Summary
The glucocorticoid receptor (GR), like many signaling proteins, depends on the Hsp90 molecular chaperone for in vivo function. Although Hsp90 is required for ligand binding in vivo, purified apoGR is capable of binding ligand with no enhancement from Hsp90. We reveal that Hsp70, known to facilitate client delivery to Hsp90, inactivates GR through partial unfolding, while Hsp90 reverses this inactivation. Full recovery of ligand binding requires ATP hydrolysis on Hsp90 and the HOP and p23 cochaperones. Surprisingly, energy from Hsp90 ATP hydrolysis appears to regulate client transfer from Hsp70, likely through a coupling of the two chaperone’s ATP cycles. Such coupling is embodied in novel contacts between Hsp90 and Hsp70 in the GR:Hsp70:Hsp90:HOP complex imaged by cryoEM. While GR released from Hsp70 is aggregation prone, release from Hsp90 protects GR from aggregation and enhances its ligand affinity. Together, this illustrates how coordinated chaperone interactions can enhance stability, function and regulation.
Introduction
Hsp90 is a ubiquitous ATP-dependent molecular chaperone required for activation of numerous client proteins, many involved in the progression of cancer and other diseases (Taipale et al., 2010). While most canonical chaperones, such as Hsp70, recognize unfolded proteins, Hsp90 is unique as it acts in the later stages of folding by binding to partially folded intermediates (Jakob et al., 1995). Extensive studies have revealed a complex ATP hydrolysis cycle in which Hsp90 undergoes dramatic conformational rearrangements (Krukenberg et al., 2011). Hsp90 is a high affinity homodimer that is dimerized in the apo state through its C-terminal domain (CTD) and adopts an extended V-shaped conformation (Krukenberg et al., 2008). The N-terminal domains (NTD) contain the nucleotide binding sites, and ATP binding stabilizes domain rearrangements such that the arms come together in an NTD-dimerized closed conformation (Ali et al., 2006), which promotes ATP hydrolysis (Wegele et al., 2003). While Hsp90’s ATPase activity is essential (Obermann et al., 1998), it is unclear how ATP hydrolysis promotes client folding and activation.
The Glucocorticoid Receptor (GR), a ligand activated transcription factor involved in critical biological processes (Chrousos and Kino, 2009), is a well-known obligate Hsp90 client. In vivo Hsp90 is required for GR to bind ligand and become active (Picard et al., 1990). To varying degrees, Hsp90 interacts with other steroid hormone nuclear receptors such as the progesterone receptor (PR). Like most nuclear receptors, GR’s activity is regulated through its C-terminal ligand binding domain (LBD), a mostly helical structure with the ligand binding pocket located in the core of the domain. In the absence of ligand, LBDs are thought to be dynamic, with the ligand providing both structural stability and allosteric control of the LBD’s ability to interact with co-regulator proteins to modulate transcription (Bain et al., 2007). Hsp90 interacts directly with GR through its LBD (GRLBD) (Howard et al., 1990).
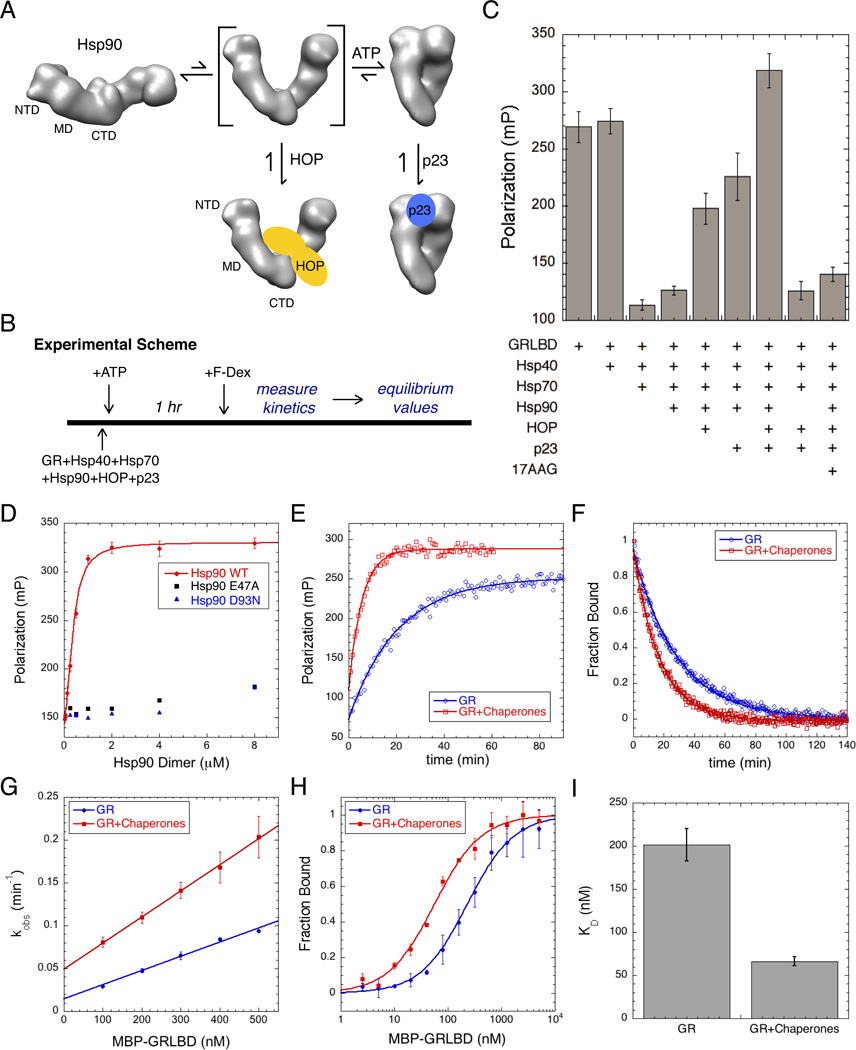
It is unclear why Hsp90 is required or how Hsp90 activates GR for ligand binding. Early reconstitution experiments with GR (Pratt et al., 2006), and with PR established that the central proteins in the maturation pathway include Hsp40, Hsp70, Hsp90, HOP, and p23 (Dittmar et al., 1996; Kosano et al., 1998). These experiments defined a general order in which proteins enter and exit the pathway (Morishima et al., 2000b), with Hsp40 and Hsp70 acting first to deliver the receptor to Hsp90 (Hernández et al., 2002; Smith et al., 1992). HOP was originally thought to act solely as an adaptor protein that facilitates delivery by providing a physical link between the two chaperones (Chen and Smith, 1998). However a recent cryo-EM reconstruction of the Hsp90:HOP complex reveals that HOP forms extensive interactions with Hsp90, pre-organizing Hsp90 NTDs for ATP hydrolysis and client binding (Southworth and Agard, 2011). The second Hsp90 cochaperone, p23, acts later in the pathway, binding to the Hsp90 complex in which GR is in its ligand binding competent state (Dittmar et al., 1997). While HOP and p23 bind to district regions on Hsp90, their binding is competitive. HOP binds to the open state, while p23 requires the closed nucleotide bound state (Figure 4A).
Figure 4. Hsp90 System Recovers GR Ligand Binding From Hsp70 Inhibition and Enhances Ligand Association.
A) Without nucleotide, Hsp90 is in an extended open state. Rotation of the NTD about the MD interface is required for dimerization of the NTDs in the ATP stabilized closed state. HOP binds the intermediate open state with rotated NTDs, and p23 binds the closed ATP bound state.
B) Experimental scheme for Figure 4.
C) Equilibrium binding of 20nM F-dex to 1 µM MBP-GRLBD with different chaperone components (±SD). Assay conditions; 50µM 17AAG, 2µM Hsp40, and 15µM Hsp70, Hsp90, HOP, and p23.
D) Saturation plot for binding of 20nM F-dex to 1 µM MBP-GRLBD with 2µM Hsp40, and 15µM Hsp70, HOP and p23, with increasing Hsp90 WT (red), and hydrolysis dead Hsp90; E47A (black) and D93N (blue) (±SD). WT Hsp90 binding curve fit to a half maximal effective concentration equation.
E) Association kinetics of 20nM F-dex to 300nM MBP-GRLBD alone (blue) and with 2µM Hsp40, and 15µM Hsp70, Hsp90, HOP, and p23 (red), fit to a single phase association.
F) Normalized dissociation kinetics of 100nM F-dex bound to 1µM MBP-GRLBD (blue) and with 15µM Hsp70, 2µM Hsp40, and 10µM Hsp90, HOP, and p23 (red). Off rates are respectively 0.041 ±0.004 min−1 and 0.059±0.002 min−1 (±SEM) (Figure S4D).
G) Average kobs vs GRLBD concentration from 3–5 separate experiments for GRLBD (blue) and with 15µM Hsp70, 2µM Hsp40, and 10µM Hsp90, HOP, and p23 (red) (±SEM). On rates determined from the slope of the linear fit to be 0.165±0.008 and 0.304±0.072 µM−1min−1 with chaperones (± weighted error of slope) (Figure S4C).
H) Normalized equilibrium binding of 20nM F-dex to GRLBD alone (blue) and with 15µM Hsp70, 2µM Hsp40, and 10µM Hsp90, HOP, and p23 (red) averaged from 2 separate experiments (±SD).
I) Average ligand KD for GRLBD with and without chaperones determined from 5 separate experiments (as in Figure 4H). KD decreases from 201±42nM to 66±12nM with chaperones (±SEM). See also Figure S3 and S4.
Like most obligate Hsp90 clients, in depth biochemical investigation of GR has been hindered by difficulty obtaining stable apo protein for in vitro investigation. As a result, previous GR studies have been carried out with proteins of variable quality and limited to basic characterizations. That said, in one of the first purely in vitro investigations, denatured and refolded GRLBD was reported to stimulate Hsp90’s hydrolysis, indicating a direct interaction between purified Hsp90 and GRLBD (McLaughlin et al., 2002).
The aim of our investigation was to determine in detail how Hsp90 promotes GR ligand binding, and in so doing, provide much needed insight into how Hsp90 activates a bona fide client. Useing highly purified recombinant proteins in an entirely in vitro system, we directly measure GRLBD ligand binding. Contrary to in vivo, our purified apo GRLBD binds ligand without aid or enhancement from Hsp90. Working under conditions where apoGR is stable, the biological dependence on Hsp90 could only be fully understood in the context of the entire chaperone system that includes Hsp70, Hsp90 and three specific cochaperones. Under these conditions, we find that the Hsp70 and Hsp90 chaperone cycles are intimately connected, with Hsp70 and Hsp90 working collaboratively to regulate and enhance GR function.
Results
Purified GRLBD Binds Ligand without Hsp90
To obtain workable quantities of GRLBD, while keeping as close as possible to wild-type, we utilized a single solubility enhancing mutation (F602S) (Ricketson et al., 2007). Although less dependent on Hsp90 for activation in vivo, this mutation allows for large-scale expression in bacteria (Bledsoe et al., 2002). GRLBD(F602S), referred to as GRLBD unless otherwise specified, was purified to homogeneity in the presence of ligand followed by extensive dialysis to remove the ligand. The GRLBD could be purified and obtained in the apo state without a solubility tag, however an N-terminal MBP tag was verified to not interfere with the shown experiments and was generally retained as it enhanced stability over long experimental time courses.
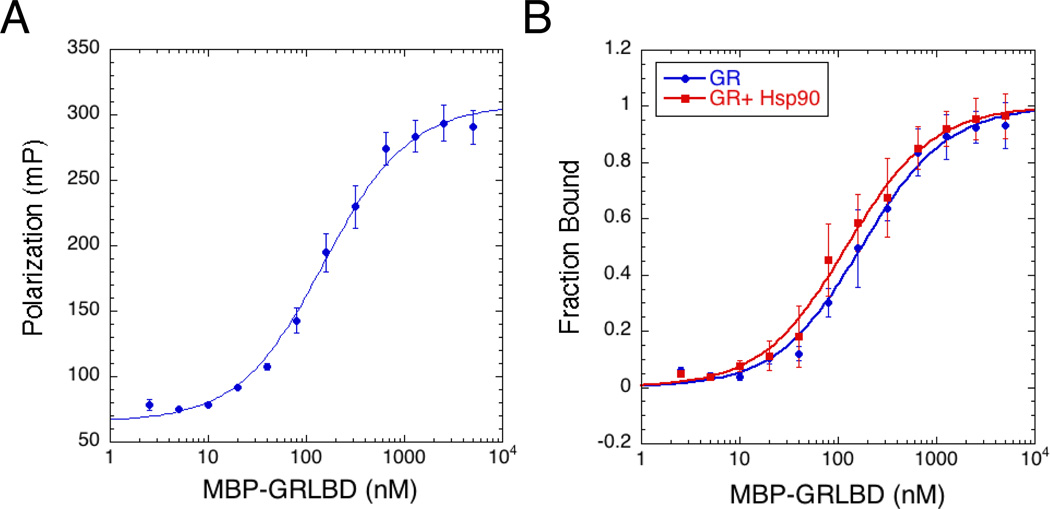
Ligand binding was monitored by measuring the increase of fluorescence polarization of fluorescein labeled dexamethasone (F-dex) as it bound to GRLBD. As previously reported, but in contrast to in vivo (Bledsoe et al., 2002), purified GRLBD can bind ligand in the absence of chaperones (Figure 1). The kinetics of F-dex binding displayed standard single-phase association and dissociation kinetics, with association taking place much faster than dissociation, indicating that our GRLBD is ligand free. Under our experimental conditions, equilibrium measurements result in a dissociation constant (KD) of 150±20 nM (Figure 1A). Given the dramatic effect of Hsp90 in vivo we were surprised that no enhancement in ligand binding by Hsp90 was detected (Figure 1B). Even with p23 and HOP, no significant Hsp90 effect was observed (not shown). In addition, we were unable to reproduce the previously reported stimulation of Hsp90’s ATPase activity by GRLBD, although we noted that the detergent used previously to stabilize GRLBD could also stimulate ATPase activity. In contrast, working with the yeast Hsp90 for which a more robust ATPase rate is observed, we confirmed the recently reported inhibition by GRLBD (Lorenz et al., 2014) (not shown).
Figure 1. GR Ligand Binding Activity In Vitro Is Hsp90 Independent.
A) Equilibrium binding of 20nM F-dex to GRLBD measured by fluorescence polarization (±SD). Binding curve fit a KD of 154±14nM (±SEM).
B) Equilibrium binding with 5mM ATP/MgCl2. GRLBD alone (blue) and with 5µM Hsp90 (red) (±SD).
Since mutations in GRLBD are known to influence GRLBD’s conformation and ligand binding properties (Pfaff and Fletterick, 2010; Ricketson et al., 2007), we wanted to ensure that the F602S mutation was not responsible for the independently functional GRLBD or lack of ATPase acceleration. From the small amount of WT GRLBD that could be purified, WT GRLBD was not only capable of binding ligand but had a higher affinity than the F602S mutant (Figure S1A), a trend consistent with a previous report (Ricketson et al., 2007). Using WT GRLBD we were still unable to detect a significant effect on Hsp90’s ATPase rate (not shown). Notably, our purified GRLBD is in a very different functional state than the denatured and refolded protein for which ATPase stimulation was observed. In addition to the nearly 300-fold difference in ligand affinities (150nM compared to 46µM), both our purified apo F602S and WT GRLBD are monomeric (Figure S2B), whereas the refolded protein was dimeric (McLaughlin and Jackson, 2002), suggesting that these two studies should not be compared. In summary, independent of the F602S mutation, our purified GRLBD can function on its own and we were unable to detect any significant high affinity functional interaction between just GRLBD and Hsp90.
Hsp70 Inhibits GR Ligand Binding by Partially Unfolding GR
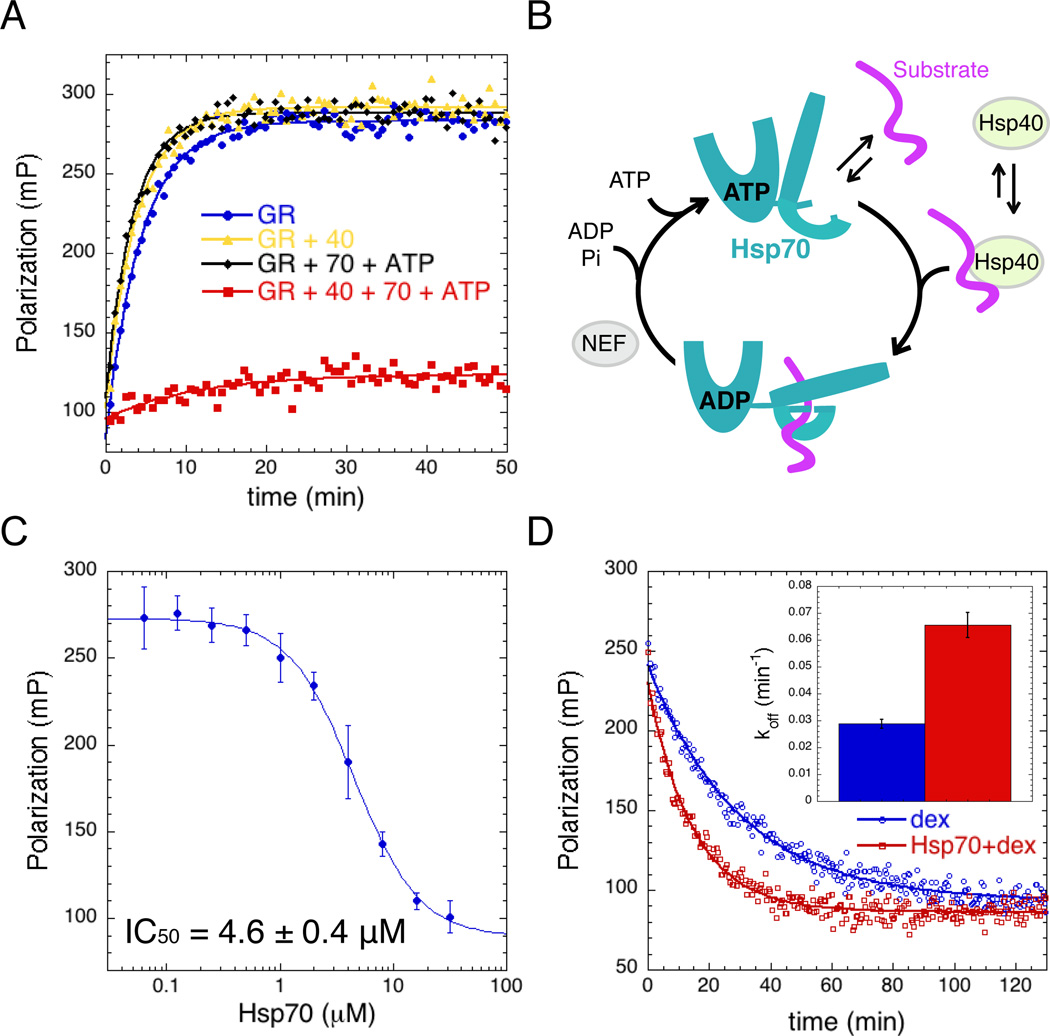
Since Hsp70 can facilitate client delivery to Hsp90, we reasoned that Hsp70 might stabilize a state of GRLBD better capable of interacting with Hsp90; therefore enhancing any effect Hsp90 might have on ligand binding. Remarkably, apo GRLBD preincubated with just the Hsp70 system resulted in complete inhibition of GRLBD ligand binding (Figure 2A). Based on previous studies (Dittmar et al., 1998), we used sub-stoichiometric amounts of yeast Hsp40 (Ydj1) to accelerate Hsp70 ATP hydrolysis. With ATP, the Hsp70 substrate binding lid is open and hydrolysis triggers allosteric changes that lead to closure of the lid, promoting stable high affinity substrate binding and protection from aggregation (Figure 2B) (Mayer and Bukau, 2005). Hsp40 has no effect on its own, but is required for GRLBD inhibition (Figure 2A) and to promote stable binding of Hsp70 to GRLBD (Figure S1D). ATP hydrolysis is also essential, as no GRLBD inhibition is observed with AMPPNP or ADP (Figure S1C). The concentration dependence of Hsp70 shows a cooperative mode of inhibition with an IC50 of 4.6±0.6µM and a Hill coefficient of 1.6±0.6 (Figure 2C). The F602S mutation had no effect on the IC50 (Figure S1E).
Figure 2. Hsp70 inhibits GR Ligand Binding.
A) Association kinetics of F-dex to GRLBD (blue), with Hsp40 (yellow), with Hsp70 and ATP (black), and with Hsp40, Hsp70 and ATP (red). Assay conditions: 5mM ATP/MgCl2, 50nM F-dex, 1µM MBP-GRLBD, 2µM Hsp40 and 15µM Hsp70.
B) Chaperone cycle of Hsp70. ATP bound NBD opens the SBD such that substrate binding is weak. Substrate and Hsp40 stimulate ATP hydrolysis, resulting in a high affinity substrate-bound state with the lid latched down. Substrate release occurs upon ADP:ATP exchange promoted by nucleotide exchange factors (NEF).
C) Hsp70 concentration dependence of equilibrium binding of 1 µM MBP-GRLBD to 20nM F-dex (with 2µM Hsp40) (±SD). Fitting a cooperative competitive binding model yields an IC50 of 4.6±0.4 µM and a Hill coefficient of 1.6±0.4 µM (±SEM).
D) Dissociation of 100nM F-dex from MBP-GRLBD (with 2µM Hsp40) initiated with excess (100µM) unlabeled dex (blue), and with 15µM Hsp70 (red), fit to a single exponential decay. Inset shows average off rate; 0.029±0.002 min−1 and 0.066±0.005 min−1 with Hsp70 (±SEM). See also Figure S1.
While this data shows that Hsp70 can bind to apo GRLBD and prevent ligand binding, we wanted to know if Hsp70 actively displaces ligand from an already ligand bound GRLBD. To investigate this, GRLBD was pre-equilibrated with Hsp40 and F-dex and ligand dissociation initiated by excess unlabeled dex with and without Hsp70 (Figure 2D). We found that Hsp70 accelerated F-dex dissociation more than 2-fold, revealing that Hsp70 can directly catalyze ligand removal from GRLBD.
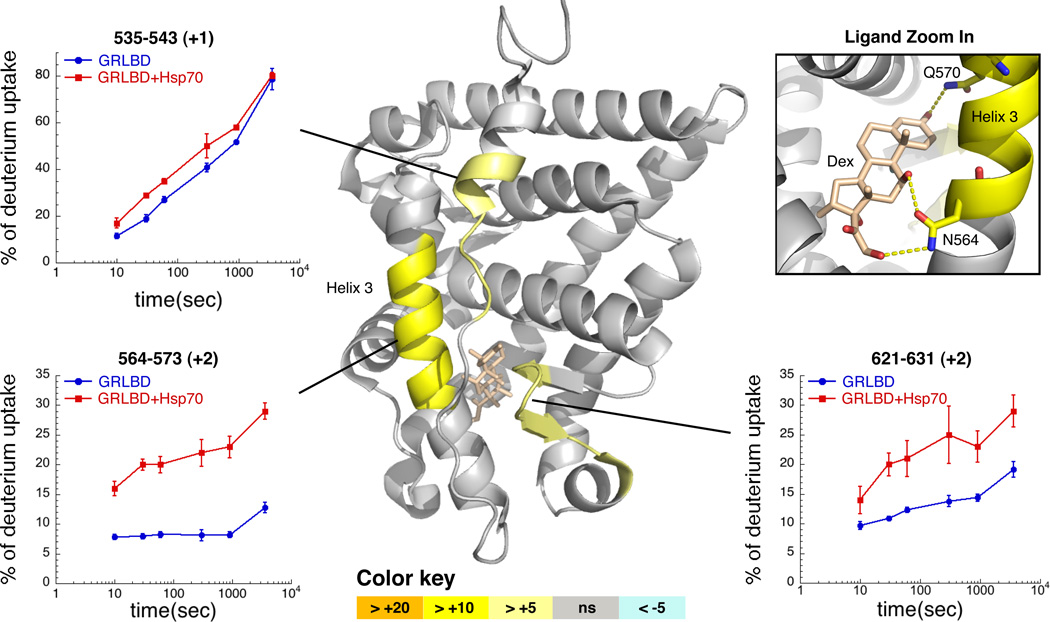
From the above, we hypothesized that Hsp70 is actively promoting GRLBD unfolding, thereby disrupting the conformation required for ligand recognition. Our expectation was that Hsp70 would significantly shift the equilibrium towards a completely unfolded GRLBD. However, limited proteolysis showed an increased sensitivity in only a single region (helix 3,Figure S2A). Hsp70-induced unfolding was more quantitatively investigated by hydrogen deuterium exchange coupled with mass spectrometry (HDX-MS). While Hsp40 alone hand no effect (not shown), there was a significant increase in H/D exchange with both Hsp70 and Hsp40 in three limited GRLBD regions (Figure3 and Figure S2). The most extensive increase was on helix 3 (residues 564–573) and then the β-sheet region (residues 621–631). This correlates with the limited proteolysis, confirming that Hsp70 induces only local unfolding. Interestingly, in addition to disrupting local structure and important hydrophobic contacts with the ligand, the region on helix 3 that was most effected contains residues N564 and Q570, which form three of the six hydrogen bonds between GRLBD and the ligand (Figure 3). This indicates that while the degree of overall unfolding is minimal, it is located for optimal disruption of ligand binding.
Figure 3. Hydrogen Exchange Mass Spectrometry Detects Partial Unfolding of GRLBD by Hsp70.
Differential HDX between GRLBD and Hsp70 bound GRLBD mapped onto the dex and coactivator peptide (not shown) bound crystal structure of GRLBD (1M2Z). Differences in the average HDX are represented as percentage change and colored according to the key with orange being faster with Hsp70. Number within parentheses is the standard deviation between 3 replicates. Corresponding deuterium build-up curves for the regions that undergo the most significant conformational change are shown (±SD). Top right shows zoom in on dex, and the three hydrogen bonds formed with helix 3. See also Figure S2.
Hsp90 Recovers GR Ligand Binding
Given that Hsp90 is essential in vivo for ligand binding, and our finding that Hsp70 holds GRLBD in an inactive state, we explored Hsp90’s ability to liberate GRLBD from Hsp70 inhibition. Including Hsp90 in the preincubation with Hsp70 and Hsp40 resulted in minimal recovery of ligand binding (Figure 4B and 4C). By contrast, significant recovery occurred with Hsp90 and either HOP or p23. Full recovery required both cochaperones. The concentrations used for HOP and p23 were saturating in their effect; thus partial recovery was not due to insufficient protein (not shown). Ligand binding recovery was entirely dependent on Hsp90, as no recovery was seen without Hsp90 or with the specific Hsp90 inhibitor, 17AAG. Thus, Hsp90 is the active component required for recovery of ligand binding, recapitulating its in vivo requirement. Interestingly, the Hsp90 concentration dependence of GRLBD recovery saturates at stoichiometric amounts of Hsp90 dimer and GRLBD (Figure 4D). This suggests a 1:1 interaction between monomeric GRLBD and dimeric Hsp90 and indicates tight binding of Hsp90 and cochaperones to the Hsp70:GRLBD complex, with an estimated KD on the order of 200nM or less.
Release of Hsp70 Inhibition Requires ATP Hydrolysis by Hsp90
Inhibition by 17AAG, which binds to Hsp90’s ATP binding pocket and prevents nucleotide binding, indicates the importance of ATP for recovery of GRLBD ligand binding. To explore further, we utilized two previously characterized hydrolysis dead mutations in Hsp90: D93N, which cannot bind nucleotide, and E47A, which can bind nucleotide and close but cannot hydrolyze ATP (Obermann et al., 1998). Neither mutant was able to recover ligand binding (Figure 4D), indicating that both nucleotide binding and hydrolysis are required to reverse Hsp70 inhibition.
Pull down studies show that in the presence of the chaperones an Hsp90:Hsp70:HOP:GRLBD complex forms and that neither 17AAG nor the Hsp90 mutations prevent formation of this large complex (Figure S3). By contrast, inhibition of ATP hydrolysis caused a loss in p23 association. Binding of p23 and HOP appear weaker than the other components, and were additionally probed by western blot. This confirmed the presence of HOP and revealed no detectable incorporation of p23 with either Hsp90 mutant or 17AAG (Figure S3). For D93N, this is expected as this mutant cannot bind ATP which is required to stabilize the closed state to which p23 binds (Ali et al., 2006). By contrast, the significant loss in p23 binding to the GR complex with the E47A mutant was unexpected since E47A has been shown to support the binary interaction between p23 and Hsp90 (Johnson et al., 2007). The loss of p23 binding to the E47A mutant in the context of the full system indicates that p23 association is inhibited by some component of the Hsp90:GR complex prior to hydrolysis.
This, in combination with the inability to restore ligand binding, clearly illustrates that hydrolysis on Hsp90 facilitates an essential step in the pathway required for the release of Hsp70’s inhibition, and that blocking hydrolysis results in a stalled inactive intermediate complex with Hsp70, Hsp90 and HOP but not p23. Consequently, p23’s role in the receptor maturation pathway must occur on a post hydrolysis state of Hsp90.
The Coordinated Action of Hsp70 and Hsp90 Enhances GR Ligand Affinity
When investigating ligand binding recovery in response to the full chaperone system, we noticed a slight enhancement in the amount of GRLBD bound to ligand (Figure 4C). This enhancement is not due to activating a previously inactive portion of the GRLBD population, as the entire chaperone system did not change the specific activity of GRLBD (Figure S4A). Instead, the full chaperone system accelerates the ligand association rate in an Hsp90 concentration-dependent manner (Figure S4B). Comparing the association and dissociation rates of F-dex to GRLBD alone or GRLBD pre-equilibrated with saturating amounts of the entire chaperone system (Figure 4B) revealed acceleration in both the association (Figure 4E) and dissociation rates (Figure 4F). The dissociation rate seen was about ~1.5 fold faster, similar to that measured with just the Hsp70 system (Figure 2C). On rates were determined from the slope of the linear fit of the kobs measured at different GRLBD concentrations (Figure 4G), revealing ~2 fold faster on rate with chaperones (Figure S4C). Additionally, equilibrium measurements show ~3 fold enhancement in the KD with the chaperones (Figures 4H, 4I and S4E). These data indicate that within the full chaperone system, Hsp90 holds GRLBD in a higher ligand affinity conformation than GRLBD alone, and thus suggests that the chaperone-bound complex is directly capable of binding ligand.
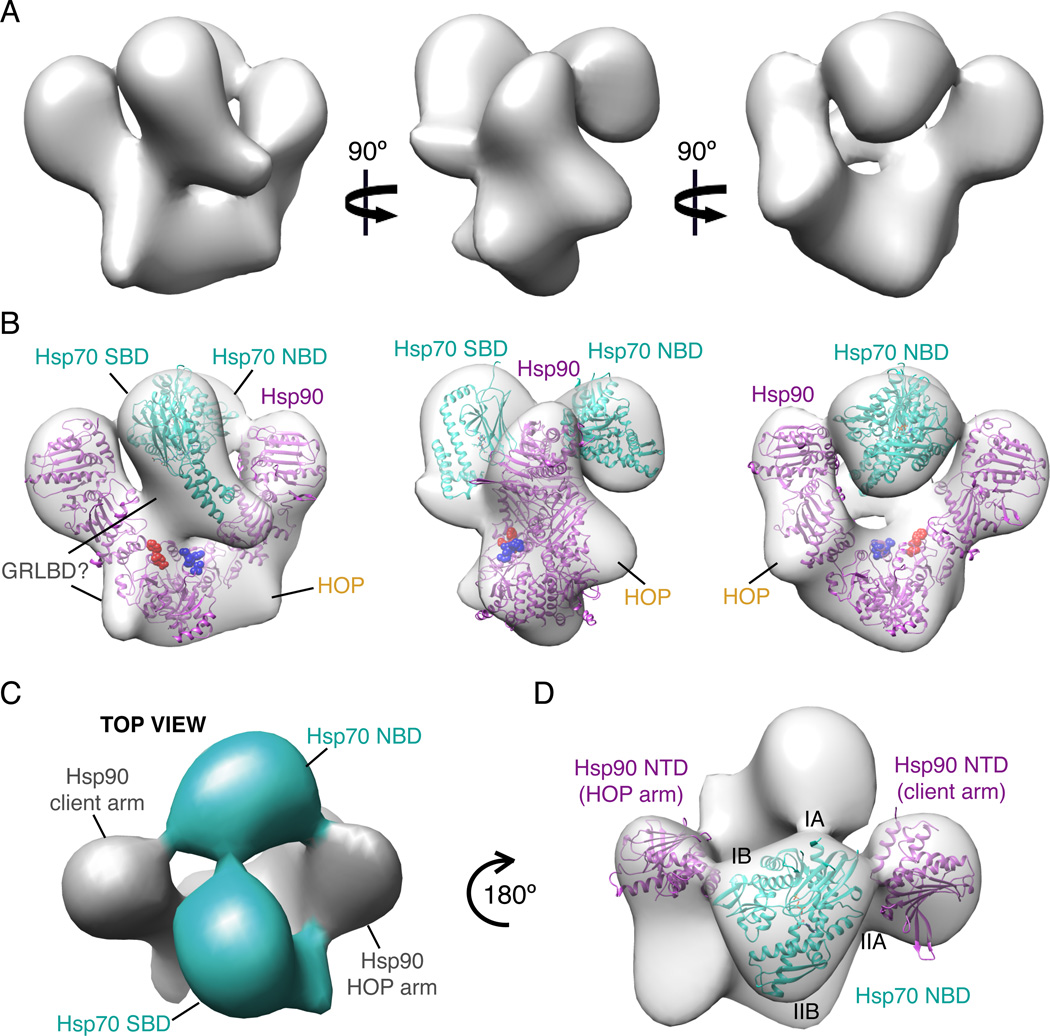
Cryo EM Map of Hsp90:HOP:Hsp70:GR Complex Supports Coupling of Hsp90 and Hsp70 ATP Hydrolysis Cycles
To provide a structural framework for understanding the hand off of GR between Hsp70 and Hsp90, we purified the intermediate complex having one Hsp90 dimer and one copy each of Hsp70, HOP and GRLBD (Figure S5), and utilized single particle cryo EM to obtain a 3D reconstruction. The assembly reactions and running buffers were used with the intention of capturing the GRLBD bound ADP state of Hsp70 and the apo HOP bound state of Hsp90. While resolution of the reconstruction is estimated to be 38Å and likely limited by sample heterogeneity, the size and distinct shape of the individual Hsp90 and Hsp70 domains allowed the general organization of these components in the complex to be determined with high confidence (Figure 5). Hsp90 is in a V-shaped state similar to the previous Hsp90-HOP EM structure (Southworth and Agard, 2011), however the overall Hsp90 organization is distinctly asymmetric with only one monomer having the fully rotated N-terminal domain (NTD) orientation seen in the HOP complex and in the Hsp90 closed ATP state crystal structure (Ali et al., 2006). To fit our density, the other Hsp90 NTD needs to be rotated ~23° outwards about the NTD-middle domain (MD) interface (Figure S6A). The opposing Hsp90 monomer that fits the HOP induced conformation has two regions of extra density centered on the MD, which likely corresponds to HOP domains shown to be necessary and sufficient for GR-activation (TPR2A, TPR2B and DP2)(Schmid et al., 2012).
Figure 5. Cryo-EM Reconstruction of Hsp90, Hsp70, HOP, GRLBD Complex.
A) Cryo-EM reconstruction of Hsp90, Hsp70, HOP, GRLBD complex filtered to 38Å.
B) Cryo-EM reconstruction as in A with placement of Hsp70 NTD (3ATU) and SBD (1DKX) in cyan, and Hsp90 NTD rotated HOP model (Southworth and Agard, 2011) in magenta. Client binding site residues for E. coli Hsp90; E466, W467, N470 shown in red and M546, M550, L553, F554 in blue (Genest et al., 2013).
C) Top view shows density for the linker between Hsp70 domains (cyan).
D) Top view showing positioning of Hsp70 NTD between Hsp90 NTDs. See also Figure S5 and S6.
The most distinguishable feature of the complex is the Hsp70 substrate binding domain (SBD), which has extra density from the substrate binding site on Hsp70 extending to the MD and C-terminal domain (CTD) interface of Hsp90, suggesting that GRLBD is simultaneously bound to both Hsp90 and Hsp70. Additionally, Hsp70 appears to be delivering GRLBD directly to the client binding site originally identified on the E. coli Hsp90 (Genest et al., 2013). When tested in yeast, mutations in this region had a significant impact on GR signaling in vivo. Indeed, the portion of the binding site mapped to the CTD amphipathic helix had been suggested to make direct contact with apo GRLBD (Fang et al., 2006).
As expected for Hsp70 in the ADP state, the Hsp70 SBD is separated from its nucleotide binding domain (NBD). Not only is there a distinct orientation between the two domains, but connecting density is visible (Figure 5C). A surprising aspect of the reconstruction is that the Hsp70 NBD is nestled between the two Hsp90 NTDs. A direct contact between Hsp90 and Hsp70 was unexpected, as in vitro binding between Hsp90 and Hsp70 is only detected in the presence of HOP, which binds both independently through separate TPR domains. This led to the conclusion that HOP was a passive linker, and that the Hsp70:Hsp90 interaction was purely through the physical connection to HOP. While the Hsp70 NBD seems to contact both Hsp90 NTDs, it makes a stronger connection with the ATPase domain of the HOP bound Hsp90 monomer. Formation of this connection requires the Hsp90 conformation induced by HOP, indicating that not only does HOP pre-organize Hsp90 for receiving clients, but also for interaction with Hsp70.
Utilizing the Hsp70 domain linker as a restraint, Hsp70 NTD best fits into the EM density such that the IB lobe is in proximity to the HOP arm of Hsp90, and the base of the IIA lobe is in proximity to the opposing Hsp90 NTD (Figure 5D). Intriguingly, this orientation suggests that the lid of the Hsp90 nucleotide binding pocket is in the region making contacts with Hsp70 NBD. The functional implications of this potential interaction are significant, as the direct contact between the ATPase domains of Hsp70 and Hsp90 provide a structural basis for the coupling of the two hydrolysis cycles suggested by our ligand binding studies.
Hsp90 is an Hsp70 Release Factor with Chaperone Function
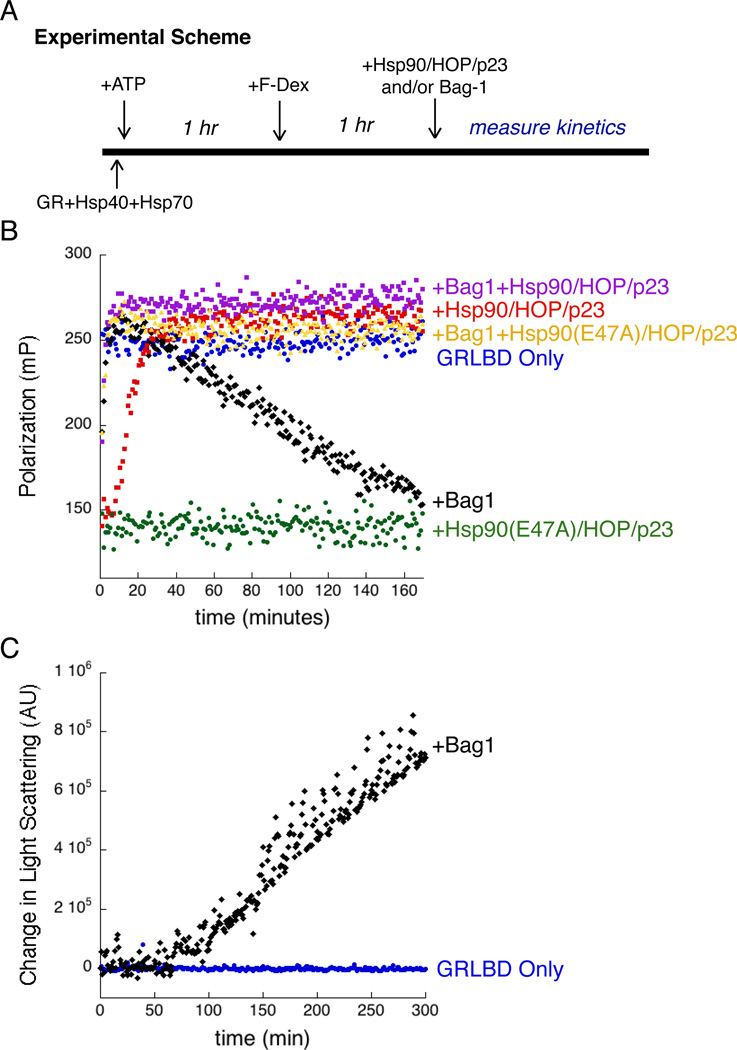
The experiments above suggest a model in which Hsp90 reverses the Hsp70 inhibition in a process that requires ATP hydrolysis by Hsp90, likely by promoting Hsp70 release from GRLBD. In the absence of other factors, substrate release from Hsp70 is slow, but is facilitated by nucleotide exchange factors that accelerate ADP release, allowing ATP rebinding, and opening of the substrate binding site and lid (Mayer and Bukau, 2005). We sought to directly contrast Hsp90 with a traditional Hsp70 exchange factor in the GR ligand binding assay. We chose the well characterized Bag-1 due to its ability to stimulate Hsp70 ADP and client release (Sondermann et al., 2001), and evidence that Bag-1 acts in the GR degradation pathway (Demand et al., 2001).
Bag-1 induced GRLBD release from Hsp70 was confirmed by pull down (Figure S1D). For functional comparison, we pre-formed the inhibited Hsp70-GRLBD complex, and then monitored the kinetics of F-dex binding resulting from Hsp70 release stimulated by either Bag-1, or Hsp90 plus HOP and p23 (Figure 6A). Under these conditions, the Hsp90 system showed a lag phase for ligand binding recovery (Figure 6B and Figure S7A). The slow recovery can be explained as the time required for the Hsp70 – Hsp90 client hand off given its dependence on the slow Hsp90 hydrolysis rate. Indeed, previous results indicated that Hsp90 ATP hydrolysis is the rate limiting step in the cycle (Morishima et al., 2001). With Hsp90, full ligand binding is restored by 40 minutes and maintained for the duration of the experiment. Bag-1 functioned very quickly with a fast recovery of ligand binding (<10min). The ligand association rate observed by release with Bag-1 is ~2 fold faster than the basal association of F-dex to GRLBD on its own, however this is followed by a steady decrease in ligand binding. This decrease is relatively slow and occurs linearly with time until almost all ligand binding is lost by 3 hours. Increase in light scattering demonstrated that the loss of function correlates with aggregation (Figure 6C). It should be noted that GRLBD alone, without chaperones, was able to maintain full ligand binding during the course of the experiment and shows no aggregation at this temperature by light scattering. This shows that GRLBD directly released from Hsp70, but not from the Hsp90 system, is in an aggregation prone state, implying that an additional GRLBD folding likely occurs on Hsp90. This indicates two distinct functions of Hsp90; one being the release of GRLBD from Hsp70, and the other being a chaperone function.
Figure 6. Hydrolysis Independent Chaperone Function of Hsp90 is Important for Maintaining an Active GR Population.
A) Experimental scheme for B. GRLBD was pre-inhibited by Hsp70 and Hsp40, and equilibrated with F-dex. Ligand binding is initiated with either Bag-1 or the Hsp90 system.
B) Ligand binding initiated with,Bag-1 (black), Hsp90 with HOP and p23 (red), Hsp90(E47A) with HOP and p23 (green), Bag-1 plus Hsp90 with HOP and p23 (purple), and Bag-1 with Hsp90(E47A) with HOP and p23 (yellow). Also shown, GRLBD alone preincubated with F-dex (blue). Assay conditions: 50nM F-dex, 1µM MBP-GRLBD, 2µM Hsp40, 15µM Hsp70, Hsp90, HOP, p23 and Bag-1.
C) Time course of light scattering for GRLBD (blue), and for GRLBD preincubated with Hsp40 and Hsp70 as for (A), with time course initiated with Bag-1 (black). Same conditions as B but without F-dex. See also Figure S7.
While Hsp90’s ability to reverse the Hsp70 inhibition is dependent on ATP hydrolysis (Figure 4B), we wanted to determine if the chaperone function is as well. To this end, we investigated the combined effects of Bag-1 and Hsp90 (with Hop and p23). When added together, a fast Bag-1 like recovery is seen, but then the full activity is maintained by the Hsp90 system (Figure 6B). This indicates that in the context of Hsp70 release by Bag-1, Hsp90 with HOP and p23 can function to prevent GRLBD loss of function. To truly decouple the Hsp70 release from the chaperone function, Bag-1 was added with hydrolysis dead Hsp90 E47A (with HOP and p23). With Bag-1, the Hsp90 E47A is just as capable as the wild type Hsp90 at preventing GRLBD loss of function (Figure 6B), clearly showing that the chaperone function is independent of ATP hydrolysis. This correlates with early work which showed that Hsp90’s ability to interact with unfolded intermediates and prevent aggregation is ATP independent (Jakob et al., 1995).
Further investigation into the combined effect with Bag-1 and Hsp90 revealed that the absence of HOP and p23 resulted in only partial suppression of the loss of function, indicating that both cochaperones are required to fully maintain GRLBD function (Figure S7B). Equivalent loss of function was seen without p23 as with the D93N mutant of Hsp90 with p23, indicating that accessing the closed state with p23 is an important aspect of Hsp90’s chaperone function (Figure S7C). Together, this suggests that Bag-1 acts within the pathway to promote nucleotide exchange on Hsp70 in the stalled complex with Hsp90 and HOP. Further evidence for this is seen when Bag-1 is added to the GRLBD pull down with the Hsp90 system. Addition of Bag-1 results in reduced levels of Hsp70 and HOP and significantly enhances incorporation of p23 (Figure S3).
Discussion
Under conditions where recombinant GRLBD is stable, recapitulating the in vivo requirement for Hsp90 activation entailed looking up stream in the pathway to the Hsp70 system. Here we showed that association with Hsp70 locally unfolds GRLBD resulting in inactivation. From this inhibited state, Hsp90 promotes recovery of ligand binding, explaining the in vivo requirement of Hsp90 for GR function. Instead of tenuous interactions in a brief handoff, there is tight coordination of an entire set of cochaperones with Hsp90 and this biochemical coordination is matched by an integral physical coordination seen in the cryo EM reconstruction of the Hsp90:Hsp70:HOP:GRLBD complex. In this complex, the interconnection between Hsp90 and Hsp70 manifests in the positioning of Hsp70 substrate binding region near the MD:CTD client binding site on Hsp90 and the direct interaction between the two ATPase domains. This provides a structural explanation for the coupling of the two hydrolysis cycles observed by recovery in ligand binding.
ATP hydrolysis by Hsp90 is essential for the reversal of the Hsp70 inhibition. This is surprising given the expectation that energy from hydrolysis is focused on driving client conformational rearrangements that promote activation. Instead, our results show that hydrolysis is utilized to regulate client transfer and Hsp70 release. Our work also provides new insight into how Hsp90 inhibitors affect the chaperone system. Hsp90 inhibitors that block the ATP binding pocket do not prevent the association of Hsp90 to the intermediate complex with Hsp70, HOP, and GR (Whitesell and Cook, 1996), but as shown here, they block reversal of the Hsp70-mediated client inhibition. In vivo, inhibition of Hsp90’s ATPase results in the proteolytic degradation of GR, as well as other Hsp90 clients, revealing a direct link between failure of client hand off, and degradation that proceeds through Hsp70 pathways (Stankiewicz et al., 2010). Hand off from Hsp70 to Hsp90 is thus a crucial regulation point in which a client’s fate is determined.
In addition to the Hsp70 release function, comparison of Hsp90 with the Hsp70 nucleotide exchange factor, Bag-1, reveals that Hsp90 also provides a chaperone function required for maintaining client stability upon Hsp70 release. Our work provides the first direct evidence of functional enhancements provided by the Hsp90 system, and moreover explains the constitutive requirement for Hsp90 throughout the functional lifetime of GR and not just during initial folding.
Hsp70 Binds to and Induces Partial Unfolding of GRLBD
Genetic studies have shown that deletion of Hsp40 in vivo leads to elevated GR signaling in both the absence and presence of hormone (Kimura et al., 1995). The Hsp40 dependent Hsp70 inhibition reported here explains this correlation. The elevated GR signaling in the absence of hormone suggests a further role of inactivation by Hsp70 beyond ligand binding inhibition, and points to Hsp70 as a crucial component that holds GR in the inactive, non-DNA binding state in the absence of ligand.
The observation that Hsp70 facilitates ligand release is surprising. It implies that Hsp70 can bind to a ligand bound form of GRLBD and actively promote ligand dissociation, presumably by unfolding. The minimal degree of unfolding observed in GRLBD was also surprising as Hsp70 is known to bind proteins in an unfolded state. In the crystal structure, Hsp70’s lid latches down over the stretch of unfolded polypeptide, which would require significant unfolding of the bound client (Zhu et al., 1996). However, there is evidence that Hsp70 can bind to some proteins while in more native states (Wawrzynow et al., 1995). Of these, σ32 was similarly shown to be only partially unfolded upon Hsp70 binding (Rodriguez et al., 2008). In this case, recent evidence revealed that the Hsp70 lid was not in a fully locked down position (Schlecht et al., 2011), suggesting that Hsp70 could accommodate protein substrates with varying degrees of tertiary structure, such as a partially folded GRLBD. In fact, the extended lid conformation fits better into the EM density for our complex than the bent conformation seen in the peptide bound structure (Figure S6C). Since most of the gain in substrate affinity in the ADP state comes from conformational changes within the substrate binding site (Mayer et al., 2000), it is plausible that GRLBD is still securely bound.
More generally, Hsp70’s ability to accelerate release of tightly bound GR ligand suggests an unappreciated mechanism whereby catalyzed unfolding can allow a more rapid response to sudden decreases in cellular ligand concentrations. Chaperones have been proposed to be involved in disassembly of transcription complexes (Freeman and Yamamoto, 2001), however, most of the attention has been centered around Hsp90 and p23 (Freeman and Yamamoto, 2002). Our findings suggest that the role of Hsp70 in promoting disassembly of GR transcription complexes has been over looked.
Modulation of GR’s Folded State by the Hsp70:Hsp90 System Results in Enhanced GR Stability, Function and Regulation
While Hsp70’s role in GR delivery to Hsp90 had been studied by Pratt and coworkers, the inactivation function of Hsp70 and therefor the role of Hsp90 in the reversal of the inhibition was not appreciated. This may be because the GR used in these studies was in an inactive state to begin with. It is worth noting that in related studies, purified PR while on ice, could bind ligand, and chaperones were only required to maintain ligand binding under elevated temperatures (Smith, 1993). Similarly, while our functional apoGRLBD could be maintained at 25°C, thermally induced aggregation of GRLBD without the solubility-enhancing MBP tag was detected at temperatures as low as 30°C (not shown). This indicates that under our experimental conditions, the apo receptor was only marginally stable. In addition, the full length wild type constructs used by Pratt and coworkers were likely more unstable, and were likely misfolded or had already formed small aggregates such that both Hsp70 and Hsp90 were required to cooperatively unfold and then refold the receptor in order to gain ligand binding. This highlights the unstable nature of GR and the genuine need for chaperone interactions in vivo.
Pratt and coworkers previously proposed that Hsp90 is holding the ligand binding cleft open (Pratt et al., 2008). In their model, Hsp70 is required to “prime” the receptor, while Hsp90 opens the binding cleft in an ATP dependent manner. Our data shows that the “priming” by Hsp70 is opening of the binding pocket, and that the crucial event resulting from ATP hydrolysis on Hsp90 is the release of Hsp70. In either case, the end result is GRLBD bound to Hsp90 with an open binding pocket (hence our faster ligand on rate).
With this in mind, the nuclear hormone receptors likely evolved a regulatory dependence on the Hsp90:Hsp70 chaperones because they are a class of proteins where partial unfolding is potentially beneficial. As represented in our model (Figure 7), we propose that the LBD has to partially unfold to allow for ligand entry and release. Unfolding by Hsp70 is thus a crucial step in opening the binding pocket. The mechanism by which Hsp70 achieves this is unclear. The cooperativity in the equilibrium inhibition could be the result of two Hsp70 binding events acting simultaneously, however with a multi state equilibrium of GRLBD conformations, it could also be the result of different binding affinities between one Hsp70 and the different GRLBD folded states, manifesting as an apparent cooperativity. Kinetic modeling could not rule out either model. Along these lines, a previous report suggested that either one Hsp70 interactively cycles on GR or two Hsp70s act sequentially in the Hsp70 priming step (Morishima et al., 2001). While the exact mechanism is still unknown, it is clear that while bound to Hsp70, GRLBD lacks the essential structural determinants for ligand binding, which can only be gained once folding completes after Hsp70 release by either a NEF or the Hsp90 system. While GRLBD released in a partially unfolded state by Bag-1 binds ligand more rapidly than GRLBD alone, without the influence of Hsp90 it is prone to aggregation, such that a fraction of the GRLBD population is lost during every cycle of Hsp70 binding and release.
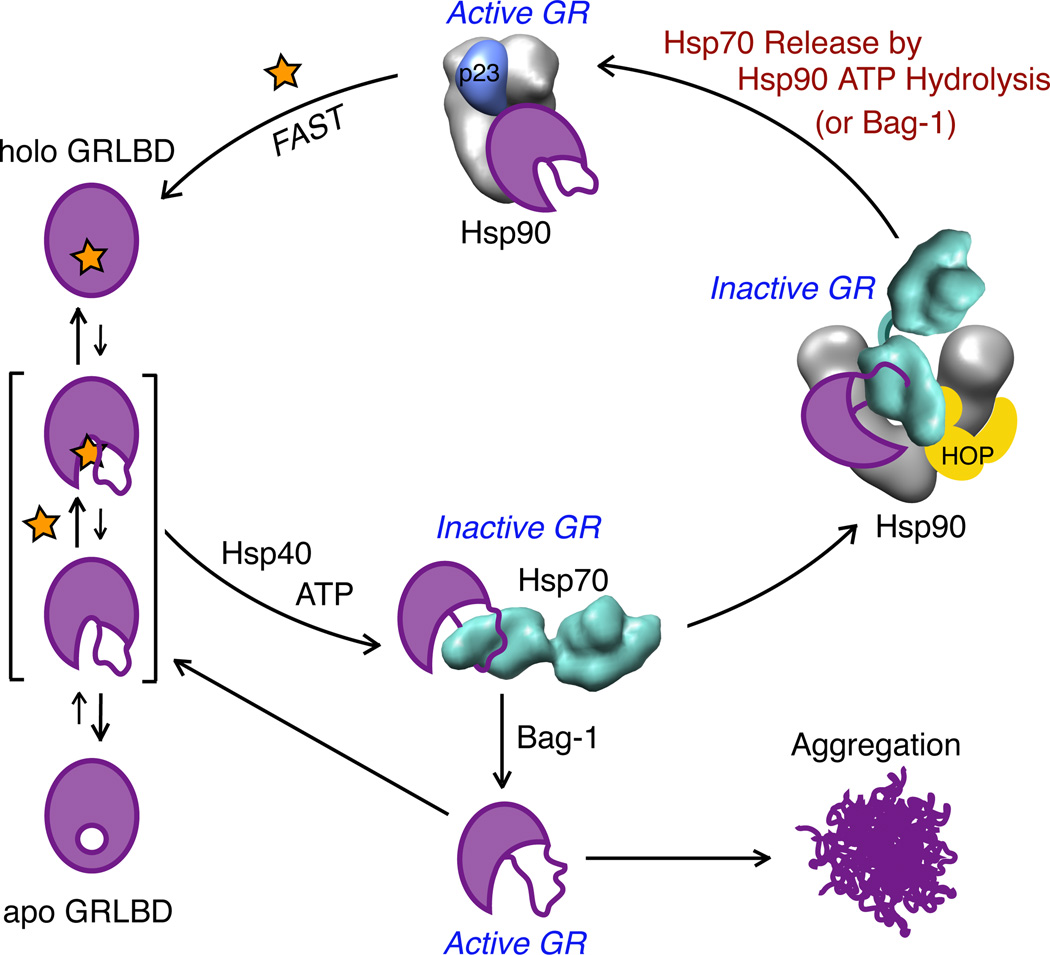
Figure 7. Model for GR Ligand Binding.
Apo GRLBD on its own is mostly folded and transiently samples a more unstructured state where the binding pocket at the core of the protein is accessible, allowing ligand association and dissociation. In a process that requires ATP hydrolysis and Hsp40, Hsp70 binds to GRLBD and promotes ligand release by stabilizing a partially unfolded inactive state. When GRLBD release from Hsp70 is stimulated by the NEF, Bag-1, GRLBD is rapidly released in a partially unfolded state that while able to bind ligand, is prone to aggregation. In the presence of Hsp90, GRLBD bound Hsp70 is brought together with Hsp90 by HOP, to form an inactive complex. Subsequent ATP hydrolysis on Hsp90 is required to release Hsp70 (and HOP), allowing progression to the closed state, and p23 binding. Together, Hsp90 and Bag-1 cooperate to promote Hsp70 release such that Hsp90 ATP hydrolysis is not required. Progression to the matured complex with Hsp90 and p23 results in high affinity ligand binding.
In the presence of the Hsp90 system, HOP recruits the GRLBD:Hsp70 complex such that GRLBD is delivered in proximity to Hsp90’s client binding site. The EM density suggests that GRLBD is making direct contact with Hsp90 while still bound to Hsp70. From this inhibited state, Hsp70 release requires hydrolysis on Hsp90. HOP mediates this interaction by promoting the NM rotation required to make contact between Hsp70 and Hsp90 ATPase domains. In the absence of HOP, Hsp90 can likely access this conformation, although with lower efficiency, thus explaining the partial ligand binding recovery seen without HOP. This is consistent with previous findings that also showed ~50% GR activation without HOP (Morishima et al., 2000a), and that HOP facilitated an Hsp70-Hsp90 interaction that can take place without HOP, but with low efficiency (Morishima et al., 2000b).
While we do not provide direct evidence that Hsp90 promotes ADP release from Hsp70, it is highly suggestive based on the direct contacts between Hsp90 and Hsp70 in the EM reconstruction. In the case of the catalytically dead Hsp90, this complex is stalled and GR activation cannot be achieved. However, ATP hydrolysis on Hsp90 is not required for GR activation in the presence of Bag-1. Our data suggests a model in which Bag-1 works within the Hsp90 pathway. Furthermore, in the EM structure, the Bag-1 (as well the bacterial exchange factor GrpE) binding site on Hsp70 is accessible. This suggests that Hsp90 and nucleotide exchange factors can work synergistically to promote nucleotide release on Hsp70. This provides an explanation for GrpE’s ability to work synergistically with bacterial Hsp90 and Hsp70 to refold denatured proteins (Genest et al., 2011), and further suggests that our findings extend to the Hsp70:Hsp90 system at a fundamental level.
Together, this work illustrates how Hsp70 and Hsp90 function as the yin and yang of protein folding in the cell. While the unfolding/inactivation by Hsp70 and the refolding/reactivation by Hsp90 might seem wastefully contradictory, their combination can be complementary. Constant rounds of Hsp70-mediated unfolding/ligand release and Hsp90-mediated refolding/ligand binding facilitates GR’s ability to provide both rapid and subtle responses to changing hormone levels while also maintaining apoGR in a non-aggregating, high affinity state. Moreover this provides opportunities for regulatory control and to coordinate signaling with protein homeostasis. Additionally, the reliance of both chaperones on cochaperones and numerous post-translational modifications enables additional levels of fine-tuning. Repeated cycles of unfolding and refolding have the additional benefit of allowing the chaperones to ensure a functional proteome by preventing the buildup of nonfunctional misfolded states and utilizing refoldability as a metric for targeting damaged proteins for degradation. These results provide clear benefits and reasons beyond stability for why many signaling proteins have evolved a dependence on the Hsp70:Hsp90 chaperone system.
Experimental Procedure
Protein Expression and Purification
Human Hsp90α, Hsp70, HOP and p23, and yeast YDJ1 were expressed in the pET151 bacterial expression plasmid with a cleavable 6× His-tag, and purified as previously described (Southworth and Agard, 2011). Human Bag-1 was expressed in a pET28a vector with a cleavable 6× His tag and purified as above. For cryo-EM, Hsp70 was purified from Sf9 cells as previously described (Southworth and Agard, 2011). For GRLBD, the LBD(F602S) (521–777) was cloned into a pMAL-c2X derivative with a N-terminal cleavable 6× His-MBP tag and a pACYCDuet derivative with a cleavable 6× His tag. Briefly, GRLBD was expressed in BL21 star (DE3) at 16°C in the presence of dexamethasone, and purified by Ni-affinity chromatography and ion exchange in the presence of ligand. Ligand was then removed by overnight dialysis, and excluded from size-exclusion chromatography (SEC). Purification was finished by extensive dialysis into ligand free buffer. See Supplemental Information for details.
Fluorescence Polarization Assays
Fluorescence polarization of fluorescein-labeled dexamethasone (Life Technologies) was measured on a SpectraMax M5 plate reader (Molecular Devices) with excitation/emission waves lengths of 485/538 nm, temperature control set at 25°C, and in 30mM HEPES pH7.5, 150mM KCl, and 2mM DTT. See Supplemental Information for details.
Hydrogen Deuterium Exchange Mass Spectrometry
Solution-phase amide HDX was carried out with a fully automated system (Goswami et al., 2013). GRLBD and Hsp70 was mixed at 1:1.2 molar ratio with 2µM Hsp40 and 5mM ATP/ MgCl2 and incubated for 1h at room temperature before HDX. For HDX, 5µl of 10µM GRLBD or the complex (GRLBD and Hsp70) was diluted to 25µl with D2O-containing HDX buffer and incubated at 4 °C for 10s, 30s, 60s, 900s or 3,600s. Following exchange, the protein was denatured by dilution to 50µl with 0.1% (v/v) TFA in 3 M urea and 50 mM TCEP and subjected to online pepsin digestion and electrospray ionization directly coupled to a high resolution (60,000 at m/z 400) Orbitrap mass spectrometer (LTQ Orbitrap XL with ETD, Thermo Fisher). In-house software was used for analysis (Pascal et al., 2012). For back exchange correction, an average of 70% recovery was estimated.
Aggregation Assay
Aggregation was monitored with a Jobin Yvon FluoroMax-3 fluorescence spectrophotomer with a temperature controlled jacket set to 25°C. Right angle light scattering was measured with excitation and emission wavelengths set to 500 nm with 1.4 nm slit widths.
Assembly of Hsp90-Hsp70-GR Loading Complex
Hsp90, HOP, Hsp70, MBP-GRLBD, and Hsp40 were incubated with ATP for 1 hour at room temperature. The complex was purified by SEC with an Ettan LC (GE Healthcare). Fractions were cross-linked with 0.02% gluteraldehyde for 20 minutes at room temperature and quenched with 20mM Tris HCl pH 7.5. Cross-linked fractions were analyzed by SDS-PAGE. Fractions containing the full complex were used for cryo-EM data collections. See Figure S5 for details.
Electron Microscopy and Three Dimensional Reconstructions
Cryo-EM data was collected as previously described (Southworth and Agard, 2011). 10,149 particles were picked from 214 images with a 2.2–4 µm defocus range. Defocus values were determined using CTFFIND (Mindell and Grigorieff, 2003) and corrected using a Weiner filter. An initial model was built from a featureless sphere with 2D class averages by projection matching in SPIDER (Frank et al., 1996). The 3D reconstruction was preformed using EMAN with ten rounds of refinement with 15° angular increments (Ludtke et al., 1999). The resolution was determined by the even-odd test in EMAN.
Supplementary Material
Highlights.
-
-
Hsp70 promotes GR ligand release and inactivation by inducing partial unfolding
-
-
Subsequent Hsp90 binding and ATP hydrolysis on Hsp90 reactivates GR ligand binding
-
-
Interaction with the Hsp90/Hsp70 chaperone systems result in enhanced GR function
-
-
Hsp90:Hop:Hsp70:GR complex reveals novel interactions between Hsp70 and Hsp90
Acknowledgements
We thank M. Liao, and Y. Cheng for help with the initial model for EM refinement, and members of the Agard lab for helpful discussions. Support for this work was provided by the PSI-Biology grant U01 GM098254 (D.A.), the UCSF Quantitative Biosciences Consortium (E.K.) and the Howard Hughes Medical Institute.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali MMU, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90–nucleotide–p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear Receptor Structure: Implications for Function. Annu. Rev. Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM. Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode of Receptor Dimerization and Coactivator Recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- Chrousos GP, Kino T. Glucocorticoid Signaling in the Cell. Annals of the New York Academy of Sciences. 2009;1179:153–166. doi: 10.1111/j.1749-6632.2009.04988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demand J, Alberti S, Patterson C, Höhfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- Dittmar KD, Banach M, Galigniana MD, Pratt WB. The role of DnaJ-like proteins in glucocorticoid receptor.hsp90 heterocomplex assembly by the reconstituted hsp90.p60.hsp70 foldosome complex. J. Biol. Chem. 1998;273:7358–7366. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- Dittmar KD, Demady DR, Stancato LF, Krishna P, Pratt WB. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. The role of p23 is to stabilize receptor.hsp90 heterocomplexes formed by hsp90.p60.hsp70. J. Biol. Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- Dittmar KD, Hutchison KA, Owens-Grillo JK, Pratt WB. Reconstitution of the steroid receptor.hsp90 heterocomplex assembly system of rabbit reticulocyte lysate. J. Biol. Chem. 1996;271:12833–12839. doi: 10.1074/jbc.271.22.12833. [DOI] [PubMed] [Google Scholar]
- Fang L, Ricketson D, Getubig L, Darimont BD. Unliganded and hormone-bound glucocorticoid receptors interact with distinct hydrophobic sites in the Hsp90 C-terminal domain. Proc. Natl. Acad. Sci. U.S.a. 2006;103:1847–18492. doi: 10.1073/pnas.0609163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjimi M, Leith A. SPIDER and WEB: Processing and Visualization of Images in 3D Electron Microscopy and Related Fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. Continuous recycling: a mechanism for modulatory signal transduction. Trends in Biochemical Sciences. 2001;26:285–290. doi: 10.1016/s0968-0004(01)01834-5. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- Genest O, Hoskins JR, Camberg JL, Doyle SM, Wickner S. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proceedings of the National Academy of Sciences. 2011;108:8206–8211. doi: 10.1073/pnas.1104703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, Masison DC, Wickner S. Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast; Mol. Cell. 2013;49:464–473. doi: 10.1016/j.molcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D, Devarakonda S, Chalmers MJ, Pascal BD, Spiegelman BM, Griffin PR. Time window expansion for HDX analysis of an intrinsically disordered protein. J. Am. Soc. Mass Spectrom. 2013;24:1584–1592. doi: 10.1007/s13361-013-0669-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández PM, Chadli A, Toft DO. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. Journal of Biological Chemistry. 2002;277:11873–11881. doi: 10.1074/jbc.M111445200. [DOI] [PubMed] [Google Scholar]
- Howard KJ, Holley SJ, Yamamoto KR, Distelhorst CW. Mapping the HSP90 binding region of the glucocorticoid receptor. J. Biol. Chem. 1990;265:11928–11935. [PubMed] [Google Scholar]
- Jakob U, Lilie H, Meyer I, Buchner J. Transient Interaction of Hsp90 with Early Unfolding Intermediates of Citrate Synthase. Journal of Biological Chemistry. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Halas A, Flom G. Nucleotide-Dependent Interaction of Saccharomyces cerevisiae Hsp90 with the Cochaperone Proteins Sti1, Cpr6, and Sba1. Mol. Cell. Biol. 2007;27:768–776. doi: 10.1128/MCB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Ichiro Y, Lindquist S. Role of the Protein Chaperone YDJ1 in Establishing Hsp90-Mediated Signal Transduction Pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- Kosano H, Stensgard B, Charlesworth MC, McMahon N, Toft D. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J. Biol. Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- Krukenberg KA, Förster F, Rice LM, Sali A, Agard DA. Multiple Conformations of E. coli Hsp90 in Solution: Insights into the Conformational Dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Quart. Rev. Biophys. 2011;44:229–255. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz OR, Freiburger L, Rutz DA, Krause M, Zierer BK, Alvira S, Cuellar J, Valpuesta JM, Madl T, Sattler M, et al. Modulation of the Hsp90 Chaperone Cycle by a Stringent Client Protein; Mol. Cell. 2014;53:941–953. doi: 10.1016/j.molcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. CMLS, Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Schröder H, Rüdiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- McLaughlin SH, Jackson SE. Folding and stability of the ligand-binding domain of the glucocorticoid receptor. Protein Science. 2002;11:1926–1936. doi: 10.1110/ps.5000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J. Mol. Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Morishima Y, Kanelakis KC, Murphy PJ, Shewach DS, Pratt WB. Evidence for iterative ratcheting of receptor-bound hsp70 between its ATP and ADP conformations during assembly of glucocorticoid receptor.hsp90 heterocomplexes. Biochemistry. 2001;40:1109–1116. doi: 10.1021/bi002399+. [DOI] [PubMed] [Google Scholar]
- Morishima Y, Kanelakis KC, Silverstein AM, Dittmar KD, Estrada L, Pratt WB. The Hsp organizer protein hop enhances the rate of but is not essential for glucocorticoid receptor folding by the multiprotein Hsp90-based chaperone system. J. Biol. Chem. 2000a;275:6894–6900. doi: 10.1074/jbc.275.10.6894. [DOI] [PubMed] [Google Scholar]
- Morishima Y, Murphy PJ, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J. Biol. Chem. 2000b;275:18054–18060. doi: 10.1074/jbc.M000434200. [DOI] [PubMed] [Google Scholar]
- Obermann WMJ, Sondermann H, Russo A, Pavletich N, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. The Journal of Cell Biology. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. HDX Workbench: Software for the Analysis of H/D Exchange MS Data. J. Am. Soc. Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff SJ, Fletterick RJ. Hormone binding and co-regulator binding to the glucocorticoid receptor are allosterically coupled. Journal of Biological Chemistry. 2010;285:15256–15267. doi: 10.1074/jbc.M110.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y. The Hsp90 Chaperone Machinery Regulates Signaling by Modulating Ligand Binding Clefts. Journal of Biological Chemistry. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Murphy M, Harrell M. In Molecular Chaperones in Health and Disease. Berlin/Heidelberg: Handbook of Experimental Pharmacology; 2006. Chaperoning of Glucocorticoid Receptors; pp. 111–138. [DOI] [PubMed] [Google Scholar]
- Ricketson D, Hostick U, Fang L, Yamamoto KR, Darimont BD. A Conformational Switch in the Ligand-binding Domain Regulates the Dependence of the Glucocorticoid Receptor on Hsp90. J. Mol. Biol. 2007;368:729–741. doi: 10.1016/j.jmb.2007.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Arsène-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones; Mol. Cell. 2008;32:347–358. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Schlecht R, Erbse AH, Bukau B, Mayer MP. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 2011;18:345–351. doi: 10.1038/nsmb.2006. [DOI] [PubMed] [Google Scholar]
- Schmid AB, Lagleder S, Gräwert MA, Röhl A, Hagn F, Wandinger SK, Cox MB, Demmer O, Richter K, Groll M, et al. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. Embo J. 2012;31:1506–1517. doi: 10.1038/emboj.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol. Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- Smith DF, Stensgard BA, Welch WJ, Toft DO. Assembly of progesterone receptor with heat shock proteins and receptor activation are ATP mediated events. J. Biol. Chem. 1992;267:1350–1356. [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Höhfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 Complex: Convergent Functional Evolution of Hsp70 Nucleotide Exchange Factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- Southworth DR, Agard DA. Client-Loading Conformation of the Hsp90 Molecular Chaperone Revealed in the Cryo-EM Structure of the Human Hsp90:Hop Complex; Mol. Cell. 2011;42:771–781. doi: 10.1016/j.molcel.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz M, Nikolay R, Rybin V, Mayer MP. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS Journal. 2010;277:3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Banecki B, Wall D, Liberek K, Georgopoulos C, Zylicz M. ATP hydrolysis is required for the DnaJ-dependent activation of DnaK chaperone for binding to both native and denatured protein substrates. J. Biol. Chem. 1995;270:19307–19311. doi: 10.1074/jbc.270.33.19307. [DOI] [PubMed] [Google Scholar]
- Wegele H, Muschler P, Bunck M, Reinstein J, Buchner J. Dissection of the contribution of individual domains to the ATPase mechanism of Hsp90. J. Biol. Chem. 2003;278:39303–39310. doi: 10.1074/jbc.M305751200. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Molecular Endocrinology. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural Analysis of Substrate Binding by the Molecular Chaperone DnaK. Science, New Series. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.