Summary
Multiprotein complexes govern virtually all cellular processes. Their 3D structures provide important clues to their biological roles, especially through structural correlations among protein molecules and complexes. The detection of such correlations generally requires comprehensive searches in databases of known protein structures by means of appropriate structure-matching techniques. Here, we present a high-speed structure search engine capable of instantly matching large protein oligomers against the complete and up-to-date database of biologically functional assemblies of protein molecules. We use this tool to reveal unseen structural correlations on the level of protein quaternary structure and demonstrate its general usefulness for efficiently exploring complex structural relationships among known protein assemblies.
Graphical Abstract
Highlights
-
•
Tool enables efficient exploration of structure similarities among protein assemblies
-
•
Multiprotein complexes show structural correlations concealed on the level of chains
-
•
Hexamer of unknown function from Salmonella matches Hcp3 protein from Pseudomonas
-
•
Strikingly similar DNA clamp complexes assemble from highly different subunits
Wiederstein et al. introduce a fast protein structure search tool that scans the complete database of multiprotein complexes. It reveals structural correlations among multiprotein complexes that are concealed on the level of single chains and supports the characterization of newly determined protein structures.
Introduction
Molecular complexes of interacting protein chains are fundamental for virtually all biological processes. Their role as functional and evolutionary units is evident since the early days of structural biology, when the first 3D structures of proteins uncovered initial examples of how multiple protein chains associate (Perutz et al., 1960; Bolton and Perutz, 1970; Birktoft and Blow, 1972). Advances in structure determination techniques and the pipelines of structural genomics projects have promoted the acquisition of atomic coordinates of macromolecular assemblies, providing the community with a plethora of structures of ever-growing size and complexity (Dutta and Berman, 2005; Berman et al., 2013; Furnham et al., 2013; Wagner and Chiu, 2013). Among the results of these efforts are acclaimed structure determinations of multiprotein complexes like that of RNA polymerase (Cramer et al., 2001; Gnatt et al., 2001) or the ribosome (Carter et al., 2000; Ban et al., 2000; Schluenzen et al., 2000), and it is fair to expect that many more of the ambitious targets of structural biology will be resolved (Bhattacharya, 2009).
Knowledge of the structure of a protein complex to atomic scale is generally of high value for understanding its biological role. However, it is usually of limited use to investigate the coordinates of a structure without interpretation against the background of other known structures. Indeed, comparison and classification of protein structures frequently reveal information on the biological roles, chemical functions, and evolutionary relationships of proteins that is difficult to obtain from experiment. The detection of structure matches on the level of whole-protein complexes is particularly informative, because proteins generally assemble to multichain complexes that act and evolve as functional units. Consequently, tools to efficiently and accurately compare multiprotein complexes against all known structures are essential for the investigation of protein structure, function, and evolution (Sippl and Wiederstein, 2012).
Database searches of proteins are customarily carried out either on the sequence level (Altschul et al., 1997; Remmert et al., 2012; Frank et al., 2010) or on the level of single-chain structures (Hasegawa and Holm, 2009). Both strategies are only partly capable of detecting structural correlations among multichain complexes. Sequence search methods, although fast, struggle with the fact that highly similar structures may have virtually no detectable sequence similarity (Flaherty et al., 1991), and with the indeterminacy of chain order in oligomers. Most importantly, the relative spatial orientation of the chains cannot be captured by sequence search methods, a problem that is obviously shared with single-chain structure comparison methods. Recently, we reported on a tool for the efficient pairwise comparison of large macromolecular complexes (Sippl and Wiederstein, 2012). Here we extend this tool and present a structure search engine that efficiently sorts all known biological assemblies of protein structures according to their structural similarity to a given query. We exemplify a number of key features of the presented method and illustrate its application in the structure-based characterization of multiprotein complexes. In particular, we search for structures matching a protein of unknown function from the pathogen Salmonella typhimurium and find significant matches on the level of quaternary structure that are concealed on the level of tertiary structure. Furthermore, we apply the presented technique in the comparative analysis of DNA clamps and report hitherto undetected structural correlations.
Results
A Structure Search on the Level of Multiprotein Complexes Reveals Significant Structural Correlations that Are Undetectable on the Level of Single Chains
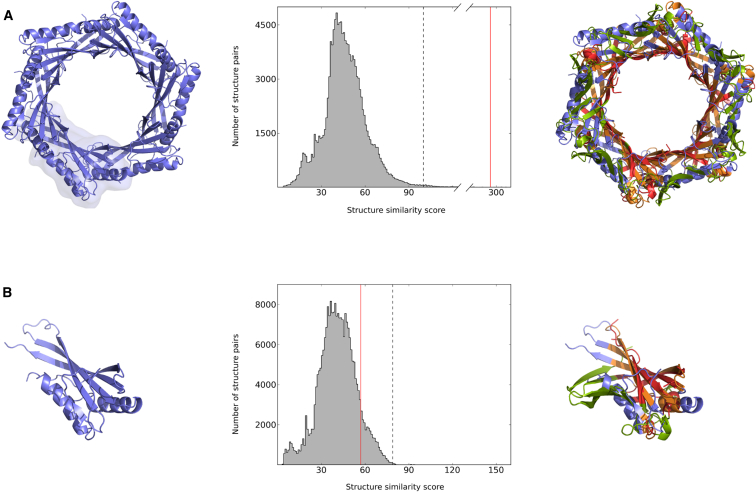
The identification of structure similarities among proteins frequently reveals important relationships on the level of chemical function, biological role, and evolutionary kinship. Structural matches of biological assemblies provide particularly relevant information, because they imply similarities between the biologically active forms of the respective proteins. We first illustrate the use of biological assemblies in the structure-based characterization of newly determined protein structures, using the example of a cytoplasmic protein of unknown function from the pathogenic bacterium S. typhimurium. The structure of this protein has been determined by X-ray crystallography and deposited in the Protein Data Bank (PDB) (ID code: 2GJV) in the course of the Protein Structure Initiative. The authors of this structure, assisted by the PISA software (Krissinel and Henrick, 2007), determined two ring-like biological assemblies for this protein, a homohexameric assembly (2gjv@1, the “@1” postfix denotes the first assembly listed in the PDB file; see Experimental Procedures) and a homododecameric assembly (2gjv@2).
The main result of the search procedure described here is a ranked target list that represents the complete repertoire of known structures ordered in terms of similarity to the query structure. On the level of biological assemblies, a structure search with hexameric 2gjv@1 identifies about 780 targets with a structure similarity score, S, above the threshold of S+ = 100.0 (Experimental Procedures). The top ranked of these targets are either from bacteriophages or bacteria and assemble to hexameric rings. Based on the high similarity of these rings, an intriguing structure/function relationship between proteins from the bacterial type VI secretion system and tail proteins of bacteriophages has been described previously (Kanamaru, 2009; Leiman et al., 2009; Pell et al., 2009; Sippl and Wiederstein, 2012). Notably, the high similarities among the multichain rings do not necessarily coincide with high similarities of the respective constituent subunits. For instance, 2gjv@1 perfectly matches the inner ring of secretory protein Hcp3 from the pathogenic bacterium Pseudomonas aeruginosa (3he1@1; Osipiuk et al., 2011; Figure 1A). Hcp3 is paralogous to Hcp1, which is part of the bacterium’s type VI secretion system. The match yields a structure similarity score S of 292.2, which clearly stands out from the bulk of random correlations (Figure 1A, middle). In contrast, the structure alignment of the individual monomers that build up the respective assemblies produces a score of 56.7, which reports a rather insignificant similarity when related to the entirety of scores (S+ = 79.0). In fact, a structure search with chain A of 2gjv against all known protein chains shows that there are tens of thousands of hits with a similarity S > 56.7 (Figure 1B, middle). Only a few of them form ring-like hexamers, and their detection is only possible with a structure search on the level of biological assemblies.
Figure 1.
A Structure Search on the Level of Multiprotein Complexes Reveals Significant Structural Correlations that Are Undetectable on the Level of Single Chains
(A and B) In order to identify similarities to other protein structures, a cytoplasmic protein of unknown function from S. typhimurium (PDB ID code: 2GJV; to be published) is compared to all proteins in PDB. (A) Left: structure of homohexameric 2gjv@1, with chain A contoured. Middle: distribution of structure similarity scores obtained from a search of 2gjv@1 against all known biological assemblies (138,294 items; November 7, 2013). Extensive structure similarity (S = 292.2, red vertical line) is found between 2gjv@1 and homohexameric secretory protein Hcp3 from P. aeruginosa (3he1@1; Osipiuk et al., 2011). Note that a score of 292.2 is far above the threshold of S+ = 100.0 (dashed line; Experimental Procedures) and close to the end of the distribution’s right tail; only 17 hits have a score S > 292.2. Right: superposition of 2gjv@1 (blue) and 3he1@1 (green), with matching parts in orange (2gjv@1) and red (3he1@1). (B) Left: structure of chain A of 2gjv. Middle: distribution of structure similarity scores obtained from a search of chain A of 2gjv against all known protein chains (242,925 items; November 7, 2013). As indicated by the red vertical line, the structure similarity S = 56.7 to monomeric Hcp3 (chain A of 3he1) is hardly distinguishable from random matches and quite below the threshold of S+ = 79.0 (dashed line). More than 25,000 other protein chains yield a score S > 56.7 when compared to chain A of 2gjv. Right: superposition of chains A of 2gjv and 3he1, respectively. Colors as in (A).
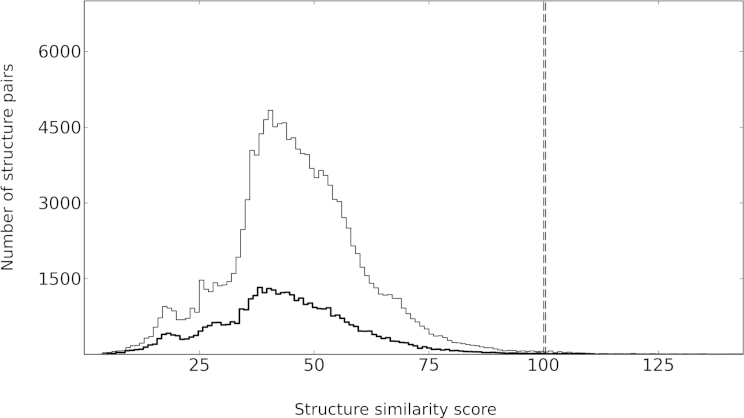
In the structure searches of the previous example, the search database comprised the complete set of known protein structures. As delineated in the Experimental Procedures, search time can be saved by using a nonredundant set of representative structures instead. However, we have to ensure that this strategy does not entail any loss of significant structure matches. Figure 2 compares the distributions of structure similarity scores obtained from an exhaustive search and a search against a subset of representative structures. A choice of Sr = 90% (Experimental Procedures) leads to a 3-fold reduction of search database size and, consequently, to a considerable gain in search speed. Importantly, the shapes and characteristics of the distributions are rather robust with respect to the removal of redundancy. In particular, their mean values and SDs are practically identical, resulting in a proper assignment of S+. Thus, all key information for the identification of significant structure matches is retained. Test searches with 20,000 randomly chosen query structures confirmed that this is a general feature of the resulting distributions and independent of any specifics of the queries.
Figure 2.
A Search against a Representative Subset of Structures Keeps the Main Information Retrieved from an Exhaustive Search
The plot shows two distributions of structure similarity scores S obtained from one-against-n structure database searches. Searches were done for a cytoplasmic protein of unknown function from S. typhimurium (2gjv@1) against all known biological assemblies (thin line, n = 138,294 items; November 7, 2013) and against a representative subset of them (bold line, n = 41,325 items, Sr = 90%; Experimental Procedures). The dashed vertical lines mark the thresholds above which S is considered to be significant at the 3σ level (Experimental Procedures). Note that these thresholds are practically identical for both distributions.
Comparative Structure Analysis of DNA Clamps
DNA clamps are oligomeric components of the DNA polymerase holoenzyme that serve as processivity-promoting factors in DNA replication. Despite different subunit stoichiometries and high sequence diversity, their ring-shaped structures are highly conserved throughout all kingdoms of life (Kuriyan and O’Donnell, 1993; Bruck and O’Donnell, 2001; Indiani and O’Donnell, 2006; Sippl and Wiederstein, 2012). Our aim here is to take a typical bacterial DNA clamp as a starting point from which we explore the structural correlations to all other protein structures presently known.
Our query structure is the dimeric β subunit of polymerase III from Escherichia coli (PDB ID code: 2POL; Kong et al., 1992). The active molecule is a ring-shaped homodimer that is fully represented by the biological assembly 2pol@1. Each monomer contains three domains. The 3D structures of these domains are similar, but their sequences are uncorrelated (Sippl and Wiederstein, 2012). The ring has exact two-fold symmetry as a consequence of dimer assembly and an approximate six-fold symmetry that is due to the individual domains.
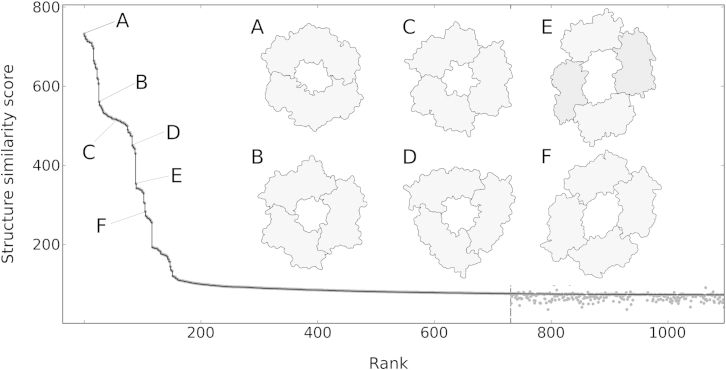
Figure 3 plots the ranks and S scores for the top 1,100 hits of a structure search with 2pol@1 and shows schematic drawings of the query and five high-scoring targets that are discussed in more detail below. The structure similarities of the top hits rapidly decrease to a score of S ∼ 100 and then stay rather constant. For this search, the structure similarity threshold S+ is 75.1, resulting in around 820 targets that have a score above S+.
Figure 3.
A Structure Search with a Dimeric Bacterial DNA Clamp against All Known Biological Assemblies Reveals Different Types of Ring-like DNA Polymerase Subunits
The set of all biological assemblies (138,602 items; November 19, 2013) is searched for structures similar to a dimeric bacterial DNA clamp (2pol@; Kong et al., 1992). Structure similarity scores S and ranks are plotted for the top 1,100 hits (black line). Structure similarities below the threshold of S+ = 75.1 (dashed vertical line) are approximate (gray dots; Experimental Procedures). The dimeric bacterial clamp matches various ring-like assemblies, six of which are schematically shown and linked to their respective positions in the hit list. (A) 2pol@1 (homodimer; Kong et al., 1992). (B) 1plr@1 (homotrimer; Krishna et al., 1994), a proliferating cell nuclear antigen from Saccharomyces cerevisiae. (C) 3hi8@1 (homotrimer; Morgunova et al., 2009), a proliferating cell nuclear antigen from Haloferax volcanii. (D) 1czd@1 (homotrimer; Moarefi et al., 2000), a DNA polymerase accessory protein from Enterobacteria phage T4. (E) 3aiz@1 (heterotetramer; Kawai et al., 2011), a DNA polymerase sliding clamp from Sulfolobus tokodaii. (F) 2z0l@8 (homotetramer; Murayama et al., 2009), a DNA polymerase processivity factor from human herpesvirus 4.
The top 45 ranks are exclusively occupied by dimeric bacterial polymerase III β subunits whose close evolutionary distance is reflected by their extensive structure similarities to 2pol@1 (Figure 3A). Beyond, we find a mixture of eukaryotic and archaeal DNA clamps, suggesting that DNA clamps of eukaryotes and archaea are more similar to each other than to bacterial DNA clamps. The superposition of bacterial and eukaryotic (or archaeal) DNA clamps immediately reveals that the entire ring structures are equivalent (Sippl and Wiederstein, 2012), although bacterial DNA clamps are dimers, whereas eukaryotic and archaeal DNA clamps are trimers (Figures 3B and 3C). Several viral DNA clamps like the polymerase accessory protein of bacteriophage T4 (rank 105; Figure 3D) are also trimers, and in terms of their basic architecture, they seem to be more closely related to the homotrimeric archaeal and eukaryotic DNA clamps than to the homodimeric bacterial clamps.
When going further down in the hit list, a sharp drop in similarity is observed, demarcating the end of the ranks occupied by full-ring matches to the query. The DNA clamps that follow match only part of the ring. Generally, this is due to one of the following reasons: either the definition of the respective biological assembly covers only part of the asymmetric unit, or experimental structure determination was confined to subunits of the ring (e.g., to one or two monomers). Moreover, DNA clamps that are assembled from four subunits turn up in the hit list. For example, at rank 112 we find a crenarchaeal sliding clamp forming an elliptic heterotetrameric complex (Figure 3E). Because the central channel of this complex is considerably larger than that of the dimeric and trimeric clamps spotted so far, only two-thirds of the query can be superimposed with the tetramer. A similar situation is encountered with (C-terminally truncated) early antigen protein D from human herpesvirus 4, another DNA processivity factor following shortly after in the hit list (Figure 3F). Although this protein is reported to be dimeric in solution (Murayama et al., 2009), tetrameric ring formation is observed in the asymmetric unit, and the authors deposited both dimeric and tetrameric biological assemblies in PDB. Indeed, in a follow-up study, the authors speculate that tetrameric ring formation might be required for virus replication in vivo (Nakayama et al., 2010).
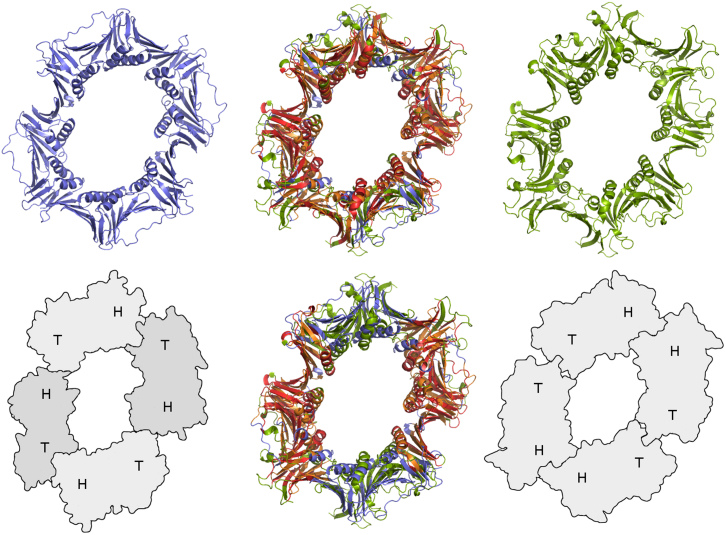
The two tetrameric complexes identified thus far differ significantly in several aspects. First, the crenarchaeal structure is a heterotetramer, whereas the viral structure is a homotetramer. Second, the sequence identity between the archaeal monomers and the viral monomer is below 10%, respectively, reflecting a large evolutionary distance between the corresponding genes. Third, although the monomers of the archaeal complex associate in a “head-to-tail” manner, the monomers of the viral complex associate in a “head-to-head” manner (Figure 4, bottom). Given these differences it is quite astounding that the quaternary structures of these two proteins are highly similar, as revealed by pairwise structure comparison (Figure 4, top). The superposition shows that large parts of the structures accurately match and that the complexes share the size and shape of the central channel as well as the twist observed between the heterodimeric planes of the archaeal structure. It has been hypothesized that the central channel of the archaeal complex can accommodate two stacked DNA duplexes and thus may clamp a Holliday junction (Kawai et al., 2011). To the viral processivity factor, a multifunctional role in virus replication has been ascribed (Sugimoto et al., 2011; Kawashima et al., 2013). The remarkable structural correlation detected between the archaeal and the viral tetramer suggests that the latter may also operate as a Holliday junction clamp. Clearly, this structural correlation motivates further investigation of the functional consequences implied by this type of ring-like assembly.
Figure 4.
Pairwise Structure Comparison of Two Tetrameric DNA Processivity Factors Reveals High Similarity in Quaternary Structure Despite Differences in the Constituent Subunits and Their Association
(Left) 3aiz@1 (Kawai et al., 2011), a heterotetrameric DNA polymerase sliding clamp from S. tokodaii.
(Right) 2z0l@8 (Murayama et al., 2009), a homotetrameric DNA polymerase processivity factor from human herpesvirus 4.
(Middle) Superposition of 3aiz@1 and 2z0l@8. Structurally equivalent parts are shown in orange and red.
(Bottom) Schematic views of the tetramers show the locations of N-terminal (H, “head”) and C-terminal (T, “tail”) domains. Due to different chain associations (“head-to-head” versus “head-to-tail” association), the full structural equivalence can only be seen if permutations are enabled in the structure comparison procedure (Sippl and Wiederstein, 2012); without permutations only two of the four chains can be aligned, respectively (bottom middle).
We conclude the exploration of our example with a note on the lower-ranking structure matches in the hit list. In the search strategy presented, most of the pairwise structure comparisons that would be covered by a truly exhaustive search are skipped, and only an appropriate subset of them actually is processed (Experimental Procedures). This does not imply, however, that the resulting hit list is incomplete. In fact, we can give approximations for all structure similarity scores that are not available from directly calculated alignments. To investigate the loss of accuracy arising from this strategy, we compare the hit list of a truly exhaustive search (i.e., a search where all pairwise alignments are calculated) to the hit list obtained in part by approximation. As shown in Figure 3, the error for approximated structure similarity scores is negligible. In particular, the error is zero for all hits expected to reveal significant matches, and only marginal for all other targets (Figure 3, gray dots).
Discussion
A frequent task in structural biology is the characterization of newly determined protein structures in terms of structure similarities to other proteins. The detection of common structural features and correlations among proteins not only points to important biological connections, but also allows for an appropriate judgment of the novelty of newly determined structures. Finding such correlations, preferably in a comprehensive manner, commonly turns out to be a demanding exercise, because the information on structure similarities is to a large part implicit, hidden, or inaccessible for biological research. The structure search engine presented here provides a means for the efficient and reliable detection of structure similarities among biological assemblies of proteins, i.e., among those structural units thought to represent the functional form of protein molecules.
As demonstrated by the examples discussed above, the detection of structure similarities on the level of protein oligomers is highly informative in that it can reveal correlations that are concealed on the level of individual chains. Of particular interest are situations where remarkably similar supramolecular structures are assembled from distinct sets of structural building blocks. In the case of the homohexameric assemblies compared in Figure 1, the respective building blocks (chains) are not only considerably different on the level of structure, but also they have extremely low (∼10%) pairwise sequence identity. Consequently, the intra- and interchain interactions associated with the respective assemblies arise from largely different groups of amino acids, and it is an intriguing question as to which constraints result in the conservation (or, alternatively, convergent evolution) of such structures of multisubunit complexes. We emphasize that without proper structure-matching tools, structural correlations of this kind remain unrecognized, and we encourage the interested reader to explore the examples discussed in the previous section as well as other structures of choice using the accompanying web service TopSearch (see Accessibility).
Although this work is focused on multiprotein complexes, the methods developed here are seamlessly accessible for other molecular objects like individual protein chains or asymmetric units. Updates of the underlying structure databases are done on a weekly basis so that TopSearch comprises all structures available in PDB. Moreover, using the upload facility of TopSearch, any set of protein structure coordinates in PDB format can be used as a query for a search against the complete structure repository. With this facility we particularly address the X-ray and nuclear magnetic resonance (NMR) communities and their need for instant and reliable characterization of the novelty of experimentally determined structures before releasing them to PDB. Beyond that, the presented tool opens a broad range of possibilities for comparative studies of protein structures, including the structure-based identification of all complexes known to contain a particular protein chain, the investigation of quaternary structure resemblances (Fenn et al., 2013; Kofler et al., 2014), the exploration of fold space (Sippl, 2009), and the evaluation of structures resulting from modeling efforts (e.g., from protein-protein docking; Aloy et al., 2005). In the process of digesting and organizing the vast amount of structures provided by experimental and computational methods, we expect a multitude of novel and unexpected structural correlations yet to be discovered.
Experimental Procedures
Database of Multiprotein Complexes
A comprehensive list of multiprotein complexes is obtained by extracting all biological assemblies from all protein structures available from the Research Collaboratory for Structural Bioinformatics (RCSB) PDB (Berman et al., 2000). In PDB, a biological assembly (or “biological unit”) is defined as a specific macromolecular assembly that is known or believed to be one of the functional forms of a molecule (Dutta et al., 2009; Dutta and Berman, 2005). A particular biological assembly may correspond to a single protein chain, or it can be as large as a complete ribosome or virus capsid, containing many individual protein chains. A PDB entry is usually accompanied by transformation matrices (rotational and translational) that are used to generate the full set of coordinates of a biological assembly.
Biological assemblies are generally derived from crystal structures. The assignments are either supplied by the crystallographers who solved the structures, or they are defined in an automated manner by specialized programs (Henrick and Thornton, 1998; Krissinel and Henrick, 2007). There are many cases where a single PDB entry contains two or more definitions of biological assemblies so that the number of putative biological assemblies is considerably larger than the number of solved structures. Currently, the PDB holds approximately 95,000 protein structure files but more than 140,000 biological assemblies.
In this work, a particular biological assembly is addressed by its four-letter PDB code followed by the @ sign and the number of the assembly as defined in REMARK 350 of the PDB file. For proteins with no biological assembly defined (mostly NMR structures), we take all coordinates listed in the respective PDB file (the first model in case of NMR structures). These entries are identified by the PDB code followed by @0 (e.g., 1nmr@0).
Some technical limitations come from the PDB file format, which allows only 99,999 atoms and 62 unique chains. Examples are virus capsids such as that of poliovirus (2plv) or simian virus 40 (1sva). For these files we include the coordinates of the asymmetric unit and, again, add @0 to the PDB code to identify them. Presently, limitations of such kind affect around 390 files. It is expected that these limitations will become obsolete with mmCIF format (Westbrook and Fitzgerald, 2003; Westbrook et al., 2005; Dutta and Berman, 2005).
Pairwise Structure Comparison
The pairwise alignment of multichain complexes presents several challenges to structure comparison methods. One challenge is that macromolecular assemblies are generally much larger than single-chain structures, implying considerably increased computing time for finding optimal matches between two multichain complexes. Furthermore, in the simultaneous alignment of multiple chains, the relative order of chains is arbitrary, and in order not to miss a solution, the algorithm has to be able to handle permutations in the construction of alignments (see, for example, Figure 4). Here, we use the structure comparison tool TopMatch (Sippl and Wiederstein, 2012) to efficiently compute accurate pairwise alignments between two proteins or protein complexes, query (Q) and target (T).
TopMatch calculates several parameters that describe the structural relationship of Q and T. In particular, the length (L) of an alignment between Q and T and the corresponding spatial deviation of Cα atoms after optimal superposition are important to quantify the structure similarity of Q and T. Both aspects are readily combined by
where is calculated from the superimposed coordinates xi (Q) and yi (T), and σ is a scaling parameter that controls the weights of individual distance errors (ri) (Sippl and Wiederstein, 2012). Accordingly, 0 ≤ S ≤ L, where a perfect match of all structurally equivalent residue pairs yields S = L, while S approaches 0 with increasing spatial deviation of Q and T.
Database Search
The most straightforward way to find proteins with high structure similarity to a given query is a pairwise structure comparison for each entry of a structure database and then to select those pairs that have high similarity scores. However, this costs considerable computing time. For example, such a search takes about 9 hr for a comparison of a hemoglobin tetramer (∼570 amino acid residues) to all ∼140, 000 biological assemblies currently available in PDB, as carried out using TopMatch on a single present-day desktop CPU.
In this procedure much time is wasted for the alignment of structures that are highly dissimilar to the query. For essentially all query structures, it can be expected that matches on the level of tertiary and quaternary structure will only be found to a small set of target structures in the database and that the bulk of target structures show little or no structure similarity to the query anyway. For instance, in the one-against-all search of hemoglobin sketched above, more than 90% of all structures in the database match, if at all, only a tiny part (<10%) of the query. In general, this means that most of the pairwise alignments done during an exhaustive structure search will be dismissed after calculation, and only a small fraction of hits that show considerable structure similarity to the query will be further analyzed.
In addition, redundancy of structures in the search database implies many dispensable pairwise structure comparisons in an exhaustive search (Holm et al., 2008). For instance, thousands of highly similar structures of globins have been deposited to PDB, and in order to find out whether a query structure matches a globin to a considerable degree, it is not necessary to compare the query to each member of a structurally highly homogeneous group of globins when a single comparison with a representative structure taken from this group tells almost the same.
Thus, we use the following strategy in our structure search procedure. We cluster the complete set of structures in the search database into groups of structurally similar molecules (Sippl, 2009). From each group we select one structure to represent all members of the group. The clustering procedure ensures that for all members of a group the structure similarity to the representative reaches at least a certain threshold. We then align the query structure to all representative structures only, thereby obtaining a nonredundant list of pairwise structure similarities. The degree of redundancy removed (and search time saved) in this way is controlled by the threshold used to build the clusters. Here, we express this threshold by the relative structure similarity Sr = 2S/(QL + TL), where QL and TL are the numbers of residues in Q and T, respectively (Sippl, 2008).
The similarities calculated between query and all representatives provide a sample of the distribution of similarities in the complete search database. Thus, they can be used to identify and sort out all structures that are expected to have only marginal similarity to the query. More specifically, let φ(S) be the distribution of structure similarity scores, S, over the complete set of structures in the search database. We approximate φ(S) by the distribution of scores obtained from the sample of representative structures. Both φ and will, in general, be dominated by rather low similarity scores, arising from the bulk of structures that only match small parts of the query (e.g., a pair of helices or less). Significant matches on the level of tertiary or quaternary structure will be comparably rare and clearly separated from the bulk, typically by several standard deviations (σ) above the average score, . Here, we use a score as threshold to distinguish representatives with high similarity to the query from insignificant similarities. We select all clusters whose representatives have a similarity score to the query greater than S+. We then calculate pairwise structure alignments between the query and all members of these clusters.
The procedure just described guarantees exact similarity scores to all structures in the database that are (1) representatives with a score above S+, or (2) in the same cluster as such a representative. These structures cover the interesting part of the hit list, in the sense that their similarities to the query are significantly above the marginal matches of the bulk. All other structures are skipped from direct pairwise alignment because extensive similarity is not expected. Nevertheless, we can efficiently approximate their similarities to the query using metric properties of the TopMatch scores: because we know the exact similarities to all representatives, we can determine a minimal structure similarity to the query for all members of the respective clusters (Sippl, 2008). In this way, no entry of the search database can get lost, the resulting hit list is complete and can finally be ordered by decreasing S to identify the best matching targets.
Accessibility
The structure search tool presented here is implemented as web service called TopSearch and can be accessed at https://topsearch.services.came.sbg.ac.at. Search results are provided as a web page that lists all target structures by decreasing structural similarity to the query structure. Queries are specified by PDB code or by upload of coordinate files in PDB format. The structure similarities found can be analyzed in detail by clicking on the respective entry in the hit list. This triggers a pairwise structure comparison of query and target with TopMatch (Sippl and Wiederstein, 2012), including a 3D visualization of the superimposed structures with Jmol (Hanson, 2010). Each target is annotated with various attributes that help to interpret the results, such as source organism, ligands, release date, and resolution. In addition, a condensed view of the target list can be selected that displays groups of structurally similar targets, thereby removing redundant entries from the target list and focusing on the structural diversity of the hits.
The repository of structures accessible in TopSearch is updated regularly with the weekly releases of the RCSB PDB. Every structure newly released by the PDB enters the structure search pipeline presented above and is processed and integrated into TopSearch within days after release. As a result, if the TopSearch query is specified by PDB code, access to the structural relations between query and all other structures in PDB is instantaneous. If the query is specified by upload of a coordinate file, the hit list is usually available within several hours.
The figures shown in this paper are prepared with the UCSF Chimera package (Pettersen et al., 2004) and PyMOL (Schrödinger).
Acknowledgments
This work was supported by Austrian Science Fund (FWF): P21294-B12. F.M. acknowledges support from FONDECYT Chile (1110400) and Iniciativa Cientifica Milenio (ICM) Chile (P09-016-F). Computations were carried out in part at the Centre for High-Performance Computing (CHPC), University of Salzburg.
Footnotes
This is an open access article under the CC BY-NC-SA license (http://creativecommons.org/licenses/by-nc-sa/3.0/).
References
- Aloy P., Pichaud M., Russell R.B. Protein complexes: structure prediction challenges for the 21st century. Curr. Opin. Struct. Biol. 2005;15:15–22. doi: 10.1016/j.sbi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Coimbatore Narayanan B., Di Costanzo L., Dutta S., Ghosh S., Hudson B.P., Lawson C.L., Peisach E., Prlić A., Rose P.W. Trendspotting in the Protein Data Bank. FEBS Lett. 2013;587:1036–1045. doi: 10.1016/j.febslet.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A. Protein structures: Structures of desire. Nature. 2009;459:24–27. doi: 10.1038/459024a. [DOI] [PubMed] [Google Scholar]
- Birktoft J.J., Blow D.M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J. Mol. Biol. 1972;68:187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Bolton W., Perutz M.F. Three dimensional fourier synthesis of horse deoxyhaemoglobin at 2.8 Angstrom units resolution. Nature. 1970;228:551–552. doi: 10.1038/228551a0. [DOI] [PubMed] [Google Scholar]
- Bruck I., O’Donnell M. The ring-type polymerase sliding clamp family. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-1-reviews3001. reviews3001.1–reviews3001.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell D.A., Kornberg R.D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Dutta S., Berman H.M. Large macromolecular complexes in the Protein Data Bank: a status report. Structure. 2005;13:381–388. doi: 10.1016/j.str.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Dutta S., Burkhardt K., Young J., Swaminathan G.J., Matsuura T., Henrick K., Nakamura H., Berman H.M. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- Fenn S., Schiller C.B., Griese J.J., Duerr H., Imhof-Jung S., Gassner C., Moelleken J., Regula J.T., Schaefer W., Thomas M. Crystal structure of an anti-Ang2 CrossFab demonstrates complete structural and functional integrity of the variable domain. PLoS ONE. 2013;8:e61953. doi: 10.1371/journal.pone.0061953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K.M., McKay D.B., Kabsch W., Holmes K.C. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc. Natl. Acad. Sci. USA. 1991;88:5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K., Gruber M., Sippl M.J. COPS benchmark: interactive analysis of database search methods. Bioinformatics. 2010;26:574–575. doi: 10.1093/bioinformatics/btp712. [DOI] [PubMed] [Google Scholar]
- Furnham N., Laskowski R.A., Thornton J.M. Abstracting knowledge from the protein data bank. Biopolymers. 2013;99:183–188. doi: 10.1002/bip.22107. [DOI] [PubMed] [Google Scholar]
- Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Hanson R.M. Jmol - a paradigm shift in crystallographic visualization. J. Appl. Crystogr. 2010;43:1250–1260. [Google Scholar]
- Hasegawa H., Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Henrick K., Thornton J.M. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- Holm L., Kääriäinen S., Rosenström P., Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C., O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- Kanamaru S. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc. Natl. Acad. Sci. USA. 2009;106:4067–4068. doi: 10.1073/pnas.0901205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A., Hashimoto H., Higuchi S., Tsunoda M., Sato M., Nakamura K.T., Miyamoto S. A novel heterotetrameric structure of the crenarchaeal PCNA2-PCNA3 complex. J. Struct. Biol. 2011;174:443–450. doi: 10.1016/j.jsb.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Kawashima D., Kanda T., Murata T., Saito S., Sugimoto A., Narita Y., Tsurumi T. Nuclear transport of Epstein-Barr virus DNA polymerase is dependent on the BMRF1 polymerase processivity factor and molecular chaperone Hsp90. J. Virol. 2013;87:6482–6491. doi: 10.1128/JVI.03428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler S., Ackaert C., Samonig M., Asam C., Briza P., Horejs-Hoeck J., Cabrele C., Ferreira F., Duschl A., Huber C., Brandstetter H. Stabilization of the dimeric birch pollen allergen Bet v 1 impacts its immunological properties. J. Biol. Chem. 2014;289:540–551. doi: 10.1074/jbc.M113.518795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X.P., Onrust R., O’Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Krishna T.S., Kong X.P., Gary S., Burgers P.M., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., O’Donnell M. Sliding clamps of DNA polymerases. J. Mol. Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- Leiman P.G., Basler M., Ramagopal U.A., Bonanno J.B., Sauder J.M., Pukatzki S., Burley S.K., Almo S.C., Mekalanos J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moarefi I., Jeruzalmi D., Turner J., O’Donnell M., Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- Morgunova E., Gray F.C., Macneill S.A., Ladenstein R. Structural insights into the adaptation of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii to a high-salt environment. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1081–1088. doi: 10.1107/S0907444909029321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama K., Nakayama S., Kato-Murayama M., Akasaka R., Ohbayashi N., Kamewari-Hayami Y., Terada T., Shirouzu M., Tsurumi T., Yokoyama S. Crystal structure of epstein-barr virus DNA polymerase processivity factor BMRF1. J. Biol. Chem. 2009;284:35896–35905. doi: 10.1074/jbc.M109.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S., Murata T., Yasui Y., Murayama K., Isomura H., Kanda T., Tsurumi T. Tetrameric ring formation of Epstein-Barr virus polymerase processivity factor is crucial for viral replication. J. Virol. 2010;84:12589–12598. doi: 10.1128/JVI.01394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J., Xu X., Cui H., Savchenko A., Edwards A., Joachimiak A. Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. J. Struct. Funct. Genomics. 2011;12:21–26. doi: 10.1007/s10969-011-9107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell L.G., Kanelis V., Donaldson L.W., Howell P.L., Davidson A.R. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. USA. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz M.F., Rossmann M.G., Cullis A.F., Muirhead H., Will G., North A.C. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Sippl M.J. On distance and similarity in fold space. Bioinformatics. 2008;24:872–873. doi: 10.1093/bioinformatics/btn040. [DOI] [PubMed] [Google Scholar]
- Sippl M.J. Fold space unlimited. Curr. Opin. Struct. Biol. 2009;19:312–320. doi: 10.1016/j.sbi.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Sippl M.J., Wiederstein M. Detection of spatial correlations in protein structures and molecular complexes. Structure. 2012;20:718–728. doi: 10.1016/j.str.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A., Kanda T., Yamashita Y., Murata T., Saito S., Kawashima D., Isomura H., Nishiyama Y., Tsurumi T. Spatiotemporally different DNA repair systems participate in Epstein-Barr virus genome maturation. J. Virol. 2011;85:6127–6135. doi: 10.1128/JVI.00258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G., Chiu W. Exploring new limits in complex biological structures. Curr. Opin. Struct. Biol. 2013;23:704–706. doi: 10.1016/j.sbi.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Westbrook J.D., Fitzgerald P.M.D. The PDB format, mmCIF, and other data formats. Methods Biochem. Anal. 2003;44:161–179. [PubMed] [Google Scholar]
- Westbrook J., Ito N., Nakamura H., Henrick K., Berman H.M. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]