gastric organoids and in vivo tumorigenesis.
(a) In vitro culture of wild-type neonatal gastric organoids. Top row. Stereomicroscopy reveals progressive growth of primary gastric organoids at days 7, 14 and 21 of air-liquid interface culture. Scale bars, 1 mm. Middle row. Left, Histologic characterization of gastric organoids reveals a well-differentiated simple cystic epithelium with peripherally oriented enveloping fibroblasts, H&E, day 7. Scale bar, 25 μm. Right, middle. Phase contrast and fluorescence microscopy of adeno Cre-GFP infection of wild-type gastric organoids. Scale bar, 200 μm. Bottom Row. Left, middle. H+/K+-ATPase-expressing parietal cells (immunofluorescence) and mucin-secreting cells (PAS), day 30, 100× magnification. Right, Immunofluorescence demonstrates E-cad+ epithelium, PCNA+ proliferative cells and SMA+ stromal cells. Scale bars, 20 μm. (b) Transformation of primary gastric organoid cultures in vitro. Gastric organoids from neonatal mice with LSL KrasG12D and/or p53flox/flox alleles were cultured with adenovirus Cre-GFP (K, P, KP) at d0 of primary plating. Scale bars, 5 mm. Left column. Stereomicroscopy of K, P and KP organoids after 20 days of primary culture and an additional 30 days of secondary passage (50 days total). Middle column. H&E staining for the indicated genotypes at day 50. Scale bar, 50 μm. As opposed to wild-type organoids, P organoids exhibited focal areas of nuclear stratification and moderate nuclear enlargement, K displayed high-grade dysplasia with markedly enlarged, pleomorphic nuclei and significant nuclear stratification, and KP organoids presented with extensive invasion, highly pleomorphic and enlarged nuclei and regional complex cribriform growth patterns associated with necrosis. Right column. Enlargements of the yellow boxed fields. (c) Transformed gastric organoids display in vivo tumorigenicity. Top row. In vivo tumor growth of organoids of the indicated genotypes, day 30 after s.c. implantation into NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac mice (NOG) mice. Arrows mark the tumor border. 2nd row. H&E staining of tumors. Scale bar, 100 μm. P and KP tumors were hypercellular with enlarged, hyperchromatic nuclei, variable gland formation, numerous mitotic figures, and eosinophilic cytoplasm. The tumor cells infiltrate into adjacent soft tissue. 3rd row. Enlarged view of glandular regions of K and KP. Scale bar, 10 μm. Bottom row. Tumor glandular epithelium expresses E-cadherin and CK19. PCNA-positive proliferative cells and SMA-positive stromal cells are present. Scale bar, 50 μm.
To model gastric cancer, we generated primary organoids from the glandular stomach of K, P or compound KP mice, modeling the TP53 loss and Kras mutations that are present in approximately 50% and 5% of gastric cancer, respectively. Inclusion of Ad Cre-GFP at the time of primary plating resulted in efficient adenoviral infection (Fig. 2a), accompanied by epithelial overexpression of KrasG12D in K and KP organoids, as well as loss of epithelial P53 immunoreactivity in P organoids (Supplementary Fig. 3a).
Similar to the pancreatic organoids, oncogene-transformed K, P and KP gastric organoids could be serially passaged (> 6 passages, longest time evaluated) as opposed to Ad Fc controls (Supplementary Fig. 3b). After 50 days of culture, K, P and KP gastric organoids all exhibited in vitro histologic dysplasia which was most profound with KP, characterized by invasive tumor cells containing highly pleomorphic and enlarged nuclei and regional complex cribriform growth patterns associated with necrosis. K and P gastric organoids exhibited relatively milder degrees of in vitro dysplasia, with cellular stratification, nuclear enlargement and pleiomorphism (Fig. 2b). These alterations represented approximately ~30–40% of the epithelium for K, 10% for P, and ~80% for KP despite uniform KrasG12D expression and p53 loss (Supplementary Fig. 3a). Also similar to pancreatic organoids, K, P and KP gastric organoids were readily infected with lentivirus and retrovirus, in marked contrast to wild type controls (Supplementary Fig. 3c and data not shown).
In 96-well air-liquid interface proliferation and invasion assays, KP organoids exhibited significantly increased proliferation and invasion (P < 0.05 for KP vs. K or P) (Supplementary Fig. 4a). Subcutaneous transplantation of K, P and KP organoid-dissociated cells all induced in vivo tumorigenicity within 30 days of inoculation into immunodeficient NOG mice (Fig. 2c, Supplementary Fig. 4b). All produced high-grade invasive carcinomas composed of malignant cells with enlarged, hyperchromatic nuclei, numerous mitotic figures, and variable amounts of eosinophilic cytoplasm. KP tumors were higher grade than K or P and exhibited decreased glandular architecture, more nuclear pleomorphism and absence of identifiable intracytoplasmic mucin vacuoles. Regions of glandular differentiation for all genotypes were E-cadherin- and CK19-positive (Fig. 2c and data not shown), but non-glandular regions were E-cadherin-negative (Supplementary Fig. 4c). These results indicated the successful modeling of oncogenic loci in primary gastric organoids with robust in situ histologic readouts in vitro and neoplastic transformation in vivo, accurately recapitulating numerous aspects of human gastric cancer progression.
Recapitulation of multi-hit colorectal tumorigenesis in primary colon organoids
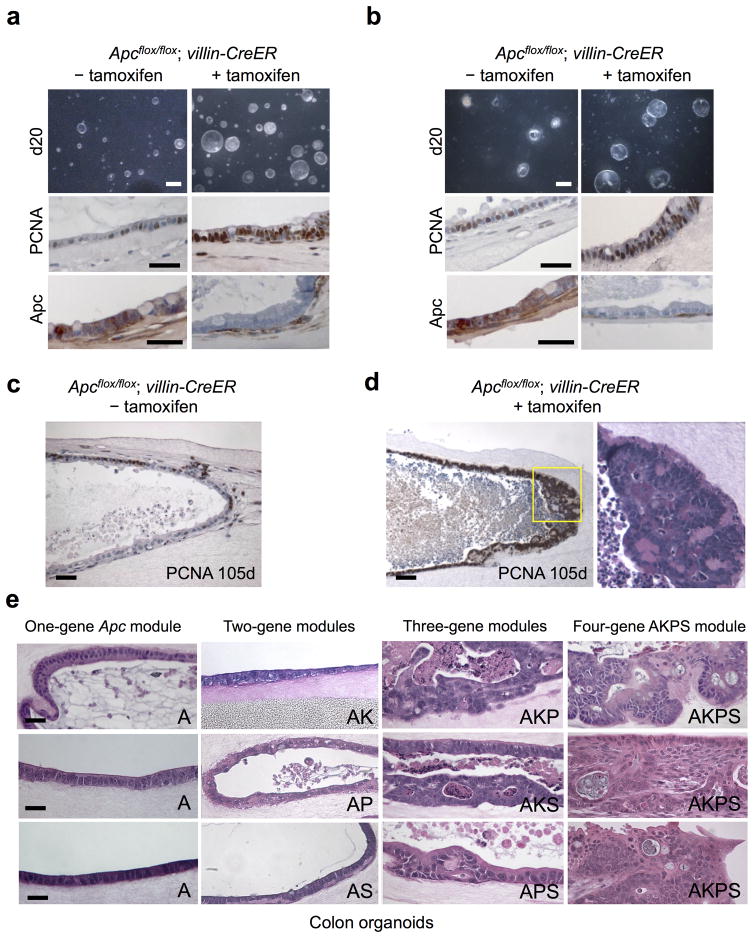
During colorectal cancer (CRC), polyposis is typically initiated by baseline APC mutations, with subsequent accumulation of additional mutations (c.f. KRAS, P53, SMAD4) required to develop metastatic adenocarcinoma. Additional signaling pathways undergoing frequent alteration in CRC include EGFR, BRAF and PI3K16–18. We applied the ability to propagate long-term epithelial/mesenchymal intestinal organoid cultures9 to CRC modeling. Accordingly, small intestine or colon organoids from Apcflox/flox; villin-CreER mice19,20 were treated with tamoxifen in vitro, inducing epithelial-specific Apc deletion and marked hyperproliferation without significant dysplasia or invasion over a 20 day period (Fig. 3a,b). Over extended time points (>100 days), in situ polyposis within the wall of Apc-null small intestinal organoids was observed, with proliferation of closely spaced tubules lined by mildly enlarged, crowded nuclei that recapitulated human tubular adenomatous polyps (Fig. 3c,d).
Figure 3. Systematic evaluation of oncogene modules of increasing complexity in primary colon organoids.
(a,b) In vitro organoid Apc deletion. Primary small intestine (a) or colon (b) organoids from neonatal Apcflox/flox; villin-CreER mice were treated with tamoxifen (2 μM, 7 days) followed by analysis at day 20. Tamoxifen-mediated Apc deletion induced overall growth, proliferative index (middle, PCNA) and APC deletion (bottom, APC IHC). Scale bars, top, 5 mm; middle, 50 μm; bottom, 50 μm. (c,d) In situ polyposis within long-term intestinal organoid cultures. Small intestine organoids from neonatal Apcflox/flox; villin-CreER mice induced without (c) or with (d) tamoxifen followed by culture for 105 days and analysis by PCNA. An in situ tubular adenomatous polyp within the organoid wall in (d) exhibits a polypoid proliferation of PCNA-positive closely spaced tubules lined by enlarged, crowded nuclei. H&E staining of the yellow-boxed region is depicted. Scale bars, 50 μm. (e) one to four oncogene CRC modules were created by infection of tamoxifen-treated Apcflox/flox; villin-CreER adult colon organoids with appropriate combinations of control LMP retrovirus (with GFP cassette) or retroviruses encoding KrasG12D, LMP p53 shRNA/GFP or LMP Smad4 shRNA/GFP followed by H&E staining at day 50 post-infection. The 1-gene Apc (“A”) and 2-gene colon modules (Apc−/−/KrasG12D (AK), Apc−/−/p53 shRNA (AP) or Apc−/−/Smad4 shRNA (AS)) only exhibited minimal dysplasia. High-grade focal dysplasia was exhibited by the 3-gene module Apc−/−/KrasG12D/p53 shRNA (AKP). The four-gene module Apc−/−/KrasG12D/p53 shRNA/Smad4 shRNA (AKPS) induced adenocarcinoma characterized by atypia, confluent sheets of cells, cribriform growth patterns, luminal necrosis, jagged infiltrating growth patterns and frank invasion. Scale bars, 50 μm.
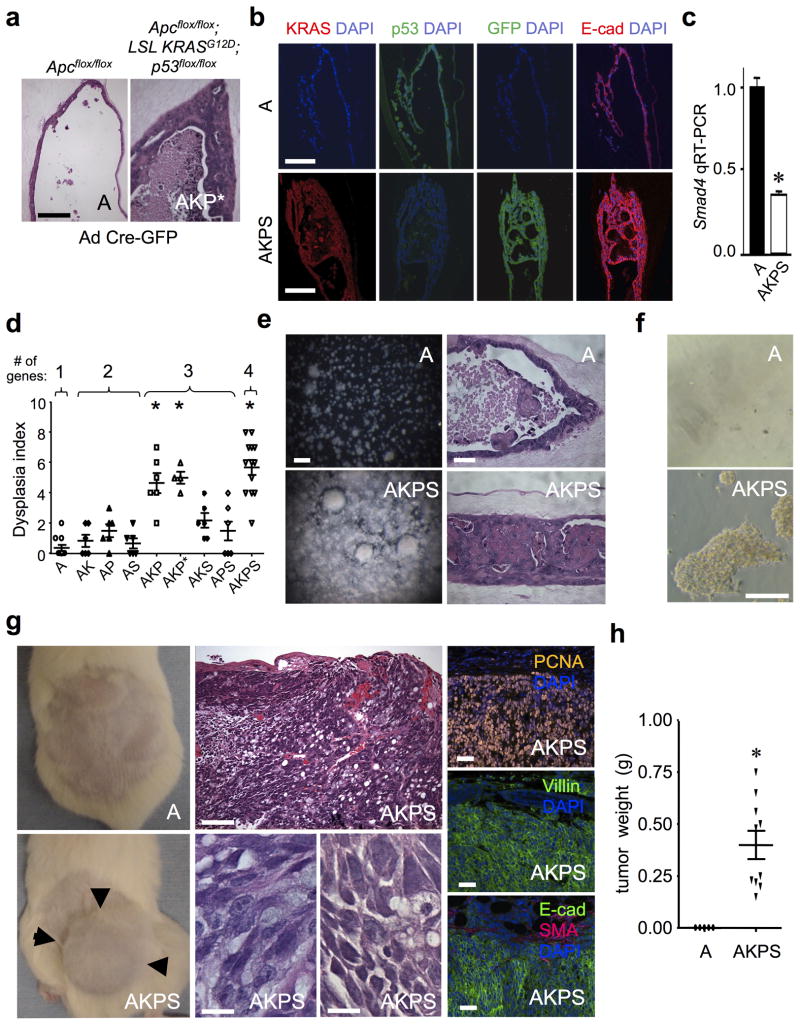
We examined the multi-hit requirement for CRC development in primary colon organoids by systematically comparing one, two, three and four oncogene modules engineered by combinations of CreER-mediated Apc deletion (“A”) and ecotropic retroviruses encoding KrasG12D (“K”), p53 shRNA/GFP (“P”) and/or Smad4 shRNA/GFP (“S”) (Fig. 3e). Adult Apc-null colon organoids (Apcflox/flox; villin-CreER + tamoxifen) are extremely permissive for retroviral or adenoviral infection (Supplementary Fig. 5). Both one-gene Apc-null (A) or two-gene modules Apc/KrasG12D (AK), Apc/p53 shRNA (AP) and Apc/Smad4 shRNA (AS) elicited only minimal dysplasia in colon organoids even after 50 days of culture, predominantly exhibiting a well-organized, stereotyped epithelial monolayer organization (Fig. 3e). However, amongst three-gene modules with different combinations of Apc, KrasG12D, p53 shRNA or Smad4 shRNA, the AKP module was notable for high-grade focal dysplasia with nuclear pleiomorphism and necrosis, either upon retroviral engineering (Fig. 3e) or adenovirus Cre-GFP infection of organoids from Apcflox/flox; lox-stop-lox (LSL) KrasG12D; p53flox/flox mice 21 (AKP*) (Fig. 4a); lesser dysplasia was observed with AKS or APS. The Apc/KrasG12D/p53 shRNA/Smad4 shRNA four-gene module (AKPS) produced an even more severe transformation, ranging from confluent sheets of cells to cribriform growth patterns with luminal necrosis and jagged infiltration typical of human colorectal adenocarcinoma (Fig. 3e and Supplementary Fig. 6).
Figure 4. Serial passage and in vivo transplantation of colon four-gene AKPS organoids.
(a) A four-gene AKP module created by adenovirus Cre-GFP infection of adult Apcflox/flox; LSL KrasG12D; p53flox/flox; organoids (AKP*) exhibited high grade dysplasia and invasion, day 50. Scale bar, 100 μm. (b) Characterization of p53 knockdown, KrasG12D expression, proliferation (PCNA) and epithelial dysplasia (E-Cadherin IF) in adult AKPS organoids. Scale bars, 50 μm. (c) Smad4 knockdown confirmed by FACS sorting and qRT-PCR of EpCAM+/GFP+ cells from AKPS organoids. n=3 experiments, mean +/− SE. * P < 0.05. (d) Dysplasia index from blinded pathologic analysis of 1–4 oncogene module-infected colon organoids at day 50. Each individual data point represents n=4–6 microscopic fields containing viable organoids. The four-gene AKPS module exhibited pronounced dysplasia exceeding one-, two- or four-gene modules (P < 0.05). Mean +/− SE. * = P values: all 1- and 2 gene modules, AKS or APS vs. AKPS, P < 0.0004. AKP or AKP* vs. AKPS, P < 0.13. (e) Serially passaged adult AKPS colon organoids exhibit robust growth (day 17, passage 4, stereomicroscopy, scale bar, 5 mm) and a frequent solid tumor mass morphology (day 10, passage 4, H&E, scale bar, 50 μm) versus cystic Apc-null 1-gene organoids (“A”). (f) Growth and focus formation of Apc-null versus AKPS cells on tissue culture plastic (day 14, passage 4, phase contrast images). Scale bar, 100 μm. (g) AKPS but not A cells demonstrated robust in vivo tumorigenicity following s.c. in vivo transplantation into NOG mice (day 45 post-implantation, 500,000 cells, passage 4). Arrows mark the tumor border. H&E staining demonstrated poorly differentiated adenocarcinoma with epithelial clusters and glands infiltrating the surrounding stroma. Tumor cells with enlarged, hyperchromatic, irregular nuclei, prominent nucleoli and occasional intracytoplasmic mucin are present. Tumors exhibited extensive PCNA-positivity and expressed epithelial markers villin and E-cadherin but not SMA. Scale bars, middle top, 100 μm; middle bottom, 10 μm; right, 50 μm. (h) AKPS versus Apc-null (“A”) tumor weight, day 45 after s.c. implantation, n=10 tumors/genotype. * = P < 0.0001.
Transformed colon AKPS foci represented ~10–20% of the total epithelium, versus ~5% for other genotypes and required ~50 days to manifest. KrasG12D expression, p53/Smad4 knockdown and E-cadherin+ epithelial origin were confirmed (Fig. 4b,c). A dysplasia index incorporating blinded pathologic examination of proliferation, nuclear atypia, invasion and cellular stratification indicated a direct correlation in colon organoids between oncogene module complexity and degree of transformation (Fig. 4d). Thus, colon organoids accurately recapitulate classical multi-hit CRC models requiring multiple mutations for invasive carcinoma16.
In contrast to colon, small intestine organoids exhibited more rapid dysplasia (~ 20 day onset) and stronger transformation sensitivity. High-grade dysplasia was observed even with the small intestine two-gene modules AK and AP and the four-gene AKPS module exhibited histology consistent with invasive adenocarcinoma (Supplementary Figs. 7 and 8), paralleling the well-established increased susceptibility of mouse small intestine versus colon to Apc-mediated tumorigenesis22. This relative promiscuity of small intestinal organoids to transformation indicated that colon organoids represent a more optimal in vitro method to model multi-hit transformation requirements of human CRC.
The transformation of four-gene module (AKPS) colon organoids was further characterized. Upon organoid dissociation and replating, Apc-null organoids (A) serially passaged with retention of cystic morphology, but AKPS organoids exhibited massive expansion frequently as solid tumor masses (Fig. 4e). AKPS but not A organoids grew in 2D on tissue culture plastic and exhibited focus formation (Fig. 4f). Further, AKPS but not A cells exhibited robust in vivo tumorigenicity within 50 days of s.c. transplantation (10/10 versus 0/10 mice, respectively), indicating full oncogenic transformation (Fig. 4g,h). These transplanted tumors expressed the epithelial markers villin and E-cadherin, were highly proliferative, and represented poorly differentiated adenocarcinoma with occasional mucinous features (Fig. 4g).
miR-483 is the predominant oncogene in the 11p15.5 CRC amplicon
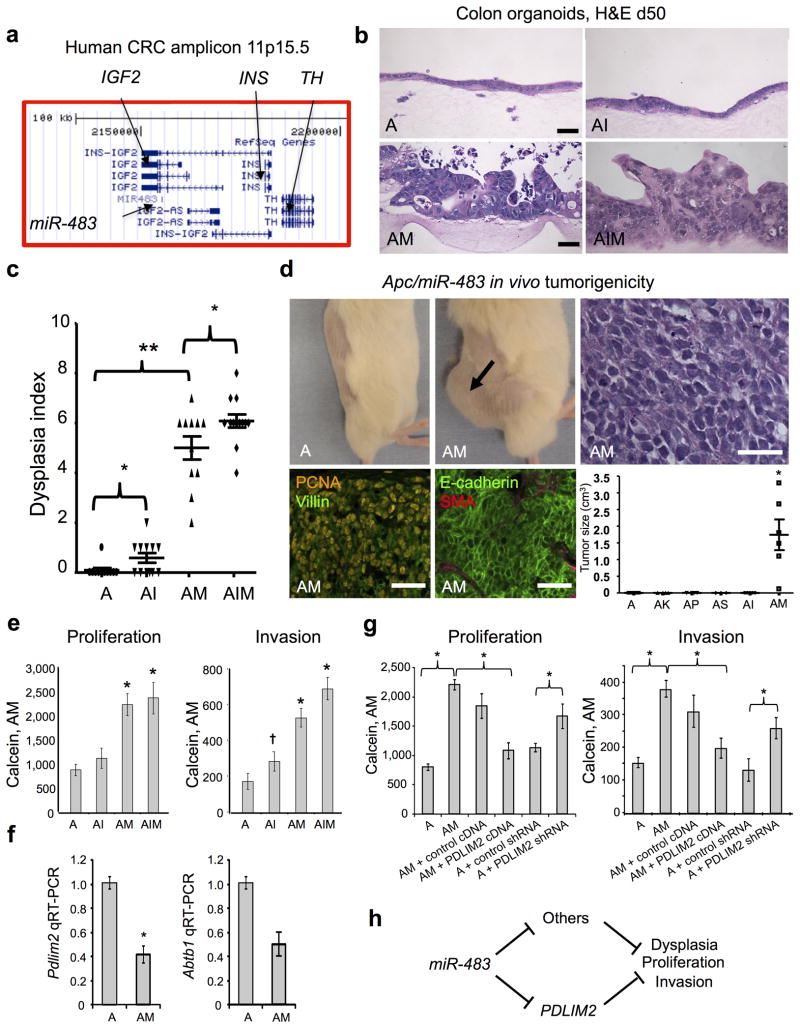
To demonstrate the utility of the primary colon organoid system for oncogene discovery applications, we functionally validated a putative colorectal cancer (CRC) locus from the TCGA CRC survey. The 11p15.5 amplicon containing IGF2, its intronic microRNA miR-483, INS and TH occurs in 7% of CRC TCGA cases12 (Fig. 5a) and is typically co-mutated with Apc but not Kras, p53 or Smad4 (www.cbioportal.org). Since only IGF2 and miR-483 are co-overexpressed from this amplicon12, we performed contextual modeling overexpressing either Igf2, miR-483, both in Apc-null colon organoids. Surprisingly, overexpression of miR-483 (AM) but not Igf2 (AI) elicited high-grade dysplasia of Apc-null organoids with nuclear pleiomorphism and epithelial stratification (Fig. 5b and Supplementary Fig. 9a). Prominent epithelial dysplasia was further confirmed by E-cadherin immunofluorescence and was only present with miR-483 but not Igf2 overexpression (Supplementary Fig. 9b). miR-483 strongly increased the dysplasia index of A organoids (Fig. 5c, A vs. AM, P < 0.001), versus the lack of AK, AP, AS 2-gene module effects on dysplasia index (Fig. 4d) and consistent with particularly robust Apc/miR-483 transforming synergy in organoid culture. Dysplasia was induced, to a lesser degree, by Igf2 overexpression (Fig. 5c, A vs. AI, P < 0.036). Further, Igf2/miR-483 oncogene cooperation was suggested by the dysplasia index of the Apc/Igf2/miR-483 3-gene module (AIM) versus Apc/miR-483 (AM) (Fig. 5c, AM vs. AIM, P = 0.0487). Notably, miR-483 exhibited potent transforming activity in vivo as colon AM organoids robustly gave rise to tumors upon s. c. transplantation, in contrast to two gene modules containing Apc and either IGF2, KrasG12D, p53 shRNA or Smad4 shRNA (AI, AK, AP, AS) from which tumors did not arise by 50 days (Fig. 5d).
Figure 5. Comparative functional validation of miR-483 and Igf2 transforming activity in primary Apc-null colon organoid culture.
(a) Schematic of human 11p15.5 CRC amplicon indicating the position of miR-483 within IGF2 (UCSC Genome Browser). (b) Functional analysis of miR-483, Igf2 or both in Apc-null colon organoids. Tamoxifen-treated Apcflox/flox; villin-CreER adult colon organoids were infected with appropriate combinations of control LMP retrovirus, Igf2 cDNA-IRES-GFP or lentivirus miR-483/GFP to generate Apc-null (A), Apc-null/Igf2 (AI), Apc-null/miR-483 (AM) or Apc-null/miR-483/Igf2 (AIM) organoids. Significant high-grade epithelial dysplasia with nuclear pleiomorphism was only observed with miR-483-overexpressing organoids (AM and AIM) but not A or AI. H&E, day 50 post-infection. Scale bars, 50 μm. (c) Dysplasia index from blinded pathologic analysis reveals marked transformation of miR-483-containing modules (AM, AIM) versus Apc-null or Apc/Igf2 (AI or AIM vs. A, ** = P < 0.0001) versus Igf2 (AI vs. A; AIM vs. AM, * = P < 0.05). Each individual data point represents n=4–6 microscopic fields containing viable organoids. Day 50 post-infection. Mean +/− SE. (d) AM but not A cells demonstrated robust in vivo tumorigenicity following s.c. transplantation into NSG mice (day 60 post-implantation, 500,000 cells, passage 5). H&E staining demonstrated poorly differentiated adenocarcinoma, scale bar, 50 μm. Tumors exhibited PCNA-positive high mitotic index and expressed epithelial markers villin and E-cadherin but not the stromal marker smooth-muscle actin (SMA), scale bars, 50 μm. (bottom, right), AM versus Apc-null (“A”) or other two gene combinations, Tumor weight examined at day 60 post s.c. organoid implantation, n=10 tumors/genotype. * = P < 0.0001. (e) 96-well invasion and proliferation analyses. FACS-sorted EpCAM+ disaggregated cells from day 14 organoids of the indicated genotypes were replated at 10,000 cells/well into 96-well air-liquid interface culture to form secondary organoids. After 14 days of additional culture, proliferation and invasion in A, AI, AM and AIM organoids was determined by Calcein Red-Orange, AM fluorescence signal. N=12 wells/genotype, mean+/−SE. For proliferation * = P < 0.00003 and for invasion * = P < 1 × 10−7, † = P <0.03, all vs. the Apc-null “A” module. (f) MiR-483 represses Pdlim2 and Abtb1 RNA in FACS-sorted EpCAM+/GFP+ cells from Apc-null colon organoids 10 days after infection with lentivirus encoding miR-483 or negative control miR. N=3 experiments, mean +/− SE, * = P<0.05. (g) PDLIM2 rescue experiments. AM vs. A organoids were assayed after 14d of culture in 96-well format for invasion and proliferation with either PDLIM2 cDNA overexpression or Pdlim2 shRNA knockdown. * = P < 0.05. N=12, mean +/− SE. (h) Schematic of mediation of miR-483-induced dysplasia by Pdlim2 and other potential targets.
In the multiplexed proliferation and invasion assay, miR-483 increased invasion of Apc-null cells (P < 1 × 10−7) versus weaker but significant effects from Igf2 (P < 0.03) (Fig. 5e). In addition, miR-483 (P < 0.00003) but not Igf2 significantly increased proliferation of Apc-null organoids (Fig. 5e). In contrast to the dysplasia index, the Igf2/miR-483 combination did not additively stimulate proliferation (P = 0.66) or invasion (P = 0.07) (Fig. 5e, AM vs. AIM).
To identify potential miR-483-repressed targets that could mediate its transforming effects in colon cancer, we identified 122 genes with a false discovery rate (FDR) <0.05 that were repressed in miR-483-amplified TCGA human colorectal cancers by searching for an inverse correlation between gene expression and copy number in 191 matched patients from the COAD/READ datasets12. This list was further restricted to genes containing computationally-predicted miR-483 seed sequences, yielding 11 loci (ABTB1, ACBD4, APBA2, CREBL2, KREMEN2, LTBP4, METRN, PDLIM2/SLIM, PORCN, RUSC1 and ZCWPW1). Filtering further for seed sequence matches that were conserved between mouse and human yielded 2 loci (ABTB1 and PDLIM2) that were selected for investigation (Supplementary Fig. 10a). Lentiviral overexpression of miR-483 repressed Pdlim2 and Abtb1 in colon organoids (Fig. 5g). Further, overexpression of PDLIM2 but not ABTB1 reversed miR-483-induced proliferation and invasion in Apc/miR-483 colon organoids (AM), while shRNA knockdown of Pdlim2 but not Abtb1 was sufficient to induce proliferation and invasion of Apc-null colon organoids (A) (Fig. 5f and Supplementary Fig. 10b,d,e); miR-483 expression strongly repressed mouse and human PDLIM2-3′ UTR-luciferase biosensor constructs (Supplementary Fig. 10c). These data support a model where PDLIM2, encoding a E3 ubiquitin ligase that represses STAT function23, is a physiologically-relevant mediator of miR-483 transforming effects in CRC (Fig. 5h).
DISCUSSION
These studies describe the general application of a single primary organoid culture method without modification to the in vitro modeling of diverse pancreatic, gastric and colon gastrointestinal malignancies. Notably, this air-liquid interface methodology robustly supports organoid growth as epithelial/mesenchymal hybrids without exogenous growth factor supplementation, and for the first time allows in vitro cancer modeling in a more physiologic milieu than previously achieved using transformed cell lines or exclusively epithelial primary cultures. We also extend our previous work9,10 to the successful long-term culture of wild-type pancreatic epithelial/mesenchymal organoids as opposed to epithelium-only5–8,13,14 or short-term embryonic pancreatic organoid cultures24,25.
One decided advantage of this method is the generation of extremely detailed in situ histologic endpoints for dysplasia and transformation in the 3D organoid context. Such profound in vitro histologic changes have not been reported upon oncogene manipulation in previous epithelial-only organotypic modeling of pancreatic or intestinal cancer26,27 or 3D cultures of immortalized breast or lung cell lines26,28. The ability to model and histologically document malignant progression completely in vitro without reliance on in vivo transplantation represents a substantial advance for cancer investigation.
This highly tractable system greatly facilitates the ability to engineer combinatorial oncogene modules in primary cultures through the use of mouse floxed oncogenic alleles superimposed upon efficient transduction with a variety of viral vectors. Combinatorial gene manipulation is highly relevant given the frequency of co-mutated loci in cancer genomes12 and the use of ecotropic retroviruses further affords a safety factor for manipulation of oncogenic loci. For both pancreatic and gastric organoids, pronounced histologic dysplasia was observed with combinatorial Kras and p53 mutations. In colon organoids, progressive transformation was observed with increasing complexity of the introduced gene modules, accurately recapitulating multi-hit models of human CRC development12,16 and culminating with pronounced histologic transformation via the 4-gene AKPS module. In contrast, the promiscuous transformation susceptibility of small intestinal organoids in our studies, even with 2-gene modules, strongly suggests that prior attempts at 3D modeling of CRC in small intestine epithelial organoids27 are (1) less than optimal compared with our present colon-based approaches and (2) are consonant with the well-documented increased susceptibility of the mouse small intestine versus colon to Apc-mediated tumorigenesis in vivo 22. To our knowledge, the AKPS module represents the first in vitro conversion of primary colon tissue to colon adenocarcinoma as judged by multiple criteria including histologic transformation, focus formation and robust in vivo tumorigenicity upon transplantation.
The oncogenic transformation of diverse gastrointestinal organoids to a fully malignant phenotype, as in the current studies, is strongly supported by in vivo tumorigenicity. Transplanted pancreatic and gastric organoids readily generated invasive adenocarcinoma with admixtures of glandular and less differentiated non-glandular areas, consistent with mouse transgenic and iPS-based approaches15,29. The colon AM and AKPS modules exhibited poorly differentiated high-grade tumors, and it will be interesting to assess if more-differentiated colon tumors could arise from 1–3 gene colon modules over extended time points. Since the organoids were in a hybrid background, the tumor transplantation studies occurred in immunodeficient mice, which are likely a permissive setting for tumorigenesis.
The substantial catalogue of genomic aberrations in malignant cells revealed by unbiased genome-scale surveys of cancer12 have highlighted the need for robust methods for large-scale, systematic functional validation of putative cancer driver loci30. Our functional studies with miR-483 provide proof-of-principle for primary organoid validation of putative drivers, with the potential to combine the histologic accuracy of in vivo studies31,32 with the experimental tractability of transformed cell lines33,34 and to complement both of these techniques. The unexpected identification of miR-483 and not Igf2 as the predominant driver within the recurrent CRC 11p15.5 amplicon 12,35 is supported by multiple criteria, including histologic transformation, proliferation and invasion. At the same time, the notion of Igf2 transforming activity is supported by blockade of Igf2-mediated tumorigenesis by a soluble Igfr2 receptor ectodomain36, we have observed modest miR-483/IGF2 cooperativity which could be more profound with higher levels of overexpression.
Prior imprinting and Igf2/Apc cooperation studies have attributed IGF2 locus oncogenicity in CRC to IGF2 itself 37,38 although these studies may be confounded by the location of miR-483 within IGF2 intron 2 (Supplementary Fig. 11 and Supplementary Discussion). MiR-483 and IGF2 are tightly linked via co-overexpression in human CRC12,35 and in 92% of non-hypermutated IGF2-overexpressing TCGA CRC cases (22.4% overall frequency)12 likely reflecting their common transcript-of-origin. MiR-483 knockdown modestly increases the spontaneous apoptosis rate of cultured HCT116 cells and the tumorigenicity of HepG2 cells more effectively than Igf2 knockdown in a liver cancer metastasis model35. However, neither the de novo transforming potential of miR-483 gain-of-function/overexpression nor the comparative oncogenicity of Igf2 versus miR-483 in CRC have been previously demonstrated before our studies. Interestingly, miR-483 induced potent transformation and in vivo tumorigenicity compared with KrasG12D, or p53 or Smad4 knockdown in Apc-null organoids, suggesting particularly strong transforming activity possibly originating in the ability of miRNAs to modulate diverse targets. The increased proliferation and invasion of primary colon organoids upon PDLIM2 shRNA knockdown agrees with observations of decreased tumorigenicity upon PDLIM2 overexpression in CRC cell lines39 and is overall consistent with PDLIM2 tumor suppressor function (Supplementary Discussion). Given the promiscuity of miRNA action, however, numerous other described and yet-undescribed targets likely contribute to miR-483 oncogenicity.
Overall, these studies describe the successful in vitro oncogenic reprogramming of diverse gastrointestinal primary tissues in epithelial/mesenchymal organoids. This single methodology should greatly facilitate cancer modeling and oncogene discovery, and complement existing in vitro cell line, iPS and in vivo screening methodologies29,31–34. The continued utilization of such models incorporating both tumor and stroma for in vitro “cancer engineering” approaches11 could certainly be extended to embryonic stem cell-based epithelial/mesenchymal organoids40 to investigate cancer biology and therapy. It will be further interesting to further exploit the mesenchymal elements of organoids to model stromal regulation of cancer; we have previously documented endogenous paracrine Wnt and Notch signaling in intestinal organoids9. Finally, the general utility of primary organoid approaches to oncogene discovery, as demonstrated herein, may be further generalized to other tissues and to screening approaches for therapeutic agents.
ONLINE METHODS
Mouse Colonies
Apcflox/flox mice which carry conditional alleles with loxP sites flanking exon 14 of Apc were kindly provided by Dr. Bart Williams at Van Andel Research Institute20 and crossed to Villin-CreER mice19 to generate Apcflox/flox; villin-CreER mice. Apcflox/flox and Apcflox/flox; LSL KrasG12D; p53 flox/flox mice were previously described21. LSL KrasG12D; p53 flox/flox mice20. NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) mice were obtained from Taconic Farms, Inc. All animals studies were performed in accordance with the National Institutes of Health guidelines for use and care of live animals and were approved by the Stanford University Institutional Animal Care and Use Committee A3213-01.
Virus Constructs
Retroviral constructs: The mutant G12D-activated Kras allele from LSL KrasG12D (Addgene Plasmid #11585) 27,41 was cloned into pBabe-puro to generate pBabe-puro-KrasG12D. MSCV/LTRmiR30-PIG ΔRI (LMP) expressing p53 shRNA within a miR-30 context and a GFP cassette (p53.1224) and the control empty vector LMP were kind gifts from Ross Dickins and Scott Lowe42. Murine Smad4 shRNAs were obtained from Open Biosystems, Thermo Scientific. Mouse Igf2 cDNA was obtained from OriGene (Plasmid #MG201606). LMP or pBabe-puro-IRES-EGFP was used as the backbone vector for Smad4 shRNA, Igf2 cDNA, or respectively. Lentiviral constructs pCDH-pre-miR-483 and pCDH-CMV-scramble-EF1-copGFP (control scramble miRNA) were from System Biosciences. Human Abtb1 and Pdlim2 cDNA were obtained from the Broad ORFeome library and cloned into pCDH-CMV-MCS -EF1-copGFP. GIPZ-Abtb1 and GIPZ-Pdlim2 mouse shRNAs and GIPZ-control shRNA were obtained from Thermo Scientific.
Retrovirus and lentivirus production and titering
For retrovirus, retroviral plasmids were cotransfected with pCL-Eco (James Chen, Stanford University) into 293T cells by Lipofectamine2000 (Invitrogen). Lentiviral plasmids were cotransfected with pLP1, pLP2 and pLP/VSVG or pMD.ecoEnv (a kind gift from Richard Mulligan and Jeng-Shin Lee, Harvard) into 293T cells by Lipofectamine2000. Retroviral or lentiviral supernatants were collected 48 hrs and 72 hrs post-transfection and concentrated by PEG-it virus solution (System Biosciences). Where appropriate, viruses were titered by infection of NIH3T3 cells and FACS analysis of GFP positive cells 48 hrs post-infection.
Three-dimensional air-liquid interface primary gastrointestinal organoid culture
Pancreas, stomach, colon or small intestine from neonatal or adult mice of appropriate genotypes was dissected lengthwise and washed in cold PBS to remove all luminal contents. We minced a 0.5–1 cm segment per dish extensively on ice and embedded the minced tissues in a 3D collagen gel using a double-dish culture system as previously described 9. Where appropriate, primary organoids from Apcflox/flox; villin-CreER mice were induced with tamoxifen in the medium for 7 days (Sigma, 2 μM in ethanol) on the day of initial plating into air-liquid interface culture. Fresh media (Ham’s F12, 20% FCS, gentamicin 50 μg ml−1) was applied every week. In general, growth of wild-type organoids was most optimal with neonatal tissue. For colon, Cre activation elicited vigorous growth of adult Apcflox/flox organoids with or without latent alleles of Kras or p53. Colon, pancreatic and gastric organoids with single mutation of K or P required neonatal tissue, although adult KP organoids from these tissues grew robustly. Gender did not affect the efficacy of culture. Oncogene-containing organoids were passaged every 20–50 days by dissociation with 200 unit ml−1 collagenase IV (Worthington) at 37 °C for 30 min, followed by 3 × 5 min wash with 100% FBS and replating at 1:4 split into new air-liquid interface collagen gels.
Viral infection of organoids
For adenoviral infection of pancreatic, gastric or colon organoids, air-liquid interface cultures were plated, and adenovirus Cre, Cre-GFP or Fc (108 pfu) in 500 μl culture medium was directly added to the top of the inner dish; organoids containing various combinations of LSL KrasG12D, p53 flox/flox or Apcflox/flox were infected in this manner. For lentiviral or retroviral infection of oncogene-containing pancreatic, gastric or colon organoids (K, P, KP, A), infection was carried out at the time of serial passage to maximize epithelial exposure to virus. Accordingly, for retrovirus/lentivirus infection of these “secondary” organoids, primary organoids at 14–20 d of growth were recovered from collagen gel by collagenase IV (Worthington) incubation followed by 0.05% trypsin/EDTA incubation to dissociate organoids into a single cell suspension. Following extensive washing with 10% FBS to inactivate collagenase/trypsin, cells were pelleted by centrifugation and incubated with retroviral particles encoding GFP, KrasG12D, p53 shRNA, Smad4 shRNA. Igf2, or lentiviral particles of pCDH-pre-hsa-miR-483-EF1-copGFP, pCDH-CMV-scramble-EF1-copGFP, pCDH-CMV-Abtb1-EF1-copGFP, pCDH-CMV-Pdlim2-EF1-copGFP, GIPZ-Abtb1 shRNA, GIPZ-Pdlim2 shRNA and GIPZ-control shRNA (moi 50 particles/cell) in the presence of growth medium and TransDux (System Biosciences) at 37 C for 30 min before serial replating into 3D collagen gel air-liquid interface culture. In general, wild-type pancreatic, gastric or colon organoids were infected robustly by adenovirus but inefficiently with lentivirus or retrovirus using these methods. Alternatively, for retrovirus infection by microinjection, retroviruses encoding KrasG12D, p53 shRNA and/or Smad4 shRNA (1 μl of 108 pfu ml−1) virus per organoid, depending on the size of particular organoid) were microinjected into the primary intestinal organoids at day 7 post-plating using an MPPI-2 pressure injector and glass-pulled capillaries (World Precision Instruments). Equivalent infection efficiency was obtained by either method.
Immunohistochemistry and immunofluorescence staining
Organoids were fixed with 4% paraformaldehyde overnight, paraffin-embedded and sectioned (4–5 μM) as previously described9. Sections were deparaffinized and stained with H&E for initial histology analysis. For further immunohistochemistry analysis, we used antibodies to the following proteins: PCNA (1:300; Invitrogen,133940), APC (1:100; Abcam), E-cadherin (1:300; BD Biosciences Pharmingen, 610182), GFP (1:100; Abcam, ab290), p53 (1:100; Santa Cruz, sc-1311), Kras (1: 100; Abcam, ab16907), Villin (1:100; NeoMarkers, MS-1499-P1), SMA (1:200; Abcam, ab5694), Pdx1 (1:100; Abcam, ab47267), Amylase (1:100; Santa Cruz, sc-31869), Somatostatin (1:200; Abcam, ab103790), Insulin (1:100; Abcam, ab63820), Glucagon (1:100; Cell Signaling, #2760), PanCK (1:200; Sigma, C2931), CK19 (1:200; Abcam, ab52625). Secondary antibodies used were: Cy3- or Dylight488-conjugated Streptavidin (1:500; Jackson ImmunoResearch Laboratories), Alexa Fluor 488 goat anti-mouse (1:1,000; Invitrogen), Alexa Fluor 488 goat anti-rabbit (1:1,000; Invitrogen), FITC-conjugated Affinipure Donkey anti-goat IgG (1:500; Jackson ImmunoResearch Laboratories)
Organoid disaggregation and FACS
Organoids were recovered and dissociated from collagen gel by collagenase IV incubation followed by 0.05% trypsin/EDTA incubation. Following extensive washing with 100% FBS, cells were filtered with 40 μM cell strainers (BD Falcon) and pelleted by centrifugation at 200 × g. Pellets were resuspended with FACS staining solution (5% FCS in PBS) and incubated with APC conjugated anti-mouse EpCAM (1:300; eBioscience) for 45 min at 4 C. Stringent wash was applied using ice cold PBS and propidium iodide (1:300; Boehringer Mannheim Corp) was used to mark dead cells followed by isolation of EpCAM+ GFP+ virus-infected epithelium using an Aria II cell sorter (BD).
Quantitative real-time PCR
For miR-483 expression, EpCAM+ GFP+ virus-infected epithelium was sorted into Ham’s F-12/20% FCS, pelleted and processed and analyzed using the TaqMan MicroRNA Cells-to CT kit (Ambion). Looped RT primer-based TaqMan was used to detect processed hsa-miR-483-5p and expression was normalized to that of the control miRNA Rnu6b; primers were obtained from Applied Biosystems and used following the manufacturer’s instructions. For all other qPCR assays, EpCAM+ GFP+ virus-infected epithelium was sorted into RLT buffer (Qiagen) and determined using the SYBR Green Quantitect PCR Kit (Qiagen) normalized to Gapdh. Assays were performed in triplicate and results from at least three independent experiments are presented. mSmad4 primers: 5′-GTTAGTGAAGGATGAGTACG-3′, 5′-CACAGGAATGTTGGGGAAGT-3′. mIgf2 primers: 5′-GTACCAATGGGGATCCCAGT-3′, 5′-TGAAGTAGAAGCCGCGGTCC-3′. mGapdh: 5′-ACTTGAAGGGTGGAGCCAAA-3′ 5′-TTCCACAATGCCAAAGTTGTCA-3′. mAbtb1: 5′- GAGTTGGCCTACGATGTTCTGAG -3′ and 5′-CTTCTCTATGACTTTGGCCAT -3′. mPdlim2: 5′-TCCCTGAGGACACACCGTGA -3′, 5′-CTACAGCCA ATCATATCCTC -3′
Proliferation and migration assay
Primary organoids of the appropriate genotypes were recovered from collagen gel by collagenase IV digestion yielding a single cell suspension as described above at 10 days following viral infection. The cell suspension was FACS sorted (Aria II, BD) for EpCAM+ epithelium (BD Biosciences) and reseeded to collagen in 96-well air-liquid interface culture (BD FluoroBlok) at 10,000 cells per well, n=12 wells/genotype. Calcein red-orange, AM (Invitrogen, Molecular Probes) was used to stain cells at day 14 following replating (following the manufacturer’s instruction). The plate was analyzed by Flexstation II 384 using 577 nm excitation and 590 nm emission filters, respectively with the sum of top and bottom reads to quantitate proliferation and the bottom read only for invasion.
Primary organoid transplantation
For colon, AKPS vs. Apc-null organoids from passage 4 or 2-gene modules versus Apc-null from passage 6 were recovered from air-liquid interface collagen gels by disaggregation with collagenase VI and collected as described above. For pancreas and stomach, cells from passage 3 day 14 K, P, and KP organoids were collected. Dissociated cells were pelleted by centrifugation and resuspended with Matrigel (50% Matrigel (BD), 10% FCS, 40% F12, in a total volume of 100 μl containing 500,000 cells) and injected s.c. into flanks of NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) mice.
Dysplasia index
Histology scores were determined blindly based on levels of 4 histological characteristics. Nuclear grade (0 = minimal nuclear changes; 1 = mild to moderate nuclear enlargement with irregularities, variably prominent nucleoli; 2 = enlarged nuclei with diffuse membrane irregularities, and prominent nucleoli ); Stratification ( 0 = 2–3 cells; 1 = 4–5 cells; 2 = >5 cells ); Mitoses ( 0 = absent; 1 = scattered; 2 = numerous ); Invasion ( 0 = absent; 1 = focal (1–2 foci); 2 = prominent >2 foci). The sum of values from these 4 categories was used as the final dysplasia index score for each sample. Each data point indicates a completely independent organoid culture. The dysplasia index was evaluated by all microscopic fields containing viable organoids, 4–6 fields per sample (N).
Bioinformatics analysis and prediction of miR-483 targets
Colorectal patient data from The Cancer Genome Atlas (TCGA) COAD and READ datasets were analyzed to identify potential miR-483 targets. Gene expression and copy number were extracted for 191 matched patients. Genes with a significant inverse correlation between the gene expression and copy number of miR-483 were filtered, resulting in 122 genes with FDR <0.05. MicroCosm version 5 was applied to computationally predict targets of miR-483 43. MicroCosm lists 1552 genes for the 3p and 5p version of the microRNA. Eleven genes overlap between both analyses. These genes have a significant inverse correlation with the copy number of mir-483 and are computationally predicted targets of mir-483. Further filtration was applied by identifying seed sequence matches that were conserved between mouse and human (MicroCosm).
Biosensor assays
The 3′UTR of mus Pdlim2 (~1kb), hsa Pdlim2 (~1kb) and corresponding mus Pdlim2* and hsa Pdlims2* with deletions of miR-483 binding site were synthesized by IDT and cloned downstream of a Renilla luciferase reporter vector pRL-tk (Promega). 293T cells in 96-well plates were transfected with 1 ng/well of firefly luciferase, 0.2 ng/well of 3′UTR Renilla reporter construct, and either 0, 10 or 50 ng/well of miR-483 expression vector pCDH-pre-hsa-miR-483-EF1-copGFP. 50, 40 or 0 ng/well empty vector pCDNA3.1 was added to provide a total of 51.2 ng of DNA per transfection. 72 hrs after transfection, Renilla/firefly luciferase was measured using the Dual Reporter Luciferase Kit (Promega).
Statistical analysis
P-values were determined using a two-tailed Student’s t-test assuming unequal variances. A P-value of 0.05 was considered significant.
Supplementary Material
Acknowledgments
We are grateful to members of the Kuo laboratory, M. Amieva, R. Honaker, T. Jacks, C. Chartier and for helpful discussion, to P. Chu and S. Michie for assistance with histology, to S. Lowe and R. Dickins for LMP-based retroviruses, and B. Williams for mouse strains, and R. Mulligan and J. Chen for lentiviral and retroviral reagents. X. L. and J.-T. N. were supported by the Dean’s Fellowship at Stanford University, A. O. by a Physician-Scientist Fellowship from the California Institute for Regenerative Medicine, J.-T. N. by an American Cancer Society Postdoctoral Fellowship, C. W-M. C. by the Croucher Foundation and T. Y. by the Royal College of Surgeons. L. N. was supported by an American Society for Clinical Oncology Young Investigator Award and U.S. National Institutes of Health grant K08CA166512. C-Z. C. was supported by grants from U.S. National Institutes of Health (1R01AI073724 and Pioneer Award DP1CA174421) and the W. M. Keck Foundation. We thank the generous support from the Ranzetta, Wang and Krishnan Family Gift Funds to C. J. K. This work was also supported by U.S. National Institutes of Health grants K08DK078033 and U01CA084301 to K. E. H.; U.S. National Cancer Institute Integrative Cancer Biology Program grant U01CA151920, U.S. National Cancer Institute Cancer Target Discovery and Development grant 1U01CA168424 and a Fidelity Foundation grant to C. J. K. and H. P. J; and U.S. National Institutes of Health Transformative R01DK085720 and U.S. National Institute of Diabetes and Digestive and Kidney Diseases Intestinal Stem Cell Consortium grant U01DK085527 to C. J. K.
Footnotes
AUTHOR CONTRIBUTIONS
X. L., L. N., A. O., D. C. C., M. A. C., P. G. R. and J.-T. N. designed and executed organoid experiments. R. K. P. performed blinded histologic interpretation. O. G. and X. G. performed bioinformatic analyses. C. W-M C., T. Y., J. Y. and J. W. performed organoid experiments and produced viruses. S. R., L. D. A. and K. E. H. provided reagents. K. E. H, C-Z. C., S. K. P., H. P. J. and C. J. K. designed experiments. X. L., L. N., D. C. C. and C. J. K wrote the manuscript.
References
- 1.Hynds RE, Giangreco A. Concise review: the relevance of human stem cell-derived organoid models for epithelial translational medicine. Stem Cells. 2013;31:417–422. doi: 10.1002/stem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 3.Stelzner M, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Pylayeva-Gupta Y, Lee KE, Bar-Sagi D. Microdissection and culture of murine pancreatic ductal epithelial cells. Methods Mol Biol. 2013;980:267–279. doi: 10.1007/978-1-62703-287-2_14. [DOI] [PubMed] [Google Scholar]
- 6.Jin L, et al. Colony-forming cells in the adult mouse pancreas are expandable in Matrigel and form endocrine/acinar colonies in laminin hydrogel. Proc Natl Acad Sci U S A. 2013;110:3907–3912. doi: 10.1073/pnas.1301889110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smukler SR, et al. The adult mouse and human pancreas contain rare multipotent stem cells that express insulin. Cell Stem Cell. 2011;8:281–293. doi: 10.1016/j.stem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. Embo J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katano T, et al. Establishment of a long-term three-dimensional primary culture of mouse glandular stomach epithelial cells within the stem cell niche. Biochem Biophys Res Commun. 2013;432:558–563. doi: 10.1016/j.bbrc.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 11.Ghajar CM, Bissell MJ. Tumor engineering: the other face of tissue engineering. Tissue Eng Part A. 2010;16:2153–2156. doi: 10.1089/ten.tea.2010.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wescott MP, et al. Pancreatic ductal morphogenesis and the Pdx1 homeodomain transcription factor. Mol Biol Cell. 2009;20:4838–4844. doi: 10.1091/mbc.E09-03-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rovira M, et al. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 17.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sansom OJ, et al. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci U S A. 2006;103:14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 20.Shibata H, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 21.Hung KE, et al. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc Natl Acad Sci U S A. 2010;107:1565–1570. doi: 10.1073/pnas.0908682107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taketo MM, Edelmann W. Mouse models of colon cancer. Gastroenterology. 2009;136:780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Attali M, et al. Control of beta-cell differentiation by the pancreatic mesenchyme. Diabetes. 2007;56:1248–1258. doi: 10.2337/db06-1307. [DOI] [PubMed] [Google Scholar]
- 25.Duvillie B, Heinis M, Stetsyuk V. In vivo and in vitro techniques to study pancreas development and islet cell function. Endocr Dev. 2007;12:46–54. doi: 10.1159/000109604. [DOI] [PubMed] [Google Scholar]
- 26.Lee KE, Bar-Sagi D. Oncogenic KRas suppresses inflammation-associated senescence of pancreatic ductal cells. Cancer Cell. 2010;18:448–458. doi: 10.1016/j.ccr.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onuma K, et al. Genetic reconstitution of tumorigenesis in primary intestinal cells. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1221926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung CT, Brugge JS. Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature. 2012;482:410–413. doi: 10.1038/nature10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3:2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.March HN, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43:1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veronese A, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 2010;70:3140–3149. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harper J, et al. Soluble IGF2 receptor rescues Apc(Min/+) intestinal adenoma progression induced by Igf2 loss of imprinting. Cancer Res. 2006;66:1940–1948. doi: 10.1158/0008-5472.CAN-05-2036. [DOI] [PubMed] [Google Scholar]
- 37.Timp W, Levchenko A, Feinberg AP. A new link between epigenetic progenitor lesions in cancer and the dynamics of signal transduction. Cell Cycle. 2009;8:383–390. doi: 10.4161/cc.8.3.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan AB, Howell JA. Insulin-like growth factor II supply modifies growth of intestinal adenoma in Apc(Min/+) mice. Cancer Res. 2000;60:1070–1076. [PubMed] [Google Scholar]
- 39.Qu Z, et al. DNA methylation-dependent repression of PDZ-LIM domain-containing protein 2 in colon cancer and its role as a potential therapeutic target. Cancer Res. 2010;70:1766–1772. doi: 10.1158/0008-5472.CAN-09-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasai K, et al. Oncogene-mediated human lung epithelial cell transformation produces adenocarcinoma phenotypes in vivo. Cancer Res. 2011;71:2541–2549. doi: 10.1158/0008-5472.CAN-10-2221. [DOI] [PubMed] [Google Scholar]
- 42.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.