Abstract
Carboxylesterases (CES, EC 3.1.1.1) are members of a superfamily of serine hydrolases that hydrolyze ester, amide, and carbamate bonds. Several different CES genes exist in mammalian species with evidence of multiple gene duplication events occurring throughout evolutionary history. There are five CES genes reported in the Human Genome Organization database, although CES1 and CES2 are the two best characterized human genes. An emerging picture of the CES family suggests that these enzymes have dual roles in the metabolism of xenobiotic and endobiotic compounds. Pesticides, such as the pyrethroids, are important xenobiotic substrates that are metabolized by CES, whereas cholesteryl esters, triacylglycerols, and 2-arachidonoylglycerol are examples of endobiotics known to be substrates for CES. Functional studies using selective chemical inhibitors, siRNA, and gene knockout models are providing valuable insights into the physiological functions of CES, and suggest that CES may be a novel target for the treatment of diseases such as diabetes and atherosclerosis. This review will examine the known physiological functions of CES, the interactions between xenobiotics (primarily pesticides) and lipids that occur with CES enzymes, and where possible the implications that these findings may have in terms of health and disease.
Keywords: carboxylesterase, pyrethroids, organophosphates, cholesteryl ester hydrolase, macrophage, xenobiotic biotransformation
Introduction
Carboxylesterases (CES, EC 3.1.1.1) are members of the α,β-serine hydrolase multigene family and are widely expressed in multiple mammalian tissues.1) CES are multifunctional enzymes that catalyze the hydrolysis of substrates containing ester, amide, and thioester bonds. They have a very broad substrate specificity, which is attributed to a large conformable active site that permits entry of numerous structurally diverse substrates 2), including endobiotics and xenobiotics. 3,4) Organophosphate (OPs), carbamate, and pyrethroid insecticides are metabolized by CES.5) These compounds have essential roles in agriculture and public health, and are important for the optimization of crop yields and the prevention of disease. However, CES are irreversibly inhibited by OPs during attempted catalytic turnover of these substrates, or reversibly inhibited by carbamates due to slow decarbamoylation rates.5) The bio-persistent organochlorine insecticides widely used during the mid 20th century were replaced by OPs, carbamates, and pyrethroids because these compounds were more environmentally labile and did not accumulate in the food chain. Pyrethroids were extremely attractive compounds because they exhibited greater selective lethality toward insects over mammals than both the OPs and carbamates.6) The quantities of pyrethroid insecticides used are rapidly increasing, with subsequent declines of OP compounds due to phase-outs and restrictions.7) Therefore, pyrethroids now rank second only to organophosphates in terms of total insecticide usage in U.S. agricultural practices.8) Thus humans are presently, and will continue to be, occupationally, environmentally, and domestically exposed to high levels of these compounds. Importantly, CES mediated metabolism of pyrethroids is a major route of their elimination in humans.9,10) Thus knowledge of CES function is important for classifying the risk posed by pyrethroids, and pesticides in general, on human health.
The role of CES in the metabolism of xenobiotics is firmly established, whereas a role for these enzymes in lipid metabolism was more recently identified. CES appears to be an example of a ‘catalytically promiscuous’ enzyme in that they seem to have acquired adventitious secondary activities apart from their main activity.73) Thus, CES appear to be dual-function enzymes and probably did not evolve for the sole purpose of metabolizing xenobiotics, although the importance of CES in the detoxication of ester-containing xenobiotics cannot be overstated. Nevertheless, recent studies have clarified roles for CES in lipid biochemistry. For example, human CES1 was shown to exhibit cholesteryl ester hydrolase activity in human macrophages and hepatocytes11), murine Ces3 exhibited triacylglycerol hydrolase activity in both mouse hepatocytes and adipocytes12), and bacterial CES was found to efficiently hydrolyze the endocannabinoid 2-arachidonoylglycerol13). These findings suggest that CES have important physiological functions in vivo. The purpose of this review will be to examine the physiological roles of CES, the interactions between xenobiotics (focusing primarily on pesticides) and lipids that occur with CES enzymes, and where possible the implications that these findings may have in terms of disease pathology and treatment will be discussed.
Genetics and enzymology of carboxylesterases
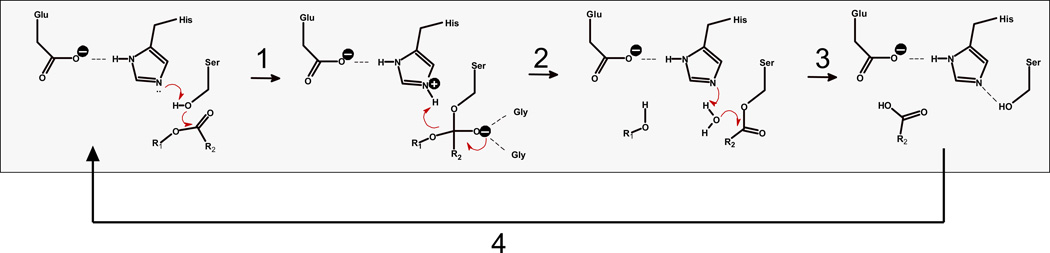
CES are a family of hydrolytic enzymes found within the serine hydrolase superfamily. Satoh and Hosokawa14) classified mammalian CES enzymes into five groups (CES 1-5), based on their amino acid homology. The majority of identified CES enzymes fall within the CES1 and CES2 sub-families. CES contain a catalytic triad of amino acids in their active site [Ser-His-Glu (or Asp)], which are essential for catalytic activity. The nucleophilic serine residue at the base of the active site can attack the carbonyl carbon of ester-containing substrates. This reaction is catalyzed by a general acid-base mechanism, utilizing His and Glu (or Asp) catalytic residues via a charge-relay complex15) (Figure 1). CES are widely distributed in several tissues, including liver, small intestine, lung, kidney, adipose tissue, testis, and macrophages; typically in the endoplasmic reticulum of cells but also in the cytoplasm.4) The hepato-intestinal axis is of particular importance for CES expression because of the high concentrations of ester-containing toxins that can be ingested orally. There are five CES genes reported in the Human Genome Organization database, although CES1 and CES2 are the two best characterized human genes.71) CES1, CES2, and CES3 encode ~60 kDa glycoproteins.16) CES1 actually represents two separate but nearly identical genes (CES1A1, CES1A2), with only slight nucleotide differences in the promoter and signal peptide regions, yet produce an identical mature protein.71) CES1 protein is expressed in several tissues with highest levels found in liver, whereas CES2 protein is expressed in a more tissue specific manner with highest levels in the intestine and liver. The abundance of CES1 in human liver was underscored by the recent findings of the Human Liver Proteome project, which determined that CES1 was the tenth most abundant protein (out of >6,000) expressed in the human liver.17) In contrast, CES3 appears to have limited expression; highest levels were reported in trachea71) and, although detected by northern blot in liver, it probably does not have a major role in xenobiotic metabolism.
Figure 1.
Catalytic cycle of CES-mediated hydrolysis of ester substrates. Step 1. Nucleophilic attack of serine Oγ on carbonyl carbon of ester substrate R2-C(=O)-OR1 and formation of tetrahedral oxyanion intermediate, which is stabilized by the oxyanion hole. Serine Oγ is activated by charge relay complex established between the Glu-His-Ser catalytic triad residues. Step 2. Collapse of oxyanion intermediate and formation of acyl-enzyme covalent intermediate and alcohol product, R1OH. Step 3. Acyl-enzyme intermediate is attacked by a water molecule, which is activated by the catalytic His residue, thereby liberating the carboxylic acid product, R2-C(=O)-OH. Step 4. Catalytic triad hydrogen bond network re-establishes itself and cycle repeats.
In terms of amino acid sequence homology, human CES1 and CES2 proteins are 48% identical, thus they are placed in two separate CES classes (class 1 and 2, respectively). Crystal structures for human CES1, but not CES2, have been obtained and indicate that 3 ligand binding sites are present (active site, side door, and Z site).72) In contrast to the human situation, the number of CES genes found in the murine genome is much higher. For instance, there are at least nine different CES1 family genes found in the murine genome, and these genes exhibit 65–77% sequence homology with human CES1 based on amino acid homology. Murine Ces1 and Ces3 are members of the mouse CES1 gene family and the corresponding enzymes have the highest homology to human CES1, exhibiting 73% and 77% amino acid sequence homology to hCES1, respectively. The redundancy of CES1 genes in the mouse genome suggests that multiple gene duplication events occurred during the evolutionary history of Mus musculus. The high sequence homology between murine CES1 family members makes biochemical characterization difficult because there is not a specific substrate that can distinguish each isoform. Recently, a CES1 gene knockout (Ces3−/− or Tgh−/−) mouse was reported by Wei et al.18), which should help characterize the biological functions of CES. Indeed, it was shown that deletion of Ces3 (also called TGH for triacylglycerol hydrolase) from mice caused decreased levels of plasma triacylglycerols (TGs), apolipoprotein B, and fatty acids when compared to wildtype mice. Furthermore, Ces3−/− mice also exhibited improved insulin sensitivity and glucose tolerance, which suggested that CES might be an attractive target for the treatment of type 2 diabetes. Lack of a CES knockout animal had previously hampered study of CES function. Indeed, CES were often described in the literature as enzymes with “unknown function”, or stated another way the CES enzymes were considered functionally unannotated. However, generation of a Ces3 knockout mouse should provide new knowledge regarding the physiological functions of this enzyme class, and help to assign bona fide physiologic substrates for Ces3.
Selective chemical inhibitors for carboxylesterases
The continued development of selective chemical inhibitors that block CES activity in cells, tissues, and organisms will be highly valuable for the elucidation of CES function in vivo. For example, Potter and Wadkins15) have used high-throughput enzyme assays and in silico approaches using recombinant rabbit and human CES proteins to guide development of highly potent and CES isoform-selective small molecule inhibitors. They have shown that the diphenylethane-1,2-dione chemotype (benzil) is an excellent scaffold for selectively inhibiting the CES family of enzymes. The 1,2-dione moiety of benzil is crucial for enzyme inhibition and the phenyl rings improve the potency of the inhibitors. The selectivity towards different CES isoforms is dependent upon substitutions within these rings. Although benzil inhibits human CES1 and CES2 with near equal potency in the sub-nanomolar range19), it has also been shown that it does not inhibit acetylcholinesterase (AChE), butyrylcholinesterase (BChE)19), and bile salt-stimulated carboxyl ester lipase (CEL)20), which is an enzyme secreted from the pancreas into the gut lumen. Our laboratory has further shown that benzil does not inhibit the endocannabinoid hydrolases, monoacylglycerol lipase (MAGL) and fatty acid amide hydrolase (FAAH) (unpublished data). Collectively, these results indicate that benzil has good selectivity toward the CES enzyme family, and related serine hydrolases (i.e., AChE, BChE, CEL, MAGL, and FAAH) are unlikely to be off-targets in vivo. Selective inhibitors will be highly useful for studying the physiological function of CES. Furthermore, since insects utilize CES to detoxify pesticides, these inhibitors might be specifically tailored to inhibit insect CES and be formulated with pesticides to reduce the prevalence of insecticide resistance in crop-damaging pests.19)
Role of carboxylesterases in xenobiotic metabolism
Several environmental pollutants, pharmaceutical agents, and illicit drugs possess ester bonds, which has the effect of increasing compound lipophilicity and bioavailability. Examples include pesticides (pyrethroid insecticides), plasticizers (phthalate esters), anti-thrombotic drugs (aspirin, clopidogrel), pro-drugs (irinotecan), and narcotics (cocaine, heroin). These compounds are inactivated (or bioactivated in the case of irinotecan) by the hydrolytic actions of CES, thus rendering water-soluble carboxylic acid and alcohol containing metabolites that are excreted from the organism, often following phase II conjugation reactions. Metabolism of pyrethroids by CES has been a focus of several laboratories because of their extensive use in agriculture21) and public health.22) CES have an important role in the detoxification of pyrethroids, which was demonstrated early in rodent studies.23) Moreover, inhibition of this metabolic pathway had significant consequences for pyrethroid mediated toxicity. For example, when OP insecticides were administered to mice prior to dosing with a pyrethroid (trans-permethrin), robust inhibition of liver CES activity resulted, which in turn increased the plasma levels of pyrethroid and subsequent neurotoxic action of trans-permethrin in vivo.23) Metabolism studies conducted in rodents also demonstrated that pyrethroids were degraded rapidly to a large number of polar metabolites via oxidative and hydrolytic enzymes.24,25) The enzymes responsible for pyrethroid biotransformation include both the cytochrome P450 (CYP) and carboxylesterase (CES) families. More recent studies using recombinant enzymes have identified the specific P450 and CES isozymes that are responsible for these biotransformation reactions.10,26–28) CYP and CES isozymes are expressed abundantly in mammalian liver29), and their efficient activities account in part for the reason that pyrethroids are relatively non-toxic in mammalian species. However, despite the attractive metabolic features of pyrethroids in mammals, insects can often establish resistance against pyrethroid insecticides following mutation and/or overexpression of genes that encode CYP and CES catabolic enzymes.30)
Interindividual variation in the rates of pyrethroid metabolism by CES in humans have been shown to be substantial9,28), which may influence toxicities caused by these compounds in the human population. CES1 is the most abundant CE isoform expressed in human liver (~50-fold greater than CES2)27), and the level of CES1 protein expression in the human population, as determined by western blotting, appears to be strikingly invariable among individuals 28) (Xie and Ross, unpublished data). In contrast, the enzymatic activity of CES1 in these same individuals varies considerably. Thus, a simple correlation between CES1 enzyme activity and CES1 protein levels in the human population is not apparent. Therefore, post-translational regulation of CES1 enzyme activity in the liver may be a major factor that accounts for the variable pharmacokinetic behavior of ester-containing xenobiotics, such as pyrethroids, observed in human populations. Alternatively, heretofore unidentified SNPs in the CES1 gene may also contribute to this variation.
The substrate specificities of pure human and rodent CES proteins toward pyrethroids reveal substantial differences. Several type II pyrethroids, which are distinguished from type I pyrethroid compounds by the presence of a cyano functionality on the alcohol moiety of the pyrethroid, were hydrolyzed by recombinant human CES1 at markedly faster rates than by a rat CES1 ortholog.26) For example, the Vmax for human CES1 catalyzed hydrolysis of deltamethrin was ~5-fold higher than the Vmax for the rat CES1 ortholog (Hydrolase A). Thus, hydrolytic metabolism appears to be the primary metabolic pathway that clears the type II pyrethroid deltamethrin in human liver, whereas CYP-mediated oxidation of deltamethrin is a minor pathway.26) This finding is in contrast to the rat liver, where CYP-mediated oxidation of deltamethrin is the most quantitatively important pathway of metabolism, whereas CES-mediated hydrolysis is slow and of relatively minor importance. Furthermore, the type I pyrethroids trans-permethrin and bioresmethrin were hydrolyzed by rat and human CES1 at comparable rates.28) Thus, rats are a good model species to study the pharmacokinetics of type I pyrethroids, yet they may not be appropriate for studying type II pyrethroids because of the much slower hydrolytic rates observed in this species.
OP insecticides, such as chlorpyrifos and parathion, are oxidized by hepatic CYPs to electrophilic metabolites (termed oxons), which are highly potent non-specific inhibitors of serine hydrolases (e.g., AChE and CES). Inhibition of AChE, which produces a rapid and lethal cholinergic crisis, is the primary mechanism by which bioactive metabolites of OPs elicit their toxicological effects.31) Nevertheless, CES are also targets of reactive oxons, which may elicit unique toxicities that have heretofore been unexamined. Our laboratory has shown that the oxons of chlorpyrifos and parathion insecticides are potent inhibitors of human CES1 with IC50 values in the sub-nanomolar range.20) Therefore, because of their abundance in liver, CES are an important scavenger of OP reactive metabolites. Moreover, development of CES1-based protein therapeutics is being pursued by the Redinbo and Potter groups with the goal of preventing OP nerve agent toxicity and protecting against the long-term side effects of these agents, which could be released on populations in a terrorist attack, during warfare, and following deliberate or accidental exposure to OP agrochemicals.32)
Carboxylesterase gene regulation
The mechanisms involved in the regulation of mammalian CES gene expression have become better clarified, yet much needs to be learned. Promoter regions have been characterized in human, rat, and mouse CES1 and CES2 genes.33) A TATA box was not found in any of the CES promoters; however, they do share several common binding sites for transcription factors (e.g., Sp1 and C/EBPα), suggesting that the orthologous CES genes have evolutionally conserved transcriptional regulatory mechanisms. The CpG methylation state of the human CES1 promoter appeared to be a major determinant of tissue-specific expression since the marked difference in CES1 gene expression in the human kidney (low) and liver (high) may arise from a difference in DNA methylation levels in the promoter region.34)
With respect to liver-enriched ligand activated transcription factors, the murine CES2 family gene Ces6 was recently shown to be regulated in vivo by the pregnane X receptor (PXR) and constitutive androstane receptor (CAR), two members of the nuclear receptor superfamily, when mice were treated with prototypical murine PXR- and CAR-specific ligands, pregnenolone 16α-carbonitrile and 1,4-bis[2-(3,5-dichloro-pyridyloxy)]benzene, respectively.35) Although it was previously shown that pyrethroids can activate human PXR and CAR, it is not apparent that pyrethroids have any effect on the levels of CES1 and CES2 mRNA in human primary hepatocytes following insecticide treatment.36) However, PXR-responsive CYP3A4 mRNA was found to be significantly induced in the pyrethroid-treated hepatocytes. Similar results were obtained when protein amounts in primary hepatocytes were examined by western blotting; CES1 and CES2 protein levels were unchanged by pyrethroid treatment, whereas CYP3A4 protein amounts were substantially increased.36) It has also been observed that pyrethroids had no effect on reporter activity when a human CES1 proximal promoter-luciferase reporter and human PXR expression vector were transiently transfected into HepG2 cells and the transfected cells treated with pyrethroids (Streit and Ross, unpublished data). No significant enhancement in luciferase reporter activity was observed by any of the compounds tested; even rifampicin, a well characterized PXR ligand, had no effect. However, when a CYP3A4 promoter-luciferase reporter was used in a similar series of experiments, luciferase reporter activity was robustly activated by the pyrethroid treatment.37) These results indicate that human CES1 gene expression is not regulated by ligand-activated PXR and that pyrethroids likely have no influence on CES1 gene expression in human populations, although murine CES (Ces6) does appear to be inducible in a PXR-dependent manner both in vitro and in vivo.35) Further evidence of a species difference with respect to nuclear receptors and CES gene expression has been observed with the liver X receptor (LXR). Like PXR and CAR, LXR α and β are also members of the nuclear receptor superfamily and are involved in the regulation of genes that control lipid metabolism.38) A transcriptomic study in wildtype and LXRαβ double knockout mice showed that a synthetic ligand for LXR caused the down-regulation of the murine CES gene Es31 by at least 2-fold in liver39), whereas other data indicated that the same LXR synthetic ligand could activate a human CES1 proximal promoter-luciferase reporter 2.6-fold in cells overexpressing LXRα.11) Thus opposing effects of LXR activation on CES gene expression may be apparent when comparing mice and humans, revealing that complex regulatory mechanisms are at work.
Role of carboxylesterases in lipid metabolism
It is now clearly apparent that CES have a role in lipid metabolism and that targeting this enzyme may impact disease phenotype, such as diabetes and atherosclerosis.11,12,18) For example, human CES1 and its murine ortholog Ces3 are responsible for mobilizing cytosolic TG pools and their subsequent assembly into very low-density lipoproteins in hepatocytes, which are subsequently trafficked out of the cells and into the circulation.40) Further, CES controls in part the lipolysis of TGs in mouse adipose tissues41), which if unregulated can result in high levels of fatty acids in the circulation and lipotoxicity, a clinical manifestation of diabetes. As mentioned earlier, CES also have a role in cholesterol metabolism in macrophages42), and the cholesteryl ester hydrolyzing activity of CES1 in human macrophages may be an important mechanism to prevent cells from becoming engorged with cholesteryl esters and thus turning into inflammatory foam cells, which accelerates atherosclerosis development.20,43) Further, data from our laboratory suggests that an endogenous cannabinoid is efficiently metabolized by CES.13) The endocannabinoids are arachidonoyl derivatives produced on demand in cells. These bioactive lipids are involved in multiple biological processes in both central and peripheral tissues.44) The pathophysiologies of obesity, diabetes, and atherosclerosis each have aspects that involve endocannabinoids.45) The two best studied of these lipid mediators are 2-arachidonoylglycerol (2-AG) and N-arachidonoyl-ethanolamide (AEA). Both compounds bind and activate the G-protein coupled receptors known as cannabinoid receptors (CB1 and CB2). CB1 is abundantly expressed in the central nervous system and to a lesser degree in peripheral tissues, such as macrophages, while CB2 receptors are predominantly expressed in macrophages and other immune cells and to a lesser extent in the CNS.46) Recent findings in animal models of disease suggest that CB1 receptor antagonism using rimonabant reduces the extent of atherosclerosis47), whereas activation of CB2 receptors by a cannabinoid agonist (Δ9-THC) protects against atherosclerosis by possibly down-regulating the production of inflammatory mediators by immune cells.48) Furthermore, activation of peripheral CB1 receptors by endogenous cannabinoids has been shown to contribute to diet-induced obesity in mice and to stimulate hepatic lipogenesis.49) Thus, endocannabinoids have important functions in controlling metabolism and regulation of their tissue levels is crucial for maintaining physiological homeostasis. Therefore, in addition to MAGL and FAAH50), CES also appears to hydrolyze 2-AG. An intriguing future direction in endocannabinoid research will be to perturb CES function using chemical inhibitors, siRNA methods, and knockout mice and determine whether this will affect endocannabinoid tone and disease phenotype.
CES1 inhibition and esterolytic metabolism of cholesteryl esters
Cardiovascular disease (CVD) continues to be the leading cause of death in the United States.51) Atherosclerosis is the pathologic process responsible for CVD and inflammation plays a major role in the development of atherosclerosis.52) During vascular damage and inflammation, blood monocytes migrate into the arterial intima where they differentiate into macrophages and secrete pro-inflammatory cytokines and chemotactic factors. Intimal macrophages scavenge oxidized low-density lipoproteins (oxLDL), of which cholesteryl esters are a major component, via scavenger receptors SR-A and CD36.53,54) Following uptake of oxLDL, cholesteryl esters are hydrolyzed to free cholesterol and fatty acids within lysosomes.55) Excess free cholesterol is toxic to cells thereby requiring intracellular free cholesterol levels to be tightly regulated.56) Consequently, free cholesterol is re-esterified by acyl coenzyme A: cholesterol acyltransferase 1 (ACAT-1) rendering highly water insoluble cholesteryl esters, which are stored in cytoplasmic lipid droplets.57) Intimal macrophages are transformed into foam cells when they accumulate excessive amounts cholesteryl esters, which is apparent under a microscope from the large number of lipid droplets found in the cytoplasm. Mobilization of cholesterol for export is initiated by the hydrolysis of the stored cholesteryl esters. This activity is catalyzed by a neutral cholesteryl ester hydrolase and is the rate-limiting step in macrophage cholesterol mobilization.58) Free cholesterol is transported out of the macrophages via ABCA1 and ABCG1 cholesterol transporters to extracellular acceptors (ApoA-I and HDL, respectively), which are then transported via HDL to the liver for disposal in the bile. Transport of cholesterol from vessel wall macrophages to liver is termed macrophage reverse cholesterol transport (RCT) and is considered to be the mechanism by which regression of atherosclerotic lesions occurs.59)
One of the candidate enzymes responsible for the cholesteryl ester hydrolase activity in human macrophages is CES1, which was originally cloned from a human THP-1 monocyte/macrophage cell line.60) In support of this, overexpression of CES1 in COS-7 cells yielded elevated amounts of cholesteryl ester hydrolytic activity in cell lysates compared to mock transfected cells.61) CES1 was also found to be localized in the cytoplasm of human macrophage cells and to be associated with the cholesteryl ester lipid droplet within the cytoplasm.43) Convincing evidence of a role for CES1 in cholesterol metabolism came from studies in which overexpression of CES1 in THP-1 macrophages was shown to markedly increase the rate of cholesterol efflux in vitro.43) Moreover, Zhao et al.62) demonstrated that over-expression of human CES1 in macrophages caused increased rates of macrophage RCT and decreased atherosclerosis in Western diet-fed Ldlr−/− mice. Furthermore, macrophage-specific hCES1 transgenic Ldlr−/− mice also exhibited improved glucose and insulin tolerance and had reduced inflammatory mediator profiles compared to their non-transgenic controls when placed on a Western diet.63) These findings were possibly due to attenuated NFκB and AP-1 activities in adipose tissue macrophages that overexpress hCES1, which may be linked to reduced cellular cholesterol levels in these cells. Thus, both in vitro and in vivo studies point to an essential role for CES1 in cholesteryl ester hydrolysis in macrophages, which may have important implications for the development of metabolic syndrome disease and atherosclerosis.
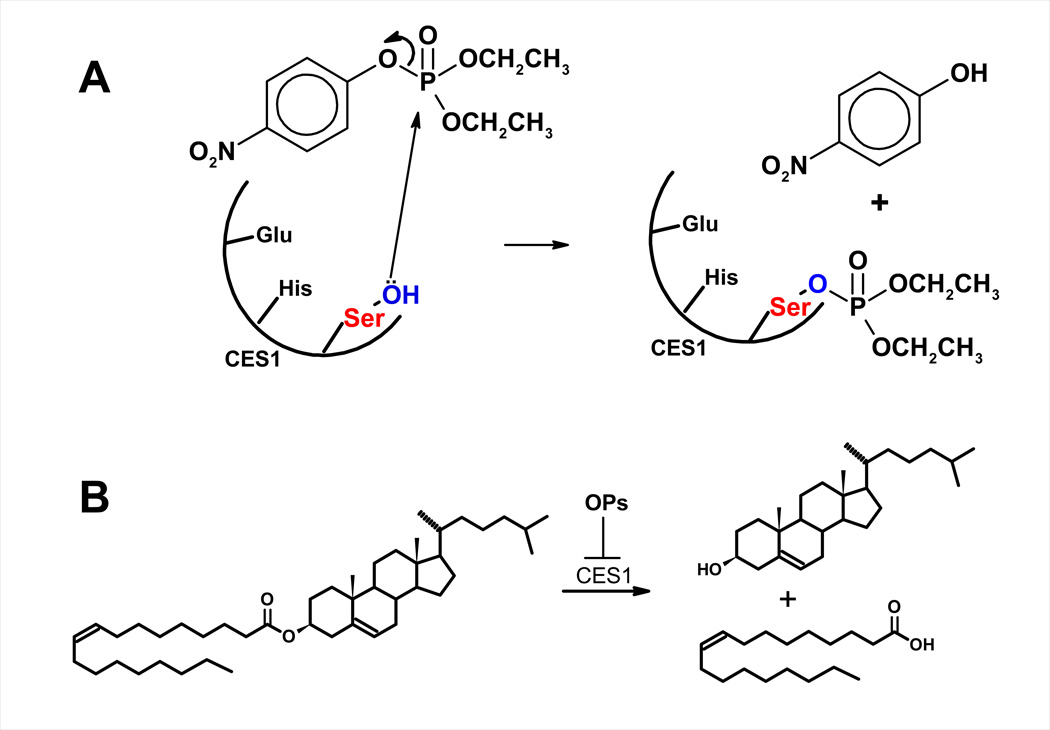
The findings described above strongly support the proposal that hydrolysis of intracellular cholesteryl esters is the rate-limiting step in macrophage RCT. If CES1 is responsible for macrophage cholesteryl ester hydrolysis, then inhibition of CES1 activity would be predicted to inhibit cholesterol efflux from macrophages. Our laboratory has been interested in studying whether environmental pollutants may exacerbate pathological foam cell formation. Toward this goal, we treated cultured human THP-1 foam cells with OP bioactive metabolite (paraoxon) or CES1-specific inhibitor (diphenylethane-1,2-dione; benzil) and found that these chemical treatments caused significant retention of cholesteryl esters in the cultured macrophages under conditions that promote lipoprotein-dependent cholesterol efflux.20) Therefore, it is conceivable that inhibition of CES1 activity may exacerbate the macrophage foam cell phenotype, i.e., make macrophages more prone to become foam cells (Figure 2).
Figure 2.
(A) Inhibition of CES by paraoxon, the bioactive metabolite of the insecticide parathion. (B) Hydrolysis of cholesteryl oleate by CES1 and inhibition of this activity by bioactive metabolites of OP insecticides.
The hypothesis that inhibition of cholesteryl ester hydrolase activity causes an excess accumulation of intracellular cholesteryl esters is not without precedence. In steroidogenic cells a neutral cholesteryl ester hydrolase cleaves cholesteryl esters to generate free cholesterol for steroid hormone biosynthesis. Following treatment of female rats with either tricresyl phosphate or butylated triphenyl phosphate, both organophosphate compounds, cholesteryl esters in the adrenal glands and ovaries were found to accumulate when compared to control animals.64) This study demonstrated that the level of cholesteryl ester accumulation correlated with the degree of inhibition of a neutral cholesteryl ester hydrolase, strongly suggesting that this inhibition was responsible for the cholesteryl ester accumulation. Further, examples of pesticides that can inhibit CES activity are not only limited to OPs. For instance, molinate is an herbicide used on rice that is bioactivated in vivo to electrophilic sulfoxide and sulfone metabolites that covalently react with rat Hydrolase A (the rat ortholog of human CES1) in a highly specific manner. This modification leads to markedly reduced CES activity in rat liver and testis, which in the case of Leydig cells could inhibit the mobilization of cholesterol esters required for testosterone biosynthesis.74) Moreover, the hypothesis that reduced cholesteryl ester hydrolase activity can increase atherogenesis also has precedence. Several examples of decreased macrophage cholesteryl ester hydrolase activity have been associated with an increased risk of atherosclerosis.(65–68) Increasingly, environmental toxicants are being recognized as etiological agents that contribute to atherosclerosis.69) Our results have clearly shown that oxons of OP insecticides are able to affect CES1-catalyzed hydrolytic activities in THP-1 monocytes/macrophages. Taken together, these findings suggest that exposure to oxons may adversely affect macrophage cholesterol efflux and contribute to the development of atherosclerosis.
In addition to man-made chemicals such as OP insecticides, it was also shown that the CES activity of recombinant CES1, THP-1 cell lysates, and intact THP-1 cells could be potently inhibited by naturally occurring lipids found in cells, such as 27-hydroxycholesterol (27-HC) and arachidonic acid.70) For example, 27-HC is a partially noncompetitive inhibitor of recombinant CES1 (Kiapp=10 nM) and could impair intracellular CES1 activity when 27-HC was added to culture medium containing intact THP1 monocytes. Therefore, elevated levels of oxysterols and arachidonic acid in cells may alter the ability of CES1 to both detoxify environmental pollutants and metabolize endogenous compounds in vivo. In order to establish the physiological relevance of this finding, our laboratory is currently determining whether the in situ generation of these natural lipids in macrophages can impede the activity of CES1.
Future outlook
Hydrolysis of ester-containing substrates in mammalian species is mainly catalyzed by the CES superfamily, which has been largely conserved throughout evolutionary history. In humans, two isoforms, CES1 and CES2, are widely distributed in organs and tissues, with particularly high expression in the hepato-intestinal axis where exposure to high concentrations of xenobiotics arises. A majority of the reports on the function of the CES enzymes have focused on their ability to metabolize drugs, pesticides, and environmental pollutants. However, it is now becoming clear that the CES family has important roles in lipid metabolism, and CES may be an attractive target for the treatment of diseases such as metabolic syndrome and atherosclerosis. The continued development of selective chemical inhibitors and gene knockout models will provide essential tools for the study of CES function in biological systems. Therefore, the outlook for CES research is exciting and many new discoveries with potential benefits for agriculture, human health, and homeland security are eagerly anticipated.
Acknowledgement
Research support to M.K.R. was provided by NIH 1R15ES015348-01A1 and 3R15ES015348-01A1S1 (competitive supplement).
References
- 1.Cygler M, Schrag JD, Sussman JL, Harel M, Silman I, Gentry MK, Doctor BP. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bencharit S, Morton CL, Hyatt JL, Kuhn P, Danks MK, Potter PM, Redinbo MR. Crystal structure of human carboxylesterase 1 complexed with the Alzheimer's drug tacrine. From binding promiscuity to selective inhibition. Chem. Biol. 2003;10:341–349. doi: 10.1016/s1074-5521(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 3.Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu. Rev. Pharmacol. Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 4.Ross MK, Crow JA. Role of carboxylesterases in xenobiotic and endobiotic metabolism. J. Biochem. Mol. Toxicol. 2007;21:187–196. doi: 10.1002/jbt.20178. [DOI] [PubMed] [Google Scholar]
- 5.Sogorb MA, Vilanova E. Enzymes involved in the detoxification of organophosphorus, carbamate and pyrethroid insecticides through hydrolysis. Toxicol. Lett. 2002;128:215–228. doi: 10.1016/s0378-4274(01)00543-4. [DOI] [PubMed] [Google Scholar]
- 6.Casida JE JE, Quistad GB. Golden age of insecticide research: Past, present, or future? Annu. Rev. Entomol. 1998;43:1–16. doi: 10.1146/annurev.ento.43.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Epstein L, Bassein S. Patterns of pesticide use in California and the implications for strategies for reduction of pesticides. Annu. Rev. Phytopathol. 2003;41:351–375. doi: 10.1146/annurev.phyto.41.052002.095612. [DOI] [PubMed] [Google Scholar]
- 8.Soderlund DM, Clark JM, Sheets LP, Mullin LS, Piccirillo VJ, Sargent D, Stevens JT, Weiner ML. Mechanisms of pyrethroid neurotoxicity: implications for cumulative risk assessment. Toxicology. 2002;171:3–59. doi: 10.1016/s0300-483x(01)00569-8. [DOI] [PubMed] [Google Scholar]
- 9.Wheelock CE, Wheelock AM, Zhang R, Stok JE, Morisseau C, Le Valley SE, Green CE, Hammock BD. Evaluation of alpha-cyanoesters as fluorescent substrates for examining interindividual variation in general and pyrethroid-selective esterases in human liver microsomes. Anal. Biochem. 2003;315:208–222. doi: 10.1016/s0003-2697(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 10.Nishi K, Huang H, Kamita SG, Kim IH, Morisseau C, Hammock BD. Characterization of pyrethroid hydrolysis by the human liver carboxylesterases hCE-1 and hCE-2. Arch. Biochem. Biophys. 2006;445:115–123. doi: 10.1016/j.abb.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh S, Zhao B, Bie J, Song J. Macrophage cholesteryl ester mobilization and atherosclerosis. Vascul. Pharmacol. 2010;52:1–10. doi: 10.1016/j.vph.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolinsky VW, Gilham D, Alam M, Vance DE, Lehner R. Triacylglycerol hydrolase: role in intracellular lipid metabolism. Cell Mol. Life Sci. 2004;61:1633–1651. doi: 10.1007/s00018-004-3426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streit TM, Borazjani A, Lentz SE, Wierdl M, Potter PM, Gwaltney SR, Ross MK. Evaluation of the ‘side door’ in carboxylesterase-mediated catalysis and inhibition. Biol. Chem. 2008;389:149–162. doi: 10.1515/BC.2008.017. [DOI] [PubMed] [Google Scholar]
- 14.Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu. Rev. Pharmacol. Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 15.Potter PM, Wadkins RM. Carboxylesterases - detoxifying enzymes and targets for drug therapy. Curr. Med. Chem. 2006;13:1045–1054. doi: 10.2174/092986706776360969. [DOI] [PubMed] [Google Scholar]
- 16.Satoh T, Taylor P, Bosron WF, Sanghani SP, Hosokawa M. La Du BN Current progress on esterases: from molecular structure to function. Drug Metab. Dispos. 2002;30:488–493. doi: 10.1124/dmd.30.5.488. [DOI] [PubMed] [Google Scholar]
- 17.Sun A, Jiang Y, Wang X, Liu Q, Zhong F, He Q, Guan W, Li H, Sun Y, Shi L, Yu H, Yang D, Xu Y, Song Y, Tong W, Li D, Lin C, Hao Y, Geng C, Yun D, Zhang X, Yuan X, Chen P, Zhu Y, Li Y, Liang S, Zhao X, Liu S, He F. Liverbase: a comprehensive view of human liver biology. J. Proteome Res. 2010;9:50–58. doi: 10.1021/pr900191p. [DOI] [PubMed] [Google Scholar]
- 18.Wei E, Ben Ali Y, Lyon J, Wang H, Nelson R, Dolinsky VW, Dyck JR, Mitchell G, Korbutt GS, Lehner R. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 2010;11:183–193. doi: 10.1016/j.cmet.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Wadkins RM, Hyatt JL, Wei X, Yoon KJ, Wierdl M, Edwards CC, Morton CL, Obenauer JC, Damodaran K, Beroza P, Danks MK, Potter PM. Identification and characterization of novel benzil (diphenylethane-1,2-dione)analogues as inhibitors of mammalian carboxylesterases. J. Med. Chem. 2005;48:2906–2915. doi: 10.1021/jm049011j. [DOI] [PubMed] [Google Scholar]
- 20.Crow JA, Middleton BL, Borazjani A, Hatfield MJ, Potter PM, Ross MK. Inhibition of carboxylesterase 1 is associated with cholesteryl ester retention in human THP-1 monocyte/macrophages. Biochim. Biophys. Acta. 2008;1781:643–654. doi: 10.1016/j.bbalip.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgson E, Levi PE. Pesticides: an important but underused model for the environmental health sciences. Environ. Health Perspect. 1996;104:97–106. doi: 10.1289/ehp.96104s197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takken W. Do insecticide-treated bed nets have an effect on malaria vectors? Trop. Med. Int. Health. 2002;7:1022–1030. doi: 10.1046/j.1365-3156.2002.00983.x. [DOI] [PubMed] [Google Scholar]
- 23.Gaughan LC, Engel JL, Casida JE. Pesticide Interactions: Effects of organophophorus pesticides on the metabolism, toxicity, and persistence of selected pyrethroid insecticides. Pest Biochem. Physiol. 1980;14:81–85. [Google Scholar]
- 24.Casida JE, Gammon DW, Glickman AH, Lawrence LJ. Mechanisms of selective action of pyrethroid insecticides. Annu. Rev. Pharmacol. Toxicol. 1983;23:413–438. doi: 10.1146/annurev.pa.23.040183.002213. [DOI] [PubMed] [Google Scholar]
- 25.Casida JE, Ruzo LO. Metabolic chemistry of pyrethroid insecticides. Pestic. Sci. 1980;11:257–269. [Google Scholar]
- 26.Godin SJ, Scollon EJ, Hughes MF, Potter PM, DeVito MJ, Ross MK. Species differences in the in vitro metabolism of deltamethrin and esfenvalerate: Differential oxidative and hydrolytic metabolism by humans and rats. Drug Metab. Dispos. 2006;34:1764–1771. doi: 10.1124/dmd.106.010058. [DOI] [PubMed] [Google Scholar]
- 27.Godin SJ, Crow JA, Scollon EJ, Hughes MF, DeVito MJ, Ross MK. Identification of rat and human cytochrome P450 isoforms and a rat serum esterase that metabolize the pyrethroid insecticides deltamethrin and esfenvalerate. Drug Metab. Dispos. 2007;35:1664–1671. doi: 10.1124/dmd.107.015388. [DOI] [PubMed] [Google Scholar]
- 28.Ross MK, Borazjani A, Edwards CC, Potter PM. Hydrolytic metabolism of pyrethroids by human and other mammalian carboxylesterases. Biochem. Pharmacol. 2006;71:657–669. doi: 10.1016/j.bcp.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson A. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 2001;6:133–224. [Google Scholar]
- 30.Heidari R, Devonshire AL, Campbell BE, Dorrian SJ, Oakeshott JG, Russell RJ. Hydrolysis of pyrethroids by carboxylesterases from Lucilia cuprina and Drosophila melanogaster with active sites modified by in vitro mutagenesis. Insect Biochem. Mol. Biol. 2005;35:597–609. doi: 10.1016/j.ibmb.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem. Res. Toxicol. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- 32.Hemmert AC, Otto TC, Wierdl M, Edwards CC, Fleming CD, MacDonald M, Cashman JR, Potter PM, Cerasoli DM, Redinbo MR. Human carboxylesterase 1 stereoselectively binds the nerve agent cyclosarin and spontaneously hydrolyzes the nerve agent sarin. Mol. Pharmacol. 2010;77:508–516. doi: 10.1124/mol.109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosokawa M, Furihata T, Yaginuma Y, Yamamoto N, Koyano N, Fujii A, Nagahara Y, Satoh T, Chiba K. Genomic structure and transcriptional regulation of the rat, mouse, and human carboxylesterase genes. Drug Metab. Rev. 2007;39:1–15. doi: 10.1080/03602530600952164. [DOI] [PubMed] [Google Scholar]
- 34.Hori T, Hosokawa M. DNA methylation and its involvement in carboxylesterase 1A1 (CES1A1) gene expression. Xenobiotica. 2010;40:119–128. doi: 10.3109/00498250903431794. [DOI] [PubMed] [Google Scholar]
- 35.Xu C, Wang X, Staudinger JL. Regulation of tissue-specific carboxylesterase expression by pregnane×receptor and constitutive androstane receptor. Drug Metab. Dispos. 2009;37:1539–1547. doi: 10.1124/dmd.109.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang D, Wang X, Chen YT, Deng R, Yan B. Pyrethroid insecticides: isoform-dependent hydrolysis, induction of cytochrome P450 3A4 and evidence on the involvement of the pregnane X receptor. Toxicol. Appl. Pharmacol. 2009;237:49–58. doi: 10.1016/j.taap.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das PC, Streit TM, Cao Y, Rose RL, Cherrington N, Ross MK, Wallace AD, Hodgson E. Pyrethroids: cytotoxicity and induction of CYP isoforms in human hepatocytes. Drug Metab. Drug Interact. 2008;23:211–236. doi: 10.1515/dmdi.2008.23.3-4.211. [DOI] [PubMed] [Google Scholar]
- 38.Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stulnig TM, Steffensen KR, Gao H, Reimers M, Dahlman-Wright K, Schuster GU, Gustafsson JA. Novel roles of liver X receptors exposed by gene expression profiling in liver and adipose tissue. Mol. Pharmacol. 2002;62:1299–1305. doi: 10.1124/mol.62.6.1299. [DOI] [PubMed] [Google Scholar]
- 40.Gilham D, Ho S, Rasouli M, Martres P, Vance DE, Lehner R. Inhibitors of hepatic microsomal triacylglycerol hydrolase decrease very low density lipoprotein secretion. FASEB J. 2003;17:1685–1687. doi: 10.1096/fj.02-0728fje. [DOI] [PubMed] [Google Scholar]
- 41.Wei E, Gao W, Lehner R. Attenuation of adipocyte triacylglycerol hydrolase activity decreases basal fatty acid efflux. J. Biol. Chem. 2007;282:8027–8035. doi: 10.1074/jbc.M605789200. [DOI] [PubMed] [Google Scholar]
- 42.Ghosh S. Cholesteryl ester hydrolase in human monocyte/macrophage: cloning, sequencing and expression of full length cDNA. Physiol. Genomics. 2000;2:1–8. doi: 10.1152/physiolgenomics.2000.2.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Zhao B, Song J, St Clair R, Ghosh S. Stable over-expression of human macrophage cholesteryl ester hydrolase (CEH) results in enhanced free cholesterol efflux from human THP1-macrophages. Am. J. Physiol. Cell Physiol. 2007;292:C405–C412. doi: 10.1152/ajpcell.00306.2006. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 2009;49:123–150. doi: 10.1146/annurev.pharmtox.011008.145616. [DOI] [PubMed] [Google Scholar]
- 45.Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nat. Rev. Drug Discov. 2008;7:438–455. doi: 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- 46.Pacher P, Steffens S. The emerging role of the endocannabinoid system in cardiovascular disease. Semin. Immunopathol. 2009;31:63–77. doi: 10.1007/s00281-009-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dol-Gleizes F, Paumelle R, Visentin V, Marés AM, Desitter P, Hennuyer N, Gilde A, Staels B, Schaeffer P, Bono F. Rimonabant, a selective cannabinoid CB1 receptor antagonist, inhibits atherosclerosis in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2009;29:12–18. doi: 10.1161/ATVBAHA.108.168757. [DOI] [PubMed] [Google Scholar]
- 48.Steffens S, Veillard NR, Arnaud C, Pelli G, Burger F, Staub C, Karsak M, Zimmer A, Frossard JL, Mach F. Low dose oral cannabinoid therapy reduces progression of atherosclerosis in mice. Nature. 2005;434:782–786. doi: 10.1038/nature03389. [DOI] [PubMed] [Google Scholar]
- 49.Osei-Hyiaman D, DePetrillo M, Pacher P, Liu J, Radaeva S, Bátkai S, Harvey-White J, Mackie K, Offertáler L, Wang L, Kunos G. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J. Clin. Invest. 2005;115:1298–305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem. Rev. 2008;108:1687–707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.American Heart Association. Heart Disease and Stroke Statistics-2008 Update. 2008 [Google Scholar]
- 52.Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111:3481–3488. doi: 10.1161/CIRCULATIONAHA.105.537878. [DOI] [PubMed] [Google Scholar]
- 53.Babaev VR, Gleaves LA, Carter KJ, Suzuki H, Kodama T, Fazio S, Linton MF. Reduced atherosclerotic lesions in mice deficient for total or macrophage-specific expression of scavenger receptor-A. Arterioscler. Thomb. Vasc. Biol. 2000;20:2593–2599. doi: 10.1161/01.atv.20.12.2593. [DOI] [PubMed] [Google Scholar]
- 54.Linton MF, Babaev VR, Gleaves LA, Fazio S. A direct role for the macrophage low density lipoprotein receptor in atherosclerotic lesion formation. J. Biol. Chem. 1999;274:19204–19210. doi: 10.1074/jbc.274.27.19204. [DOI] [PubMed] [Google Scholar]
- 55.Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. J. Biol. Chem. 1975;250:8487–8795. [PubMed] [Google Scholar]
- 56.Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J. Clin. Invest. 2002;110:905–911. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang TY, Chang CC, Lin S, Yu C, Li BL, Miyazaki A. Roles of acyl-coenzyme A:cholesterol acyltransferase-1 and-2. Curr. Opin. Lipidol. 2001;12:289–296. doi: 10.1097/00041433-200106000-00008. [DOI] [PubMed] [Google Scholar]
- 58.Rothblat GH, Llera-Moya MDL, Favari E, Yancey PG, Kellner-Weibel G. Cellular cholesterol flux studies: methodological considerations. Atherosclerosis. 2002;163:1–8. doi: 10.1016/s0021-9150(01)00713-4. [DOI] [PubMed] [Google Scholar]
- 59.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ. Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- 60.Ghosh S. Cholesteryl ester hydrolase in human monocyte/macrophage: cloning, sequencing and expression of full length cDNA. Physiol. Genomics. 2000;2:1–8. doi: 10.1152/physiolgenomics.2000.2.1.1. [DOI] [PubMed] [Google Scholar]
- 61.Zhao B, Natarajan R, Ghosh S. Human liver cholesteryl ester hydrolase: cloning, molecular characterization, and role in cellular cholesterol homeostasis. Physiol. Genomics. 2005;23:304–310. doi: 10.1152/physiolgenomics.00187.2005. [DOI] [PubMed] [Google Scholar]
- 62.Zhao B, Song J, Chow WN, St. Clair RW, Rudel LL, Ghosh S. Macrophage-specific transgenic expression of cholesteryl ester hydrolase significantly reduces atherosclerosis and lesion necrosis in Ldlr−/−mice. J. Clin. Invest. 2007;117:2983–2992. doi: 10.1172/JCI30485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bie J, Zhao B, Song J, Ghosh S. Improved insulin sensitivity in high-fat high-cholesterol fed LDLR−/− mice with macrophage-specific transgenic expression of cholesteryl ester hydrolase: Role of macrophage inflammation and infiltration into adipose tissue. J. Biol Chem. 2010;285:13630–13637. doi: 10.1074/jbc.M109.069781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Latendresse JR, Azhar S, Brooks CL, Capen CC. Pathogenesis of cholesteryl lipdoisis of adrenocortical and ovarian interstitial cells in F344 rats caused by tricresyl phosphate and butylated triphenyl phosphate. Toxicol. Appl. Pharmacol. 1993;122:281–289. doi: 10.1006/taap.1993.1197. [DOI] [PubMed] [Google Scholar]
- 65.Mathur SN, Field FJ, Megan MB, Armstrong HL. A defect in mobilization of cholesteryl esters in macrophages. Biochim. Biophys. Acta. 1985;834:48–57. doi: 10.1016/0005-2760(85)90175-4. [DOI] [PubMed] [Google Scholar]
- 66.Ishii I, Oka M, Katto N, Shirai K, Saito Y, Hirose S. Beta-VLDL-induced cholesterolester deposition in macrophages may be regulated by neutral cholesterol esterase activity. Arterioscler. Thomb. 1992;12:1139–1145. doi: 10.1161/01.atv.12.10.1139. [DOI] [PubMed] [Google Scholar]
- 67.Hakamata H, Miyazaki A, Sakai M, Suginohara Y, Sakamoto Y, Horiuchi S. Species differences in cholesteryl ester cycle and HDL-induced cholesterol efflux from macrophage foam cells. Arterioscler. Thomb. 1994;14:1860–1865. doi: 10.1161/01.atv.14.11.1860. [DOI] [PubMed] [Google Scholar]
- 68.Yancey PG, St. Clair RW. Mechanism of the defect in cholesteryl ester clearance from macrophages of atherosclerosis-susceptible White Carneau pigeons. J. Lipid Res. 1994;35:2114–2129. [PubMed] [Google Scholar]
- 69.O'Toole TE, Conklin DJ, Bhatnagar A. Environmental risk factors for heart disease. Rev. Environ. Health. 2008;23:167–202. doi: 10.1515/reveh.2008.23.3.167. [DOI] [PubMed] [Google Scholar]
- 70.Crow JA, Herring KL, Xie S, Borazjani A, Potter PM, Ross MK. Inhibition of carboxylesterase activity of THP1 monocytes/macrophages and recombinant human carboxylesterase 1 by oxysterols and fatty acids. Biochim. Biophys. Acta. 2010;1801:31–41. doi: 10.1016/j.bbalip.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanghani SP, Sanghani PC, Schiel MA, Bosron WF. Human carboxylesterases: an update on CES1, CES2 and CES3. Protein Pept. Lett. 2009;16:1207–1214. doi: 10.2174/092986609789071324. [DOI] [PubMed] [Google Scholar]
- 72.Redinbo MR, Potter PM. Mammalian carboxylesterases: from drug targets to protein therapeutics. Drug Discov. Today. 2005;10:313–325. doi: 10.1016/S1359-6446(05)03383-0. [DOI] [PubMed] [Google Scholar]
- 73.Copley SD. Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 2003;7:265–272. doi: 10.1016/s1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 74.Jewell WT, Miller MG. Identification of a carboxylesterase as the major protein bound by molinate. Toxicol. Appl. Pharmacol. 1998;149:226–234. doi: 10.1006/taap.1998.8381. [DOI] [PubMed] [Google Scholar]