Abstract
Precise regulation of axon branching is crucial for neuronal circuit formation, yet the mechanisms that control branch formation are not well understood. Moreover, the highly complex morphology of neurons makes them critically dependent on protein/membrane trafficking and transport systems, although the functions for membrane trafficking in neuronal morphogenesis are largely undefined. Here we identify a kinesin adaptor, Calsyntenin-1 (Clstn-1), as an essential regulator of axon branching and neuronal compartmentalization in vivo. We use morpholino knockdown and a Clstn-1 mutant to show that Clstn-1 is required for formation of peripheral but not central sensory axons, and for peripheral axon branching in zebrafish. We used live imaging of endosomal trafficking in vivo to show that Clstn-1 regulates transport of Rab5-containing endosomes from the cell body to specific locations of developing axons. Our results suggest a model in which Clstn-1 patterns separate axonal compartments and define their ability to branch by directing trafficking of specific endosomes.
Keywords: axon branching, calsyntenin, endosome, polarity, trafficking, zebrafish
Introduction
Development of complex neuronal morphology and functional circuits requires precise regulation of axon formation and branching. Axon branching allows innervation of multiple targets, and tight control over where branches form is essential for circuit formation, plasticity, and regeneration after injury. Although several molecular signals are known to influence branching (Schmidt and Rathjen, 2010; Gallo, 2011; Gibson and Ma, 2011), the mechanisms controlling axon branch formation remain poorly understood.
Many neurons extend multiple axons or branches that navigate to distinct targets. For example, vertebrate sensory neurons must extend separate axons to targets in the periphery and the CNS for proper sensory circuit formation. Peripheral and central sensory axons follow distinct pathways and have a different capacity for branching (Andersen et al., 2011). In some cases, these axons have been shown to respond differently to guidance signals (Liu and Halloran, 2005). Moreover, intrinsic activity of LIM homeodomain transcription factors specifically regulates formation and branching of zebrafish peripheral but not central sensory axons (Segawa et al., 2001; Becker et al., 2002; Andersen et al., 2011; Tanaka et al., 2011). Nonetheless, the molecular mechanisms regulating compartmentalization of separate axons from one neuron are largely unknown.
Calsyntenins are cadherin superfamily transmembrane proteins first identified as synaptic proteins (Vogt et al., 2001; Hintsch et al., 2002). Vertebrates have three calsyntenins (Clstn-1, -2, and -3) expressed predominantly in the nervous system (Hintsch et al., 2002). A human clstn-2 allele is associated with increased episodic memory ability (Preuschhof et al., 2010), and a Caenorhabditis elegans ortholog is required for learning (Ikeda et al., 2008; Hoerndli et al., 2009). Clstn-3 regulates synapse development in mice (Pettem et al., 2013; Um et al., 2014), suggesting that developmental roles may underlie learning and memory functions. Clstn-1 is a kinesin-1 adaptor that regulates trafficking and processing of amyloid precursor protein (Konecna et al., 2006; Araki et al., 2007; Steuble et al., 2012; Vagnoni et al., 2012). Calsyntenin protein levels are reduced in human Alzheimer's disease brains (Ringman et al., 2012; Vagnoni et al., 2012) and accumulate in dystrophic neurites (Araki et al., 2003; Uchida et al., 2013). Nonetheless, how Clstn-1 influences neuronal function and how its trafficking function may relate to roles in learning and memory are not known. Clstn-1 is found in growth cones of cultured neurons (Steuble et al., 2010). However, roles for Clstn-1 in development have not been demonstrated.
Here we identify Clstn-1 as a critical regulator of axon branching and compartmentalization during development of vertebrate sensory axon arbors. We use live imaging in vivo to show that Clstn-1 is required specifically for formation and branching of peripheral but not central sensory axons in zebrafish. Moreover, we develop a method for high-speed, high-resolution, in vivo imaging of endosomal trafficking in neurons. We find that specific endosomal populations display different dynamics in different neuronal compartments, and we find that endosome trafficking is important for peripheral axon branching. Finally, we show that Clstn-1 regulates endosomal dynamics and the transport of endosomes from the cell body into axons and branch points during neuronal morphogenesis in vivo.
Materials and Methods
Animals.
Adult zebrafish (Danio rerio) were kept in a 14/10 h light/dark cycle. Embryos were maintained at 28.5°C and staged as hours postfertilization (hpf) as described previously (Kimmel et al., 1995). Wild-type AB strain or transgenic Tg(−3.1ngn1:GFP-CAAX) (Andersen et al., 2011) embryos of either sex were used for all experiments. All animals were handled in accordance with guidelines from the Institutional Animal Care and Use Committee and the National Institutes of Health.
In situ hybridization.
Zebrafish clstn1 cDNA in pME18S-FL3 vector was obtained from Open Biosystems. A T7 promoter site was added to the clstn1 cDNA via PCR using the following primers: forward 5′-GGATGTTGCCTTTACTTCTA-3′, and reverse 5′-TAATACGACTCACTATAGGGAGACGACCTGCAGCTCGAGCACA-3′.
A digoxigenin-labeled riboprobe for clstn1 mRNA was synthesized by in vitro transcription using T7 RNA polymerase (Roche) and hydrolyzed to an average size of 200–500 bases by alkaline hydrolysis (Cox et al., 1984). Whole-mount in situ hybridization was performed as previously described (Halloran et al., 1999).
Immunohistochemistry.
Embryos were fixed overnight in 4% PFA and labeled with monoclonal anti-HNK1 antibody (ZN-12, 1:250; Zebrafish International Resource Center) for wild-type and mutant embryos, or with mouse or rabbit anti-GFP antibody (1:1000; Invitrogen) for transgenic embryos. Antibody detection was performed with a Vectastain IgG ABC detection kit (Vector Laboratories), or for fluorescent labeling, with AlexaFluor-488-conjugated secondary anti-mouse or anti-rabbit antibodies (4 μg/ml; Invitrogen).
DNA constructs, morpholinos (MOs), and injection.
DNA expression constructs were made using Multisite Gateway Cloning System (Invitrogen) into Tol2 vectors (Kwan et al., 2007). Constructs were cloned behind a cis-regulatory element of the neurogenin1 gene (−3.1ngn1) (Blader et al., 2003) to drive expression in RB neurons as described previously (Andersen et al., 2011). The Clstn-1 intracellular domain or mCherry (as a control) were connected to prenylated GFP with a self-cleaving 2A peptide. Zebrafish Rab5c, constitutively active form of Rab5c (CA-Rab5c; Q81L), Rab7, and Rab11a in Gateway pDONR vectors (Clark et al., 2011) were obtained from Brian Link (Medical College of Wisconsin) and cloned behind the −3.1ngn1 promoter with N-terminal GFP or photoactivatable (PA) GFP fusions. To mosaically label RB cells, 25 pg DNA was injected into one-cell stage embryos. For photoactivation experiments, 11 pg of −3.1ngn1:TagRFP-CAAX (Andersen et al., 2011) and 11 pg of −3.1ngn1:PA-GFP-Rab5c DNA constructs were coinjected into one-cell stage embryos.
Translation blocking and splice blocking MOs against Clstn-1 were synthesized by Gene Tools. The translation blocking MO sequence, with the start codon in italics, is 5′-GATCCGCATCGCGTCTTCGTAATAA-3′. The splice blocking MO was designed to the exon5/intron5 boundary, and its sequence is 5′-GAGTTTTGGCAGTCACTTACCATCC-3′. A scrambled standard control MO was used as an injection control: 5′-CCTCTTACCTCAGTTACAATTTATA-3′. MOs were injected at 750 μm in 1 nl volume into one-cell stage embryos.
We tested the efficacy of the splice-blocking MO with RT-PCR on mRNA collected from embryos at 24 hpf using the following primers: RT primer in exon 10, 5′-ACAAACAAAGTCACTGCCGGGAAC-3′; forward PCR primer spanning the exon1–2 boundary, 5′-GCGGTGGATGCAGCCAAAGTAAAT-3′; reverse PCR primer in exon 6, 5′-GTTTGCAGGTGGGCTTGATGTTGA-3′.
Bright-field and fixed sample imaging.
Bright-field images were captured on a Nikon TE300 inverted microscope with a Spot RT camera (Diagnostic Instruments) and processed with MetaMorph Software (Universal Imaging). Fluorescent images of fixed embryos were captured with an Olympus FV1000 laser-scanning confocal microscope with 40× (UPlan FLN air, NA 0.75) or 60× (UPlanSApo oil-immersion, NA 1.35) objectives.
Time-lapse imaging.
For live confocal imaging, embryos were anesthetized in 0.02% tricaine and mounted in 1% low melting agarose in 10 mm HEPES E3 medium as described previously (Andersen et al., 2010). Images of Tg(−3.1ngn:GFP-CAAX) embryos were collected on an Olympus FV1000 laser-scanning confocal microscope using a 40× objective (UPlan FLN, NA 0.75). Imaging was started between 17 and 19 hpf, when peripheral RB axons are extending out of the spinal cord. Optical sections (1 μm) covering a total of 18–27 μm thickness were captured every 35–196 s. Imaging duration ranged from 1.5–6 h.
Photoactivation and imaging of PA-GFP-Rab5 were performed on the Olympus FV1000 confocal using a 60× objective (UPlanSApo oil-immersion, NA 1.35). An ROI in the region of the cell body or a branch point was activated with a 405 nm laser for a duration of 1 s. z-stacks of 1 μm optical sections were captured at 34–100 s time intervals, for durations ranging from 39 to 192 min.
Live high-speed imaging of endosomal trafficking was performed on a Swept-Field confocal microscope (Bruker Nano Surfaces FM) equipped with a Nikon CFI Plan Apo VC 60× oil-immersion objective (NA 1.40). Embryos were 23 hpf at the beginning of the experiment, and 1–20 1-μm-thick optical sections were captured every 2 s for a total duration of 400 s.
Quantification and data analysis.
Fluorescent images and movies were processed and quantified with Volocity Software (PerkinElmer). Axon lengths and growth rates were calculated as described previously (Andersen et al., 2011). Peripheral axon branching was quantified by counting the number of axon branches crossing the horizontal myoseptum in somites 8–13 of 22 hpf embryos labeled with HNK-1 antibody. In individually labeled neurons, branching was also quantified by counting the total number of peripheral axon branch tips. Growth cone bifurcations and interstitial branching were quantified by counting new branching events/min from time-lapse movies. Cell number was measured as the number of RB cell bodies in one side of the spinal cord along somites 8–13 of 22 hpf embryos. Growth cone volume was measured in Volocity software with an ROI drawn around the growth cone.
Travel distance of photoactivated Rab5c vesicles was measured in Volocity as the greatest distance from the cell body at time points ranging from 1 to 30 min after photoactivation. Comparisons were made with two-way ANOVA. To measure changes in vesicle accumulations, an ROI was drawn around the vesicle accumulation and average fluorescence intensity was measured at 3 min and 30 min after photoactivation. The change in signal is reported as percentage of original intensity (intensity at 30 min divided by intensity at 3 min). To measure retention of vesicles photoactivated at branch points, the volume of GFP signal at the branch point was measured at 2 min and 20 min after photoactivation, and the change is reported as the percentage of original volume remaining at 20 min.
Endosomal trafficking movies were built in Volocity and corrected for drift in ImageJ (Schneider et al., 2012). Endosomal speeds were measured in Volocity from kymographs made in ImageJ. Speeds were measured during the fastest run per vesicle in a 400 s imaging period. To determine endosomal direction, we measured the net direction of each individual punctum over 400 s, and categorized vesicle movement as either anterograde (away from the cell body), retrograde (toward the cell body), or no net movement.
All statistical analyses were done using Prism 5.0 (GraphPad Software). Errors are reported as SEM.
Transcription Activator-like Effector Nuclease (TALEN) mutant generation.
TALENs were designed to target exon 2 of clstn1 as described previously (Dahlem et al., 2012) using the TALEN Golden Gate Assembly System (Cermak et al., 2011). Plasmids with repeat-variable di-residue (RVD) repeats were obtained from Addgene, and RVD repeat arrays were assembled as described previously (Cermak et al., 2011). RVD repeat arrays were cloned into pCS2TAL3DD and pCS2TAL3RR to generate left and right TALEN constructs (Dahlem et al., 2012).
pCS2TAL3DD and pCS2TAL3RR TALEN plasmid templates were linearized with NotI, and 5′-capped RNA was transcribed in vitro (mMESSAGE mMACHINE SP6 Kit, Ambion). Equal amounts of left and right TALEN mRNA (200 pg total) were injected into one-cell stage embryos.
Genomic DNA was extracted from individual 2 d postfertilization (dpf) embryos as described previously (Dahlem et al., 2012). Embryos were incubated in extraction buffer containing 500 μg/ml proteinase K at 55°C, and the reaction was terminated by incubation at 100°C for 5 min.
High-resolution melt analysis (HRMA) was used to detect TALEN-induced mutations (Dahlem et al., 2012). The following primers were used to generate a 150 bp amplicon that included the TALEN target site in exon 2: forward 5′-ttcaccttcacagtaaataagcaca-3′, reverse 5′-atggaggatcttacctgcgta-3′. HRMA was performed in a 20 μl reaction containing 40–400 ng embryonic genomic DNA, 1× MeltDoctor HRM Master Mix (Applied Biosystems) and 300 nm of each primer. Thermocycling conditions were as follows: enzyme activation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 15 s, annealing/extension at 60°C for 1 min; denaturation at 95°C for 10 s; annealing at 60°C for 1 min; high resolution melting at 95°C for 15 s; annealing at 60°C for 15 s. Three replicates were done for DNA samples from each embryo. HRMA data were collected with StepOnePlus Real-Time PCR System (Applied Biosystems) and analyzed with High Resolution Melt Software version 3.0 (Invitrogen).
Mutant clstn-1uw7 embryos at 22 hpf were genotyped after immunohistochemistry. Genomic DNA was extracted from individual embryos with hot sodium hydroxide (Meeker et al., 2007), followed by PCR amplification of the mutated region. Derived cleaved amplified polymorphic sequence analysis (Neff et al., 2002) was used to identify mutants. The following PCR primers were used to generate 158 bp amplicons: forward 5′-acgtatcatggtatcgcgac-3′; reverse 5′-gtttcagaaagcatgaggaagc-3′. PCR was performed in a 50 μl reaction containing 20–60 ng embryonic genomic DNA, 10 μm of each primer, and GoTaq DNA polymerase (Promega). PCR products were digested with Hpy99I restriction enzyme (New England Biolabs).
Results
clstn1 is expressed in the developing CNS and in sensory neurons
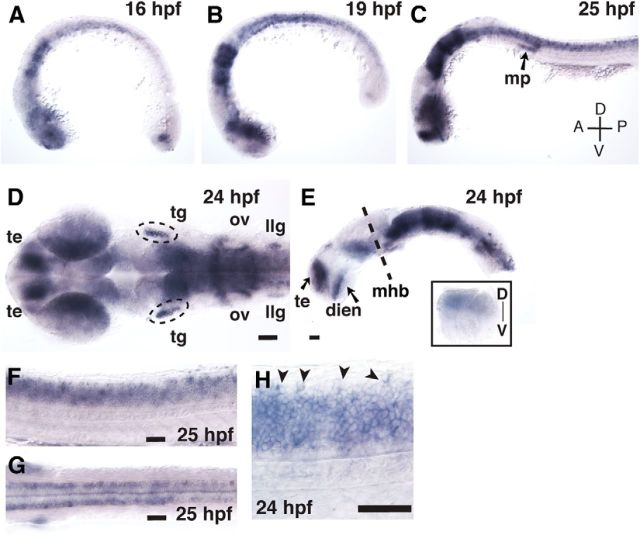
We initially identified zebrafish clstn1 in a microarray experiment (part of a separate study) as a gene regulated by LIM homeodomain transcription factors, suggesting it may function in differential development of sensory axons. We examined clstn1 mRNA expression during neural development stages and found expression in sensory neurons as well as other regions of the CNS (Fig. 1). At 16 hpf, when primary neurons first begin extending axons, clstn1 is expressed in the spinal cord and in regions of the forebrain, retina, and hindbrain (Fig. 1A). Expression increases from 16 to 24 hpf (Fig. 1B,C), and at 24 hpf, clstn1 is expressed strongly in the telencephalon, ventral forebrain, dorsal retina, ventral midbrain, hindbrain, and spinal cord (Fig. 1D,E,F). clstn1 is also expressed in the cranial sensory ganglia and in the migrating primordium of the lateral line during the period of sensory ganglia axon formation (Fig. 1C,D). In the spinal cord, clstn1 appears to be expressed in most differentiated neurons, including Rohon-Beard (RB) sensory neurons in the dorsal spinal cord, motor neurons in the ventral spinal cord, and interneurons in intermediate regions (Fig. 1F–H). Thus, clstn1 is found in the developing nervous system during stages of axon outgrowth, suggesting it may function in axon development.
Figure 1.
clstn1 mRNA is expressed in the developing nervous system. A–C, Lateral views of whole-mount embryos showing in situ hybridization for clstn1 mRNA at stages indicated. D, E, Dorsal (D) and lateral (E) views of expression in the head at 24 hpf. E, Inset, Dorsal expression in the eye. F, G, Lateral (F) and dorsal (G) views of the trunk at 25 hpf showing expression in the spinal cord. H, Higher-magnification view showing expression in the RB cell bodies (arrowheads) at 24 hpf. mp, Migrating primordium; te, telencephalon; tg, trigeminal ganglion; ov, otic vesicle; llg, lateral line ganglion; dien, diencephalon; mhb, midbrain-hindbrain boundary; D, dorsal; V, ventral; A, anterior; P, posterior. Scale bars, 40 μm.
Clstn-1 knockdown reduces peripheral sensory axons
To test roles for Clstn-1 in sensory axon development, we first used MOs to knock down Clstn-1. We designed two MOs: one that binds the region of the start codon to block translation and another that binds the exon5–intron5 boundary to block mRNA splicing. We tested the efficacy of the splice-blocking MO by amplifying the region from exon 2 to exon 6 with RT-PCR. We were able to amplify the expected product from wild-type but not from Clstn-1 MO-injected embryos, suggesting that Clstn-1 knockdown causes an intronic insertion in the mRNA product. Standard control MO-injected embryos and uninjected wild-type were used as controls.
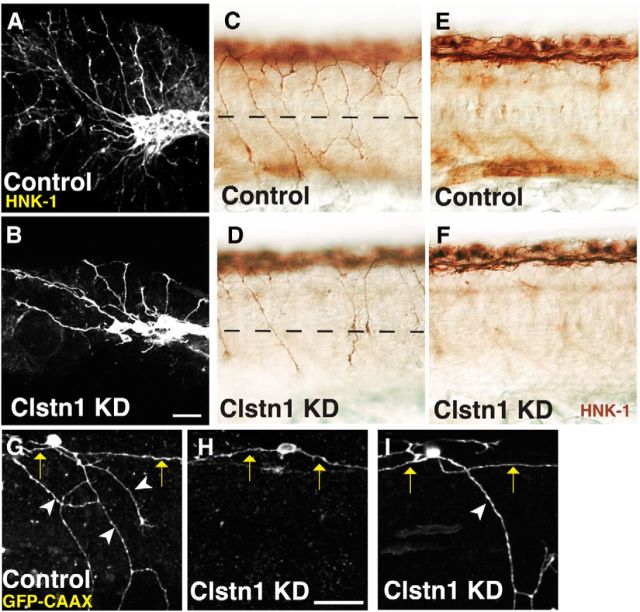
We first analyzed the effects of Clstn-1 knockdown by labeling cranial trigeminal neurons and spinal RB neurons with anti-HNK1 antibody. Clstn-1 knockdown caused a marked reduction in peripheral axons of both trigeminal and RB neurons (Fig. 2A–D). We quantified the RB axon phenotype by counting the number of peripheral axon branches that cross the horizontal myoseptum at 22 hpf and found significantly fewer crosses in Clstn-1 MO-injected embryos compared with either uninjected wild-type or standard control MO-injected embryos (Table 1). We found no significant difference between wild-type and standard control MO (p = 0.15, unpaired t test). Thus, for some measurements (indicated in Table 1), we pooled these controls. The number of RB cells (Table 1) and the integrity of the central axon fascicle (Fig. 2E,F) were not affected, suggesting that Clstn-1 has a specific function in peripheral axon growth or branching. Both MOs produced similar phenotypes (Table 1), suggesting that the phenotype is specific to Clstn-1 knockdown. We used the splice blocking MO for the remaining experiments described here.
Figure 2.
Clstn-1 knockdown reduces branches in peripheral sensory axons. A, B, Lateral views of trigeminal ganglia labeled with anti-HNK-1 in uninjected (A) and Clstn-1 knockdown (KD) (B) embryos showing reduced branching of trigeminal axons. C, D, Lateral views of RB neurons labeled with anti-HNK-1 showing a reduction in peripheral axons in Clstn-1 KD (D) versus control MO-injected embryos (C). E, F, The focal plane of central axons in the same embryos showing no effect on the central fascicle. G–I, Single-labeled RB cells expressing GFP-CAAX show a reduction of peripheral outgrowth (H) and peripheral branching (I) in Clstn-1 KD embryos. Arrows indicate central axons; arrowheads indicate peripheral axons. Scale bars, 40 μm.
Table 1.
Peripheral axon branching is reduced by Clstn-1 knockdowna
| Axons crossing myoseptum RB population (somites 8–13) | No. of RB neurons (somites 8–13) | Branches crossing myoseptum (individual RBs) | No. of peripheral axon tips (individual RBs) | |
|---|---|---|---|---|
| Controls | — | 15.6 ± 0.5* | — | — |
| Wild-type | 9.5 ± 1.2b | — | 1.6 ± 0.4h | 10.7 ± 1.4j |
| Standard control MO | 11.0 ± 1.8c | — | — | — |
| ATG MO | 5.4 ± 0.6d | 14.4 ± 0.5f | ND | ND |
| Splice MO | 6.5 ± 0.7e | 16.1 ± 0.7g | 0.3 ± 0.2i | 3.9 ± 0.9k |
aAll comparisons made with two-tailed, unpaired t test. ND, Not determined.
bn = 12 embryos.
cn = 21 embryos.
dn = 29 embryos; p = 0.0012 compared with wild-type; p = 0.03 compared with standard control MO.
en = 17 embryos; p < 0.0001 compared with wild-type; p = 0.0002 compared with standard control MO.
fn = 25 embryos; p = 0.1 compared with pooled controls.
gn = 7 embryos; p = 0.5 compared with pooled controls.
hn = 17 cells in 14 embryos.
in = 13 cells in 12 embryos; p = 0.005 compared with wild-type.
jn = 15 cells in 13 embryos.
kn = 8 cells in 7 embryos; p = 0.003 compared with wild-type.
*Pooled uninjected wild-type and standard control morpholino controls; n = 15 embryos.
To further investigate whether Clstn-1 knockdown affects central axons and to ask whether the reduction in peripheral axons is the result of defects in axon formation or in branching, we labeled individual RB neurons. We injected DNA encoding membrane-targeted GFP, driven by regulatory elements from the neurogenin-1 gene (Blader et al., 2003) into embryos at the one-cell stage. This method results in mosaic labeling of RB cells and allows visualization of the entire axon morphology of individual neurons. The peripheral RB axon forms as a branch from a central axon in the close vicinity of the cell body or directly from the cell body (Andersen et al., 2011). We found that 50% of RB neurons (n = 18 cells in 14 embryos) did not extend a peripheral axon after Clstn-1 knockdown (Fig. 2H), compared with 13% (n = 23 cells in 18 embryos) in control embryos without MO (Fig. 2G), a significant difference (p = 0.02, Fisher's Exact test). Moreover, peripheral axons that did extend showed reduced branching (Fig. 2I). Quantification of axon branches revealed a significant decrease in peripheral branches crossing the horizontal myoseptum, as well as a reduction in peripheral axon branches on individually labeled RB neurons in Clstn-1 knockdown embryos (measured as number of peripheral axon branch tips; Table 1). These data suggest that Clstn-1 mediates both the initial branching of the peripheral axon from the central axon as well as secondary branching in the formed peripheral axon. In contrast, central axon length was unaffected by Clstn-1 knockdown. Central axons were on average 389 ± 32 μm long (n = 11 axons in 8 embryos) in control embryos and 380 ± 41 μm (n = 16 axons in 9 embryos) in Clstn-1 knockdown embryos (p = 0.12, unpaired t test), further supporting the idea that Clstn-1 functions in the differential behavior of peripheral versus central axons.
Generation of Clstn-1 mutants with TALENs
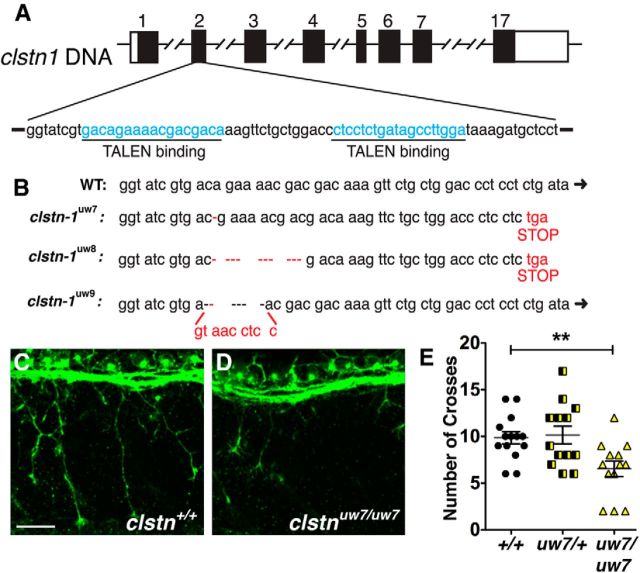
To further investigate Clstn-1 function and verify the knockdown phenotype, we generated a Clstn-1 mutant using TALENs (Dahlem et al., 2012). Two splice variants of Clstn-1, which differ in the presence of exon 3, are found in zebrafish. To generate mutations that affect both splice forms, we designed two TALEN pairs targeting the second exon (Fig. 3A). TALEN RNA was injected into one-cell stage wild-type embryos, and the efficacy of TALEN pairs in generating mutations was tested with HRMA of the embryos at 2 dpf (Dahlem et al., 2012). Short amplicons (150 bp) from genomic DNA of TALEN-injected embryos were generated with primers spanning the TALEN binding region. The amplicons undergo denaturation and renaturation, and HRMA detects changes in thermostability of heteroduplexes that result from mutations. We detected mutations in 10 of 10 TALEN-injected embryos tested with HRMA. The siblings of these embryos were raised to adulthood to generate mutant lines.
Figure 3.
TALEN generation of Calsyntenin-1 mutants. A, TALENs were designed against the second exon of Clstn-1. B, clstn-1uw7and clstn-1uw8 result in nucleotide deletions with a frame shift and predicted premature stop. clstn-1uw9 has a 6 bp deletion and 9 bp insertion, which does not result in a frame shift. C, D, RB neurons labeled with HNK-1 antibody in wild-type (C) and clstn1uw7/uw7 mutant (D) embryos from incross of clstn1uw7/+ heterozygous fish. E, Quantification of axon branches crossing the horizontal myoseptum in wild-type (+/+), clstn1uw7/+ heterozygous, and clstn1uw7/uw7 homozygous embryos. **p = 0.004 (unpaired, two-tailed t test). Scale bar, 40 μm.
We tested for germline transmission of mutations using HRMA on F1 embryos and found that 20% of the founder fish tested produced F1 embryos with mutations. We raised F1 fish and sequenced their DNA to determine the nature of the mutations. We detected three independent mutations in the F1 generation (Fig. 3B). Allele clstn1uw7 has a single nucleotide deletion, which results in a frame shift and a premature stop after amino acid 54 (of 954 amino acids total). The resulting protein would lack most of the extracellular cadherin domains and all of the transmembrane and intracellular domains, including the calcium and kinesin binding regions, suggesting a likely null. Allele clstn1uw8 has a 10 nucleotide deletion and a similar frame shift and stop. Allele clstn1uw9 has a 6 nucleotide deletion and 9 nucleotide insertion, which does not result in a frame shift. Interestingly, we observed mutation clstn1uw7 in multiple offspring of a single founder, suggesting that the mutation may have occurred early in the process of founder gametogenesis.
We analyzed the Clstn-1 mutant phenotype by crossing heterozygous carriers of clstn1uw7. Embryos showed no gross morphological defects at 1 dpf, similar to MO-injected embryos. To assess sensory axon development, we labeled embryos with HNK-1 antibody. We compared age-matched siblings and found a marked reduction in RB peripheral axon branching in mutant embryos (Fig. 3C–E). These data strongly support the MO results and indicate that the Clstn-1 knockdown phenotype is specifically caused by Clstn-1 loss of function. Interestingly, at 5 dpf, 22% of the embryos from crosses of heterozygous clstn1uw7 carriers (n = 530 embryos total) showed defects, including small or missing eyes, moderate to severe pericardial and periorbital edema, abnormal body curvature, and visibly smaller heads, suggesting multiple potential roles for Clstn-1 in embryonic development. However, because there is no gross morphological phenotype that allows us to identify living mutant embryos at early stages (1 dpf), we use MO knockdown for the live imaging experiments described below.
Live imaging of RB axon growth reveals Clstn-1 role in branch formation
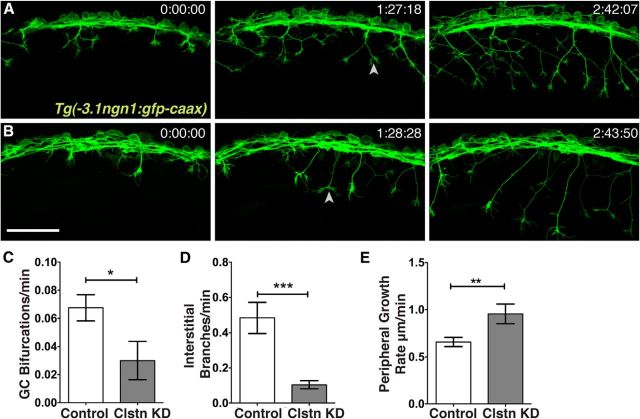
Zebrafish RB axons branch by two mechanisms: growth cone bifurcation and interstitial back branching (Andersen et al., 2011). To investigate the specific motile processes and type of branching regulated by Clstn-1, we used live imaging of axon behavior. We imaged RB axons using a transgenic line in which all RB neurons are labeled with GFP-CAAX (Tg(−3.1ngn1:GFP-CAAX)) (Fig. 4A,B; Movies 1 and 2). We found that the number of filopodial protrusions and interstitial branches that formed along axons was significantly reduced in Clstn-1 MO-injected embryos compared with standard control MO-injected embryos (Fig. 4D), suggesting that Clstn-1 is required for the initial formation of branches. We also found a significant reduction in growth cone bifurcation events (Fig. 4C), suggesting that Clstn-1 functions in a step common to both branching mechanisms. Moreover, we measured peripheral RB growth cone volume and found that growth cones in Clstn-1 knockdown embryos were on average 206 ± 19 μm3 (n = 24 growth cones in 5 embryos) compared with 307 ± 49 μm3 for control growth cones (n = 20 growth cones in 6 embryos), a significant reduction (p = 0.038, unpaired t test). This result suggests that Clstn-1 may be important for the cytoskeletal dynamics mediating growth cone size or shape. Interestingly, the rate of peripheral axon extension was increased in Clstn-1 knockdown embryos (Fig. 4E), indicating that Clstn-1 knockdown does not generally impair the ability of axons to grow but specifically their ability to branch.
Figure 4.
Live imaging reveals reduced peripheral branching activity in Clstn-1 knockdown. A, B, Time-lapse images (lateral views, anterior left, z-projections) from Tg(−3.1ngn1:gfp-caax) embryos injected with standard control MO (A) or Clstn-1 MO (B). Arrowheads indicate bifurcation events. C–E, Quantification of axon branching and growth rate. *p = 0.03 (unpaired, two-tailed t test). ***p = 0.0008 (unpaired, two-tailed t test). **p = 0.009 (unpaired, two-tailed t test). Scale bar, 100 μm. Time is in hours:minutes:seconds.
RB axon development in control embryo. Time-lapse movie of axons in a Tg(−3.1ngn1:GFP-CAAX) embryo. Images were acquired every 34 s and are played back at 20 frames per second, for a total movie duration of 1:54:48 (hours:minutes:seconds). Movie is 680× faster than real time. Scale bar, 30 μm.
RB axon development after Clstn-1 knockdown. Time-lapse movie of −3.1ngn1:GFP-CAAX-expressing neurons in Clstn-1 MO-injected embryos. Images were acquired every 196.5 s and are played back at 4 frames per second, for a total movie duration of 2:30:43 (hours:minutes:seconds). Movie is 786× faster than real time. Scale bar, 30 μm.
Clstn-1 intracellular domain inhibits peripheral axon outgrowth
Clstn-1 binds kinesin light chain via its intracellular domain (ICD) and can serve as a molecular adaptor between kinesin-1 and vesicular cargo (Konecna et al., 2006; Araki et al., 2007; Ludwig et al., 2009; Vagnoni et al., 2011, 2012). The Clstn-1 ICD has been shown in cultured cells to act as a dominant-negative and interfere with normal kinesin binding and Clstn-mediated trafficking (Araki et al., 2007; Ludwig et al., 2009). To test whether Clstn-1 regulates axon branching via its trafficking function, we expressed the Clstn-1 ICD in sensory neurons (Fig. 5A–C). We injected DNA encoding the ICD linked to GFP-CAAX by the self-cleaving 2A peptide, which allows us to label the expressing neurons without directly tagging the ICD protein. We found that 43% of cells expressing the ICD (n = 14 cells in 12 embryos) failed to extend a peripheral axon by 24 hpf (Fig. 5C), compared with 13% of control neurons labeled with only GFP-CAAX (n = 15 cells in 11 embryos). Furthermore, axons that did form showed less branching. On average, control neurons had 11.8 ± 1.3 peripheral axon tips per neuron (n = 10), whereas Clstn-1 ICD-expressing neurons had 6.2 ± 1.4 peripheral axon tips per neuron (n = 11; p = 0.01, unpaired t test). Individual Clstn-1 ICD neurons had fewer branches crossing the horizontal myoseptum (0.79 ± 0.2, n = 14 cells in 11 embryos), compared with control cells (1.8 ± 0.3, n = 14 cells in 10 embryos) (p = 0.015, unpaired t test). These effects are similar to those seen with Clstn-1 knockdown, supporting the idea that the Clstn-1 ICD acts as a dominant-negative and prevents interaction of endogenous Clstn-1 with kinesin-1.
Figure 5.
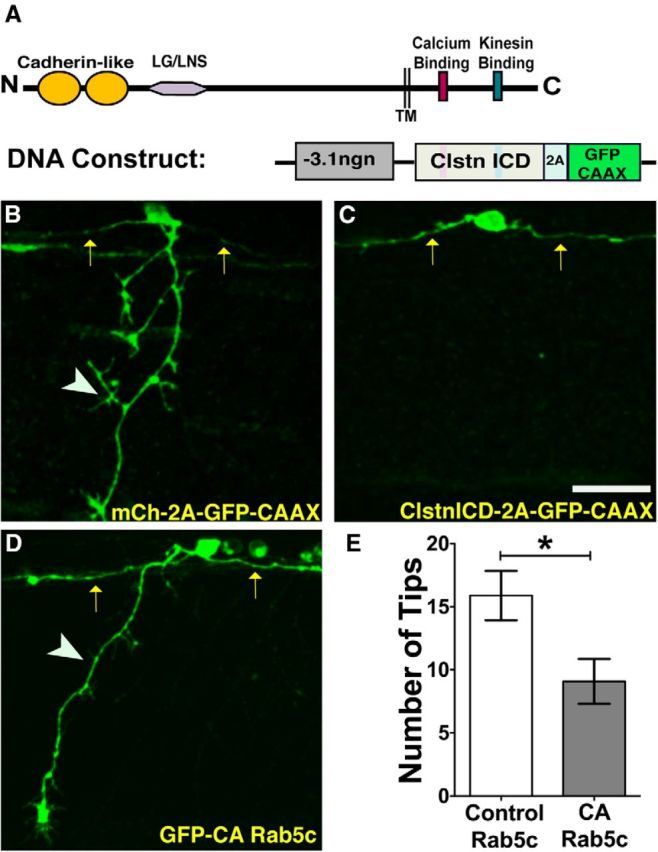
Clstn-1 intracellular domain or CA-Rab5c expression reduces peripheral RB branching. A, Schematic showing structure of Clstn-1 and a DNA construct used to express Clstn-1 ICD in RB neurons. TM, Transmembrane domain; 2A, self-cleaving peptide. B, C, Individually labeled RB neurons expressing either GFP-CAAX (B) or Clstn-1 ICD (C) showing failure of peripheral axon formation in Clstn-1 ICD-expressing cell. Arrows indicate central axons; arrowheads indicate peripheral axons. D, RB neuron expressing constitutively active Rab5c (CA-Rab5c) shows reduced branching. E, Quantification of peripheral axon tips per neuron. *p = 0.02 (unpaired, two-tailed t test). Scale bar, 40 μm.
Disruption of endosomal trafficking causes peripheral axon defects
We focused on endosomes as potential cargos because Clstn-1 is associated with endosomes in cultured neurons (Steuble et al., 2010). To test whether endosomal trafficking is necessary for sensory axon development, we first disrupted trafficking with a constitutively active (CA) form of Rab5c (Rab5c Q81L) (Clark et al., 2011). CA-Rab5 has been shown in other systems to cause enlargement of early endosomes and prevent conversion to late endosomes, which is critical for integrity of the endosomal network and for recycling of Rab5 to nascent endosomes (Rink et al., 2005). We expressed CA-Rab5c in RB neurons and quantified effects on peripheral axon formation and branching. We found that CA-Rab5c does not affect peripheral axon outgrowth; however, it did cause a reduction in peripheral branching compared with neurons expressing unmutated Rab5c (Fig. 5D,E). These data indicate that a properly functioning endosomal trafficking system is critical for peripheral axon branching.
Clstn-1 regulates endosomal transport from the cell body to axons and branch points
In cultured cells, Clstn-1 has been shown to recruit kinesin to post-TGN carriers and regulate cargo transport from the TGN into endosomal carriers (Ludwig et al., 2009). However, whether Clstn-1 controls endosomal trafficking in neuronal processes is unknown. Because Clstn-1 colocalizes with a subpopulation of Rab5-labeled endosomes in hippocampal neurons (Steuble et al., 2010), we tracked movements of Rab5-containing vesicles during the process of sensory axon formation and branching. We used a previously described zebrafish Rab5c construct (Clark et al., 2011) to label these endosomes in vivo. To determine whether Clstn-1 regulates vesicular transport from the cell body to specific axonal locations, we labeled Rab5c with photoactivatable GFP (PA-GFP). We coinjected −3.1ngn1:PA-GFP-Rab5c with −3.1ngn1:TagRFP-CAAX DNA to label individual RB neurons. We photoactivated the GFP in the cell body at stages when central axons have formed and are extending rapidly, and peripheral axons are extending out of the spinal cord. We then imaged the progressive accumulation of labeled vesicles moving into central and peripheral axons (Fig. 6). To quantify vesicle transport into axons, we measured the furthest distance the PA-GFP signal moved into axons over time after photoactivation. In wild-type embryos, PA-GFP-Rab5c vesicles originating in the cell body move rapidly from the cell body into both central and peripheral axons (Fig. 6A; Movie 3), reaching an average distance of 19.1 ± 4.6 μm into central axons and 19.0 ± 6.3 μm into peripheral axons within 2 min of photoactivation (Fig. 6A,C,D). Over the course of ∼30 min, vesicles progressively moved further along the length of the central axons and the newly forming and branching peripheral axon. In contrast, Clstn-1 knockdown embryos showed a significant reduction in PA-GFP-Rab5c vesicle movement into both peripheral and central axons, with PA-GFP signal moving on average 15.6 ± 1.8 μm into central axons and 10.6 ± 2.1 μm into peripheral axons within 2 min after photoactivation (Fig. 6B–D; Movie 4). Although peripheral axon length was variable at these stages, we analyzed cells with peripheral axon lengths in equivalent ranges (14–42 μm in control and 10–43 μm in Clstn-1 knockdown), and the mean axon length was not significantly different between groups.
Figure 6.
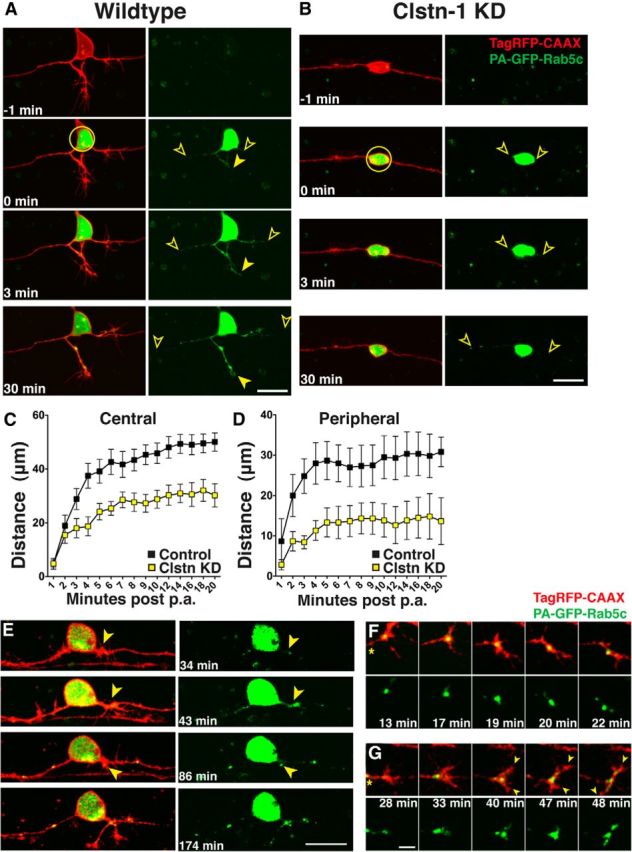
Rab5 endosomes move into developing axons and Clstn-1 knockdown inhibits transport of early endosomes from the cell body into axons. A, B, Time-lapse images of PA-GFP-Rab5c-labeled endosomes in TagRFP-CAAX-labeled RB cells in control (A) and Clstn KD (B) RB neurons. Photoactivation was in the cell body (yellow circle) and spread of photoactivated Rab5c vesicles into axons was measured as the farthest vesicle from the cell body (open arrowheads). C, Quantification of Rab5 vesicle movement into central axons. Vesicles advance more slowly in Clstn-1 knockdown compared with wild-type. p = 0.0003 (two-way ANOVA). D, Rab5 vesicles move more slowly into peripheral axons in Clstn-1 knockdown embryos compared with wild-type. p = 0.03 (two-way ANOVA). E, Time-lapse images of peripheral axon initiation in neuron in which PA-GFP-Rab5 was photoactivated in the cell body. GFP-Rab5 accumulation is present at peripheral axon initiation site (arrowhead) and fills forming growth cone. F, Time-lapse images of peripheral growth cone turning. Growth cone is initially growing up, but little Rab5 enters the upper protrusion, which retracts. Growth cone redirects to area of large Rab5 accumulation. The accumulation moves into lower branch, which extends. *Axon shaft. G, Time-lapse images of bifurcating peripheral growth cone. Rab5 moves into both branches (arrowheads) of bifurcating growth cone. Scale bars: A, B, E, 20 μm; F, G, 5 μm. Time is minutes after photoactivation.
PA-GFP-Rab5c in a control RB neuron. Time-lapse movie of a −3.1ngn1:Tag-RFP-CAAX and −3.1ngn1:PA-GFP-Rab5c-expressing neuron. PA-GFP activated in the cell body. Images were acquired every 34 s and are played back at 10 frames per second, for a total movie duration of 34 min. Movie is 340× faster than real time.
PA-GFP-Rab5c in a Clstn-1 knockdown neuron. Time-lapse movie of a −3.1ngn1:Tag-RFP-CAAX and −3.1ngn1:PA-GFP-Rab5c-expressing neuron in a Clstn-1 MO-injected embryo. PA-GFP activated in the cell body. Images were acquired every 62.7 s and are played back at 5 frames per second, for a total movie duration of 33 min, 25 s. Anterior to the right. Movie is 314× faster than real time.
To explore the relationship between vesicle accumulation and formation of axons and branches, we photoactivated PA-GFP-Rab5c in the cell body of wild-type neurons at early developmental stages and imaged cells over extended time periods. In cells where we captured peripheral axon formation from the earliest stages (n = 4), Rab5c vesicles accumulated at the peripheral axon initiation site at the time of initiation and moved into the forming peripheral axon as it emerged (Fig. 6E). Similarly, during peripheral growth cone turning or bifurcation, Rab5c vesicles accumulated in the regions of the growth cone that were maintained and subsequently extended (Fig. 6F,G) in all cases where we captured such events (n = 30 events in 8 cells in 8 embryos). These observations suggest that delivery of Rab5c-containing endosomes is important for axon or branch formation.
To further investigate vesicle movement to branch points and the role of Clstn-1 in this process, we analyzed vesicle accumulations in neurons photoactivated during stages when peripheral axons are actively growing and branching. In wild-type embryos, PA-GFP-Rab5c vesicles appeared to accumulate in puncta along the axons and showed particularly robust accumulations at the peripheral axon initiation point, at secondary branch points, and in growth cones within minutes after photoactivation (Fig. 7A). In Clstn-1 knockdown neurons that did form peripheral axons, vesicles did not show robust accumulations at branch points (Fig. 7B), nor did vesicles accumulate on central axons in neurons that failed to form peripheral axons (Fig. 6B). To quantify vesicle accumulations at branch points, we compared the fluorescence intensity of individual accumulations at 30 min after activation to the original intensity at 3 min after activation (Fig. 7C). In control neurons, fluorescence intensity of vesicle accumulations increased during this period, suggesting a continued supply of vesicles from the cell body. In contrast, intensity decreased in Clstn-1 knockdown embryos, suggesting either that fewer vesicles are delivered to axon branch points or that they are not retained at branch points. To test retention, we photoactivated the GFP at branch points and measured the percentage of PA-GFP signal remaining at the branch point after 20 min (Fig. 7D,E). We found that vesicles dispersed from the branch point to an equal degree in wild-type versus Clstn-1 knockdown neurons (Fig. 7F). These results suggest that Clstn-1 influences Rab5c vesicle delivery from the cell body to branch points but not retention of vesicles at branch points. Overall, our results show that Rab5c-labeled endosomal carriers from the cell body move rapidly into developing axons and may deliver cargo important for branch formation to specific axonal locations. Moreover, Clstn-1 is required for vesicle transport into axons and for robust accumulation of endosomes at branch points.
Figure 7.
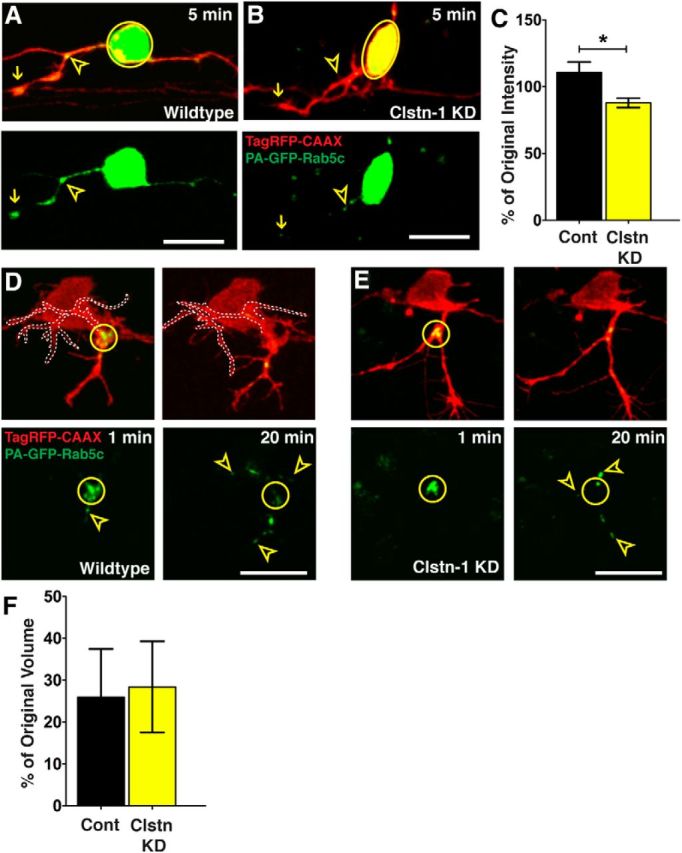
Clstn-1 knockdown reduces Rab5 vesicle delivery to branch points. A, Endosomes accumulate at branch points (open yellow arrowheads) and in growth cones (arrows) of peripheral axons in control embryos. B, Clstn-1 knockdown embryos show reduced accumulations at branch points. C, Quantification of GFP accumulation intensity change at branch points (intensity at 30 min after activation divided by intensity at 3 min after activation). *p = 0.03 (unpaired, two-tailed t test). D, E, Photoactivation in branch point (yellow circle) of control neuron (D) or Clstn-1 KD neuron (E). Signal is largely dispersed from the branch points by 20 min after PA. Arrowheads indicate vesicles that moved from branch point into axon branches. Left branch in D is outlined for clarity. F, Quantification of change in vesicle accumulation at branch points (volume at 20 min divided by volume at 2 min) shows no difference between control and Clstn-1 KD (p = 0.885). Scale bars, 20 μm. Time is minutes after photoactivation.
Live imaging of endosome dynamics shows differential trafficking in RB axons
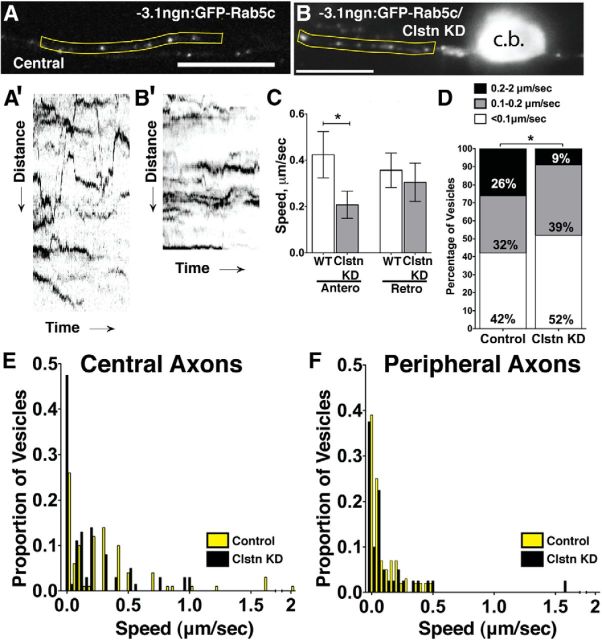
To further investigate dynamics of individual endosomes within specific axon compartments and characterize roles for Clstn-1 in endosome dynamics, we performed high temporal resolution imaging of endosome movements at 24 hpf, a stage when central and peripheral axons are established. In general, very little is known about endosomal dynamics in neurons, especially in vivo, largely because of the challenges involved in imaging very rapid vesicular movements in 3D axon arbors in vivo. We used swept field confocal microscopy (Castellano-Muñoz et al., 2012), which provides sufficient acquisition rates, signal-to-noise, and spatial resolution to image endosomal movements in vivo. We used GFP-tagged zebrafish Rab5c, Rab7, and Rab11a constructs to label subpopulations of early, late, and recycling endosomes, respectively (Clark et al., 2011). We first characterized endosome dynamics in wild-type RB neurons and compared their behavior in central and peripheral axons (Fig. 8). We found that all three endosome types can move both directions in both axon types, although they vary considerably in directional preference and speed. Rab5c- and Rab11a-labeled vesicles show an equal preference for anterograde and retrograde directions (Fig. 8A′,D; Movie 5). In contrast, GFP-Rab7-labeled endosomes move preferentially in the retrograde direction (Fig. 8C′,D; Movie 6), consistent with described roles in retrograde transport in vitro (Deinhardt et al., 2006; Harrington and Ginty, 2013). We quantified speed at which endosomes move along axons by generating kymographs and calculating the speed of the fastest run during a 400 s imaging period. We found that endosomal populations behave differently from one another and showed differences in central and peripheral RB neurons (Fig. 8E). GFP-Rab5c-labeled early endosomes move the fastest and on average are significantly faster in central axons than peripheral axons. GFP-Rab11a-labeled recycling endosomes are the slowest but also show differential rates between central and peripheral axons. GFP-Rab7-labeled late endosomes do not exhibit differential transport speeds between the central and peripheral axons. Together, these data show that different endosome populations have distinct transport profiles in sensory neurons, and suggest that differential protein trafficking may underlie the different behaviors of peripheral versus central axons.
Figure 8.
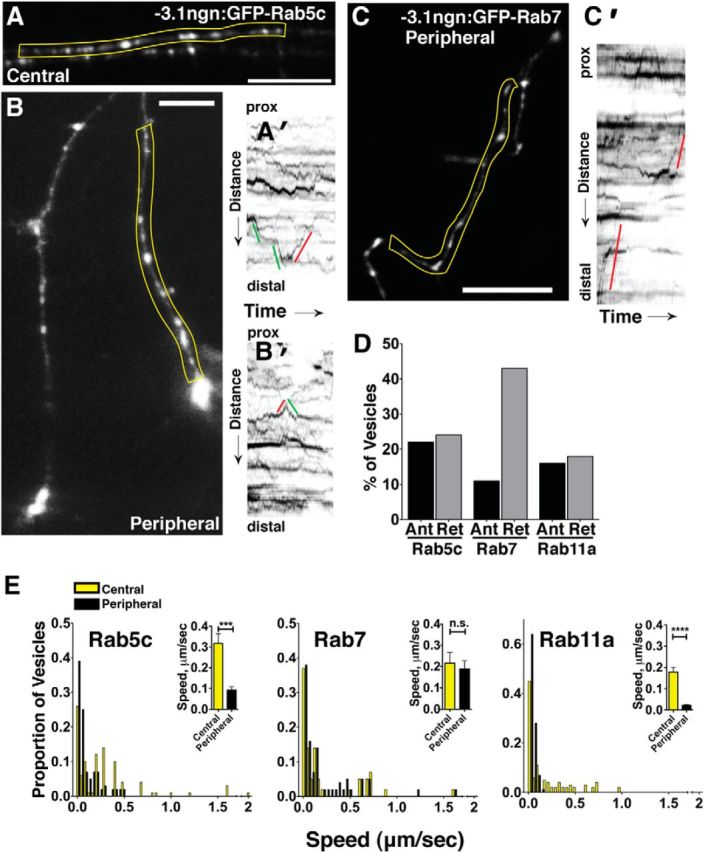
Characterization of endosome dynamics in RB axons. A, B, GFP-Rab5c-labeled endosomes in central (A) and peripheral (B) axons. A′, Kymograph of yellow region in A with lines indicating anterograde (green) or retrograde (red) movement, with reference to the cell body. prox, Proximal. B′, Kymograph of peripheral Rab5c endosomes in yellow region from B. C, Peripheral RB with GFP-Rab7-labeled late endosomes. C′, Kymograph of yellow region in C showing retrograde (red) movements. D, Directionality of GFP-Rab5c-, GFP-Rab7-, and GFP-Rab11-labeled endosomes in RB cells. Vesicles with no net movement during the imaging period are not shown in the graph. E, Speed distributions of GFP-Rab5c-, GFP-Rab7-, and GFP-Rab11-labeled endosomes in central (yellow) and peripheral (black) axons. Insets, Mean speeds in central and peripheral axons. ***p = 0.001 (unpaired, two-tailed t test). ****p < 0.0001 (unpaired, two-tailed t test). n.s., Not significant. Scale bar, 20 μm.
Rab5c-labeled early endosomes in central RB axons. Time-lapse movie of −3.1ngn1:gfp-Rab5c-expressing neurons. Images were acquired every 2 s and are played back at 10 frames per second, for a total movie duration of 398 s. Movie is 200× faster than real time. Scale bar, 10 μm.
Rab7-labeled late endosomes in a peripheral RB axon. Time-lapse movie of a −3.1ngn1:gfp-Rab7-expressing neuron. Images were acquired every 2 s and are played back at 10 frames per second, for a total movie duration of 148 s. Movie is 200× faster than real time. Time displayed as hours:minutes:seconds. Scale bar, 10 μm.
Clstn-1 is required for rapid endosomal movement in vivo
Our PA-GFP-Rab5 analysis showed that Clstn-1 is required for vesicle transport from the cell body to axons. To ask whether Clstn-1 influences vesicle transport along axons, we directly measured endosome speeds and directionality in Clstn-1 knockdown embryos. We found that movement of Rab5c-containing vesicles was significantly affected by Clstn-1 knockdown (Fig. 9). Clstn-1 knockdown embryos had fewer vesicles moving at the fastest rates in central axons (Fig. 9D), and this effect was selective for vesicles moving anterogradely (Fig. 9C), suggesting an effect on kinesin-based transport. In contrast, we did not detect differences in the fast vesicle speed category in peripheral axons. However, our PA-GFP-Rab5c analysis above (Fig. 6), performed over longer time periods, did reveal significant effects on the ability of Rab5-containing vesicles to move into peripheral axons. We also did not see a significant difference in speeds of Rab7- or Rab11-labeled endosomes after Clstn-1 knockdown (data not shown). Together, these data suggest that Clstn-1 affects a distinct subpopulation of early endosomes and is required for rapid movement of Rab5 endosomes along axons.
Figure 9.
Clstn-1 knockdown reduces early endosomal speeds. A, B, Single frames from time-lapse movies of GFP-labeled Rab5c in central RB axons of control (A) and Clstn-1 KD (B) embryos at 24 hpf. c.b., Cell body. A′, B′, Kymographs from yellow regions of A and B of vesicle movement in control (A′) and Clstn-1 knockdown (B′) central RB axons, showing reduced movement in Clstn-1 knockdown embryos. C, Speeds of vesicles moving in the anterograde directions were significantly reduced. *p = 0.05 (unpaired, two-tailed t test). D, Percentage of vesicles moving at specific speeds shows fewer early endosomes traveling at fast speeds in central axons of Clstn-1 knockdown embryos. *p = 0.03 (χ2 test). E, F, Speed distributions of endosomes in central (E) and peripheral (F) axons of control (yellow) and Clstn-1 knockdown (black) embryos.
Discussion
In this study, we uncover a novel molecular player important for axon branching and compartmentalization. We demonstrate that Clstn-1 is critical for growth and branching of peripheral sensory axons and differentially affects the behavior of separate axons from one neuron. Furthermore, our data indicate that Clstn-1 acts in part through a trafficking role and controls the transport of endosomal carriers. Our ability to image live endosome dynamics as vertebrate neurons develop complex axon arborizations in vivo has revealed new insight into Clstn-1 function and into differential endosome dynamics in specific axon compartments. Our results suggest that regulated trafficking of early endosomes from the cell body to specific axon locations is crucial for neuronal compartmentalization and axon branching.
Precise control over axon branching is essential for neuronal circuit formation. Many factors have been shown to influence axon branching, including extracellular cues, intracellular signaling molecules, and cytoskeletal regulators (Schmidt and Rathjen, 2010; Gallo, 2011; Gibson and Ma, 2011; Lewis et al., 2013). Nonetheless, the mechanisms by which these and other factors drive axon branch formation at correct locations remain poorly understood. Accurate localization of branching signals likely requires tight control of protein trafficking within the neuron, but how traffic regulators contribute to branching patterns is not known. Our results uncover novel roles for a kinesin adaptor, Clstn-1, in control of axon branching. Adaptor molecules that mediate cargo binding to kinesin motors are numerous and diverse (Hirokawa et al., 2010; Namba et al., 2011), and their functions are just beginning to be understood. Another kinesin adaptor, c-Jun N-terminal kinase (JNK)-interacting protein 3 (JIP3), was found to restrict branching of cerebellar neurons via GSK3β signaling (Bilimoria et al., 2010) but whether its kinesin adaptor function is involved is not known. Our results show that Clstn-1 controls branching in part through its function as a director of membrane trafficking. Interestingly, we found that Clstn-1 is important for both interstitial branching and growth cone bifurcation. Little is known about mechanisms of growth cone bifurcation, although it may involve mechanisms different from interstitial branching in some cases (Schmidt and Rathjen, 2010; Gibson and Ma, 2011). Our results show that these two types of branching share some molecular mechanisms in sensory neurons, in that both require Clstn-1. Moreover, our finding that growth cone size is reduced by Clstn-1 knockdown suggests that Clstn-1 also influences processes controlling growth cone motility. Multiple studies have demonstrated shared mechanisms between growth cone guidance and axon branching (Dent et al., 2003; Gibson and Ma, 2011). Clstn-1 may be responsible for delivering signals that mediate both these processes, such as regulators of the cytoskeleton or regulators of membrane turnover processes, both of which are key components of axon growth and guidance (Dent and Gertler, 2003; Hall and Lalli, 2010; Tojima et al., 2011).
Our results also reveal new insight into the roles for endosomal transport in regulation of axon branching. Disruption of trafficking with CA-Rab5 inhibited peripheral axon branching, suggesting that an intact endosomal trafficking system is necessary for branch formation. In addition, our imaging of endosome dynamics shows that at least some of the important cargo for Clstn-1-mediated branching is carried by Rab5-containing endosomes. PA-GFP-Rab5 imaging revealed that Rab5-containing vesicles move rapidly from the cell body to both the central and peripheral axons during early stages of axon outgrowth in wild-type embryos. Moreover, PA-GFP-Rab5 vesicles accumulate at branch points, and these accumulations persist and even increase over extended imaging periods, suggesting a continued supply of activated GFP-Rab5 vesicles from the cell body. Our finding that Clstn-1 is required for Rab5 vesicle transport and for accumulation of vesicles at branch points supports the idea that Clstn-1 functions to direct endosome traffic from the cell body to axons. Together, our data suggest that a subpopulation of Rab5-containing endosomes deliver cargo important for the branching process. These results complement previous studies showing emerging roles for endosomal signaling in proper neuronal branching and circuit formation. For example, dynein-mediated transport of Rab5-containing endosomes is required for dendrite branching in Drosophila sensory neurons (Satoh et al., 2008). Proper acidification of endosomal vesicles is critical for TrkB endosomal signaling and therefore branching of hippocampal axons and dendrites (Ouyang et al., 2013). Rab11-dependent trafficking also regulates BDNF-induced TrkB trafficking and hippocampal dendrite branching (Lazo et al., 2013). Our study extends these findings by showing that rapid movement of Rab5 endosomes into developing axons is critical for axon branching in vivo, and identifies a specific kinesin adaptor that sorts Rab5 vesicles into axons. Recent studies showing that mitochondria recruitment to axon branch sites promotes branch formation (Courchet et al., 2013; Spillane et al., 2013; Tao et al., 2014) and that synaptotagmin and vesicle cycling are important mediators of axon branching (Granseth et al., 2013; Greif et al., 2013) also support the idea that tight regulation of axonal transport and membrane trafficking are critical components of axon branching.
Our finding that Clstn-1 affects anterograde and not retrograde transport suggests Clstn-1 associates with a select subpopulation of early endosomes. This result is consistent with the known kinesin-binding activity of Clstn-1 (Konecna et al., 2006; Araki et al., 2007) and with a previous study showing that Clstn-1 moves anterogradely in axons of cultured neurons (Konecna et al., 2006). Moreover, we found that Clstn-1 knockdown inhibits anterograde movement of some, but not all, Rab5-labeled vesicles. Although there are no unique markers to label distinct subpopulations of Rab5 early endosomes, prior studies showed that Clstn-1 selectively colocalizes with amyloid precursor protein in a subset of early endosomes in hippocampal neurons (Steuble et al., 2010), supporting the idea that it mediates transport of a select subpopulation of Rab5-containing endosomes. Interestingly, another study showed that the kinesin adaptor JIP3 can also bind dynein and mediate retrograde transport of JNK and lysosomes in zebrafish lateral line axons (Drerup and Nechiporuk, 2013). Our work supports a model in which Clstn-1 functions predominantly as a kinesin adaptor to deliver early endosomes containing branching factors to specific axonal locations.
Our study also sheds new light on the poorly understood process of how two axons from one neuron achieve molecular compartmentalization. Axon branches from one neuron often must navigate separate pathways to distinct targets. This process likely requires that axons have molecular differences to allow distinct responses to guidance cues. We previously showed that central and peripheral RB axons have differential responses to, and requirements for, guidance cues (Liu and Halloran, 2005), suggesting differential localization of receptors or signaling components. In C. elegans sensory neurons, targeting of netrin receptors to one axon branch confers netrin responsiveness to that branch (Hao et al., 2010). Even within one axon, guidance receptors can be localized to specific axon membrane segments, and such intra-axonal patterning depends at least partly on intrinsic sorting mechanisms (Katsuki et al., 2009). However, the molecular mechanisms controlling axon patterning differences are largely unknown. Our results show that Clstn-1-mediated endosomal trafficking is one mechanism important for conferring compartmentalization of two axons from one neuron. Interestingly, we found that specific endosome populations (Rab5 and Rab11 vesicles) show different dynamics in central versus peripheral axons, suggesting that these endosomes contain cargos that help define the distinct molecular character and behavior of the two axon types.
An important question raised by our results is why Clstn-1 regulates endosome movement in peripheral and central axons when it appears to function specifically for the formation and branching of peripheral axons. One possibility is that it regulates central axon transport of different cargo proteins, which affect aspects of central axon function not apparent with our analyses. Alternatively, more complex trafficking mechanisms might be in play, in which cargos are trafficked through the central axon compartment before being delivered to the peripheral axon. Such traffic routes could be similar to transcytosis mechanisms shown in cultured neurons, in which receptor or adhesion molecules destined for the axon are first trafficked to the somatodendritic membrane and then are selectively endocytosed and trafficked to axons (Wisco et al., 2003; Ascaño et al., 2009; Winckler and Mellman, 2010).
Our results show that, in the context of axon branching, Clstn-1 acts in a large part through its kinesin adaptor and trafficking functions. However, it is possible that other domains or functions of the Clstn-1 protein, such as the extracellular cadherin or LG domains, or the intracellular Ca2+ binding domain, contribute to its roles in neuronal development. Interestingly, the C. elegans calsyntenin ortholog is required for learning and its mutant phenotype can be rescued by expression of the cleaved extracellular domain in neurons, suggesting that the extracellular domain is important for learning (Ikeda et al., 2008). Also, mammalian Clstn-3 regulates synapse formation through extracellular domain interaction with neurexin (Pettem et al., 2013; Um et al., 2014). Our Clstn-1 mutant embryos show gross morphological defects in head morphology at later developmental stages, indicating that there may be multiple important functions for Clstn-1 in development. Further investigation of the mutant phenotype will be important to reveal all the functions of Clstn-1.
Footnotes
This work was supported by National Institutes of Health Grants R01NS042228 and R56NS086934 to M.C.H., Grant F31NS074606 to O.Y.P., and Grant R44MH065724 to K.W.E. We thank Brian Link for Rab constructs, David Grunwald and Tim Dahlem for help with generating TALENs, Julie Last for assistance with swept field confocal microscopy, Erik Dent for comments on the manuscript, Namrata Asuri for experiments that inspired this work, and Bill Davis and Andrew Goelz for fish care and technical support.
The authors declare no competing financial interests.
References
- Andersen E, Asuri N, Clay M, Halloran M. Live imaging of cell motility and actin cytoskeleton of individual neurons and neural crest cells in zebrafish embryos. J Vis Exp. 2010;36:1726. doi: 10.3791/1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EF, Asuri NS, Halloran MC. In vivo imaging of cell behaviors and F-actin reveals LIM-HD transcription factor regulation of peripheral versus central sensory axon development. Neural Dev. 2011;6:27. doi: 10.1186/1749-8104-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Tomita S, Yamaguchi H, Miyagi N, Sumioka A, Kirino Y, Suzuki T. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta-protein precursor metabolism. J Biol Chem. 2003;278:49448–49458. doi: 10.1074/jbc.M306024200. [DOI] [PubMed] [Google Scholar]
- Araki Y, Kawano T, Taru H, Saito Y, Wada S, Miyamoto K, Kobayashi H, Ishikawa HO, Ohsugi Y, Yamamoto T, Matsuno K, Kinjo M, Suzuki T. The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J. 2007;26:1475–1486. doi: 10.1038/sj.emboj.7601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascaño M, Richmond A, Borden P, Kuruvilla R. Axonal targeting of Trk receptors via transcytosis regulates sensitivity to neurotrophin responses. J Neurosci. 2009;29:11674–11685. doi: 10.1523/JNEUROSCI.1542-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Ostendorff HP, Bossenz M, Schlüter A, Becker CG, Peirano RI, Bach I. Multiple functions of LIM domain-binding CLIM/NLI/Ldb cofactors during zebrafish development. Mech Dev. 2002;117:75–85. doi: 10.1016/S0925-4773(02)00178-8. [DOI] [PubMed] [Google Scholar]
- Bilimoria PM, de la Torre-Ubieta L, Ikeuchi Y, Becker EB, Reiner O, Bonni A. A JIP3-regulated GSK3beta/DCX signaling pathway restricts axon branching. J Neurosci. 2010;30:16766–16776. doi: 10.1523/JNEUROSCI.1362-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blader P, Plessy C, Strähle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech Dev. 2003;120:211–218. doi: 10.1016/S0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- Castellano-Muñoz M, Peng AW, Salles FT, Ricci AJ. Swept field laser confocal microscopy for enhanced spatial and temporal resolution in live-cell imaging. Microsc Microanal. 2012;18:753–760. doi: 10.1017/S1431927612000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Winter M, Cohen AR, Link BA. Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev Dyn. 2011;240:2452–2465. doi: 10.1002/dvdy.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchet J, Lewis TL, Jr, Lee S, Courchet V, Liou DY, Aizawa S, Polleux F. Terminal axon branching is regulated by the LKB1-NUAK1 kinase pathway via presynaptic mitochondrial capture. Cell. 2013;153:1510–1525. doi: 10.1016/j.cell.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, DeLeon DV, Angerer LM, Angerer RC. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinhardt K, Salinas S, Verastegui C, Watson R, Worth D, Hanrahan S, Bucci C, Schiavo G. Rab5 and Rab7 control endocytic sorting along the axonal retrograde transport pathway. Neuron. 2006;52:293–305. doi: 10.1016/j.neuron.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/S0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- Dent EW, Tang F, Kalil K. Axon guidance by growth cones and branches: common cytoskeletal and signaling mechanisms. Neuroscientist. 2003;9:343–353. doi: 10.1177/1073858403252683. [DOI] [PubMed] [Google Scholar]
- Drerup CM, Nechiporuk AV. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 2013;9:e1003303. doi: 10.1371/journal.pgen.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G. The cytoskeletal and signaling mechanisms of axon collateral branching. Dev Neurobiol. 2011;71:201–220. doi: 10.1002/dneu.20852. [DOI] [PubMed] [Google Scholar]
- Gibson DA, Ma L. Developmental regulation of axon branching in the vertebrate nervous system. Development. 2011;138:183–195. doi: 10.1242/dev.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granseth B, Fukushima Y, Sugo N, Lagnado L, Yamamoto N. Regulation of thalamocortical axon branching by BDNF and synaptic vesicle cycling. Front Neural Circuits. 2013;7:202. doi: 10.3389/fncir.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greif KF, Asabere N, Lutz GJ, Gallo G. Synaptotagmin-1 promotes the formation of axonal filopodia and branches along the developing axons of forebrain neurons. Dev Neurobiol. 2013;73:27–44. doi: 10.1002/dneu.22033. [DOI] [PubMed] [Google Scholar]
- Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloran MC, Severance SM, Yee CS, Gemza DL, Raper JA, Kuwada JY. Analysis of a zebrafish semaphorin reveals potential functions in vivo. Dev Dyn. 1999;214:13–25. doi: 10.1002/(SICI)1097-0177(199901)214:1<13::AID-DVDY2>3.0.CO%3B2-3. [DOI] [PubMed] [Google Scholar]
- Hao JC, Adler CE, Mebane L, Gertler FB, Bargmann CI, Tessier-Lavigne M. The tripartite motif protein MADD-2 functions with the receptor UNC-40 (DCC) in Netrin-mediated axon attraction and branching. Dev Cell. 2010;18:950–960. doi: 10.1016/j.devcel.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington AW, Ginty DD. Long-distance retrograde neurotrophic factor signalling in neurons. Nat Rev Neurosci. 2013;14:177–187. doi: 10.1038/nrn3253. [DOI] [PubMed] [Google Scholar]
- Hintsch G, Zurlinden A, Meskenaite V, Steuble M, Fink-Widmer K, Kinter J, Sonderegger P. The calsyntenins: a family of postsynaptic membrane proteins with distinct neuronal expression patterns. Mol Cell Neurosci. 2002;21:393–409. doi: 10.1006/mcne.2002.1181. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hoerndli FJ, Walser M, Fröhli Hoier E, de Quervain D, Papassotiropoulos A, Hajnal A. A conserved function of C. elegans CASY-1 calsyntenin in associative learning. PLoS One. 2009;4:e4880. doi: 10.1371/journal.pone.0004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda DD, Duan Y, Matsuki M, Kunitomo H, Hutter H, Hedgecock EM, Iino Y. CASY-1, an ortholog of calsyntenins/alcadeins, is essential for learning in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:5260–5265. doi: 10.1073/pnas.0711894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuki T, Ailani D, Hiramoto M, Hiromi Y. Intra-axonal patterning: intrinsic compartmentalization of the axonal membrane in Drosophila neurons. Neuron. 2009;64:188–199. doi: 10.1016/j.neuron.2009.08.019. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Konecna A, Frischknecht R, Kinter J, Ludwig A, Steuble M, Meskenaite V, Indermühle M, Engel M, Cen C, Mateos JM, Streit P, Sonderegger P. Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol Biol Cell. 2006;17:3651–3663. doi: 10.1091/mbc.E06-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lazo OM, Gonzalez A, Ascaño M, Kuruvilla R, Couve A, Bronfman FC. BDNF regulates Rab11-mediated recycling endosome dynamics to induce dendritic branching. J Neurosci. 2013;33:6112–6122. doi: 10.1523/JNEUROSCI.4630-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TL, Jr, Courchet J, Polleux F. Cell biology in neuroscience: cellular and molecular mechanisms underlying axon formation, growth, and branching. J Cell Biol. 2013;202:837–848. doi: 10.1083/jcb.201305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Halloran MC. Central and peripheral axon branches from one neuron are guided differentially by Semaphorin3D and transient axonal glycoprotein-1. J Neurosci. 2005;25:10556–10563. doi: 10.1523/JNEUROSCI.2710-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Blume J, Diep TM, Yuan J, Mateos JM, Leuthäuser K, Steuble M, Streit P, Sonderegger P. Calsyntenins mediate TGN exit of APP in a kinesin-1-dependent manner. Traffic. 2009;10:572–589. doi: 10.1111/j.1600-0854.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques. 2007;43:610–614. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- Namba T, Nakamuta S, Funahashi Y, Kaibuchi K. The role of selective transport in neuronal polarization. Dev Neurobiol. 2011;71:445–457. doi: 10.1002/dneu.20876. [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–615. doi: 10.1016/S0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- Ouyang Q, Lizarraga SB, Schmidt M, Yang U, Gong J, Ellisor D, Kauer JA, Morrow EM. Christianson syndrome protein NHE6 modulates TrκB endosomal signaling required for neuronal circuit development. Neuron. 2013;80:97–112. doi: 10.1016/j.neuron.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettem KL, Yokomaku D, Luo L, Linhoff MW, Prasad T, Connor SA, Siddiqui TJ, Kawabe H, Chen F, Zhang L, Rudenko G, Wang YT, Brose N, Craig AM. The specific α-neurexin interactor calsyntenin-3 promotes excitatory and inhibitory synapse development. Neuron. 2013;80:113–128. doi: 10.1016/j.neuron.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuschhof C, Heekeren HR, Li SC, Sander T, Lindenberger U, Bäckman L. KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory. Neuropsychologia. 2010;48:402–408. doi: 10.1016/j.neuropsychologia.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Ringman JM, Schulman H, Becker C, Jones T, Bai Y, Immermann F, Cole G, Sokolow S, Gylys K, Geschwind DH, Cummings JL, Wan HI. Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol. 2012;69:96–104. doi: 10.1001/archneurol.2011.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Satoh D, Sato D, Tsuyama T, Saito M, Ohkura H, Rolls MM, Ishikawa F, Uemura T. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rathjen FG. Signalling mechanisms regulating axonal branching in vivo. Bioessays. 2010;32:977–985. doi: 10.1002/bies.201000054. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–436. doi: 10.1016/S0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Spillane M, Ketschek A, Merianda TT, Twiss JL, Gallo G. Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 2013;5:1564–1575. doi: 10.1016/j.celrep.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuble M, Gerrits B, Ludwig A, Mateos JM, Diep TM, Tagaya M, Stephan A, Schätzle P, Kunz B, Streit P, Sonderegger P. Molecular characterization of a trafficking organelle: dissecting the axonal paths of calsyntenin-1 transport vesicles. Proteomics. 2010;10:3775–3788. doi: 10.1002/pmic.201000384. [DOI] [PubMed] [Google Scholar]
- Steuble M, Diep TM, Schätzle P, Ludwig A, Tagaya M, Kunz B, Sonderegger P. Calsyntenin-1 shelters APP from proteolytic processing during anterograde axonal transport. Biol Open. 2012;1:761–774. doi: 10.1242/bio.20121578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Nojima Y, Shoji W, Sato M, Nakayama R, Ohshima T, Okamoto H. Islet1 selectively promotes peripheral axon outgrowth in Rohon-Beard primary sensory neurons. Dev Dyn. 2011;240:9–22. doi: 10.1002/dvdy.22499. [DOI] [PubMed] [Google Scholar]
- Tao K, Matsuki N, Koyama R. AMP-activated protein kinase mediates activity-dependent axon branching by recruiting mitochondria to axon. Dev Neurobiol. 2014;74:557–573. doi: 10.1002/dneu.22149. [DOI] [PubMed] [Google Scholar]
- Tojima T, Hines JH, Henley JR, Kamiguchi H. Second messengers and membrane trafficking direct and organize growth cone steering. Nat Rev Neurosci. 2011;12:191–203. doi: 10.1038/nrn2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Gomi F, Murayama S, Takahashi H. Calsyntenin-3 C-terminal fragment accumulates in dystrophic neurites surrounding abeta plaques in tg2576 mouse and Alzheimer disease brains: its neurotoxic role in mediating dystrophic neurite formation. Am J Pathol. 2013;182:1718–1726. doi: 10.1016/j.ajpath.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Um JW, Pramanik G, Ko JS, Song M-Y, Lee D, Kim H, Park K-S, Sudhof TC, Tabuchi K, Ko J. Calsyntenins function as synaptogenic adhesion molecules in concert with neurexins. Cell Rep. 2014;6:1096–1109. doi: 10.1016/j.celrep.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnoni A, Rodriguez L, Manser C, De Vos KJ, Miller CC. Phosphorylation of kinesin light chain 1 at serine 460 modulates binding and trafficking of calsyntenin-1. J Cell Sci. 2011;124:1032–1042. doi: 10.1242/jcs.075168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnoni A, Perkinton MS, Gray EH, Francis PT, Noble W, Miller CC. Calsyntenin-1 mediates axonal transport of the amyloid precursor protein and regulates Abeta production. Hum Mol Genet. 2012;21:2845–2854. doi: 10.1093/hmg/dds109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt L, Schrimpf SP, Meskenaite V, Frischknecht R, Kinter J, Leone DP, Ziegler U, Sonderegger P. Calsyntenin-1, a proteolytically processed postsynaptic membrane protein with a cytoplasmic calcium-binding domain. Mol Cell Neurosci. 2001;17:151–166. doi: 10.1006/mcne.2000.0937. [DOI] [PubMed] [Google Scholar]
- Winckler B, Mellman I. Trafficking guidance receptors. Cold Spring Harb Perspect Biol. 2010;2:a001826. doi: 10.1101/cshperspect.a001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisco D, Anderson ED, Chang MC, Norden C, Boiko T, Fölsch H, Winckler B. Uncovering multiple axonal targeting pathways in hippocampal neurons. J Cell Biol. 2003;162:1317–1328. doi: 10.1083/jcb.200307069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RB axon development in control embryo. Time-lapse movie of axons in a Tg(−3.1ngn1:GFP-CAAX) embryo. Images were acquired every 34 s and are played back at 20 frames per second, for a total movie duration of 1:54:48 (hours:minutes:seconds). Movie is 680× faster than real time. Scale bar, 30 μm.
RB axon development after Clstn-1 knockdown. Time-lapse movie of −3.1ngn1:GFP-CAAX-expressing neurons in Clstn-1 MO-injected embryos. Images were acquired every 196.5 s and are played back at 4 frames per second, for a total movie duration of 2:30:43 (hours:minutes:seconds). Movie is 786× faster than real time. Scale bar, 30 μm.
PA-GFP-Rab5c in a control RB neuron. Time-lapse movie of a −3.1ngn1:Tag-RFP-CAAX and −3.1ngn1:PA-GFP-Rab5c-expressing neuron. PA-GFP activated in the cell body. Images were acquired every 34 s and are played back at 10 frames per second, for a total movie duration of 34 min. Movie is 340× faster than real time.
PA-GFP-Rab5c in a Clstn-1 knockdown neuron. Time-lapse movie of a −3.1ngn1:Tag-RFP-CAAX and −3.1ngn1:PA-GFP-Rab5c-expressing neuron in a Clstn-1 MO-injected embryo. PA-GFP activated in the cell body. Images were acquired every 62.7 s and are played back at 5 frames per second, for a total movie duration of 33 min, 25 s. Anterior to the right. Movie is 314× faster than real time.
Rab5c-labeled early endosomes in central RB axons. Time-lapse movie of −3.1ngn1:gfp-Rab5c-expressing neurons. Images were acquired every 2 s and are played back at 10 frames per second, for a total movie duration of 398 s. Movie is 200× faster than real time. Scale bar, 10 μm.
Rab7-labeled late endosomes in a peripheral RB axon. Time-lapse movie of a −3.1ngn1:gfp-Rab7-expressing neuron. Images were acquired every 2 s and are played back at 10 frames per second, for a total movie duration of 148 s. Movie is 200× faster than real time. Time displayed as hours:minutes:seconds. Scale bar, 10 μm.