Abstract
Tick gut glycoprotein, designated as Bm86, found on the luminal surface of the plasma membrane of gut epithelial cells of Boophilus microplus, which is a concealed antigen, has been used as vaccine candidate molecule for immunization against ticks. To better understand the molecular diversity of Bm86 gene in ticks, a portion of the cDNA was sequenced from an Indian isolate of B. microplus. Comparison of nucleotide sequence revealed that Indian isolate had 97 % homology (18 polymorphisms) with that of the Australian isolate and 96 % homology (20 polymorphisms) with that of the Cuban vaccine strain. Further, the Indian isolate differed from the Cuban vaccine isolate at 7 amino acid loci, including 5 substitutions (at residues 88, 94, 175, 176 and 177) and 2 deletions (at 183 and 184). However, protein prediction studies did not show any difference in the putative antigenic epitopes of the protein expressed.
Keywords: Rhipicephalus microplus, Concealed antigen, Bm86, Phylogenetic tree, Tick vaccine
Introduction
Ticks are considered as major impediment in livestock production not only as pests but also vectors of devastating diseases. The largest monetary loss in the livestock industry is attributed to the production losses resulting from tick infestation and the expenditure incurred to control both the ticks as well as the disease pathogens they transmit. Infestation caused by tick, Boophilus microplus, one-host tick, which transmits Babesiosis in cattle, accounts for huge economic loses. The most common method of tick control is the use of chemical acaricides in developing countries, but it has many disadvantages such as environmental pollution, development of resistance to acaricides and residues in meat, milk, hide, skin etc., (Nolan et al. 1989).
Alternate approaches for tick control include the use of natural host resistance and development of vaccines to induce an immunological response against tick infestations. Vaccination of cattle against the tick B. microplus by inducing an immunologic reaction against antigen in the tick gut has been reported. Micrograms of this ‘concealed’ antigen (Bm86) conferred effective protection in cattle against this tick (Willadsen and Kemp 1988).
It is well established fact that genetic variation has a direct bearing on the antigenicity of vaccines. It is essential to select as the seed material the predominant strains in a particular region for preparation of effective vaccines. Sequence variation has been reported to be one of the reasons of reduced effect of Bm86 based vaccine. The sequence variation was studied in the Bm86 locus of B. microplus strains from Australia, Mexico, Cuba, Argentina and Venezuela (Garcia-Garcia et al. 1999). Hence, a study was undertaken to determine the sequence variation of locus B. microplus strain and compared with reference strain.
Materials and methods
Collection and processing of ticks
Semi engorged B. microplus ticks were collected from naturally infested cattle in and around Chennai, Tamil Nadu, India and the ticks were dissected immediately after collection. To get 50 mg of midgut tissue for RNA isolation, 5–6 ticks were dissected. Tick dissection was carried out by as per method described by Mulenga et al. (2001). Ticks were surface sterilized in 0.1 % diethyl pyrocarbonate (DEPC) water and dissection was performed on ice under a dissection microscope. Following retraction of the dorsal exoskeleton, midgut were removed from the rest of the body and once again washed with 0.1 % DEPC water. Following dissection, the midgut was frozen in liquid nitrogen, pulverized and used for RNA isolation.
RNA isolation and reverse transcriptase-polymerase chain reaction
Total RNA was extracted from 50 mg midgut tissues of B. microplus tick by Acid Guanidium Thiocyanate Phenol Chloroform method (Chomczynski and Sacchi, 1987). Total RNA was treated with 100 Units of RNAse-free DNAse for 1 h at 37 °C, heat-inactivated at 80 °C for 10 min and then reverse transcribed. Complementary DNA (cDNA) was synthesized from total RNA using Revert Aid first strand cDNA synthesis kit, according to the manufacturer’s instructions, (MBI Fermentas, USA) for a 20 μl reaction.
Polymerase chain reaction (PCR) was carried out in the thermo cycler (Eppendorf, Germany) for amplifying 625 bp of Bm86 gene (13-638 Nucleotides) using specific primers [For 5′-CGG TGG TTC GAC GCA GTG AC-3′ and [Rev 5′-GTC CGG TCC TTT GGG GCT C-3′ (Rand et al. 1989). The amplification was performed with the following steps viz. initial denaturation at 94 °C for 2 min, 35 cycles of denaturation at 94 °C for 60 s, annealing at 54 °C for 60 s, extension at 72 °C for 1 min with a final extension at 72 °C for 5 min. The PCR products were checked in 1.5 % agarose gel electrophoresis and purified using gel extraction kit (Genei, Bangalore).
Sequence analysis
Purified PCR product was sequenced in an automated DNA sequencer (ABI prisms, Genei, Bangalore, India). The sequence, named as Chennai 1, was submitted to the Genbank. For comparison with Chennai 1 sequence, twelve nucleotide sequences were retrieved from Gen bank. The nucleotide sequences as well as the deduced amino acid sequences were analyzed using the ‘multi-align editor’ function of the Gene Tool Lite software package (Version 1.0) for putative polymorphism. Further, the sequences were aligned using Clustal W method (Thompson et al. 1994) and phylogenetic trees were constructed in the MegAlign program of the Lasergene software (DNASTAR Inc., USA).
Based on the information of 113 amino acid residues, it was investigated if there could be any putative antigenic change in epitopes in the Chennai 1 isolate, when compared to the Cuban vaccine isolate. For this, the entire amino acid sequence (650) of the vaccine strain was used as the standard and compared with the same stretch of sequences into which the 113 amino acid residues of Chennai 1 isolate was substituted. The sequences were compared using the PROPRED (http://bioinformatics.uams.edu/mirror/propred/) and PROPRED 1 (http://bioinformatics.uams.edu/mirror/propred1/) online tools (Singh and Raghava 2001).
Results
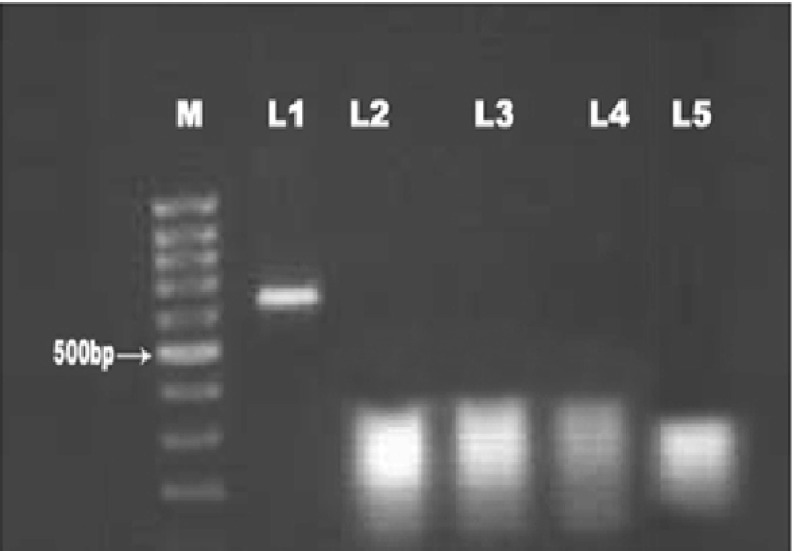
Amplification of the Bm86 gene at an expected size of 625 bp was occurred when cDNA of B. microplus was used (Fig. 1). The sequence information of 577 nucleotides of the 625 bp PCR product was obtained with fidelity. This sequence, named as Chennai 1, was assigned GenBank accession number (DQ131539). The 577 bp sequence was subjected to nucleotide BLAST (Altschul et al. 1997) to determine the similarity of this sequence to any of the previously published sequences of Bm86 gene of B. microplus. Considerable homology was observed with sequences of Bm86 from Australian (GenBank Accession No. M29321) and Cuban (GenBank Accession No. AF150891) isolates of B. microplus. When the 577 bp sequence from Chennai 1 isolate was compared with that of the Australian and Cuban isolates, using `multi-align editor’ function of the GENETOOL software, Chennai 1 isolate differed at 23 nucleotides position. The notable difference was the deletion of a 6-nucleotide region starting from nucleotide 545–550 in Chennai 1 isolate (Fig. 2). The Chennai 1 isolate had 97 % homology (18 polymorphisms) and 96 % homology (20 polymorphisms) with that of the Australian isolate and Cuban vaccine strain respectively.
Fig. 1.
Amplificationof Bm86 gene of 625 bp by RT-PCR. M—100 bp DNA ladder L1—Boophilus microplus L2—Rhipicephalus haemaphysaloides L3—Haemaphysalis bispinosa L4—Hyalomma anatolicum anatolicum L5—Negative control
Fig. 2.

Alignment of the 23 polymorphic nucleotides of the 577 bp sequence from Chennai 1, Australian and Cuban isolates. Sequences identical to Chennai 1 are indicated with a period. Alignment gaps are indicated by dashes. The numbers indicate nucleotide positions on the sequences
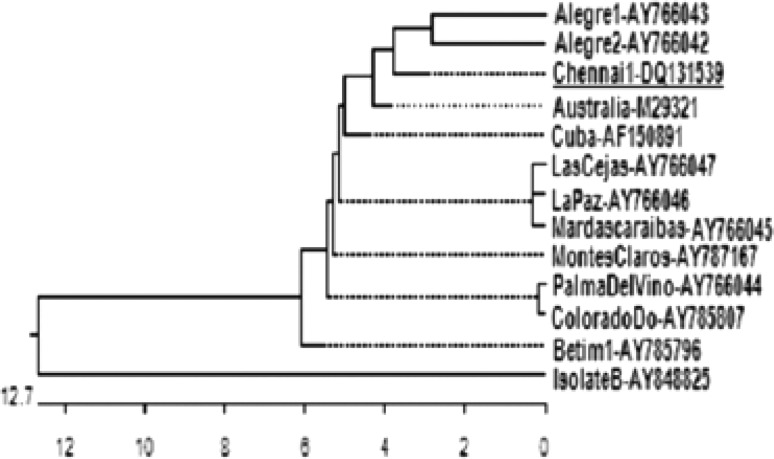
When short stretch of nucleotide sequence (342 bp i.e., from the position 236–577) from the Chennai 1 isolate was used in BLAST analysis, homology was observed with more number of Bm86 sequences that are available in the public database. This short stretch of sequence was compared with 12 other sequences of the same length from different isolates and phylogenetic tree was constructed from the nucleotide sequence comparison (Fig. 3). It can be observed that based on nucleotide sequence, the Chennai 1 isolate aligns closer to the two South American isolates (Alegre 1 and Alegre 2) rather than the Australian and Cuban isolates. The pair wise distances based on nucleotide sequences is given (Table 1). This revealed that the Chennai 1 isolate had 96.5 and 98 % homology with that of the Cuban and Australian isolates, respectively.
Fig. 3.
Phylogenetic tree derived using Clustal W from the nucleotide sequences of Bm86 gene fragments of Chennai 1 and other isolates
Table 1.
Pair distances between partial Bm 86 nucleotide sequence of Chennai 1 isolate and other sequences in public data base, derived using Clustal W
| A | B | C | D | E | F | G | H | I | J | K | L | M | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | *** | 81.6 | 97.1 | 96.8 | 96.8 | 96.6 | 96.6 | 96.6 | 96.6 | 94.5 | 96.5 | 96.8 | 96.8 | A |
| B | 22.9 | *** | 84.8 | 84.5 | 84.8 | 84.5 | 84.5 | 84.2 | 83.3 | 79.6 | 81.9 | 82.8 | 83.9 | B |
| C | 2.9 | 19.7 | *** | 99.7 | 99.7 | 99.4 | 99.4 | 99.4 | 98.6 | 93.7 | 96.8 | 98.3 | 99.1 | C |
| D | 3.2 | 19.4 | 0.3 | *** | 99.4 | 99.1 | 99.1 | 99.7 | 98.6 | 93.4 | 96.5 | 98.0 | 98.9 | D |
| E | 3.2 | 19.7 | 0.3 | 0.6 | *** | 99.1 | 99.1 | 99.1 | 98.3 | 93.4 | 96.5 | 98.0 | 98.9 | E |
| F | 3.5 | 19.7 | 0.6 | 0.9 | 0.9 | *** | 99.4 | 98.9 | 98.0 | 93.4 | 96.2 | 97.7 | 98.6 | F |
| G | 3.5 | 20.5 | 0.6 | 0.9 | 0.9 | 0.6 | *** | 98.9 | 98.0 | 93.4 | 96.2 | 97.7 | 98.6 | G |
| H | 3.5 | 19.7 | 0.6 | 0.3 | 0.9 | 1.2 | 1.2 | *** | 98.3 | 93.1 | 96.2 | 97.7 | 98.6 | H |
| I | 3.5 | 20.5 | 1.5 | 1.8 | 1.8 | 2.0 | 2.0 | 2.0 | *** | 92.5 | 95.6 | 97.1 | 97.7 | I |
| J | 5.7 | 24.5 | 6.6 | 6.9 | 6.9 | 7.3 | 7.3 | 7.3 | 7.9 | *** | 93.9 | 94.0 | 93.4 | J |
| K | 3.3 | 22.1 | 3.0 | 3.3 | 3.3 | 3.6 | 3.6 | 3.6 | 4.2 | 6.1 | *** | 97.4 | 96.5 | K |
| L | 2.9 | 21.3 | 1.7 | 2.0 | 2.0 | 2.3 | 2.3 | 2.3 | 2.9 | 6.0 | 2.4 | *** | 98.0 | L |
| M | 3.2 | 20.9 | 0.9 | 1.2 | 1.2 | 1.5 | 1.5 | 1.5 | 2.3 | 6.9 | 3.3 | 2.0 | *** | M |
| A | B | C | D | E | F | G | H | I | J | K | L | M | N |
The percent similarity is represented in the upper triangle and the percent divergence is represented in lower triangle
A = Alegre 1, B = Isolate B, C = Mar Das Caraibas, D = Palma Del Vino, E = Montes Claros, F = Las Cejas, G = La Paz, H = Colorado Do, I = Betim 1, J = Alegre 2, K = Chennai, L = Australian, M = Cuban
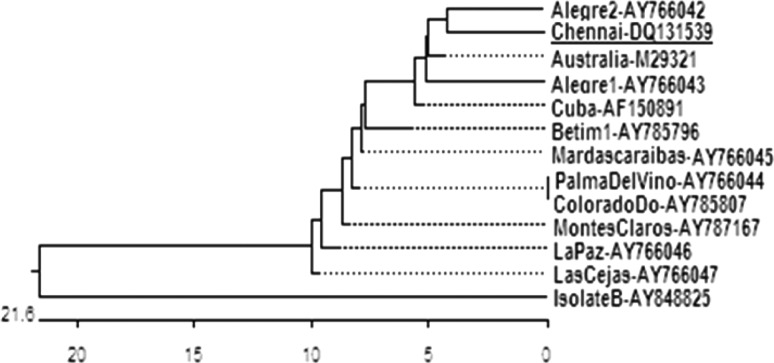
The translated amino acid sequence (113 amino acid residues from residue 82–194 of Bm86 gene) of the 342 bp fragment was also compared with those of 12 isolates retrieved from Genbank. The 113 amino acid alignment results showed that the sequence of Chennai 1 isolate differs from the Cuban vaccine isolate at 7 loci, including 5 substitutions (at residues 88, 94, 175, 176 and 177) and 2 deletions (at 183 and 184). The phylogenetic tree was constructed from the amino acid sequence (Fig. 4). It was observed that amino acid sequence of Chennai 1 isolate has greater homology with South American isolate (Alegre 2) rather than Australian and Cuban isolates. The data of pair wise distance of amino acid sequence between isolates is depicted (Table 2). The Chennai 1 isolate had 94.7 % similarity (4.6 %) with Cuban isolate, while it was much closer to the Australian isolate (97.4 % similarity).
Fig. 4.
Phylogenetic tree derived using Clustal W from the amino acid sequences of Bm86 gene fragments of Chennai 1 and other isolates
Table 2.
Pair distances between partial Bm 86 amino acid sequence of Chennai 1 isolate and other sequences in public data base, derived using Clustal W
| A | B | C | D | E | F | G | H | I | J | K | L | M | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | *** | 76.5 | 94.8 | 93.9 | 93.9 | 93.0 | 93.0 | 93.9 | 93.0 | 89.6 | 90.3 | 93.9 | 94.8 | A |
| B | 42.9 | *** | 81.7 | 81.7 | 80.9 | 80.9 | 80.9 | 81.7 | 80.0 | 76.5 | 77.9 | 79.1 | 80.0 | B |
| C | 5.4 | 34.2 | *** | 99.1 | 99.1 | 98.3 | 98.3 | 99.1 | 96.5 | 91.3 | 95.6 | 97.4 | 98.3 | C |
| D | 6.4 | 34.2 | 0.9 | *** | 98.3 | 97.4 | 97.4 | 100.0 | 95.7 | 90.4 | 94.7 | 96.5 | 97.4 | D |
| E | 6.4 | 34.2 | 0.9 | 1.8 | *** | 97.4 | 97.4 | 98.3 | 95.7 | 90.4 | 94.7 | 96.5 | 97.4 | E |
| F | 7.3 | 34.2 | 1.8 | 2.7 | 2.7 | *** | 98.3 | 97.4 | 94.8 | 90.4 | 93.8 | 95.7 | 96.5 | F |
| G | 7.3 | 1.8 | 34.2 | 2.7 | 2.7 | 1.8 | *** | 97.4 | 94.8 | 90.4 | 93.8 | 95.7 | 96.5 | G |
| H | 6.4 | 34.2 | 0.9 | 0.0 | 1.8 | 2.7 | 2.7 | *** | 95.7 | 90.4 | 94.7 | 96.5 | 97.4 | H |
| I | 7.3 | 38.4 | 3.6 | 4.5 | 4.5 | 5.4 | 5.4 | 4.5 | *** | 87.8 | 90.3 | 93.9 | 94.8 | I |
| J | 11.3 | 42.9 | 9.3 | 10.3 | 10.3 | 10.3 | 10.3 | 10.3 | 13.3 | *** | 92.0 | 92.2 | 91.3 | J |
| K | 8.4 | 37.8 | 4.6 | 5.5 | 5.5 | 6.5 | 6.5 | 5.5 | 8.4 | 8.4 | *** | 95.6 | 94.7 | K |
| L | 6.4 | 38.4 | 2.7 | 3.6 | 3.6 | 4.5 | 4.5 | 3.6 | 6.4 | 8.3 | 3.6 | *** | 97.4 | L |
| M | 5.4 | 37.0 | 1.8 | 2.7 | 2.7 | 3.6 | 3.6 | 2.7 | 5.4 | 9.3 | 4.6 | 2.7 | *** | M |
| A | B | C | D | E | F | G | H | I | J | K | L | M |
The percent similarity is represented in the upper triangle and the percent divergence is represented in lower triangle
A = Alegre 1, B = Isolate B , C = Mar Das Caraibas, D = Palma Del Vino, E = Montes Claros, F = Las Cejas, G = La Paz, H = Colorado Do, I = Betim 1, J = Alegre 2, K = Chennai, L = Australian, M = Cuban
Discussion
The ticks of different regions have undergone different evolutionary process associated with marked morphological, physiological and genetic variations. It has been documented that habitat adaptation could have resulted in strains with variations that have in fact altered the susceptibility of different strains of ticks to Bm86 based vaccines (Cobon et al. 1996; Garcia-Garcia et al. 1999, 2000). Twenty-one amino acid substitutions were observed between the Yeerongpilly (Australian) Bm86 and Bm85 from an Argentinean isolate of B. microplus (strain A), expressed in Pichia pastoris, resulted in a new Bm86 homologue, Bm95 gene (Garcia-Garcia et al. 2000).
The sequence variations found in the Chennai isolate may be due to significant alteration of the antigenic characteristics of Bm86 protein, the sequences were analyzed with PROPRED. The results indicate that the putative epitopes of the Bm86 protein from Chennai isolate of B. microplus was similar to that of the vaccine isolate. However, in this study only 113 amino acids were analyzed. A significant polymorphism was noticed in the Bm 86 fragment from Chennai 1 isolate (7 out of 113 residues examined). The Bm86 protein is considerably large (650 residues of amino acids) and it is possible that more polymorphisms have occurred during evolution of local isolates, thus making it antigenically different from the homologue used for vaccine preparation.
In sum, the full length Bm86 gene from many local Indian isolates will be sequenced in future studies and the amino acid sequences will be compared to observe the extent of polymorphism in this gene.
Acknowledgments
The authors wish to thank Tamil Nadu Veterinary and Animal Sciences University (TANUVAS), Chennai, India, for providing all the facilities necessary to carry out this work.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang Z, Miller W, Lipman DL. Gapped BLAST and PSI- BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single—Step method of RNA isolation by acid guanidinium thiocyanate- phenol-chloroform extraction. Anal Biochem. 1987;62:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Cobon GS, Moore JT, Johnston LAY, Willadsen P, Kemp DH, Sriskantha A, Riding GA, Rand KN (1996) DNA encoding a cell membrane glycoprotein of a tick gut. US patent and Trade mark office Appl. No. 325071435/240.2. Cited in Willadsen P, 2001
- Garcia-Garcia JC, Gonzalez IL, Gonzalez DM, Valdes M, Mendez L, Lamberti J, D’Agostino B, Citroni D, Fragoso H, Oritz M, Rodriguez M, de la Fuente J. Sequence variations in the Boophilus microplus Bm 86 locus and implications for immunoprotection in cattle vaccinated with this antigen. Exp Appl Acarol. 1999;23:883–895. doi: 10.1023/A:1006270615158. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia JC, Montero C, Redondo M, Vargas M, Canales M, Boue O, Rodriguez M, Joglar M, Machado H, Gonzalez IL, Valdes M, de la Mendez L, Fuente J. Control of ticks resistant to immunization with Bm 86 in cattle vaccinated with the recombinant antigen Bm 95 isolated from the cattle tick Boophilus microplus. Vaccine. 2000;18:2275–2287. doi: 10.1016/S0264-410X(99)00548-4. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Sugimoto C, Ingram G, Ohashi K, Onuma M. Characterization of two cDNAs encoding serine proteinases from the hard tick Haemaphysalis longicornis. Insect Biochem Mol Biol. 2001;31:817–825. doi: 10.1016/S0965-1748(00)00187-9. [DOI] [PubMed] [Google Scholar]
- Nolan J, Wilson JT, Green PE, Bird PE. Synthetic pyrethroid resistance in field samples in cattle tick (Boophilus microplus) Aus Vet J. 1989;66:179–182. doi: 10.1111/j.1751-0813.1989.tb09796.x. [DOI] [PubMed] [Google Scholar]
- Rand KN, Moore T, Srikantha A, Spring K, Tellam R, Willadsen P, Cobon GS. Cloning and expression of a protective antigen from the cattle tick Boophilus microplus. Proc Natl Acad Sci. 1989;86:9657–9661. doi: 10.1073/pnas.86.24.9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Raghava GPS. Propred: prediction of HLA–DR binding sites. Bioinformatics. 2001;17(12):1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willadsen P, Kemp DH. Vaccination with concealed antigen for tick control. Parasitol Today. 1988;4:196–198. doi: 10.1016/0169-4758(88)90084-1. [DOI] [PubMed] [Google Scholar]