Highlights
-
•
Waterlogging adversely affects sugarcane productivity and quality.
-
•
A subtractive cDNA library was prepared from sugarcane leaf tissue and sequenced to generate ESTs.
-
•
EST sequences were used to identify transcripts induced by waterlogging.
-
•
The sequenced clones were classified by predicted functions and stress-related genes formed the largest class.
-
•
EST sequences were also used to identify putative novel microRNAs and their targets.
Abbreviations: ABA, abscisic acid; ADF, actin depolymerizing factor; ESTs, expressed sequence tags; Hsps, heat shock proteins; PTGS, post-transcriptional gene silencing; REMs, remorins; SSA, snow pea pod senescence-associated; SUCEST, sugarcane EST database; TGS, transcriptional gene silencing
Keywords: EST, Novel miRNAs, Sugarcane, Suppression subtractive hybridization, Waterlogging stress
Abstract
Sugarcane is an important tropical cash crop meeting 75% of world sugar demand and it is fast becoming an energy crop for the production of bio-fuel ethanol. A considerable area under sugarcane is prone to waterlogging which adversely affects both cane productivity and quality. In an effort to elucidate the genes underlying plant responses to waterlogging, a subtractive cDNA library was prepared from leaf tissue. cDNA clones were sequenced and annotated for their putative functions. Major groups of ESTs were related to stress (15%), catalytic activity (13%), cell growth (10%) and transport related proteins (6%). A few stress-related genes were identified, including senescence-associated protein, dehydration-responsive family protein, and heat shock cognate 70 kDa protein. A bioinformatics search was carried out to discover novel microRNAs (miRNAs) that can be regulated in sugarcane plants subjected to waterlogging stress. Taking advantage of the presence of miRNA precursors in the related sorghum genome, seven candidate mature miRNAs were identified in sugarcane. The application of subtraction technology allowed the identification of differentially expressed sequences and novel miRNAs in sugarcane under waterlogging stress. The comparative global transcript profiling in sugarcane plants undertaken in this study suggests that proteins associated with stress response, signal transduction, metabolic activity and ion transport play important role in conferring waterlogging tolerance in sugarcane.
1. Introduction
Sugarcane, belonging to the genus Saccharum (Poaceae), is an important industrial crop accounting for nearly 75% of sugar produced worldwide [1]. Being a round the year crop, it has to face a wide range of vagaries of nature, specially biotic and abiotic stresses [2]. Incessant rains during the monsoon cause flooding and waterlogging, especially in low lying areas near the rivers. This adversely affects cane productivity and maturity. Thus, a major portion of the sugarcane production is lost, causing heavy financial loss to the farmers and millers. In sugarcane cultivation, among abiotic stresses, waterlogging is an acute problem particularly where surface drainage facilities are not adequate [3]. The extent of waterlogging injury to the plant is dependent on several factors, such as flooding depth, duration and flow of water in the field [4,5]. The need for improved productivity, coupled with limited resources for environmental modification and the availability of only a finite amount of land for expansion, points to genetic improvement as the major means to augment sugarcane yield. Understanding the physiological and molecular basis of stress tolerance/resistance is crucial in improving the performance of a crop especially in stressful environment [6].
Considerable progress has been made in understanding the molecular, biochemical and physiological basis of traits that enable plants to cope with individual stresses. The molecular improvement approach holds promise in being able to selectively incorporate specific traits into superior cultivars in a fraction of the time and reducing the effort required to achieve similar results through conventional breeding [7,8]. Genome sequencing and expressed sequence tags (ESTs) development has great potential in facilitating crop improvement for stress resistance. It is becoming realistic, for example, to combine large amounts of DNA sequence data with high-throughput molecular biology methods to identify genes that are differentially expressed under specific environmental conditions [8,9]. EST collections may be useful as direct sources of genes, but their major advantage is in allowing comparisons of expression pattern to be made between target tissues. Such evaluations have been made highly efficient by the application of array screening technologies and these have been gainfully employed in sugarcane, using both macroarray and microarray systems [10,11]. Although there is no real substitute for a complete genome sequence, cost-benefit analysis has proved that ESTs are worth their cost and form the core foundations for various genome scale experiments within the plant genomes like sugarcane that are tough to assemble due to their complexity [12]. A large amount of sugarcane EST resource has already been developed and submitted to GenBank that has been providing a very valuable means for gene discovery [13,14]. A drought-inducible gene, designated as SoDip22, has been identified in the leaves of sugarcane, using this approach [15]. Data mining of the SUCEST database with cold inducible genes from other plant species identified 33 assembled sequences that were homologous [16]. Real-time reverse transcription-PCR profiling of selected EST clusters identified several sugarcane clusters that show differential expression in response to red rot infection and drought stress conditions in sugarcane [17].
Small 20–24 nucleotides (nt) non-coding RNAs are important regulators of gene expression, causing either transcriptional gene silencing (TGS) or post-transcriptional gene silencing (PTGS) [18,19]. Several distinct size classes of small RNAs with dedicated functions have been identified, including miRNAs [20]. Deep sequencing and bioinformatics strategies offer a powerful means for quantitative and qualitative profiling of small RNA populations and it is convenient for exploring small RNAs in plant species such as sugarcane [21–23]. EST analysis has proven to be an economically feasible alternative for miRNAs discovery in species lacking a draft genome sequence [24]. We used EST analysis and DNA database analysis in detail to identify new potential miRNA genes and their targets. Since the ESTs and expressed genes come from the production of actual gene expression, our investigation provides more support and confidence in the discovery of new potential miRNAs and their targeted genes.
The information related to gene expression in sugarcane under waterlogging stress is still patchy, so the present study was undertaken to generate ESTs induced under waterlogging stress and to have a glimpse of genes expressed under such stress. In this study, we also used a bioinformatics search based on sequence similarity or the secondary structure of precursors, and have identified seven novel miRNAs expressed in water-logged sugarcane.
2. Materials and methods
2.1. Plant material
In an effort to elucidate the genes underlying plant responses to inundation, a waterlogging tolerant popular sugarcane variety, BO 91, suitable for the North Central zone in India, was grown at Indian Institute of Sugarcane Research experimental farm, under stress (in lowland area) and normal condition for preparation of subtractive cDNA library. This farm has both lowland and upland areas. This lowland area gets the runoff water from the upland and remains waterlogged during the monsoon. About 30–35 cm water level was maintained for one month in the lowland area in order to maintain the waterlogging stress.
2.2. Total and poly (A)+ RNA isolation
Sugarcane leaf tissues were collected from six month old crop at 45 days after water inundation and from the plants grown in normal condition of same age. Sampling at 45 days was decided arbitrarily to provide ample time for the resistant genotype to express the genes for water logging tolerance, and the adverse effect of the stress is pronounced and responses of the tolerant plant more visible. The sampled tissues were rinsed thoroughly with ice cold DEPC treated autoclaved water to prevent the RNA from degradation. After washing, the samples were quickly frozen in liquid nitrogen and transferred to −80 °C deep freezer for further use. Total RNA was isolated by the modified protocol using guanidine thiocyanate-phenol [25]. Approximately five-gram sugarcane tissues were grounded to fine powder in liquid nitrogen and RNA was extracted by using GTC salt buffer. RNA was selectively precipitated from the pool of nucleic acid using lithium chloride salt. Poly (A)+ RNA was isolated from 200 to 500 μg of total RNA using Oligo (dT)-cellulose through affinity chromatography. The 1% denaturing agarose gel electrophoresis was carried out to check the quality, integrity and size distribution of the RNA, using a submarine horizontal agarose slab gel. mRNA quantification was also performed by UV absorbance at A260 and A280.
2.3. Subtractive library construction
The Clontech PCR-Select cDNA Subtraction kit was used for preparation of subtractive cDNA library to identify differentially expressed transcripts in leaf of water-logged sugarcane. Double-stranded cDNA synthesis was carried out on poly (A)+ mRNA derived from the leaf tissues of waterlogged stressed (tester) and normal (driver) plants. The tester and driver cDNAs were then digested with RsaI, which yields blunt end fragments of approximately 400 bp length on an average. Both, tester and driver, cDNA populations were processed following the manufacturer’s instructions, with some modifications. The tester cDNA was aliquoted into two halves, and each halve was ligated with different cDNA adaptors. Adapter ligation was followed by two rounds of hybridization with an excess of driver cDNA as per manufacturer’s protocol. The resultant products were subjected to two cycles of PCR with adaptor targeted primers to amplify the desired differentially expressed sequences. Amplifications were performed on a MyCycler (BioRad, USA). First PCR master mix contained 10x PCR reaction buffer, 0.2 mM dNTPs, 0.4 μM PCR primer 1 and Advantage cDNA polymerase (Clontech, USA). PCR was performed under the following conditions: 94 °C (25 sec) denaturing step followed by 30 cycles each consisting of a denaturation step at 94 °C (10 sec), an annealing step at 66 °C (30 sec) and an elongation step at 72 °C (1.5 min). Before using the primary PCR products as templates for secondary PCR, these were diluted 10 fold with sterile water. The second PCR master mix contained 10x PCR reaction buffer, 0.2 mM dNTPs, 0.4 μM nested PCR primer 1, 0.4 μM nested PCR primer 2 and Advantage cDNA polymerase. PCR was run through 20 cycles each consisting of a denaturing step at 94 °C (15 sec), an annealing step at 66° (30 sec) and an elongation step at 72 °C (1.5 min). cDNA molecules were size-selected and fractions larger than 250 bp were cloned non-directionally into the pGEM-T-Easy Vector (Promega, USA). The ligated products, i.e., pGEM-T Easy vector containing insert fragments, were transformed into Escherichia coli cells by using electroporation apparatus (Pulse Controller, BioRad, USA). Commercially available electro-competent DH10B cells of E. coli (GibcoBRL Life Technologies, USA) were used as the hosts.
2.4. Template preparation
cDNA libraries were plated onto solid Luria Bertani (LB) medium with ampicillin, X-GAL and IPTG to select recombinant clones. White colonies were randomly picked to 96 well plates containing Freezing Medium (36 mM K2HPO4, 13.2 mM KH2PO4, 1.7 mM sodium citrate, 0.4 mM MgSO4, 6.8 mM (NH4)2 SO4, in LB medium and 4.4% glycerol), grown overnight and later stored at −80 °C. Recombinant plasmids were isolated using QIAprep Turbo Miniprep Kit (QIAGEN, Germany) by employing Genesis Workstation 2000 (TECAN, Switzerland) robotic system as recommended by the supplier.
2.5. Sequencing: sequence analysis and identification of novel miRNAs and their target prediction
Recombinant plasmids were single-pass sequenced from the 5′ end. Sequencing reactions were performed with 200 ng of plasmid DNA using the ABI Prism BigDye Terminator Sequencing Kit (Applied Biosystems, USA) and analyzed on ABI3700 automated DNA analyzer (Applied Biosystems, USA). Primary processing of the EST sequences was carried out with the basecaller PHRED [26], for base calling and quality trimming. Screening of vector/adapter sequences was carried out with the help of Cross_match (www.phrap.org) software. EST sequences having PHRED score >20 and longer than 299 bp were submitted to the GenBank. However, for the purpose of clustering, size exclusion was not done. Resulting high-quality cDNA sequences were assembled into contigs using Sequencher v4.6 program (Gene Codes, Ann Arbor, Mich, USA) with default parameters. Sequence similarity searches were performed against NCBI non-redundant database (www.ncbi.nlm.nih.gov/BLAST/) using BLAST internet tool keeping all the default parameters. The top scoring hits were annotated for Gene Ontology (GO) annotation according to the putative function returned by BLASTX.
EST sequences were subjected to stem-loop structure prediction using the Mfold web server (http://mfold.rna.albany.edu/?q=mfold). Predictions were made using RNA sequences containing 50–200 nucleotides on either side of the candidate miRNA. In case no apparent local foldback structure was predicted for a given sequence, larger upstream and downstream sequences were submitted for Mfold analysis. The criteria used to identify candidate-structured precursors were those suggested by Allen et al, [27]. Target genes of the miRNAs were predicted using the online tool psRNATarget, a small RNA target analysis server for plants (http://plantgrn.noble.org/psRNATarget/). This tool uses an iterative parallel Smith-Waterman algorithm and a weighted scoring scheme in which mismatched bases are penalized according to their type and location in the alignment. Mismatches to the 5′ and central regions of the miRNA were preferentially penalized compared with mismatches to the 3′ region of the miRNA.
3. Results and discussion
3.1. Waterlogging induced morphological and structural modifications in sugarcane
Sugarcane genotype BO 91 was used to prepare a subtractive library to study the genes expressed in sugarcane under waterlogged condition. The selected sugarcane genotype, BO 91, is well adapted to areas prone to waterlogging and possesses good tolerance and growth in water-logged condition [3,28]. Development of aerial roots and profuse rooting was observed under stressed condition but not in normal condition of cultivation (Fig. S1). Various morphological attributes were recorded in sugarcane variety BO 91 growing in water-logged and normal condition. Mean value of all the growth and quality parameters are furnished in Table S1. Many unfavourable deviations were observed in most of the morphological traits when the cane was grown under waterlogging condition. Considerable increase was observed in number of nodes (60.0%); internode length (17.5%) and nodes with aerial root (419.5%) – the characters supposedly conferring tolerance to waterlogging stress. Under longer duration of inundation, some morphological, anatomical, physiological and biochemical changes take place in the plant for sake of adaptation and/or survival [29]. Waterlogged sugarcane plants, like those of many species, undergo structural changes leading to cell lysis and the formation of aerial roots.
Development of adventitious roots in response to waterlogging is considered to be a tolerance mechanism to increase root aeration that allows the plant to maintain root functions during flooding and to avoid O2 deficiency [30]. These above ground roots tend to grow horizontally to remain near the water-air interface. Sugarcane root system developed a dense mat of aero-tropic, small diameter roots when flooded. Significant increase in aerial root formation was also observed in sugarcane subjected to longer duration of waterlogging [4].
Increase in internode length and decrease in cane diameter was observed when the cane was subjected to longer duration of inundation. In many cases, it is this characteristic of the shoot that contributes most strongly in securing oxygen when the usual supply-route is blocked by flooding or submergence. Achieving unusually fast rate of extension to make contact with a source of oxygen, and also light and carbon dioxide is a major feature contributing to survival in standing water stress conditions [4,5].
3.2. Construction of waterlogging-specific subtractive cDNA library
Different gene expression analyses have provided valuable insights into metabolic perturbations and readjustments in plants associated with different abiotic stresses such as salt, low/high temperatures and drought [31,32]. ESTs generated from cDNA libraries should ideally represent all the genes expressed in a target organ/tissue, at a specific developmental stage and/or in a specific environment. However, variation in expression level of different genes in a particular tissue results in mRNAs that differ in abundance, which makes it difficult to capture the mRNAs of low abundance, in cDNA libraries. Redundant sequencing of clones representing the same expressed genes is another problem that lays burden on the efficiency and cost effectiveness of EST sequencing [33,34] which hinders research laboratories with small budgets to perform EST characterization studies. To overcome this problem, different strategies based on normalized cDNA libraries have been reported in many organisms including plants [33,35]. Using the suppression subtraction hybridization method, Caturla et al. [36] identified several genes associated with submergence in adventitious root primordia of Sesbania. As the co-ordinated regulation of multiple genes appears to be an important factor in long-term flooding-stress response of plant systems [37] a suppression subtraction hybridization approach was followed in this study for understanding how genes express differentially in waterlogging tolerant sugarcane genotype BO 91. Sugarcane varieties are highly heterozygous complex polyploid hybrid species and any two varieties differ in the number of chromosomes, thus both would be differing at the genomic/functional level for hundreds of genes making it impossible to pin-point the genes of interest. A subtractive library from the same variety in the presence or absence of a stress targets the genes that are differentially expressed in response to that stress. The subtractive cDNA library was prepared by subtracting the common genes expressing in normal condition from the genes expressing specifically in waterlogged condition. A total of 96 clones were sequenced to develop water-logged specific sugarcane ESTs, out of which a total of 81 sequences were generated, 77 (80%) sequences passed quality check and were >100 bp. The average cDNA insert size was 500 bp and average good quality sequence size was 306 bp. A brief summary of waterlogging induced subtraction library is given in (Table 1). The size distribution of ESTs generated from waterlogged specific ESTs revealed all the generated 81 sequences were >100 bp and out of them 42 sequences were found >300 bp (Fig. 1A) and they were submitted to NCBI under the accession numbers (GenBank: GT757740 to GT757780). In order to remove redundancy, all the sequenced ESTs were assembled using Sequencher program. The assembled data was manually verified to remove any mis-assembled sequences. The final assembly contained a total of 53 unigene ESTs with a set of 42 reads assembled as 14 contigs, while the remaining 39 sequences were singletons (Fig. 1B).
Table 1.
Characteristics of the water-logged subtractive ESTs.
| ESTs | Count |
|---|---|
| cDNA clones sequenced (5′ end) | 96 |
| Total sequences generated | 81 |
| Sequences of good quality (>100bp) | 77 |
| Success index (%) | 80 |
| Average insert size (bp) | 500 |
| Average size of good sequences (bp) | 306 |
| Number of singletons | 39 |
| Number of contigs | 14 |
| Number of reads assembled in contigs | 42 |
| Number of unique putative transcripts (unigenes) | 53 |
| Longest unigenes (bp) | 719 |
| Shortest but >100 bp unigene (bp) | 117 |
| Number of assembled ESTs with significant BLASTX match | 49 |
| No significant BLASTX match | 4 |
| Observed redundancy (%) | 45 |
Fig. 1.
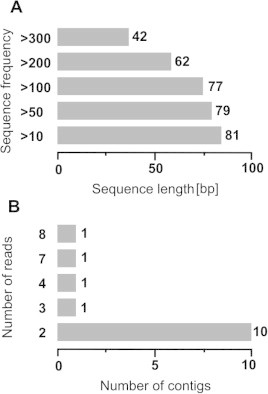
(A) Size distribution of ESTs generated from water-logged leaf cDNAs (B) Reads assembled in contigs.
3.3. Sequence analysis of cDNA fragments derived from subtractive hybridization
Unigene gene sequences were searched using the BLASTX tool from the NCBI non-redundant database to find protein similarity. Biological roles were inferred for the genes identified as differentially expressed in response to waterlogging after submitting the sequences to protein database searches (Fig. 2). Fifty-three clones identified from the subtraction library in this endeavor could be classified into 16 functional categories based on the predicted functions (Table 2). The stress related genes were the largest functional class, representing 15% of the genes identified. The second largest category was that of sequences encoding the unknown proteins. These represented 13% of the expressed genes. In this study, it was observed that approximately 21% of the identified genes in the tolerant cultivar are either unclassified, or encode unknown proteins which were much lower than unknown genes (68%) identified in rice genotypes tolerant to flooding [38]. A significant proportion of genes related to catalytic activity (13%) and cell growth (10%) were also induced during waterlogging stress.
Fig. 2.
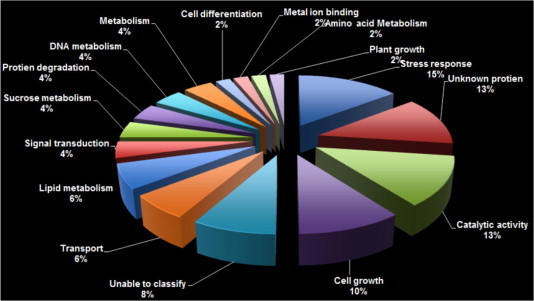
Functional characterization of water-logged induced sugarcane leaf unigenes according to the best matched BLAST search results.
Table 2.
Putative functions associated with ESTs induced during waterlogging in different functional categories.
| Functional Class | Unigene identifier⁎ | Putative function# |
|---|---|---|
| Amino acid metabolism | SUL21-001-E04-N-023.g | Methionine synthase protein |
| Catalytic activity | Contig_7 | Probable cytochrome P450 monooxygenase |
| Contig_12 | RmID substrate binding domain containing protein | |
| SUL21-001-B04-N-029.g | UDP-glucose 6-dehydrogenase | |
| SUL21-001-C02-N-006.g | Putative 2-oxoglutarate dehydrogenase E2 subunit | |
| SUL21-001-G06-N-040.g | Diaminopimelate epimerase | |
| SUL21-001-H06-N-048.g | Alcohol dehydrogenase | |
| Cell differentiation/cellulose degradation | Contig_15 | Exoglucanase precursor |
| Cell growth | Contig_6 | Alpha-tubulin |
| SUL21-001-C12-N-082.g | ||
| SUL21-001-D03-N-026.g | Actin | |
| SUL21-001-D04-N-030.g | Putative early nodulin | |
| SUL21-001-G01-N-004.g | Actin depolymerizing factor | |
| DNA metabolism | SUL21-001-B08-N-045.g | Histone H2A.Z |
| SUL21-001-H12-N-096.g | DNA methyltransferase | |
| Plant growth | SUL21-001-G02-N-008.g | Auxin-regulated protein-like |
| Lipid metabolism | SUL21-001-B06-N-045.g | Putative CLB1 protein (calcium-dependent lipid binding) protein |
| SUL21-001-D07-N-058.g | Phospholipase D alpha 1 (PLD alpha 1) (choline phosphatase 1) (phosphatidylcholine-hydrolyzing phospholipase D 1) | |
| SUL21-001-E10-N-071.g | GDSL-like lipase/acylhydrolase family protein | |
| Metabolism | SUL21-001-F01-N-011.g | Putative phosphoglycerate kinase |
| SUL21-001-F11-N-091.g | Beta-galactosidase precursor | |
| Metal ion binding | SUL21-001-A12-N-085.g | Metal ion binding |
| Protein degradation | Contig_18 | Kelch repeat-containing F-box-like |
| SUL21-001-C09-N-066.g | 26S proteasome regulatory particle triple-A ATPase subunit6 | |
| Signal transduction | Contig_4 | Putative protein kinase homolog |
| SUL21-001-C05-N-034.g | Putative receptor serine/threonine kinase PR5K | |
| Stress response | Contig_2 | Putative senescence-associated protein |
| SUL21-001-G08-N-056.g | ||
| Contig_11 | Dehydration-responsive family protein | |
| SUL21-001-D05-N-030.g | Adhesive/proline-rich protein homolog-like protein | |
| SUL21-001-G09-N-068.g | Legumain precursor | |
| SUL21-001-G12-N-088.g | Heat shock cognate 70 kDa protein | |
| SUL21-001-H10-N-080.g | DnaJ protein, putative | |
| Sucrose metabolism | Contig_5 | Sucrose synthase 2 (sucrose-UDP glucosyltransferase 2) |
| SUL21-001-H07-N-060.g | ||
| Transport | Contig_16 | Calcium-transporting ATPase 2 |
| SUL21-001-A02-N-005.g | Putative cation diffusion facilitator | |
| SUL21-001-F04-N-031.g | Clathrin assembly protein, putative | |
| Unable to classify | SUL21-001-C07-N-050.g | Hypothetical protein∼predicted by FGENESH etc. |
| SUL21-001-C11-N-082.g | Putative remorin 1 protein | |
| SUL21-001-E06-N-039.g | pnFL-2 | |
| SUL21-001-E12-N-087.g | rRNA intron-encoded homing endonuclease | |
| SUL21-001-F06-N-047.g | Expressed protein | |
Unigene (non redundant) EST sequences identity number.
The putative annotation was based on GenBank nr (BLASTX).
Plants have evolved adaptation mechanisms to sense oxygen deficiency in their environments and make coordinated physiological and structural adjustments to enhance their hypoxic tolerance [5]. It has been demonstrated that the synthesis of several proteins engaged in the glycolysis and fermentation processes is induced in plants exposed to anaerobic conditions. These anaerobic polypeptides include aldolase [39,40], pyruvate decarboxylase [41] and alcohol dehydrogenase [42–44]. Genes encoding alcohol dehydrogenase are reported in the current study (Table 2). Alcohol dehydrogenase 1 transcripts and alcohol dehydrogenase enzyme activity were analyzed intensively in maize seedlings and rice [45,46]. It has been proposed that a high level of mitochondrial ALDH2 protein under anaerobiosis conferred tolerance to submergence in rice [47].
It was interesting to observe the presence of a gene tag SUL21-001-C11-N-082.g for remorins (REMs) protein in the library. This protein family has been reported in several plants and animals with no distinct function. Specific biological roles of different remorins from the various groups remain to be investigated, but gene expression data suggest that some of these proteins might have key functions during responses to biotic and abiotic stimuli and might possibly be involved in hormone mediated responses and signal transduction. Interestingly, remorins have also been found in detergent-resistant membrane fractions, called lipid rafts [48] and these are thought to act as docking sites for specific proteins involved in many important cellular processes, including secretion, signal transduction, and the perception of pathogens [49]. The EST cluster SUL21-001-G01-N-004.g encoding for actin depolymerizing factor (ADF) was obtained in this study. The actin cytoskeleton forms a dynamic, stimulus-responsive network in eukaryotic cells and is involved in essential functions, including cell division and cytoplasmic streaming. Increasing evidence has suggested that proteins of the ADF family could be involved in the actin cytoskeleton rearrangement and signal transduction events occurring when plants are subjected to stress conditions [50]. We found that cluster SUL21-001-C05-N-034.g encoded a protein similar to a putative serine/threonine protein kinase from Oryza sativa (Table 2). Members of the protein kinase superfamily catalyzes the reversible transfer of γ-phosphate from ATP to serine, threonine, or tyrosine on target proteins. Protein kinases have been implicated in a large number of plant processes, including the development of plant reproductive organs in Petunia hybrida and Arabidopsis thaliana [51] and it is reasonable to assume that stress induced protein kinases also exist in sugarcane.
Chaperones are proteins capable of assisting the folding of other proteins in vivo [52]. Conditions of stress, such as temperature variation can induce protein aggregation inside cells and consequently, many chaperones were first identified as heat shock proteins (Hsps). Chaperones are able to bind to exposed hydrophobic residues in unfolded proteins (usually buried in the native state) and according to [53] such a mechanism prevents the incorrect folding of the protein. The expression of chaperone proteins, including heat shock and DnaJ (Table 2) may indicate their role when sugarcane plant was subjected to water logging stress. Most of the chaperone and stress-related protein genes including Hsp70, DnaJ, and heat shock transcription factors were also reported in sugarcane EST (SUCEST) database [54].
Leaf senescence is a regulated process that corresponds to the last stage in leaf development. It is characterized by dramatic changes in cellular metabolism and the sequential degeneration of cellular structures. The occurrence of leaf senescence associated proteins confers its role under waterlogging stress in sugarcane in the current study and also evidenced in other studies. In addition to ageing, multiple developmental and environmental signals are able to trigger senescence. Drought, darkness, leaf detachment, and the hormones abscisic acid (ABA) and ethylene are reported to induce leaf yellowing [55]. Thirteen snow pea pod senescence-associated (SSA) cDNAs were isolated from a 5-day stored pod cDNA library using differential screening [56]. Sequence comparison indicated that these cDNA clones encoded proteins related to cell membrane and nutrient remobilization, to disease response-related and ribosomal proteins, and to ubiquitin-conjugating enzyme.
3.4. Identification of microRNAs induced under waterlogging
The identification of miRNAs and their targets has proven to be an important tool in understanding miRNA-dependent gene regulation in many plant species including sugarcane. A large number of microRNAs and their putative targets have been identified in sugarcane subjected to drought, salt stress, and pathogen infection [21,22,57]. 26 conserved miRNA families and two putative novel miRNAs, as well as a number of trans-acting small interfering RNAs had been identified in sugarcane axillary bud growth, using bioinformatics approach [58]. Waterlogging is one of the major causes of reduced crop yield among the globe [59]. The most radical effect caused due to waterlogging is reduced access of oxygen to the rhizosphere [60]. In Arabidopsis, a widespread change has been observed in 5–10% of the transcriptome when subjected to low oxygen condition [61,62]. Co-incidentally dramatic changes have also been observed in protein synthesis and degradation with corresponding mRNA changes [63]. It has been proved now that microRNAs play important role in posttranscriptional modulation of gene expression involving in plant responses to abiotic stresses. However, the regulation of miRNA in the various morphological responses to waterlogging is not very well established in plants especially in sugarcane. In maize seedlings, a total of 61 mature miRNAs had been identified including 32 waterlogging responsive miRNAs, most of these were negatively regulated under waterlogging condition [64]. In order to understand the molecular mechanisms regulating gene expression in response to water logging we identified putative microRNAs and their targets using computational approach.
The most challenging problem in understanding plant miRNAs is identifying novel miRNAs, for which different approaches have been used in plants including forward genetics, computational prediction, and direct cloning and sequencing [19]. To uncover additional sugarcane-specific miRNA candidates within our sequenced set, in all, 7 small RNAs met our criteria as established according to Allen et al. [27] and were considered to be putative novel sugarcane miRNAs. We did find both the miRNA and the miRNA∗ sequences (Fig. 3). The detection of miRNAs∗ is a strong clue, albeit not infallible, about the existence of precursor hairpin structures, adding credence to the authenticity of the predicted candidates. Precursors of these novel miRNAs had negative folding-free energies ranging from −39 to −24 kcal mol−1, with an average of about −33 kcal mol−1 according to the Mfold RNA folding platform. These values are similar to the free energy values of other plant miRNA precursors and are much lower than the reported folding-free energies of tRNAs or rRNAs [65]. Finally, BLASTN analysis against all nucleotide sequences in the NCBI databases revealed no homologs for these miRNAs in other plant species, signifying that these newly identified putative miRNAs are specific to sugarcane species. To clarify the biological functions of the newly identified sugarcane miRNAs, we searched for putative target genes using the psRNATarget program with default parameters [66]. The target genes found for novel sugarcane miRNAs are listed in (Table S2) and most of them code for hypothetical DnaJ like proteins. The miRNA-regulation of gene expression plays a key role in the development and response to biotic and abiotic stresses [67]. To date, only 35 sugarcane miRNAs are deposited at the miRBase database current version (release 20). The identification of miRNAs and their targets is important not only to help us learn more about the roles of miRNAs in sugarcane development and physiology, but also to provide a basis for further studies on RNAi-based regulation mechanisms under waterlogging stress.
Fig. 3.

Prediction of secondary structures for novel miRNA candidates under waterlogging condition of sugarcane. The putative miRNA sequences are highlighted in red and miRNA∗ sequences are highlighted in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
In summary, the comparative global transcript profiling in sugarcane plants undertaken in this study suggests that transcripts associated with stress response, signal transduction, metabolic activity and ion transport play an important role in conferring waterlogging tolerance in the sugarcane genotype, BO 91. The sensitivity of the subtraction hybridization technique allowed the identification of factors involved in different biological functions. The result indicates that the lengths of the sequences reported in this study are good enough to retrieve hits in GenBank database. Sequences of only two unigenes showed homology to the proteins identified in sugarcane which indicates that information related to genes expressed during waterlogging stress is inadequate at global level. However, in the present study, 53 unique sequences were generated from 96 clones which may only give a glimpse of the transcriptome expressing in waterlogging tolerant sugarcane genotype under study. Further work is needed to consolidate the preliminary results and find out the battery of genes involved in transient to stable response to stress by sampling at different time intervals and various organs that perceive and respond to the stress. The cDNA clones obtained from this work need to be further characterized using simpler model systems like yeast and E. coli or higher systems, particularly gene knockouts or through ectopic over-expression of genes using a transgenics approach. miRNA analysis was undertaken with the objective to chance upon any relevant contributors to stress tolerance. Our findings provide the first miRNA discovery in sugarcane under waterlogging stress and are suggestive of the potential role of miRNA in regulatory pathways of sugarcane and other crops. Future work on these lines would definitely enhance our understanding of the genetic basis of the waterlogging stress tolerance mechanisms and provide newer directions in developing sugarcane varieties tolerant to waterlogging stress.
Acknowledgements
The authors gratefully acknowledge to Inter-disciplinary Centre for Plant Genomics (ICPG), Department of Plant Molecular Biology, UDSC, New Delhi, for cDNA library sequencing and ESTs generation. Financial support provided by Department of Biotechnology (DBT) Ministry of Science and Technology Govt. of India, New Delhi is greatly acknowledged.
Appendix A. Supplementary data
Sugarcane plant grown in water-logged condition and normal condition (a) plants growing in waterlogging condition (b) close-up (as left) (c) aerial roots produced under waterlogging condition (d) profuse root mass produced under waterlogging condition (e) plants growing in normal field condition.
Growth and yield performance of BO91 cultivated in waterlogging and normal condition.
Predicated targets of novel miRNA candidates under waterlogging condition of sugarcane.
References
- 1.Grivet L., Arruda P. Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr. Opin. Plant Biol. 2002;5:122–127. doi: 10.1016/s1369-5266(02)00234-0. [DOI] [PubMed] [Google Scholar]
- 2.Menossi M., Silva-Filho M.C., Vincentz M., Van-Sluys M.A., Souza G.M. Sugarcane functional genomics: gene discovery for agronomic trait development. Int. J. Plant Genomics. 2008;2008:458732. doi: 10.1155/2008/458732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Extension publication, (2010) Effect of Waterlogging in Sugarcane and its Management, Sugarcane Breeding Institute India. 185.
- 4.Gilbert R.A., Rainbolt C.R., Morris D.R., Bennett A.C. Morphological responses of sugarcane to long-term flooding. Agron. J. 2007;99:1622–1628. [Google Scholar]
- 5.Nishiuchi S., Yamauchi T., Takahashi H., Kotula L., Nakazono M. Mechanisms for coping with submergence and waterlogging in rice. Rice. 2012;5 doi: 10.1186/1939-8433-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson P., Robertson M., Cooper M., Hammer G. The role of physiological understanding in plant breeding; from a breeding perspective. Field Crops Res. 1996;49:11–37. [Google Scholar]
- 7.Edwards D., Batley J. Plant genome sequencing: applications for crop improvement. Plant Biotechnol. J. 2010;8:2–9. doi: 10.1111/j.1467-7652.2009.00459.x. [DOI] [PubMed] [Google Scholar]
- 8.Henry R.J. Next-generation sequencing for understanding and accelerating crop domestication. Brief Funct. Genomics. 2012;11:51–56. doi: 10.1093/bfgp/elr032. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton J.P., Buell C.R. Advances in plant genome sequencing. Plant J. 2012;70:177–190. doi: 10.1111/j.1365-313X.2012.04894.x. [DOI] [PubMed] [Google Scholar]
- 10.Carson D.L., Botha F.C. Genes expressed in sugarcane maturing internodal tissue. Plant Cell Rep. 2002;20:1075–1081. [Google Scholar]
- 11.Casu R.E., Grof C.P.L., Rae A.L., McIntyre C.L., Dimmock C.M., Manners J.M. Identification of a novel sugar transporter homologue strongly expressed in maturing stem vascular tissues of sugarcane by expressed sequence tag and microarray analysis. Plant Mol. Biol. 2003;52:371–386. doi: 10.1023/a:1023957214644. [DOI] [PubMed] [Google Scholar]
- 12.Rudd S. Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci. 2003;8:321–329. doi: 10.1016/S1360-1385(03)00131-6. [DOI] [PubMed] [Google Scholar]
- 13.Ma H.M., Schulze S., Lee S., Yang M., Mirkov E., Irvine J., Moore P., Paterson A. An EST survey of the sugarcane transcriptome. Theor. Appl. Genet. 2004;108:851–863. doi: 10.1007/s00122-003-1510-y. [DOI] [PubMed] [Google Scholar]
- 14.Vettore A.L., da Silva F.R., Kemper E.L., Souza G.M., da Silva A.M., Ferro M.I.T., Henrique-Silva F., Giglioti E.A., Lemos M.V.F., Coutinho L.L., Nobrega M.P., Carrer H., Franca S.C., Bacci M., Goldman M.H.S., Gomes S.L., Nunes L.R., Camargo L.E.A., Siqueira W.J., Van Sluys M.A., Thiemann O.H., Kuramae E.E., Santelli R.V., Marino C.L., Targon M.L.P.N., Ferro J.A., Silveira H.C.S., Marini D.C., Lemos E.G.M., Monteiro-Vitorello C.B., Tambor J.H.M., Carraro D.M., Roberto P.G., Martins V.G., Goldman G.H., de Oliveira R.C., Truffi D., Colombo C.A., Rossi M., de Araujo P.G., Sculaccio S.A., Angella A., Lima M.M.A., de Rosa V.E., Siviero F., Coscrato V.E., Machado M.A., Grivet L., Di Mauro S.M.Z., Nobrega F.G., Menck C.F.M., Braga M.D.V., Telles G.P., Cara F.A.A., Pedrosa G., Meidanis J., Arruda P. Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 2003;13:2725–2735. doi: 10.1101/gr.1532103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugiharto B., Ermawati N., Mori H., Aoki K., Yonekura-Sakakibara K., Yamaya T., Sugiyama T., Sakakibara H. Identification and characterization of a gene encoding drought-inducible protein localizing in the bundle sheath cell of sugarcane. Plant Cell Physiol. 2002;43:350–354. doi: 10.1093/pcp/pcf039. [DOI] [PubMed] [Google Scholar]
- 16.Nogueira F.T.S., De Rosa V.E., Menossi M., Ulian E.C., Arruda P. RNA expression profiles and data mining of sugarcane response to low temperature. Plant Physiol. 2003;132:1811–1824. doi: 10.1104/pp.102.017483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta V., Raghuvanshi S., Gupta A., Saini N., Gaur A., Khan M.S., Gupta R.S., Singh J., Duttamajumder S.K., Srivastava S., Suman A., Khurana J.P., Kapur R., Tyagi A.K. The water-deficit stress- and red-rot-related genes in sugarcane. Funct. Integr. Genomics. 2010;10:207–214. doi: 10.1007/s10142-009-0144-9. [DOI] [PubMed] [Google Scholar]
- 18.Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Khraiwesh B., Pugalenthi G., Fedoroff N.V. Identification and analysis of red sea mangrove (Avicennia marina) microRNAs by high-throughput sequencing and their association with stress responses. PLoS ONE. 2013;8:e60774. doi: 10.1371/journal.pone.0060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arif M.A., Frank W., Khraiwesh B. Role of RNA interference (RNAi) in the moss Physcomitrella patens. Int. J. Mol. Sci. 2013;14:1516–1540. doi: 10.3390/ijms14011516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira T.H., Gentile A., Vilela R.D., Costa G.G.L., Dias L.I., Endres L., Menossi M. MicroRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.) PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0046703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiebaut F., Grativol C., Carnavale-Bottino M., Rojas C.A., Tanurdzic M., Farinelli L., Martienssen R.A., Hemerly A.S., Ferreira P.C.G. Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics. 2012;13 doi: 10.1186/1471-2164-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanca A.S., Vicentini R., Ortiz-Morea F.A., Del Bem L.E.V., da Silva M.J., Vincentz M., Nogueira F.T.S. Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biol. 2010;10 doi: 10.1186/1471-2229-10-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B.H., Pan X.P., Wang Q.L., Cobb G.P., Anderson T.A. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II, error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 27.Allen E., Xie Z.X., Gustafson A.M., Carrington J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Kapur R., Duttamajumder S.K., Srivastava B.L., Madhok H.L., Kumar R. Harvest index and the components of biological yield in sugarcane. Indian J. Genet. Plant Breed. 2013;73:386–391. [Google Scholar]
- 29.Barclay A.M., Crawford R.M.M. Plant-growth and survival under strict anaerobiosis. J. Exp. Bot. 1982;33:541–549. [Google Scholar]
- 30.Kovar J.L., Kuchenbuch R.O. Commercial importance of adventitious rooting to agronomy. Basic Life Sci. 1994;62:25–35. [Google Scholar]
- 31.Cheong Y.H., Chang H.S., Gupta R., Wang X., Zhu T., Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Provart N.J., Gil P., Chen W.Q., Han B., Chang H.S., Wang X., Zhu T. Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol. 2003;132:893–906. doi: 10.1104/pp.103.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonaldo M.D.F., Lennon G., Soares M.B. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez P., Paniego N., Lew S., Hopp H.E., Heinz R.A. Differential representation of sunflower ESTs in enriched organ-specific cDNA libraries in a small scale sequencing project. BMC Genomics. 2003;4:40. doi: 10.1186/1471-2164-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patanjali S.R., Parimoo S., Weissman S.M. Construction of a uniform-abundance (normalized) cDNA library. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1943–1947. doi: 10.1073/pnas.88.5.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caturla M., Chaparro C., Schroeyers K., Holsters M. Suppression subtractive hybridization to enrich low-abundance and submergence-enhanced transcripts of adventitious root primordia of Sesbania rostrata. Plant Sci. 2002;162:915–921. [Google Scholar]
- 37.Dennis E.S., Dolferus R., Ellis M., Rahman M., Wu Y., Hoeren F.U., Grover A., Ismond K.P., Good A.G., Peacock W.J. Molecular strategies for improving waterlogging tolerance in plants. J. Exp. Bot. 2000;51:89–97. [PubMed] [Google Scholar]
- 38.Minhas D., Grover A. Towards developing transgenic rice plants tolerant to flooding stress. Proc. Indian Natl. Sci. Acad. 1999;65:33–50. [Google Scholar]
- 39.Kelley P.M., Freeling M. Anaerobic expression of maize glucose phosphate isomerase-I. J. Biol. Chem. 1984;259:673–677. [PubMed] [Google Scholar]
- 40.Kelley P.M., Tolan D.R. The complete amino-acid-sequence for the anaerobically induced aldolase from maize derived from cDNA clones. Plant Physiol. 1986;82:1076–1080. doi: 10.1104/pp.82.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wignarajah K., Greenway H. Effect of anaerobiosis on activities of alcohol-dehydrogenase and pyruvate decarboxylase in roots of Zea mays. New Phytol. 1976;77:575–584. [Google Scholar]
- 42.Dennis E.S., Gerlach W.L., Pryor A.J., Bennetzen J.L., Inglis A., Llewellyn D., Sachs M.M., Ferl R.J., Peacock W.J. Molecular analysis of the alcohol-dehydrogenase (Adhl) gene of maize. Nucleic Acids Res. 1984;12:3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dennis E.S., Sachs M.M., Gerlach W.L., Finnegan E.J., Peacock W.J. Molecular analysis of the alcohol dehydrogenase-2 (Adh2) gene of maize. Nucleic Acids Res. 1985;13:727–743. doi: 10.1093/nar/13.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sachs M.M., Freeling M. Selective synthesis of alcohol-dehydrogenase during anaerobic treatment of maize. Mol. Gen. Genet. 1978;161:111–115. [Google Scholar]
- 45.Andrews D.L., Cobb B.G., Johnson J.R., Drew M.C. Hypoxic and anoxic induction of alcohol-dehydrogenase in roots and shoots of seedlings of Zea mays – Adh transcripts and enzyme-activity. Plant Physiol. 1993;101:407–414. doi: 10.1104/pp.101.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie Y., Wu R. Rice alcohol-dehydrogenase genes – anaerobic induction, organ specific expression and characterization of cDNA clones. Plant Mol. Biol. 1989;13:53–68. doi: 10.1007/BF00027335. [DOI] [PubMed] [Google Scholar]
- 47.Nakazono M., Tsuji H., Li Y.H., Saisho D., Arimura S., Tsutsumi N., Hirai A. Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol. 2000;124:587–598. doi: 10.1104/pp.124.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raffaele S., Bayer E., Lafarge D., Cluzet S., Retana S.G., Boubekeur T., Leborgne-Castel N., Carde J.P., Lherminier J., Noirot E., Satiat-Jeunemaitre B., Laroche-Traineau J., Moreau P., Ott T., Maule A.J., Reymond P., Simon-Plas F., Farmer E.E., Bessoule J.J., Mongrand S. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell. 2009;21:1541–1555. doi: 10.1105/tpc.108.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajendran L., Simons K. Lipid rafts and membrane dynamics. J. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- 50.Aon M.A., Cortassa S., Casati D.F.G., Iglesias A.A. Effects of stress on cellular infrastructure and metabolic organization in plant cells. Int. Rev. Cytol. 2000;194:239–273. doi: 10.1016/s0074-7696(08)62398-0. [DOI] [PubMed] [Google Scholar]
- 51.Decroocqferrant V., Vanwent J., Bianchi M.W., Devries S.C., Kreis M. Petunia-hybrida homologs of shaggy/zeste-white-3 expressed in female and male reproductive-organs. Plant J. 1995;7:897–911. doi: 10.1046/j.1365-313x.1995.07060897.x. [DOI] [PubMed] [Google Scholar]
- 52.Ellis R.J., Hartl F.U. Protein folding in the cell: competing models of chaperonin function. FASEB J. 1996;10:20–26. doi: 10.1096/fasebj.10.1.8566542. [DOI] [PubMed] [Google Scholar]
- 53.Flynn G.C., Chappell T.G., Rothman J.E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 54.Borges J.C., Peroto M.C., Ramos C.H. Molecular chaperone genes in the sugarcane expressed sequence database (SUCEST) Genet. Mol. Biol. 2001;24:85–92. [Google Scholar]
- 55.Lim P.O., Nam H.G. The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr. Top. Dev. Biol. 2005;67:49–83. doi: 10.1016/S0070-2153(05)67002-0. [DOI] [PubMed] [Google Scholar]
- 56.Pariasca J.A.T., Sunaga A., Miyazaki T., Hisaka H., Sonoda M., Nakagawa H., Sato T. Cloning of cDNAs encoding senescence-associated genes, ACC synthase and ACC oxidase from stored snow pea pods (Pisum sativum L. var saccharatum) and their expression during pod storage. Postharvest Biol. Technol. 2001;22:239–247. [Google Scholar]
- 57.Carnavale Bottino M., Rosario S., Grativol C., Thiebaut F., Rojas C.A., Farrineli L., Hemerly A.S., Ferreira P.C. High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS ONE. 2013;8:e59423. doi: 10.1371/journal.pone.0059423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ortiz-Morea F.A., Vicentini R., Silva G.F., Silva E.M., Carrer H., Rodrigues A.P., Nogueira F.T. Global analysis of the sugarcane microtranscriptome reveals a unique composition of small RNAs associated with axillary bud outgrowth. J. Exp. Bot. 2013;64:2307–2320. doi: 10.1093/jxb/ert089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyer J.S. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 60.Agarwal S., Grover A. Molecular biology, biotechnology and genomics of flooding-associated low O(2) stress response in plants. Crit. Rev. Plant Sci. 2006;25:1–21. [Google Scholar]
- 61.Branco-Price C., Kawaguchi R., Ferreira R.B., Bailey-Serres J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation (vol 96, pg 647, 2005) Ann. Bot. 2005;96:1142. doi: 10.1093/aob/mci217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klok E.J., Wilson I.W., Wilson D., Chapman S.C., Ewing R.M., Somerville S.C., Peacock W.J., Dolferus R., Dennis E.S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey-Serres J., Voesenek L.A.C.J. Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008;59:313–339. doi: 10.1146/annurev.arplant.59.032607.092752. [DOI] [PubMed] [Google Scholar]
- 64.Zhai L.H., Liu Z.J., Zou X.L., Jiang Y.Y., Qiu F.Z., Zheng Y.L., Zhang Z.X. Genome-wide identification and analysis of microRNA responding to long-term waterlogging in crown roots of maize seedlings. Physiol. Plant. 2013;147:181–193. doi: 10.1111/j.1399-3054.2012.01653.x. [DOI] [PubMed] [Google Scholar]
- 65.Bonnet E., Wuyts J., Rouze P., Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 66.Dai X.B., Zhao P.X. PsRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khraiwesh B., Zhu J.K., Zhu J.H. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. BBA Gene Regul. Mech. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sugarcane plant grown in water-logged condition and normal condition (a) plants growing in waterlogging condition (b) close-up (as left) (c) aerial roots produced under waterlogging condition (d) profuse root mass produced under waterlogging condition (e) plants growing in normal field condition.
Growth and yield performance of BO91 cultivated in waterlogging and normal condition.
Predicated targets of novel miRNA candidates under waterlogging condition of sugarcane.


