Abstract
The first TCR-dependent checkpoint in the thymus determines αβ vs. γδ T lineage fate and sets the stage for later T cell differentiation decisions. We had previously shown that early T cells in NOD mice that are unable to rearrange a TCR exhibit a defect in checkpoint enforcement at this stage. To determine if T cell progenitors from wild type NOD mice also exhibit cell-autonomous defects in development we investigated their differentiation in the Notch-ligand presenting OP9-DL1 co-culture system, as well as by analysis of T cell development in vivo. Cultured CD4 and CD8 double negative (DN) cells from NOD mice exhibited major defects in the generation of CD4 and CD8 double positive (DP) αβT cells, while γδT cell development from bipotent precursors was enhanced. Limiting dilution and single cell experiments show that the divergent effects on αβ and γδ T cell development did not spring from biased lineage choice but from increased proliferation of γδT cells and impaired accumulation of αβT lineage DP cells. In vivo, NOD early T cell subsets in the thymus also show characteristics indicative of defective β-selection, and peripheral αβT cells are poorly established in mixed bone marrow chimeras, contrasting with strong γδT as well as B cell repopulation. Thus, NOD T cell precursors reveal divergent, lineage-specific differentiation abnormalities in vitro and in vivo from the first TCR-dependent developmental choice point, which may have consequences for subsequent lineage decisions and effector functions.
Keywords: T cell development, thymus, β-selection, γδT cells
Introduction
T cells in all jawed vertebrates can be divided into two major subsets, those that use αβ T cell receptors (TCR) and those that use γδ TCR. These two major lineages are broadly similar, both requiring successful TCR gene rearrangement to develop and utilizing many of the same effector genes. Yet accumulating evidence shows that their development is promoted by different balances of transcription factors, activating signals, and inputs from Notch-Delta interaction. While most of these effects have been seen in the artificial context of targeted mutant mice, we here present evidence that there are unexpectedly sharp, divergent abnormalities in αβT and γδT cell developmental programming in the unmanipulated genetic background of the autoimmune-prone, nonobese diabetic (NOD) mouse.
All T cell lineages arise from the few multipotent precursor cells that migrate from bone marrow to the thymus, which provides the environmental signals, including Notch ligands and cytokines, needed for these progenitor cells to carry out the process of T lineage specification and commitment. These cells undergo extensive proliferation and an ordered series of differentiation stages characterized by a progressive induction of T cell identity genes and loss of stem cell and other genes permitting alternative fate choices (1). These stages are defined by specific changes in cell surface markers. In mice, the early double negative (DN) stages lack CD4 and CD8, as well as rearranged TCR, and differentiate first from CD44+CD25−cKit+ DN1 cells (or early thymic precursors (ETPs)) to CD44+CD25+cKit+ DN2 cells and then to CD44-CD25+cKitlo DN3 cells (2, 3). T cell receptor (TCR) rearrangements are ongoing from the DN2 through the DN3 stage, and differentiation past the first T cell checkpoint at DN3 requires successful rearrangement and expression of a TCRβ chain or TCRβ and δ chains. If TCRβ molecules are expressed, they pair with invariant preTα molecules, triggering TCR signaling that results in β-selection (4-6). This is a process of rapid, major regulatory changes and extensive proliferation, coupled with effects on survival, culminating in differentiation to the CD4+CD8+ double positive (DP) stage. These DP cells undergo TCRα rearrangements and further selection and differentiation, based primarily but not solely on properties of the new αβTCR. T cells with strongly self-reactive TCRs are negatively selected and die, while cells with more moderate specificity for self-antigens differentiate into a variety of effector populations including CD4 or CD8 single positive T cells, as well as unconventional cells like regulatory T cells, NKT, and CD8αα cells, before exiting to the periphery (reviewed in (7)). Alternatively, if β and δ TCR chains are rearranged before a TCRβ chain, γδTCR expression triggers an alternative differentiation program, γδ-selection, that typically results in a lower level of proliferation and no upregulation of CD4 or CD8 (reviewed in (8)). The nature of the receptor itself does not necessarily determine whether a cell will undergo αβT or γδT cell programming, but the intensity and timing of that signal is crucial, with preTα/TCRβ typically giving weaker and γδTCR stronger signals (9, 10). Thus, β- and γδ-selection represent the first of several stages at which survival and further differentiation of a cell is dependent upon characteristics of the cell’s TCR and associated signaling apparatus. Many T cell identity genes, including those needed for TCR signaling, are turned on in the DN2-DN3 stages for first use at β- or γδ-selection (1). For this reason, any fundamental genetically-determined defect in T cells affecting signaling, survival or proliferation could alter fate choices at this stage while also having an effect on, without being complicated by, subsequent lineage choices, positive and negative selection, and peripheral activation events.
Autoimmune Type 1 diabetes (T1D) results from abnormal responses to self antigens by peripheral T cells, due to both a failure of T cell self-tolerance mechanisms and breakdown of active T cell suppression of these autoreactive cells (reviewed in (11)). A number of studies have reported that different T lineages from autoimmune diabetes-prone NOD mice have defective numbers and/or responses to various stimuli relative to cells from other non-autoimmune mouse strains, as possible factors contributing to disease. The T cell lineages implicated in T1D include CD4, CD8, NKT, and regulatory αβT cells, as well as γδT IEL cells (11-16). However, by the time mature T cells initiate and propagate an autoimmune response to peripheral tissues, they have already undergone a sequence of prior developmental choices and signaling encounters with antigen presenting cells that may have contributed cumulatively to their abnormalities. Therefore, to shed light on the possible origins of inappropriate responses of T cells in NOD mice, we have turned to the earliest stage when the TCR response machinery is first used to dictate cell fate. This is during intrathymic development, at the checkpoint where commitment to the αβ and γδ T cell lineages is established.
Although T cell populations in the thymus of NOD mice appear to be generally similar to those of other mouse strains in steady state (17, 18), the early DN populations have not been investigated in great detail. Several studies have reported abnormalities in NOD thymic development. We previously reported that thymocytes from NOD.Prkdcscid/scid and NOD-Rag1−/−mice appear to break through the β-selection checkpoint, resulting in the aberrant development of DP cells (19). NOD thymuses have also been shown to accumulate newly differentiated DP cells more slowly than conventional strains after dexamethasone-induced depletion of the DP compartment (20). Furthermore, a number of studies show defects in NOD DP cell responses to various stimuli, including responsiveness to ConA (21), apoptosis induced by glucocorticoids and β-irradiation (20) or anti-CD3ε (22), and responses to thymic selection signals (23-26). NOD mice also have defects in development of NKT cells from DP (17, 27). All of these traits point to potential alterations in TCR signaling in NOD thymic T cells, which may have consequences for mature T cell activity (12). The nature of early T cell responses is set by the cellular signaling components, which in turn have effects on the thresholds for distinct responses including positive and negative selection of αβTCR+ DP cells (reviewed in (7, 28)).
The earliest stages of T cell development are difficult to assess in the presence of ongoing TCR rearrangements because they represent only a small fraction of total thymocytes and because of the tremendous proliferation that occurs during differentiation from DN stages to DP, which can mask underlying developmental defects. In addition, steady-state proportions of thymic populations are affected by lineage decisions, cell death, and emigration (7). We have therefore used several approaches to compare the earliest stages of T cell development and lineage decisions in NOD to non-autoimmune C57BL/6 (B6) mice. To investigate the intrinsic developmental potential of NOD T cell precursors, we took advantage of the Notch-ligand expressing OP9-delta-like 1 (DL1) co-culture system, which has been used extensively to elucidate the roles of Notch and other factors on the T cell developmental process in mice and humans (29-32). We also carried out detailed flow cytometric analyses of early T cell developmental stages as well as tracking population dynamics by BrdU labeling in vivo and in mixed bone marrow chimeras. Our findings demonstrate that NOD early T cells exhibit impaired β-selection, while γδ-selection is strongly enhanced.
Materials and Methods
Mice
NOD/ShiLtJ, C57BL/6J, BALB/cJ, NOD.Rag1−/− and B6.Rag1−/− mice were obtained from Jackson Laboratories (Bar Harbor, ME) and then bred and maintained in micro-isolator cages in our specific pathogen-free mouse colony in the Caltech Animal Resources facility. (NOD X B6)F1.Rag1−/− mice used for bone marrow chimeras were bred in our facility from the parent strains. Mice were used for these studies at 4-7 weeks of age. Euthanasia and animal care followed NIH guidelines, under protocols approved by the Institute Animal Care and Use Committee.
Antibody staining, cell sorting, and flow cytometric analysis
To isolate DN populations, mice were sacrificed, their thymuses were removed and single cell suspensions made. Mature cells were depleted as previously described (33) by staining with biotinylated antibodies to CD8a, TCRγδ, TCRβ, Gr1, Ter119, CD122, NK1.1, Dx5, and CD11c, after which the cells were incubated with streptavidin coated magnetic beads and then passed through a magnetic column (Miltenyi Biotec, Auburn, CA). Eluted DN cells were either stained for direct flow cytometric analysis or for the sorting of specific DN populations into OP9 cultures. ETP (CD25−CD44hi c-Kithi), DN2 (CD25+CD44hic-Kithi), DN3a (CD25+CD44lo c-Kitlo CD27loFSClo), and DN3b (CD25+CD44lo c-Kitlo CD27hiFSChi) precursor cells, as well as γδT cells, were sorted from DN cells using a FACSAria with Diva software (Becton Dickinson Immunocytometry Systems (BDIS), Mountain View, CA). For analysis of co-cultures, cells were forcefully pipetted, filtered through nylon mesh to remove stromal cell clumps, and stained for CD45, to identify input cells, plus various combinations of lineage identifying antibodies. Cells were analyzed using a FACSCalibur (BDIS) or MACSQuant (Miltenyi) and FlowJo software (Treestar, Ashland, OR). Antibodies were purchased from eBiosciences (San Diego, CA). For intracellular staining, DN thymocytes were stained with surface markers, fixed and permeabilized using Cytofix/Cytoperm Kit (BD) and stained with conjugated antibodies to TCRβ or TCRγδ.
Cell culture
OP9-DL1 co-cultures were carried out as previously described (33, 34). For T cell development, thymic DN cells were placed on monolayers of OP9-DL1, supplemented with 2.5 ng/ml IL-7 and 5 ng/ml Flt3L. After 7 days cultures were transferred to new plates with fresh OP9-DL1 cells and the medium replaced, supplemented with 1 ng/ml IL-7. Human forms of the cytokines were used and obtained from Peprotech (Rocky Hill, NJ). Tissue culture media and fetal calf serum were obtained from Invitrogen/Gibco (Carlsbad, CA). For bone marrow cultures, cells were column-depleted of mature cells using biotinylated antibodies to CD3e, CD4, CD8, CD19, Gr-1, Ter119, CD11b, NK1.1 as described above. 105 cells were plated on OP9-DL1 monolayers in 10 cm tissue culture flasks (Corning) with the addition of 10 ng/ml IL-7 and 5 ng/ml Flt3L. After 6 days, the bone marrow-derived cells, which are predominantly DN1 and DN2 cells, were removed from the monolayers and replated on OP9-DL4 cells with 2 ng/ml IL-7. For proliferation assays, DN cell subsets were FACS sorted and then stained for 8 min at 37oC with 5 μM CFSE (Cell Trace CFSE Proliferation Kit, Invitrogen), in accordance with the manufacturer’s instructions, before they were placed in culture.
In vivo assays
For BrdU pulse-labeling of thymocytes, B6 and NOD mice were injected intraperitoneally with 1 mg Bromo-deoxyuridine (BrdU) in 0.2 ml PBS twice four hours apart. Mice were sacrificed after 1, 2 and 3 days and their thymocytes were stained with surface antibodies followed by intracellular staining for BrdU-FITC in accordance with BrdU Flow Kit instructions (BD Biosciences). For mixed bone marrow chimeras, female 8-16 week old NOD X B6)F1.Rag1−/− mice were irradiated with 400 Rads using a Cs-137 source irradiator and retroorbitally injected with 5 × 106 bone marrow cells each from NOD and B6 mice, mixed 1:1, and assayed between 5 and 9 weeks after cell injection. To reduce the risk of infection, mice were housed using autoclaved cages, bedding and food, and were treated with an antibiotic, Baytril (Bayer), in drinking water for several days before and two weeks after irradiation.
RNA extraction and quantitative RT-PCR analysis
RNA was isolated from sorted TCRγδ+ cells from B6 and NOD mouse thymuses and real time quantitative (Q)PCR carried out as previously described (19). Briefly, cells were lysed in Qiazol (Qiagen, Valencia, CA), RNA isolated using RNAeasy extraction kits (Qiagen), and cDNA reverse transcribed using random hexamers and SuperscriptIII (Invitrogen) following the manufacturer’s instructions. cDNA samples were diluted and mixed with gene-specific primers and SyBr GreenER (Invitrogen) and run on an ABI Prism 7900HT Sequence Detector (ABI, Mountain View, CA). Primers used for this study were synthesized by Eurofins MWG Operon (Huntsville, AL) and have been published previously (19, 35, 36) except for those for Sox13: Forward- GAGAAGCTGCTGTCCAGTGAC and Reverse-GACCAAAAGCTGGAGTTCCTT. Vβ1.1 and Jβ4 primer sequences were obtained from Verykokakos et al. (37). Results were calculated using the ΔCt method, normalizing all samples to Actb expression.
Results
NOD DN cells are impaired in the generation and maintenance of αβT lineage DP cells, but not γδT cells, in OP9-DL1 co-cultures
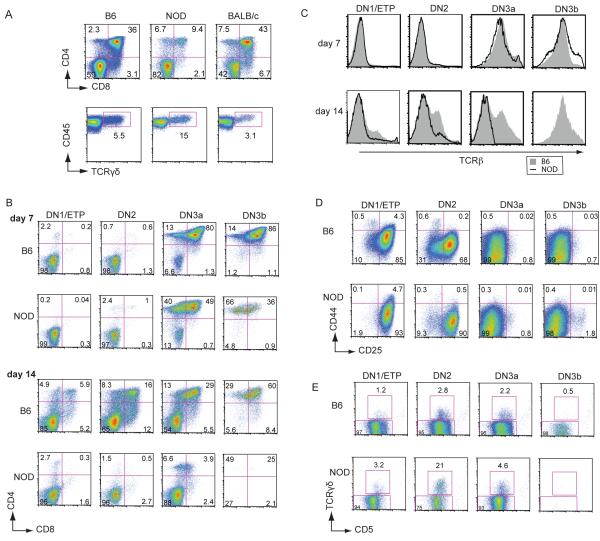
Because steady state thymic T cell populations reflect a combination of intrinsic and extrinsic factors, proliferation, cell death, and emigration, which can mask developmental differences, we wished to assess the intrinsic developmental potential of T cell precursors from B6, NOD, and BALB/c mice under the same environmental conditions using the OP9-DL1 co-culture system (30, 34). Immature DN thymocytes were purified by depletion of mature cells and placed in OP9-DL1 co-culture with IL-7 and Flt3L. When analyzed on day 12 of culture, B6 and BALB/c DN cells were found to generate and maintain high percentages of DP cells as expected, while NOD DN cells produced a much lower proportion of DP cells under the same conditions (Fig. 1A, top panels). In the same cultures, NOD DN cells produced a higher proportion of γδT cells relative to B6 and BALB/c DN cells (Fig. 1A, bottom panels).
FIGURE 1. NOD DN cells generate and maintain few DP αβT cells while retaining γδT development in vitro.
A. Flow cytometric analyses showing the development of DP and γδT cells from magnetic bead-purified DN cells from B6, NOD and BALB/c thymocytes, after 12 days of OP9-DL1 co-culture in 5 ng/ml IL-7 and Flt3L. Similar results were obtained from cultures harvested on day 8 and when IL-7 concentrations were dropped to 2 ng/ml for the last 5 days of culture or were maintained at 2 ng/ml throughout the culture period (data not shown). (B E) Development of FACs sorted DN1, DN2, DN3a, and DN3b (post-β-selection) cells from B6 and NOD thymuses cultured in 2.5 ng/ml IL-7 and 5 ng/ml Flt3L. B. Flow cytometric plots showing CD4 and CD8 DP cell development at days 7 and 14 from each progenitor population. C. Histograms showing levels of surface TCRβ expression in the same cultures for B6 and NOD cell populations at days 7 and 14 of culture. No data are shown for NOD DN3b cells on day 14 because very few cells are remaining. D. Flow cytometric plots of CD44 vs. CD25 showing differences in initial DN differentiation in the same cultures at day 7. E. γδT cell development in the same cultures at day 14. Percentages of cells in each gate are indicated. These results are representative of at least 3 independent experiments.
To assess the kinetics of stage-specific αβ vs. γδ lineage development of NOD and B6 DN cells, we tracked the development of purified intrathymic DN precursor subsets, including DN1/ETP (cKithiCD25−CD44hi), DN2 (cKithiCD25+CD44hi), and DN3a (cKitloCD25+CD44loCD27loFSClo) populations that retain both γδ and αβ T cell potential, as well as DN3b cells (cKitloCD25+CD44loCD27hiFSChi), which, as we previously showed for B6 mice, have already completed TCRβ rearrangement and pre-TCR signaling (34). Although NOD thymocytes also upregulate CD27 at β-selection, they do so to a lesser extent than B6 cells (see below), so NOD and B6 DN3b cells were enriched by sorting based upon combined expression of relatively high levels of CD27 and large size (high forward scatter (FSC), while DN3a cells were sorted based upon lower CD27 and lower FSC (38).
When precursor DN subsets from NOD and B6 thymuses were placed in OP9-DL1 co-cultures, neither B6 nor NOD DN1/ETP and DN2 cells generated DP cells by day 7 (Fig. 1B). However, by day 14, B6 DN1/ETP and DN2 cell cultures contained abundant cells that had differentiated to the DP stage while, in contrast, NOD DN1 and DN2 cells produced few if any DP cells. This is not due simply to a delay in differentiation, as NOD DP cell production did not increase with time (data not shown).
In contrast, DN3 cultures from both B6 and NOD thymuses began to express CD4 and/or CD8 by day 7 of OP9-DL1 co-culture (Fig. 1B), a time point when few DN1 or DN2 cells from either strain had turned on CD4 or CD8. Confirming the αβT cell lineage choice of the accumulated DP cells, all cultures that contained abundant DP cells, including B6 and NOD DN3a and DN3b at day 7, as well as all B6 DN subsets at day 14, clearly expressed surface TCRβ, while those NOD and B6 cultures with few DP cells express very little TCRβ (Fig. 1C). At day 7, the percentage of DP from NOD DN3a cells was moderately lower in comparison with B6 DN3a cells, while NOD DN3b cells appeared to produce fewer cells overall and a lower proportion of DP cells. By day 14 the differences were even more pronounced, with a very low proportion of DP cells in NOD DN3a cultures and very few surviving cells in the DN3b cultures. This latter result was confirmed in further experiments when the ability of individual post-β-selection DN3b cells to generate DP cells was tested. Single DN3b cells were sorted from NOD and B6 thymuses and placed in OP9-DL1 co-culture for 7 days. While all individual NOD and B6 DN3b cells proliferated and differentiated, fewer DP cells were generated in the NOD cultures (35.2 + 8.9, mean + SEM) as compared with B6 cultures (179.5 + 53.5) (p < 0.005, Mann-Whitney U Test). These results suggest a possible loss of differentiation potential, proliferation, and/or viability in the generation of DP cells from DN cells sorted both before and after β-selection in the NOD thymus.
To identify the stages at which the initial differentiation of the NOD DN cells might be blocked, the same cultures were stained with CD44 and CD25 at day 7 (Fig. 1D). Most of the cells from cultures seeded with DN3a and DN3b cells from either B6 or NOD mice had progressed to DN4, or later stages, as shown by the downregulation of CD25 and CD44 by day 7, in agreement with their predominantly DP phenotype. While both B6 and the NOD DN1 and DN2 cells differentiated successfully to the DN3 stage, the NOD DN1 and DN2 cell cultures produced a much lower proportion of DN4 and later cells (Fig. 1D), suggestive of a partial block at the β-selection checkpoint from NOD DN1 and DN2 cells.
In marked contrast to their poor production of DP cells, the DN1, DN2 and DN3a cultures from NOD thymocytes generated γδT cells very successfully (Fig. 1E). All of the NOD populations produced a higher proportion of γδT progeny than the corresponding B6 precursors, with DN2 cells producing the greatest differences: 2.8% from B6 and 21% from NOD DN2 cultures on day 14. As expected, very few γδT cells were generated from sorted DN3b cells from either strain, in agreement with our previously published results for B6 mice (34). Thus, even though NOD DN1 and DN2 cells are extremely poor at passing through the β-selection checkpoint to generate DP αβT lineage cells in vitro, they are normal or enhanced in their ability to produce γδT lineage cells.
These results show that NOD precursors exhibit a lineage specific intrinsic defect in development when placed in OP9-DL1 cultures. DN3 cells from NOD mice, even those that had undergone β-selection in the thymus, generate fewer DP cells than B6 DN3 cells, and the earliest DN1 and DN2 T cell precursors sorted from NOD thymuses are profoundly defective in their ability to go through β-selection in vitro. However, these cells retain the ability to produce abundant TCRγδ+ cells.
Prior thymic contact is not required for skewing of αβ versus γδ T cell fate from NOD cells
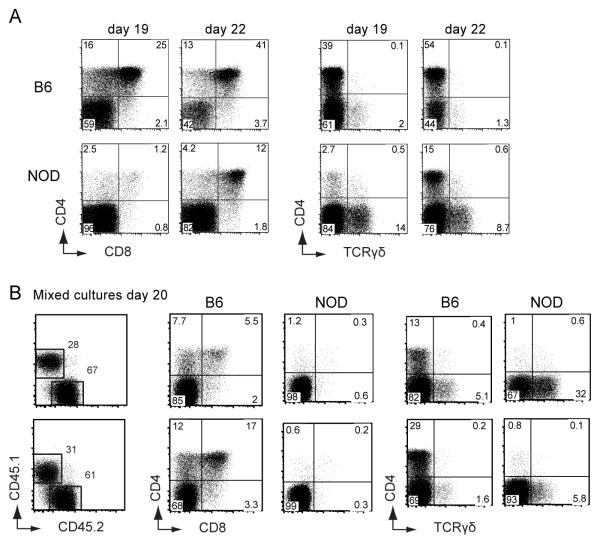
Although the highly skewed production of αβ vs. γδ lineage cells from NOD DN cells appears intrinsic to these thymic-derived cells, two concerns could be raised. First, these cells might have already encountered biasing interactions in the thymus from which we isolated them. Second, the OP9-DL1 cultures present the Delta-like 1 (DL1) Notch ligand, whereas the Notch ligand expressed in the normal thymus is DL4, which provides a somewhat different Notch signal to these cells (39-41). Therefore, we also tested the T-lineage development of prethymically derived bone marrow cell progenitors in vitro, providing Delta-like 4 (DL4) as the source of Notch ligands rather than DL1. As shown in Figure 2A, cells derived from precursor-enriched B6 and NOD bone marrow cells (BMCs) generate both DP and γδT cells on OP9-DL4 after 19-22 days of culture. However, NOD progenitor cells again exhibited a poor differentiation of DP cells (left panels) and enhancement of γδT cell generation (right panels) in comparison with those from B6 mice. Furthermore, when NOD and B6 cells were co-cultured, similar differences in αβT vs. γδT development were observed between the two strains (Figure 2B). As shown in two independent cultures, NOD cells (CD45.1+) produced a lower proportion of DP cells and more γδT cells than cultured cells from B6 mice (CD45.2+). These results confirm the profound intrinsic and lineage-specific differences in T cell developmental programming between NOD and B6 precursor cells.
FIGURE 2. NOD bone marrow progenitor cells also show attenuated in vitro development of DP αβT cells and enhanced γδT cell development.
Bone marrow cells enriched for progenitors were isolated from NOD and B6 mice and cultured on OP9-DL1 and DL4 cells as described in Materials and Methods. Flow cytometry plots show the generation of CD4/CD8 DP cells and γδT cells from B6 and NOD bone marrow cultures analyzed after 19 and 22 days (A) and two independent cultures established from mixing equal numbers of NOD and B6 bone marrow cells, analyzed after 20 days (B). Results are representative of at least 3 replicate wells from 1 of 2 independent experiments with similar results.
NOD DN2 cells generate a higher absolute number of γδT cells and fewer DP cells than B6 DN2 cells in vitro
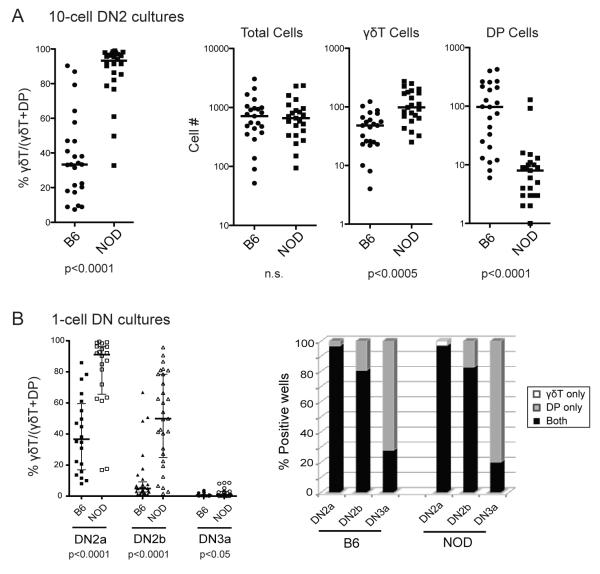
Skewed production of γδT cells relative to DP cells could result from lineage-specific alterations in proliferation or survival as well as lineage choice itself. To distinguish between these possibilities, we focused on the DN2 cells, which particularly differed between NOD and B6 in terms of their abilities to generate DP vs. γδT cells in OP9-DL1 co-cultures. First, we assessed the total cell numbers of DP and γδT cells generated from limited numbers of precursor cells in these cultures. Ten DN2 cells from B6 and NOD thymuses were sorted directly into each of 24 individual OP9-DL1 cultures and analyzed by flow cytometry on day 13. The percentage of γδT cells out of the total number of differentiated cells [calculated as 100 ×#γδT/(#γδT plus #DP)] for each well is shown (Fig. 3A, left panel) and a highly significant difference was found between cultured B6 and NOD DN2 cells. Although DN2 cells from NOD and B6 thymuses yielded comparable total numbers of CD45+ cells per well, NOD cells produced significantly higher absolute numbers of γδT cells, as well as significantly fewer DP cells (Fig. 3A, right panels). Thus, both poor DP cell differentiation and enhanced γδT cell development contribute to the NOD phenotype.
FIGURE 3. NOD and B6 DN2 cells generate more γδT and fewer DP cells in ten-cell and single cell OP9-DL1 co-cultures but retain bipotency.
A. Plots showing the relative proportion of γδT cells in comparison with DP cells generated from each individual B6 and NOD 10-cell DN2 cell culture, calculated as %γδT/(γδT + DP). The absolute numbers of CD45+ total cells, γδT cells, and DP cells generated in each of those cultures are also shown. A horizontal bar indicates the median value of individual B6 or NOD cultures. B. Plot showing the relative proportions of γδT cells in comparison to DP (calculated as %γδT/(γδT + DP)) generated from single cell cultures of sorted DN2a, DN2b and DN3a cells (left). To determine the commitment of the cells to αβ vs. γδ lineages, the lineage choices of individual cells are shown as percentages of individual wells containing γδT cells only, both γδT and DP cells, or DP cells only (right). p-values given below plots were determined using Mann-Whitney U tests. Data are representative of two independent experiments.
To determine whether or not the differences in αβT vs. γδT differentiation are the result of alterations in lineage choice, single cell differentiation analyses were performed. Because commitment to the γδT cell lineage may occur as early as the DN2 stage (42), we sought to determine if DN2 cells from either mouse strain start with a biased frequency of cells precommitted to either lineage. For this experiment single cells from two subsets of the DN2 CD44+CD25+cKithi population were tested separately based on their surface c-Kit levels, the less mature DN2a (CD44+CD25+cKit++) cells and the more mature, T lineage committed DN2b (CD44+CD25+cKit+) cells (33). Each well was scored for the presence and numbers of γδT and DP cells after 13 days of OP9-DL1 culture. Results in Fig. 3B (left panel) show again that NOD DN2 cells, both DN2a and DN2b, generally produce proportionately more γδT cells and fewer DP cells as a percentage of total differentiated cells than corresponding cells from B6 mice. However, differences between B6 and NOD were clearly not due to premature commitment in cells from either strain as they exhibited similar percentages of bipotent cells (black bars) at each stage (Fig. 3B, right panel). Almost all DN2a cells from both strains produced both γδT and DP cells. Although the majority of DN2b cells were still bipotent, individual DN2b cells from both NOD and B6 mice were more likely than DN2a stage cells to generate only DP cells, a trend which continued to the DN3a stage when few cells remained bipotent and most generated only DP cells, as shown previously for B6 mice (34). Few single cells from either strain produced only γδT cells. Thus, the high proportion of γδT cells and low proportion of DP cells in NOD cultures is not a result of increased previous γδT lineage commitment of the precursors but rather the result of enhanced γδT cell production and reduced DP cell generation after the lineage choice is made.
NOD γδT cells proliferate more rapidly than B6 γδ T cells in vitro
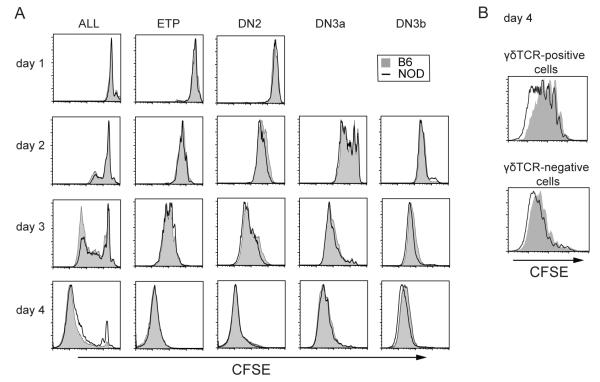
The normal preponderance of αβ-lineage DP cells over γδT cells in the thymus depends not only on the frequency of different selection events, but also on the greater extent of net proliferation that is normally associated with the αβ-lineage program after β-selection. To address the question of whether or not some of the differences in αβT vs. γδT cell development are attributable to a NOD defect in proliferation, unsorted thymocytes (“All”) as well as sorted DN1/ETP, DN2, DN3a, and DN3b populations were stained with CFSE and placed in OP9-DL1 culture. NOD DN subsets showed similar, or slightly faster, proliferation rates than B6 cells over the first four days of culture (Fig. 4A), ruling out a generalized proliferation defect. Most prominently, the γδTCR-positive cells generated in the first 4 days from NOD DN3a cultures undergo more rapid and extensive proliferation than γδTCR-positive cells from B6 cell cultures, while the γδTCR-negative cells from NOD and B6 cells proliferated more similarly (Fig. 4B). Thus, the high numbers of γδT cells generated from NOD DN cells result, at least in part, from their greater proliferation after surface TCRγδ expression in comparison with B6 γδT cells. Furthermore, the lower numbers of DP cells generated from NOD DN3 cells is not due to an initial defect in proliferation after β-selection.
FIGURE 4. B6 and NOD DN populations proliferate at similar rates in vitro, but NOD γδT cells proliferate more rapidly than B6 γδT cells.
DN cell populations were purified by cell sorting and stained with CFSE before co-culture with OP9-DL1 cells. A. Total thymocytes (All) and purified DN1, DN2, DN3a, and DN3b cells were harvested for analysis of remaining CFSE levels on days 1, 2, 3 and 4. B. Sorted DN3a cells were cultured on OP9-DL1 cells, harvested after 4 days, and stained to distinguish TCRγδ+ cells. Histograms show CFSE levels for B6 (solid gray) and NOD (black line) cells. Data are representative of 3 independent experiments with similar results.
NOD and B6 thymic γδT cells exhibit similar patterns of gene expression
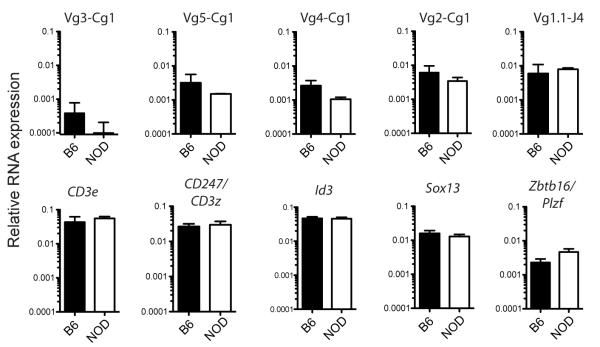
We recently reported the expression of many key T cell genes in DN populations and γδT cells from NOD and B6 mice, and found no major differences in expression of Ptcra, Notch1, Hes1, Tcf7, Runx1, and Bcl11b among others (33). However, recent evidence suggests that γδT cells may be comprised of multiple lineages, distinguished by their different dependence on specific transcription factors: Sox13, Id3 or Zbtb16/Plzf (37, 43-45). To determine whether NOD thymocytes preferentially generate all classes of γδT cells or just one particular subtype, cells were analyzed for expression of these transcription factor genes as well as for diagnostic TCR-Vβ rearrangements. We sampled Vβ rearrangements representing members of the TCR-Cβ1 cluster, which includes adult, fetal and tissue-specific receptors (reviewed in (8)). We also assayed for Vβ1.1-Jβ4 rearrangements, which mark a recently characterized population of innate γδT cells with Vβ1.1-Vδ6.3 TCRγδ receptors. These cells rapidly produce IL-4 and IFN-β, upregulate Zbtb16/PLZF, a critical transcription factor in NKT differentiation (45), and are Id3 independent (37). NOD and B6 γδT cells expressed fairly similar levels of transcripts from Vβ3, Vβ5, Vβ4, and Vβ2 to Cγ1 rearrangements, as well as Vβ1.1-Jβ4 rearrangements (Fig. 5). No differences were observed in expression of Id3 or of the CD3e, CD247 (CD3ζ, and Sox13 genes. However, Zbtb16/Plzf expression was somewhat higher in NOD γδT cells in comparison to B6 cells. Given the similarity in TCR Vβ usage, this could reflect a generally elevated state of activation (45). Thus, there appear to be few if any major differences in the types of γδ T cells produced in NOD and B6 mouse thymuses, suggesting that the differences we observe in development and rates of proliferation might be generalized to multiple γδT cell subsets.
FIGURE 5. Expression of TCR-Vγ rearrangements and other genes in sorted thymic NOD and B6 γδT cells.
QPCR results from TCRγδ+ cells sorted from NOD and B6 thymuses using primers to detect subsets of specific TCR-Vβ rearrangements and other T cell genes. Data are shown as the geometric mean +/− SD, n=2-5. For additional gene expression comparisons, see ref. 33.
NOD thymocytes exhibit multiple abnormalities around the β-selection checkpoint
A number of costimulatory and other cell surface receptors, with known effects on TCR signals, survival, and/or proliferation, are induced at β- and γδ-selection in response to TCR signaling via the newly produced pre-TCR and γδTCR, similar to events after the activation of peripheral T cells. The combinations and levels of these molecules can not only indicate the nature and intensity of the TCR signals, but also can influence further response thresholds through their modulating and enhancing roles. The degree of CD5 upregulation, for example, has been a useful indicator of the intensity of signals driving γδ-selection vs. β-selection (10), and once induced, CD5 can affect further development (10) and signal thresholds for positive and negative selection (46, 47).
We have previously shown that upregulation of the costimulatory receptor, CD27, involved in mature T cell survival and proliferation, can be used to distinguish DN3 cells that have rearranged and transcribed an intracellular (ic) TCRβ (DN3b) from those that are still TCR-negative in B6 mice (34). However, NOD DN3 cells express lower amounts of CD27 after β-selection (icTCRβ+) (Fig. 6A, top row, left panel). The costimulatory receptor CD28, which is not expressed until after β- (or γδ-) selection (48), and CD5, are also induced to a lower level in TCRβ+ NOD DN cells than B6 cells (top, middle panels). These observed differences are not due to variations in the total amount of TCRβ expressed nor to cell size, as the icTCRβ staining intensity and forward scatter (FSC) histograms are similar between the two strains (top, right panels).
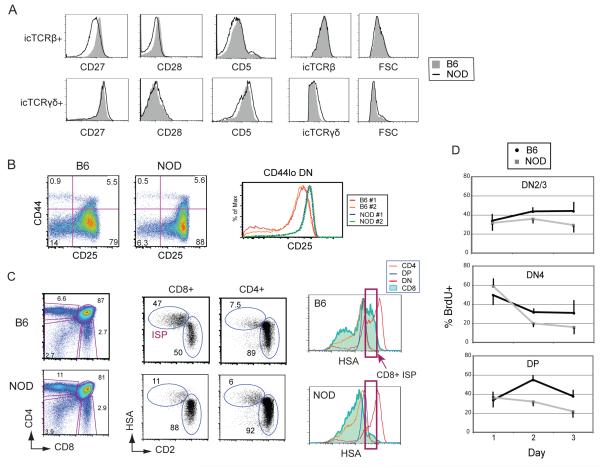
FIGURE 6. NOD thymocytes show evidence of a partial block at β-selection and abnormalities in differentiation to DP cells in vivo.
A. Histograms showing levels of activation-induced surface receptors CD27, CD28 and CD5, on freshly isolated DN thymocytes from NOD (black lines) and B6 mice (solid gray), using intracellular staining (ic) for TCRβ (top row) or TCRγδ (bottom row) to distinguish cells that have passed through β-selection or γδ-selection respectively. Data are representative of 3-4 independent experiments. B. Flow cytometric analysis showing CD44 vs. CD25 expression in DN subsets from freshly isolated NOD and B6 thymuses (left panels). Percentages of cells in each quadrant are as indicated. A histogram comparing CD25 surface expression levels between gated NOD and B6 CD44lo DN cells (DN3 and DN4 cells) from two mice of each strain. Data are representative of >10 mice of each strain. C. Flow cytometric analyses showing CD4 vs. CD8 plots for total thymocytes from NOD and B6 mice (left), CD8 and CD4 gated cells, stained to distinguish immature (ISP) cells from mature SP cells (middle panels), and histograms showing HSA levels for DN, DP, CD8+ and CD4+ cells (right) which distinguish less mature cells (HSAhi) from more mature cells (HSAlo). The red boxes indicate the intermediate HSA level for B6 CD8 ISP cells (top panel, arrow) with no corresponding major population in the NOD thymus (bottom panel, arrows). Data are representative of 3 independent experiments. D. Percentages of BrdU+ cells among DN2/3 (CD25+), DN4 (CD25−), and DP cell populations 1-3 days after in vivo BrdU pulse-labeling, showing a major difference in accumulation of BrdU+ DP cells on day 2 between B6 and NOD mice. Mean + SD values over time are plotted for B6 and NOD thymocytes.
In contrast to the results for αβT cell precursors, icTCRγδ+ cells from B6 and NOD thymuses express similar levels of surface CD27 and CD28 (Fig. 6A, bottom row, left panels). Furthermore, NOD γδ T cells express a broader range of surface CD5 levels, both higher and lower, and a higher total amount of TCRγδ than that expressed by B6 γδT cells, despite comparable cell sizes as determined by FSC profile (bottom, right panels). The higher levels of CD27 and CD5 attained by thymic γδT cells from both NOD and B6 mice reflect the higher intensity of signals generated at γδ-selection compared with those generated at β-selection (9, 10). Likewise, the lower induction of CD27, CD28 and CD5 in icTCRβ+ cells from NOD as compared to B6 mice suggests that NOD cells may experience a weaker or otherwise altered β-selection signal, while γδT cell signals are more similar between the two strains.
Depletion of subsets post-β-selection in the normal NOD thymus
B6 and NOD thymuses exhibit few obvious differences in their proportions of major populations in steady state (17, 18). However, because the OP9-DL1 co-culture system revealed major intrinsic defects in NOD differentiation at the earliest TCR dependent checkpoint, we performed a series of detailed flow cytometric analyses of freshly purified NOD and B6 thymocytes to evaluate more closely if DN cells within the NOD thymus exhibit any comparable abnormalities in vivo. We found that while all DN populations are present, as measured by CD44 and CD25 profiles, the percentages and shapes of the differentiation plots differ (Fig. 6B, left panels). Most prominently, the levels of surface CD25 are much higher in CD25+ cells from NOD than B6 cells, as seen in the density plots, as well as in histograms of all CD44lo cells (including DN3 to DN4 cells) (Fig. 6B, right panel). Furthermore, NOD thymuses consistently have a lower proportion of DN4 cells relative to DN3 cells (lower left vs. lower right quadrants). Elevated CD25 in DN3 cells and lower proportions of DN4 cells can be indicative of a partial block in β-selection as reported for thymuses from mice with mutations in the pre-TCR or Notch signaling pathways (49-52).
The generally accepted route of differentiation for B6 mouse αβT cells, after β-selection and the downregulation of CD25 leading to the DN4 stage, is the upregulation of CD8 into immature single-positive (ISP) cells prior to the expression of surface CD4 in the DP stage (7). A previous study reported low numbers of CD8+ ISP cells in NOD thymuses (17). To confirm this finding and to determine the route of differentiation from DN4 to DP in NOD thymuses, CD8 ISP cells were distinguished from mature CD8 single positive (SP) cells by their retention of high levels of surface CD24 (heat shock antigen, HSA), which then decline during differentiation. B6 thymocytes follow this pattern, expressing a high percentage of total gated CD8+ cells that are HSAhi and CD2int (ISP) cells, as compared to the mature HSA intermediate and CD2hi single positive (SP) CD8+ cells (Fig. 6C). However, NOD thymocytes have a much lower percentage of CD8 ISP cells that is only partially offset by a corresponding HSAhi CD2int CD4+ ISP population. Furthermore, none of the populations from NOD thymuses have a major subset of cells with the same level of HSA that is expressed by the B6 ISP cells, as indicated by the red boxes (Fig. 6C, right panels), suggesting a paucity of cells transiting through this stage of development.
The interpretation that there might be a reduced influx into the ISP compartment from previous stages was further supported by the kinetics of proliferation and turnover of DN and DP subsets in B6 and NOD thymuses in vivo, as determined by the fate of a cohort of BrdU pulse-labeled cells. A previous study reported an anomalous dip in BrdU+ DP cells in NOD mice after 2-3 days (18). We wished to confirm and extend this finding in the context of a β-selection defect by additional DN cell analysis. Mice were injected intraperitoneally with BrdU, sacrificed after 1, 2 or 3 days, and the thymocytes were stained for surface receptors and intracellular BrdU. One day after BrdU injection the proportions of labeled cells within the DN2/DN3 (CD25hi), DN4 (CD25− CD44−), and DP populations were similar between NOD and B6 (Fig. 6D). The DN2/3 cells were expected to accumulate BrdU+ cells due to extensive proliferation at the DN1 and DN2 stages, although most DN3 cells normally pause in proliferation before β-selection. Over 50% of the DN4 cells were pulse-labeled at this early time point, because cells that have passed through the β-selection checkpoint from DN3 undergo a rapid burst of proliferation while differentiating (Fig. 4A, DN3b cells). DP cells proliferate very little, but BrdU+ cells are expected to flow into the DP compartment and accumulate during subsequent days due to the continuing differentiation of the rapidly proliferating DN4 and ISP cells. Accordingly, on day 2 post-injection, the percentage of BrdU+ DN4 cells from both B6 and NOD declined precipitously (Fig. 6D, middle panel), as expected due to turnover and differentiation from non-labeled DN3 precursors, confirming the pulse nature of the labeling. The B6 DP compartment showed an almost two-fold increase in the proportion of BrdU+ cells over the proportion at day 1, again as expected due to continuing differentiation of the BrdU+ labeled DN4 cells from the preceding days. However, the NOD DP compartment did not show a corresponding increase in the percentage of BrdU+ cells at day 2, but rather showed a decline. The failure of BrdU-labeled NOD cells to become DP was not due to their retention in the DN4 compartment. The rapid loss of BrdU+ cells from all phenotypic compartments suggests that failure to accumulate ISP cells in NOD thymuses is not due to poor proliferation or rapid modulation of surface CD4, CD8 and HSA, but instead due to a survival failure. Taken together, our results support the notion that NOD early T cells in vitro and in vivo have a partial block around the β-selection checkpoint, leading to a reduction in DP cell accumulation.
Lineage-specific differences in the establishment of αβT vs. γδT cell and B cell populations in mixed bone marrow chimeras
To determine whether preferential development of γδT cells, as well as reduced production of αβT cells, from NOD precursors occur in vivo as well as in vitro, we used mixed bone marrow chimeras to compare the kinetics of T cell differentiation and export between NOD and B6 cells in the same animal. An equal number of NOD and B6 bone marrow cells were injected into irradiated (NOD × B6)F1.Rag1−/− mice, which were assayed for reconstitution at 5-9 weeks of age. Five weeks post-injection both NOD (CD45.1+) and B6 (CD45.2+) cells were found in the thymus (Fig. 7A, left panels) with relatively normal steady state proportions of DN, DP, SP and γδT populations established at this time. However, a lower proportion of CD45.1+ NOD cells was found in the spleen compared with CD45.2+ B6 cells (Fig. 7A, right panels). The NOD cells were predominantly CD19+ B cells with a T:B cell ratio of 2.2:85 as compared with the B6 T:B ratio of 35:56 (Fig. 7A, right top panels). This T cell repopulation defect was primarily due to disproportionately fewer TCRαβ+ T cells. In contrast, relatively normal percentages and total yields of NOD TCRγδ cells were found. Thus, in vivo as well as in vitro, there is an NOD defect in production of αβT cells, and this is lineage-specific.
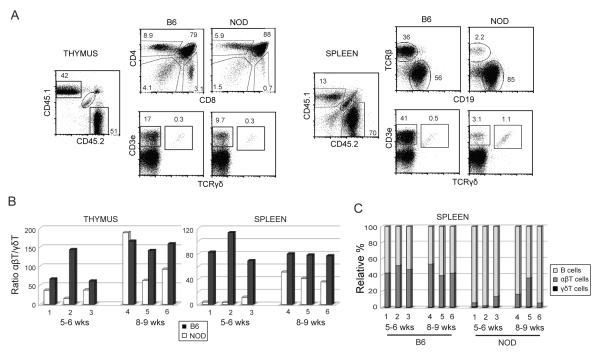
FIGURE 7. NOD cells exhibit differences in establishment of γδT vs. αβT cells in mixed bone marrow chimeras.
Bone marrow cells isolated from NOD and B6 mice and 1 × 106 cells from each strain were mixed and injected into irradiated (NOD × B6)F1.Rag1−/− recipients. Mice were sacrificed after 5-6 or 8-9 weeks and analyzed by flow cytometry for lymphocyte populations in thymus and spleen using CD45.1 (NOD) and CD45.2 (B6) to distinguish donor strains. A. Flow cytometric analysis of a representative chimeric mouse (#1) analyzed at 5 weeks post-injection. Plots on the left show thymocytes were stained for CD4 and CD8 (upper panels) and CD3ε and TCRγδ (lower panels). Splenocytes, shown on the right, were stained for αβTCR and CD19, to distinguish B cells (upper panels), and CD3ε and TCRγδ (lower panels). B, C. Summary of FACs staining results from six mixed bone marrow chimeric mice, three analyzed after 5-6 weeks (#1-3) and three after 8-9 weeks (#4-6). B. Graphs showing the ratio of TCRαβ+/TCRγδ+ cells in thymus (left) and spleen (right) generated from B6 (black bars) and NOD (white bars) donors. C. Graph showing of the relative percentages of B, αβT, and γδT lymphocytes derived from B6 and NOD donors in spleens from the same individual chimeric mice.
A summary of results from all 6 mixed bone marrow chimeric mice analyzed at 5-6 and 8-9 weeks of age is shown in Figure 7B and C. In 5/6 thymuses and 6/6 spleens from chimeric mice, NOD cells exhibited a lower ratio of TCRαβ+:TCRγδ+ cells when compared with B6 cells in the same mouse. In agreement with the steady-state phenotype of NOD mice, this effect was kinetic, not absolute. At the later time point, by 8-9 weeks post-injection (Fig. 7B, #4-5), the relative proportions of TCRαβ+:TCRγδ+ cells in the thymus and spleen became more similar between NOD and B6 than that observed at 5-6 weeks (Fig. 7B, #1-3), indicating that NOD αβT cells eventually accumulate. The defect in NOD αβT production was again shown to be lineage specific, as relatively high numbers of B cells were produced in these animals (Fig. 7C). Thus, NOD BMCs were much less efficient than B6 BMCs at producing mature αβ T cells in chimeric hosts, even though they were capable of rapidly generating γδT and B cells. These results show that the NOD genotype confers divergent effects on αβ and γδ lineage differentiation in vivo as well as in vitro.
DISCUSSION
The first TCR dependent checkpoint is the first of many stages in which the TCR signaling apparatus of the cell is used for survival, lineage choice and differentiation functions. The earliest T lineage decision, which is made at this point, is the fundamental choice between αβT and γδT cell fates, which is determined by differences in TCR signal strength and results in initiation of very different developmental programs. Furthermore, recent evidence suggests that different γδT cell lineages may be selected at this point depending on the characteristics of the rearranged TCR and transduced signal at this stage (reviewed in (43)).
The OP9-DL1 co-culture system provides the Notch ligands and other factors required by early T cells for passage through the first TCR-dependent checkpoint and differentiation into γδT or αβT cells and it is a powerful tool allowing detailed study of the T cell developmental program and lineage choice in both mice and humans (29-32, 53, 54). Using this in vitro culture system we show that NOD early DN cells are very poor at generating DP αβT cells, while the same cells under the same conditions exhibit enhanced γδT cell differentiation and proliferation. This was true of purified DN thymocytes as well as precursor enriched bone marrow cells and in the presence of DL4, the normal thymic Notch ligand, rather than DL1. Because the DN thymocytes are the progenitors of all T cell lineages, defects expressed at these earliest stages may also be present in all subsequent stages and may have a major impact on later thymic selection events, lineage choices, and peripheral developmental choices and responsiveness.
While the OP9-DL1 cell culture system may exacerbate the relatively poor DP differentiation by NOD early T cell progenitors, we have several additional lines of evidence that indicate that β-selection may also be defective in the NOD thymus. First, the heightened CD25 expression in NOD DN3 cells is similar to pre-TCR and Notch mutations that give a partial block at β-selection (49-52). Second, CD5 can be used as an indicator of the intensity of TCR signaling (10), and β-selected icTCRβ+ cells from NOD thymuses weakly upregulate coreceptor molecules CD5, as well as CD27 and CD28, as compared to the corresponding populations from B6 thymuses. The low proportion of CD8+ ISP cells found in NOD thymuses has been previously reported (17), but the paucity of HSA-intermediate cells in any corresponding population, and the poor accumulation of DP cells in BrdU-labeling studies, suggest that this may be due to an impairment of the cells differentiating past β-selection to the DP compartment rather than simply use of an alternate route of differentiation. The difference does not appear to be at the level of cell proliferation after β-selection as DN3b cells from NOD thymuses proliferate at a rate similar to B6 DN3b cells, as measured by CFSE. CD27 is associated with proliferation and survival of cells after β-selection, as well as survival of peripheral CD8 T cells (55, 56), and it is possible that the low level of CD27 upregulation upon β-selection in NOD mice may contribute to the poor accumulation of DP cells in he BrdU experiments and in OP9-DL1 culture. Overall, these results are consistent with previous reports showing that DP cells accumulate very slowly in NOD thymuses after depletion of the DP compartment with dexamethasone (20), and can be interpreted as an impairment in β-selection in NOD mice.
The preferential development of γδT cells from NOD DN subsets is not simply a concentration artifact resulting from the poor DP differentiation in these animals. There is evidence that some γδT cell precursors can become committed precociously at the DN2 stage (42, 57), before cells reach the conventional β-selection checkpoint. That does not appear to be the case here, as single DN2 cells sorted from NOD thymuses were predominantly bipotent just as in B6 thymuses, despite producing relatively higher numbers of γδT cells compared with DP cells. Thus, the skewing of NOD differentiation towards the γδT lineage is more likely to be due to survival and proliferation than to lineage choice. The major differences in αβT versus γδT cell development in vitro may result from alterations in intrinsic signaling pathways in NOD early T cells with differential effects on the two lineages. For example, stronger intrinsic IL-7 signals, lower Notch signals, weaker β-catenin/TCF1 activity, and stronger TCR signals have all been shown to favor the differentiation, survival, and/or proliferation of γδT cells over αβT cells (58-62). Although lineage choice may be normal in NOD DN cells, their ability to survive, proliferate and differentiate in the chosen lineage may differ between β- and γδ-selected T cells. Normally, in B6 and most strains examined, the β-selection program dictates much greater proliferative expansion than γδ-selection. Our results imply that this difference is greatly reduced or reversed in NOD mice.
In addition to their preferential development from NOD DN cells in OP9-DL1 culture, we found that γδT cells generated in vivo in NOD thymuses proliferate more, express a wider range of surface CD5, and upregulate all assayed activation-induced receptors very well. This suggests that NOD γδT cells may be produced as a result of unusually robust or variable signaling responses in the thymus. Thus it is possible to interpret the preferential development of γδT cells from NOD precursors as an outcome from a stronger or altered TCR signal, such that γδT cell differentiation, proliferation and survival is actively promoted (9, 10, 63, 64) while DP differentiation and survival are disfavored (54). Alternatively, the TCR signal may not be stronger in NOD cells but they may interpret any viability-promoting signal as a γδ T cell selection signal, thereby leaving many cells that undergo the β-selection program starved of viability support. Although further work is needed to resolve which intrinsic mechanisms are affecting αβ vs. γδ T cell differentiation in NOD DN cells, there is evidence that immature and mature NOD T cells may transduce abnormal TCR signals during activation (reviewed in (12)). In addition, NOD.Prkdcscid/scid and NOD.Rag−/− thymocytes fail to arrest at the β-selection checkpoint, undergoing spontaneous differentiation to DP cells without restoration of thymic cellularity (19), suggesting that NOD DN cells, even in the absence of a rearranged TCR, may undergo a spontaneous, but only partial, TCR signal. A number of factors can cause this type of breakthrough phenotype including enhanced TCR and Notch signals (6, 65). Interestingly, we previously reported data showing a higher level of Notch3 and Dtx1 expression, but not Notch1 or Hes1, in freshly isolated NOD DN3a cells in comparison with B6 DN3a cells (33), although the significance of this difference is not yet clear.
Recent studies have demonstrated that events at β-selection or even earlier can have a major effect on the characteristics and further lineage choices of αβTCR+ DP cells (66-68). Loss of a subset of differentiating cells after β-selection might result in a differentially selected DP population that, for example, transduces a stronger than average TCR signal or is more resistant to certain types of apoptosis. If aberrant NOD β-selection events alter the signaling characteristics of the final DP population, this could have major ramifications for negative and positive selection of conventional CD4 and CD8 T cells. The same could be true for the selection of γδT cells as well as “unconventional” αβT cells with self-reactive TCRs, such as NKT, CD8αα, and regulatory T cells (69). In addition, although little is known about possible roles of γδT cells in autoimmunity, γδT IELs may play an important role in peripheral tolerance and have been found to be able to control diabetes progression in NOD mice (14). γδT cells reactive to insulin peptide B:9-23 have been reported to be generated in NOD but not B6 mice (70) which provides further evidence of their potential role in autoimmune diabetes. The diversity of γδT lineages, their peripheral immune responses and their roles in disease have been a subject of great interest very recently, especially in light of the ability of particular subsets to rapidly secrete IL-17, IFN-β and IL-4 (reviewed in (43, 71)).
The results of this study show that NOD lineage T cells have fundamental and intrinsic abnormalities from the earliest stages in their responses to activating signals, either via TCR or other environmental triggers, even in the absence of MHC/peptide/APC interactions. These defects may affect lineage choices and TCR repertoires via alterations in DP selection thresholds or alter peripheral immune responses to specific stimuli, or both. Finally, whether or not these traits found in NOD early T development directly contribute to autoimmunity, these studies demonstrate that different mouse strains can show major variations in β- and γδ-selection developmental programming, which may deviate significantly from the currently accepted pathways.
Acknowledgments
We wish to thank J. C. Zuñiga-Pflücker for generously providing OP9-DL1 and OP9-DL4 cells for these experiments; Rochelle Diamond, Stephanie Adams, Diane Perez and Pat Koen from the Caltech Cell Sorting Facility for cell sorting; Natasha Bouey, Robert Butler, Ruben Bayon, and Beth Olsen for excellent animal care; Marion Duprilot for preliminary gene expression data; and Tom Taghon (University of Ghent, Belgium) and members of the Rothenberg lab, especially Rochelle Diamond, for helpful suggestions.
Footnotes
This work was supported by grants from the Juvenile Diabetes Foundation Research International and National Institutes of Health (AI64590) (to M.A.Y.), as well as the Al Sherman Foundation, The Louis A. Garfinkle Memorial Laboratory Fund, the Vanguard Charitable Endowment for Diabetes in Memory of Bently Pritsker, and the Albert Billings Ruddock Professorship (to E.V.R.).
References
- 1.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 3.Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nat Rev Immunol. 2002;2:888–897. doi: 10.1038/nri937. [DOI] [PubMed] [Google Scholar]
- 4.von Boehmer H, Aifantis I, Feinberg J, Lechner O, Saint-Ruf C, Walter U, Buer J, Azogui O. Pleiotropic changes controlled by the pre-T-cell receptor. Curr Opin Immunol. 1999;11:135–142. doi: 10.1016/s0952-7915(99)80024-7. [DOI] [PubMed] [Google Scholar]
- 5.Michie AM, Zuniga-Pflucker JC. Regulation of thymocyte differentiation: pre-TCR signals and beta-selection. Semin Immunol. 2002;14:311–323. doi: 10.1016/s1044-5323(02)00064-7. [DOI] [PubMed] [Google Scholar]
- 6.Kruisbeek AM, Haks MC, Carleton M, Michie AM, Zuniga-Pflucker JC, Wiest DL. Branching out to gain control: how the pre-TCR is linked to multiple functions. Immunol Today. 2000;21:637–644. doi: 10.1016/s0167-5699(00)01744-8. [DOI] [PubMed] [Google Scholar]
- 7.Rothenberg EV, Yui MA. Development of T Cells. In: Paul WE, editor. Fundamental Immunology. 6th ed Lippincott Williams & Wilkins; Philadelphia, PA: 2008. pp. 376–406. [Google Scholar]
- 8.Xiong N, Raulet DH. Development and selection of γδ T cells. Immunol Rev. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 9.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of γδTCR signaling efficiently diverts thymocytes to the αβ lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Hayes SM, Li L, Love PE. TCR signal strength influences αβ/γδ lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Anderson MS, Bluestone JA. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol. 2005;23:447–485. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 12.Delovitch TL, Singh B. The nonobese diabetic mouse as a model of autoimmune diabetes: immune dysregulation gets the NOD. Immunity. 1997;7:727–738. doi: 10.1016/s1074-7613(00)80392-1. [DOI] [PubMed] [Google Scholar]
- 13.Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone JA. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–440. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- 14.Locke NR, Stankovic S, Funda DP, Harrison LC. TCR γδ intraepithelial lymphocytes are required for self-tolerance. J Immunol. 2006;176:6553–6559. doi: 10.4049/jimmunol.176.11.6553. [DOI] [PubMed] [Google Scholar]
- 15.Wicker LS, Miller BJ, Mullen Y. Transfer of autoimmune diabetes mellitus with splenocytes from nonobese diabetic (NOD) mice. Diabetes. 1986;35:855–860. doi: 10.2337/diab.35.8.855. [DOI] [PubMed] [Google Scholar]
- 16.Bendelac A, Carnaud C, Boitard C, Bach JF. Syngeneic transfer of autoimmune diabetes from diabetic NOD mice to healthy neonates. Requirement for both L3T4+ and Lyt-2+ T cells. J Exp Med. 1987;166:823–832. doi: 10.1084/jem.166.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godfrey DI, Kinder SJ, Silvera P, Baxter AG. Flow cytometric study of T cell development in NOD mice reveals a deficiency in αβTCR+CD4-CD8-thymocytes. J Autoimmun. 1997;10:279–285. doi: 10.1006/jaut.1997.0129. [DOI] [PubMed] [Google Scholar]
- 18.Berzins SP, Venanzi ES, Benoist C, Mathis D. T-cell compartments of prediabetic NOD mice. Diabetes. 2003;52:327–334. doi: 10.2337/diabetes.52.2.327. [DOI] [PubMed] [Google Scholar]
- 19.Yui MA, Rothenberg EV. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J Immunol. 2004;173:5381–5391. doi: 10.4049/jimmunol.173.9.5381. [DOI] [PubMed] [Google Scholar]
- 20.Bergman ML, Penha-Goncalves C, Lejon K, Holmberg D. Low rate of proliferation in immature thymocytes of the non-obese diabetic mouse maps to the Idd6 diabetes susceptibility region. Diabetologia. 2001;44:1054–1061. doi: 10.1007/s001250100600. [DOI] [PubMed] [Google Scholar]
- 21.Zipris D, Lazarus AH, Crow AR, Hadzija M, Delovitch TL. Defective thymic T cell activation by concanavalin A and anti-CD3 in autoimmune nonobese diabetic mice. Evidence for thymic T cell anergy that correlates with the onset of insulitis. J Immunol. 1991;146:3763–3771. [PubMed] [Google Scholar]
- 22.Kwon H, Jun HS, Yang Y, Mora C, Mariathasan S, Ohashi PS, Flavell RA, Yoon JW. Development of autoreactive diabetogenic T cells in the thymus of NOD mice. J Autoimmun. 2005;24:11–23. doi: 10.1016/j.jaut.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Kishimoto H, Sprent J. A defect in central tolerance in NOD mice. Nat Immunol. 2001;2:1025–1031. doi: 10.1038/ni726. [DOI] [PubMed] [Google Scholar]
- 24.Lesage S, Hartley SB, Akkaraju S, Wilson J, Townsend M, Goodnow CC. Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J Exp Med. 2002;196:1175–1188. doi: 10.1084/jem.20020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liston A, Lesage S, Gray DH, O’Reilly LA, Strasser A, Fahrer AM, Boyd RL, Wilson J, Baxter AG, Gallo EM, Crabtree GR, Peng K, Wilson SR, Goodnow CC. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity. 2004;21:817–830. doi: 10.1016/j.immuni.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Holler PD, Yamagata T, Jiang W, Feuerer M, Benoist C, Mathis D. The same genomic region conditions clonal deletion and clonal deviation to the CD8αα and regulatory T cell lineages in NOD versus C57BL/6 mice. Proc Natl Acad Sci U S A. 2007;104:7187–7192. doi: 10.1073/pnas.0701777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner MJ, Hussain S, Mehan M, Verdi JM, Delovitch TL. A defect in lineage fate decision during fetal thymic invariant NKT cell development may regulate susceptibility to type 1 diabetes. J Immunol. 2005;174:6764–6771. doi: 10.4049/jimmunol.174.11.6764. [DOI] [PubMed] [Google Scholar]
- 28.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zuniga-Pflucker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 31.De Smedt M, Hoebeke I, Plum J. Human bone marrow CD34+ progenitor cells mature to T cells on OP9-DL1 stromal cell line without thymus microenvironment. Blood Cells Mol Dis. 2004;33:227–232. doi: 10.1016/j.bcmd.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 33.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine Slam-associated adaptor protein-dependent “innate” γδ T cells. PLoS ONE. 2010;5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, Owen MJ, Hayday AC. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 39.Koch U, Fiorini E, Benedito R, Besseyrias V, Schuster-Gossler K, Pierres M, Manley NR, Duarte A, Macdonald HR, Radtke F. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, Zuklys S, Hollander GA, Shima DT, Habu S. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med. 2008;205:2507–2513. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Besseyrias V, Fiorini E, Strobl LJ, Zimber-Strobl U, Dumortier A, Koch U, Arcangeli ML, Ezine S, Macdonald HR, Radtke F. Hierarchy of Notch-Delta interactions promoting T cell lineage commitment and maturation. J Exp Med. 2007;204:331–343. doi: 10.1084/jem.20061442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang J, Volkmann A, Raulet DH. Evidence that γδ versus αβ T cell fate determination is initiated independently of T cell receptor signaling. J Exp Med. 2001;193:689–698. doi: 10.1084/jem.193.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kreslavsky T, Gleimer M, von Boehmer H. αβ versus γδ lineage choice at the first TCR-controlled checkpoint. Curr Opin Immunol. 2010;22:185–192. doi: 10.1016/j.coi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, Kang J. Regulation of γδ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 45.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azzam HS, DeJarnette JB, Huang K, Emmons R, Park CS, Sommers CL, El-Khoury D, Shores EW, Love PE. Fine tuning of TCR signaling by CD5. J Immunol. 2001;166:5464–5472. doi: 10.4049/jimmunol.166.9.5464. [DOI] [PubMed] [Google Scholar]
- 47.Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Muller W, Killeen N, Rajewsky K. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- 48.Williams JA, Hathcock KS, Klug D, Harada Y, Choudhury B, Allison JP, Abe R, Hodes RJ. Regulated costimulation in the thymus is critical for T cell development: dysregulated CD28 costimulation can bypass the pre-TCR checkpoint. J Immunol. 2005;175:4199–4207. doi: 10.4049/jimmunol.175.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Sommers CL, Burshtyn DN, Stebbins CC, DeJarnette JB, Trible RP, Grinberg A, Tsay HC, Jacobs HM, Kessler CM, Long EO, Love PE, Samelson LE. Essential role of LAT in T cell development. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
- 51.Hinton HJ, Alessi DR, Cantrell DA. The serine kinase phosphoinositide-dependent kinase 1 (PDK1) regulates T cell development. Nat Immunol. 2004;5:539–545. doi: 10.1038/ni1062. [DOI] [PubMed] [Google Scholar]
- 52.Mao C, Tili EG, Dose M, Haks MC, Bear SE, Maroulakou I, Horie K, Gaitanaris GA, Fidanza V, Ludwig T, Wiest DL, Gounari F, Tsichlis PN. Unequal contribution of Akt isoforms in the double-negative to double-positive thymocyte transition. J Immunol. 2007;178:5443–5453. doi: 10.4049/jimmunol.178.9.5443. [DOI] [PubMed] [Google Scholar]
- 53.de Pooter RF, Zuniga-Pflucker JC. Generation of immunocompetent T cells from embryonic stem cells. Methods Mol Biol. 2007;380:73–81. doi: 10.1007/978-1-59745-395-0_5. [DOI] [PubMed] [Google Scholar]
- 54.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T cell receptor-instructed αβ versus γδ lineage commitment revealed by single-cell analysis. J Exp Med. 2008;205:1173–1186. doi: 10.1084/jem.20072425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gravestein LA, van Ewijk W, Ossendorp F, Borst J. CD27 cooperates with the pre-T cell receptor in the regulation of murine T cell development. J Exp Med. 1996;184:675–685. doi: 10.1084/jem.184.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hendriks J, Xiao Y, Rossen JW, van der Sluijs KF, Sugamura K, Ishii N, Borst J. During viral infection of the respiratory tract, CD27, 4-1BB, and OX40 collectively determine formation of CD8+ memory T cells and their capacity for secondary expansion. J Immunol. 2005;175:1665–1676. doi: 10.4049/jimmunol.175.3.1665. [DOI] [PubMed] [Google Scholar]
- 57.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential Notch dependency at the αβ and γδ T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Kang J, Coles M, Raulet DH. Defective development of β/δ T cells in interleukin 7 receptor-deficient mice is due to impaired expression of T cell receptor β genes. J Exp Med. 1999;190:973–982. doi: 10.1084/jem.190.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the αβversus γδT cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 60.Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines αβversus γδ lineage fate. J Exp Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melichar H, Kang J. Integrated morphogen signal inputs in γδ versus αβ T-cell differentiation. Immunol Rev. 2007;215:32–45. doi: 10.1111/j.1600-065X.2006.00469.x. [DOI] [PubMed] [Google Scholar]
- 62.Hayday AC, Pennington DJ. Key factors in the organized chaos of early T cell development. Nat Immunol. 2007;8:137–144. doi: 10.1038/ni1436. [DOI] [PubMed] [Google Scholar]
- 63.Hayes SM, Love PE. A retrospective on the requirements for γδ T-cell development. Immunol Rev. 2007;215:8–14. doi: 10.1111/j.1600-065X.2006.00476.x. [DOI] [PubMed] [Google Scholar]
- 64.Lauritsen JP, Haks MC, Lefebvre JM, Kappes DJ, Wiest DL. Recent insights into the signals that control αβ/γδ-lineage fate. Immunol Rev. 2006;209:176–190. doi: 10.1111/j.0105-2896.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- 65.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 66.Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H. Identification of pre- and postselection TCRαβ+ intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 67.Gallo EM, Winslow MM, Cante-Barrett K, Radermacher AN, Ho L, McGinnis L, Iritani B, Neilson JR, Crabtree GR. Calcineurin sets the bandwidth for discrimination of signals during thymocyte development. Nature. 2007;450:731–735. doi: 10.1038/nature06305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pennington DJ, Silva-Santos B, Silberzahn T, Escorcio-Correia M, Woodward MJ, Roberts SJ, Smith AL, Dyson PJ, Hayday AC. Early events in the thymus affect the balance of effector and regulatory T cells. Nature. 2006;444:1073–1077. doi: 10.1038/nature06051. [DOI] [PubMed] [Google Scholar]
- 69.Baldwin TA, Hogquist KA, Jameson SC. The fourth way? Harnessing aggressive tendencies in the thymus. J Immunol. 2004;173:6515–6520. doi: 10.4049/jimmunol.173.11.6515. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L, Jin N, Nakayama M, O’Brien RL, Eisenbarth GS, Born WK. Gamma delta T cell receptors confer autonomous responsiveness to the insulin-peptide B:9-23. J Autoimmun. 2010;34:478–484. doi: 10.1016/j.jaut.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonneville M, O’Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]