Abstract
AIM: To understand the local pathophysiological alterations and gene ontology-based functional classification of colonic biopsies into inflammatory and neoplastic diseases.
METHODS: Total RNA was extracted from frozen biopsies and amplified by T7-method. Expression profile was evaluated by Atlas Glass 1K microarrays. After microarray quality control, applicable data were available from 10 adenomas, 6 colorectal adenocarcinomas (CRCs), and 6 inflammatory bowel diseases (IBDs). Multivariate statistical and cell functional analyses were performed. Real-time RT-PCR and immunohistochemistry were used for validation.
RESULTS: Discriminant analysis of selected genes, could correctly reclassify all 22 samples using 4 parameters (heat shock transcription factor-1, bystin-like, calgranulin-A, TRAIL receptor 3). IBD samples were characterized by overregulated chemokine (C-X-C motif) ligand 13, replication protein A1, E74-like factor 2 and downregulated TNF receptor-associated factor 6, BCL2-interacting killer genes. In adenomas upregulation of TNF receptor-associated factor 6, replication protein A1, E74-like factor 2 and underexpression of BCL2-associated X protein, calgranulin-A genes were found. CRC cases had significantly increased epidermal growth factor receptor, topoisomerase-1, v-jun, TNF receptor-associated factor 6 and TRAIL receptor 3, and decreased RAD51 and RAD52 DNA repair gene, protein phosphatase-2A and BCL2-interacting killer mRNA levels. Epidermal growth factor receptor RT-PCR and immunohistochemistry, topoisomerase-1 RT-PCR confirmed the chip results.
CONCLUSION: Different histological alterations can be reclassified by functional, multivariate analysis using cDNA microarrays. Further studies with expanded sample number are needed for subclassification of pathological alterations.
Keywords: Adenoma, Biopsy samples, Colorectal cancer, Gene expression, Inflammatory bowel diseases, Microarray technology
INTRODUCTION
mRNA expression array analysis is usually performed on high volume surgery or blood samples. However, evaluation of routine biopsy specimens could yield information, as to how the local pathological processes differ from healthy counterparts. In the gastrointestinal tract, biopsy samples are routinely taken. The mRNA expression study of these samples could allow further insight into the development of inflammatory, preneoplastic and neoplastic diseases.
These specimens could not be applied previously for expression array studies, because array technology even today needs significantly more RNA than can be isolated from the tiny biopsy specimen. However, new techniques and commercial kits have recently become available for the reliable mRNA amplification without an effect on the original gene expression pattern[1].
Previous microarray analyses reported in the literature were performed predominantly from surgically resected colon adenocarcinoma samples[2], with gene expression analysis of colonic biopsies done in only two cases[3,4].
In a few studies, local pathological alterations were examined using cDNA microarrays in cells and tissue structures laser-microdissected from surgical material[4,5]. The mRNA expression patterns of tumorous and normal tissues were usually compared by cDNA microarrays[5,7-11] or oligonucleotide microarrays[12,13] containing some hundreds to several thousand fixed target sequences.
Microarray gene expression profiling of adenomas as a precancerous stage of colon adenocarcinoma is less represented in the scientific literature. Oligonucleotide or cDNA microarray-based molecular diagnostics of malignancy in colon adenoma and colorectal cancer samples was described using 10[14], 9[6] and 4[15] adenoma samples compared to adenocarcinoma and normal colonic tissues.
Inflammatory bowel diseases (IBDs) were rarely analyzed by microarrays. Lawrance et al[16] published an oligonucleotide microarray comparison between surgically resected human ulcerative colitis (UC), Crohn's disease (CD) and normal colonic tissues. Recently, Langmann et al[3] used mucosal biopsy specimens for global gene expression profiling in patients with UC and CD.
cDNA-based mid-sized (1K) commercial arrays have recently appeared, which do not need any special hybridization, washing or scanner apparatus. These open-platform arrays, if of good quality and reproducibility, can contribute to a widespread use of the array technology. Mid-sized so called “overall” microarrays with fixed target cDNAs from the most important cell processes (adhesion, apoptosis, cell cycle, DNA replication and repair, extracellular matrix remodeling, cytoskeleton, immune regulation, metabolism, stress response, oncogenesis and tumor suppression, growth factor-related cell proliferation, neuroendocrine regulation, signal transduction, transcription and transport) give us opportunity for analysis of different cell functions that may be associated with diseases.
Bioinformatical analysis of these types of arrays can then be used to detect the expression pattern differences between limited numbers of diagnostic groups. Multivariate statistical analysis methods can be applied for the development of automated classification methods like image analysis based cervical cancer screening[17,18].
In the present study we aimed to prove that normal, inflammatory, premalignant and malignant colon biopsies that can be used for mRNA expression analysis and the expression differences can be utilized in a multivariate classification system. As colorectal adenocarcinoma (CRC) frequently arises in the setting of various high-risk conditions such as adenomatous polyps and IBD, we looked for gene expression pattern-based connections between these types of diseases.
MATERIALS AND METHODS
Patients and samples
Routine biopsy specimens were collected from the pathological and normal part of the colon, placed in RNALater Stabilization Reagent. Total RNA was extracted from frozen biopsy specimens from 11 patients with adenomatous polyps, 12 with CRC and 11 with IBDs (5 UC, 6 CD). After quality control of the microarrays, applicable data were available from 10 patients with adenomatous polyps, 6 with CRC and 6 with IBD (3 UC, 3 CD).
Four male and two female patients of 53-86 years (median, 72.17 years) with CRC were involved in the study. Five patients had left side (2 rectal, 33%; 3 sigmoid , 50%) and 1 had right side (coecal, 17%) involvement. Two patients had localized disease without nodal or distant organic involvement (Dukes B stage, 33%), 1 patient had nodal involvement (17%, Dukes C stage) and 3 had liver metastases (50%, Dukes D stage).
Nine male and one female patient of 17-77 years (median, 61.2 years) with colorectal adenoma were involved in the study. Five of them had left side (1 rectal, 10%; 3 sigmoid, 30%; 1 descendent colonic, 10%), and 5 had right side (2 ascendant colonic, 20%; 1 coecal, 10%; 2 total colonic, 20%) involvement. Six patients had tubular (60%), 2 had tubulovillous (20%) and 2 had villous adenoma (20%). Only one of the villous adenoma patients' biopsy sample contained severe dysplastic alteration.
One male and five female IBD patients of 23-72 years. (42 years. median) were involved in the study. Three of them [50%; 1 UC (33%) and 2 CD (66%)] had total and three [50%; 2 UC (66%) and 1 CD (33%)] had left side colonic disease. The grade of inflammation was severe in 3 cases (50%) and moderate in 3 cases (50%).
Methods
Total RNA isolation using Qiagen RNeasy Mini Kit:Frozen biopsy samples were lysed and homogenized in a mixture of 300 μL GITC-containing lysis buffer and 3 μL β-mercaptoethanol by Polytron homogenizator for 30-40 s. The lysed samples were digested in proteinase K solution at 55°C for 10 min. After silica membrane cleaning, according to the manufacturer’s description and DNase I treatment (in order to absolutely remove genomic DNA), the total RNA was eluted in 50 μL RNase-free water. Quantity and quality of the isolated RNA was tested by measuring the UV absorbance, by using real-time RT-PCR (Light Cycler G6PDH Housekeeping Gene Set, Roche) and by agarose gel electrophoresis. The high quality, intact total RNA samples, which showed regular 18S and 28S ribosomal RNA bend pattern during the agarose gel analysis, and showed positive real-time RT-PCR reaction were used for microarray analysis.
T7 RNA amplification and labeling: Because the total RNA content of the biopsy samples was lower than 10 µg, which was not enough for one hybridization reaction- the mRNA fraction was amplified by the T7-method (MessageAmp I aRNA Kit, Ambion Inc., US), according to the manufacturer’s instructions, fluorescently labeled probes were synthesized using amino allyl UTPs and Cy-3 and Cy-5 monoreactive dyes (Amersham Biosciences Ltd., England).
Atlas Glass 1.0K microarray analysis: The mRNA expression profile was evaluated by Atlas Glass microarrays (BD Clontech Inc. US, 1081 genes). Two hundred μL fluorescently labeled probes (mixture of the appropriate Cy3- and Cy5-labelled cDNA) were mixed with the prewarmed hybridization solution and hybridized to the microarray for 16 h at 50°C. Washing steps were done at room temperature in washing solutions (containing 0.75 mmol/L DTT) according to the manufacturer’s instructions. The slides were dried by blowing with carbone-dioxyde and were scanned by Axon GenePix4000B reader on 532 nm (Cy3) and 635 nm (Cy5) wavelengths.
Data analysis: Scanned arrays were evaluated by the GenePix Pro 4.1 software. Automated spot detection using local background determination was done and feature extraction (ratio of medians, ratio of means, Cy3/Cy5) was performed. Microarrrays with nonhomogeneous background and/or incomplete housekeeping gene spot set were removed from further analysis. Ratios of medians of the detected features were applied in the normalization. Means of the ratio of medians were normalised to be 1. Results were exported into Acquity 3.1 software (Axon Inc.) and datasets were established. Dataset selection into classification categories was performed. Mean, median, SD of each parameter was calculated. Underexpression was defined as ratio of means ± SD < 0.5, overexpression as ratio of means ± SD > 2.0. One-way (group) ANOVA and multivariate exploratory techniques (discriminant analysis, factor analysis and hierarchical cluster analysis) were performed by SAS 6.12 version statistical software. Hierarchical cluster analysis was done using Ward’s method (Euclidean distances). The colonic cases were clustered according to their expression pattern based on the filtered ANOVA results. Gene annotation and functional classification were done using Atlas Gene List Version 4.0. Functional analysis and visualization of biological association network were done using Pathway assist 2.53 software.
Validation: Real-time RT-PCR was used for validation of expression microarray results (Roche LightCycler). One-step RT-PCR was carried out using the LightCycler h-β2M Housekeeping Gene Set and RNA Master Hybridization Probes kit. For the relative quantification, commercially available β-2-microglobulin (142 bp length fragment) was used as a reference, and newly designed and synthesized epidermal growth factor receptor (EGFR) (227 bp length fragment) or DNA topoisomerase I (Top1) (184 bp length fragment) were used as a target gene. The following primer and hybridization probe sequences were used: EGFR9 S primer: 5’-atcctgccggtggcatt-3’, EGFR12 A primer: 5’-gttcaggctgacgactgca-3’, EGFR-FL probe: 5’-caggacggacctccatgcctttga-3’, EGFR-LC probe: 5’-LC Red 640-cctagaaatcatacgcggcaggacc-3’ in case of EGFR, and Top1-fw primer: 5’-acatcatgcttaaccctagttcac-3’, Top1-as primer: 5’-cagagcaagcttgtcgatg-3’, TOP1-FL probe: 5’-cggatcttgtccacacattttttcagc-3’, TOP1-LC probe: 5’-LC Red 640-ccgagcagtctcgtatttctgccag-3’ in case of DNA-topoisomerase-I. Evaluation of relative ratios (diseased/normal/same patient) was prepared using RelQuant software.
EGFR immunohistochemistry: Formalin fixed paraffin embedded 4 μm thick colonic biopsy tissue sections were dewaxed and rehydrated. Antigen unmasking was carried out by nuclease free Proteinase K digestion for 20 min at room temperature. After washing twice in PBS, endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 30 min at room temperature. After washing 3 times in PBS for 3 min, a specific blocking was done with 1% BSA-PBS solution for 10 min at room temperature. Then the slides were incubated with diluted EGFR culture supernatant antibody (1 μL EGFR antibody and 40 μL PBS) (Clone: H-11, DAKO) at 37°C for 60 min in a humidified chamber. After washing 3 times in PBS, signal conversion was carried out with the LSAB2 system (DAKO) according to the manufacturer’s instructions. Haematoxylin co-staining was done.
Ethical consideration: All routine colonic biopsy specimens from the patients were taken after informed consent and ethical permission was obtained for participation in the study.
RESULTS
Identification of commonly over-and underexpressed genes in colonic diseases
Genes that were up- and downregulated in at least 2/3 of cases per sample group in colorectal cancer , in adenoma and in IBD were considered as a commonly over- and underexpressed genes. CRC cases are characterized by upregulated genes in the DNA replication (such as replication protein A1, DNA topoisomerase IIα, DNA topoisomeraseI), cell cycle (including cyclin A1, cyclin-dependent kinase 10, protein NIMA-interacting 1), extracellular matrix remodeling (like keratin 5, perlecan, enactin), transcription regulation (such as IRF5, 6, and E74-like factor 2) , oncogenesis (including v-jun, BRCA2) and growth factor related cell proliferation (EGFR, VEGFB, hepatocyte growth factor, transforming growth factor β2) cell function groups; and downregulated genes in the DNA repair (RAD51 and 52 homolog), tumor suppression and apoptosis (like BCL2-interacting killer) cell function groups. Adenoma cases in comparison showed altered gene expression data in apoptosis (such as TNF receptor-associated factor 6, BAX), growth factors, receptors and their signal transduction (like calgranulin A, KIT ligand, Ran GTPase activating protein 1), oncogenesis and tumor suppression (including growth factor receptor-bound protein 10, p53-induced protein and betaglycan) functional groups. IBD cases are featured by the gene expression changes of immune regulation (including GM-CSF2, CXCL13, MMP-3, MMP-12 and interleukin 1 receptor antagonist), transport [like transferring and solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5], and growth factor-related cell proliferation (such as B cell growth factor 1, GM-CSF).
The most differentially expressed genes between the 3 sample groups in colon-filtered results of ANOVA
We analyzed the expression differences between all 1081 genes using ANOVA. The results of ANOVA were filtered according to the number of cases. The genes without enough data from all colonic samples were removed from the analysis. 19 genes were found to be significantly differently expressed (P < 0.05) between the colonic sample groups using filtered ANOVA method (Table 1).
Table 1.
The most differentially expressed genes between the 3 sample groups in the colon
| Name | GenBank ID | Differences Between sample groups | P | Number of valid cases/sample groups | Ratio of means1 in adenoma | Ratio of means1 in CRC | Ratio of means1 in IBD | Cell function |
| Adhesion | ||||||||
| Bystin-like | L36720 | 1-3 | 0.0005 | 5, 2, 2 | 0.612 ± 0.62676 | 1.47 ± 0.789131 | 7.3375 ± 1.134906 | Other cell adhesion |
| 2-3 | 0.0008 | proteins | ||||||
| Apoptosis | ||||||||
| Tumor necrosis factor | AF016267 | 1-2 | 0.0044 | 4, 4, 5 | 0.49075 ± 0.315247 | 2.7695 ± 1.308503 | 0.5032 ± 0.270649 | Death domain |
| receptor superfamily, | 2-3 | 0.0046 | receptors | |||||
| member10c, decoy | ||||||||
| without an intracellular domain | ||||||||
| Extracellular matrix, cytoskeleton | ||||||||
| Matrix metalloproteinase | X75308 | 2-3 | 0.0395 | 10, 6, 6 | 0.8075 ± 0.406413 | 0.557 ± 0.324015 | 1.222333 ± 0.558757 | Metallo |
| 13 (collagenase 3) | proteinases | |||||||
| Metabolism, blood coagulation | ||||||||
| Ubiquitin C | M26880 | 1-3 | 0.0325 | 10, 5, 5 | 0.9329 ± 0.687387 | 0.8456 ± 0.51739 | 2.3002 ± 1.110219 | Protein turnover |
| 2-3 | 0.0227 | |||||||
| Plasminogen activator, urokinase | M15476 | 1-3 | 0.0089 | 6, 5, 4 | 0.482167 ± 0.248489 | 1.0554 ± 0.524046 | 1.513 ± 0.417319 | Serine proteases |
| Oncogenes and tumor suppression | ||||||||
| AXL receptor | M76125 | 1-2 | 0.0209 | 7, 5, 4 | 0.258571 ± 0.237985 | 2.0162 ± 1.429626 | 1.3225 ± 0.780286 | Oncogenes and |
| Tyrosine kinase | umor | |||||||
| suppressors | ||||||||
| Ras homolog gene family, | Z35227 | 1-2 | 0.0076 | 7, 5, 3 | 0.806429 ± 0.732179 | 2.736 ± 1.083434 | 1.115 ± 0.249638 | Oncogenes and |
| member H | tumor suppressors | |||||||
| RAB5A, member | M28215 | 1-3 | 0.0151 | 8, 5, 5 | 0.5745 ± 0.438766 | 0.9958 ± 1.224974 | 2.263 ± 0.863959 | General trafficking |
| RAS oncogene family | ||||||||
| Receptors and ligands | ||||||||
| Small inducible cytokine | M21121 | 1-2 | 0.0087 | 8, 4, 6 | 0.706125 ± 0.402215 | 1.94025 ± 0.799844 | 1.0615 ± 0.371365 | Growth factors, |
| A5 (RANTES) | cytokines, and | |||||||
| chemokines | ||||||||
| Pleiotrophin (heparin | M57399 | 1-2 | 0.0373 | 5, 5, 2 | 0.3474 ± 0.242632 | 2.7236 ± 1.872401 | 0.477 ± 0.166877 | Growth factors, |
| binding growth factor 8) | cytokines, and | |||||||
| chemokines | ||||||||
| Patched (Drosophila) | U43148 | 1-3 | 0.0043 | 9, 5, 5 | 0.653667 ± 0.463297 | 0.7752 ± 0.448416 | 3.8946 ± 2.578342 | Growth factor and |
| homolog | 2-3 | 0.0058 | chemokine | |||||
| receptors | ||||||||
| Stem cell growth factor; | D86586 | 1-2 | 0.01755 | 5, 5, 4 | 0.3264 ± 0.168316 | 1.6386 ± 0.957384 | 0.5585 ± 0.420226 | Ggrowth factors, |
| lymphocyte secreted | cytokines, and | |||||||
| C-type lectin | chemokines | |||||||
| Interleukin 7 receptor | M29696 | 1-2 | 0.0096 | 8, 4, 4 | 0.40775 ± 0.12511 | 1.774 ± 1.113227 | 0.32975 ± 0.133809 | Interleukin and |
| 2-3 | 0.0066 | interferon | ||||||
| receptors | ||||||||
| Signal transduction | ||||||||
| Myotonic dystrophy | Y12337 | 1-2 | 0.0253 | 10, 6, 6 | 0.7611 ± 0.296331 | 1.375167 ± 0.301345 | 1.4415 ± 0.521753 | Intracellular |
| protein kinase like protein | 1-3 | 0.0131 | kinase | |||||
| network members | ||||||||
| Transcription | ||||||||
| Heat shock | M64673 | 1-2 | 0.0136 | 9, 6, 3 | 0.732444 ± 0.421368 | 1.7565 ± 0.747317 | 0.385333 ± 0.320644 | Transcription |
| transcription factor 1 | activators | |||||||
| and repressors | ||||||||
| Immediate early protein | M62831 | 2-3 | 0.0209 | 8, 5, 5 | 0.552875 ± 0.271426 | 0.4708 ± 0.276956 | 0.989 ± 0.255144 | Basic transcription |
| factors | ||||||||
| High-mobility group | M23619 | 1-3 | 0.0003 | 9, 6, 5 | 0.734556 ± 0.324623 | 0.897667 ± 0.371581 | 1.7982 ± 0.238334 | Chromatin |
| (nonhistone chromosomal) | 2-3 | 0.0011 | proteins | |||||
| protein isoforms I and Y | ||||||||
Cy3/Cy5 (disease/normal).
Factor analysis and discriminant analysis
Factor analysis was prepared on the basis of the results of variance analysis. Factor analysis resulted in two different factor groups. Factor 1 had the most considerable explorative variance value (6.331196), but the factor 2 also showed significant explorative variance values (4.710563). The factor analysis gives information about the functional gene groups which can differentiate the observed diseases according to their different expression levels (Table 2).
Table 2.
Results of the factor analysis
| STAT. factor analysis |
Factor loadings (Varimax raw) Extraction: Principal components (Marked loadings > 0.700000) |
Cell function | |
| Factor 1 | Factor 2 | ||
| Stem cell growth factor | 0.896313 | -0.053631 | Growth factors, cytokines,and chemokines |
| Pleiotrophin (heparin binding growth factor 8) | 0.882657 | 0.006966 | Growth factors, cytokines, and chemokines |
| Small inducible cytokine A5 (RANTES) | 0.802949 | 0.169587 | Growth factors, cytokines, and chemokines |
| Interleukin 7 receptor | 0.835349 | -0.151447 | Interleukin and interferon receptors |
| Tumor necrosis factor receptor superfamily, member 10c | 0.742897 | -0.145755 | Death domain receptors |
| Signal transducer and activator of transcription 2113 kDa | 0.820315 | 0.066954 | Transcription activators and repressors |
| Heat shock transcription factor 1 | 0.745691 | 0.032620 | Transcription activators and repressors |
| Patched (Drosophila) homolog | -0.113082 | 0.768421 | Growth factor and chemokine receptors |
| RAB5A, member RAS oncogene family | -0.044311 | 0.834146 | General trafficking |
| Ubiquitin C | -0.045668 | 0.719247 | Protein turnover |
| High-mobility group protein isoforms I and Y | -0.029882 | 0.943945 | Chromatin proteins |
| Explorative variance | 6.331196 | 4.710563 | |
According to the expression changes of the following genes the three colonic disease groups can be significanty distinguished: HSF1 (P = 0.012537), bystin-like (P = 0.001027), calgranulin A (P = 0.043831), and TNFR superfamily member 10c (P = 0.037888) (Figure 1).
Figure 1.
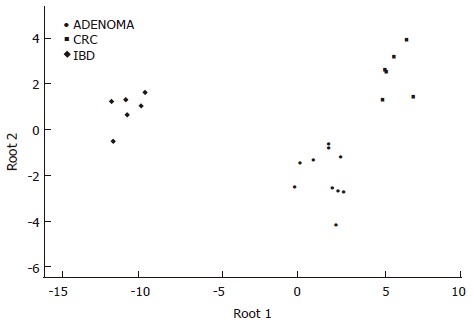
Discriminant analysis of colonic biopsy specimens. Note the clear separation of the single classification groups based on the discriminatory genes detailed in the results section.
Cluster analysis
Four different clusters can be identified: two IBD-related clusters containing one CRC case, a carcinoma group containing one adenoma and one ulcerative colitis case, and an adenoma cluster with nine adenoma cases (Figure 2). Tree diagram of 22 colonic cases showed considerable accordance with the conventional histopathological diagnoses. Excluding one case (AD1), all adenoma cases belong to a significantly distinct cluster. The cluster of severely inflammed IBD cases contained one colorectal adenocarcinoma case, as it showed a similar gene expression pattern. This observation could reflect that colorectal cancer can develop from chronic inflammation and some types of adenomas.
Figure 2.
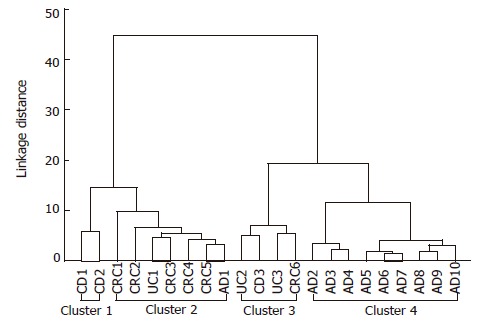
Tree diagram for 22 colonic diseases (Ward's method euclideandistances). Four different clusters can be identified: 1 and 3 are IBD-related clusters containing one CRC case; 2 is a carcinoma group containing one adenoma and one ulcerative colitis cases; 4 is an adenoma cluster with nine adenoma cases. AD: Adenoma; CD: Crohn’s disease; CRC: Colorectal cancer; UC: Ulcerative colitis.
Validation
All samples with remaining total RNA (9 CRCs, 2 adenomas and 3 CD samples), were tested using one-step EGFR and TOP1 real-time RT-PCR. Using β-2-microglobulin as a standard control, EGFR- and TOP1 mRNA levels were measured, and ratios of these concentrations to the standard were determined. Ratios for each disease group along with normal samples were compared, and the resulting relative ratios determined (diseased/normal in the same patient). The CRC samples showed higher EGFR mRNA level than the normal paired sample (relative ratio: 3.008, SD: 4.591), while there was no differential expression found in the adenoma samples (relative ratio: 0.772, SD: 0.060). Lower EGFR expression was measured in CD samples (relative ratio: 0.451, SD: 0.173) compared to the normal adjacent mucosa. Increased, 2.802-fold TOP1 mRNA level was evaluated in the CRC samples (SD: 3.884), but TOP1 level of the other sample groups was within the normal range (realtive ratio-adenoma: 0.774, SD: 0.0368; realtive ratio-CD: 0.942, SD: 0.337). The expression differences of EGFR and TOP1, found by microarray analysis, could be confirmed in most, but not in all cases. First, because of the limited total RNA amount in several of the small biopsy samples, the starting isolated total RNA was used in its entirety for probe synthesis. Secondly, differences between the two gene expression analysing method could be due to different gene sequences being amplified and detected during the real-time RT-PCR and the microarray analysis.
EGFR-immunohistochemistry
In colorectal carcinoma samples, EGFR was found to be mildly and moderately expressed in all carcinoma cells. Diffuse cytoplasmatic EGFR staining was found in all carcinoma samples. In normal samples, moderate and high EGFR expression was found in some epithelial cells, but in total amount, EGFR expression was decreased compared to carcinoma samples (Figure 3). EGFR protein expression data were in correlation with EGFR mRNA expression data from both microarray and real-time RT-PCR analysis.
Figure 3.
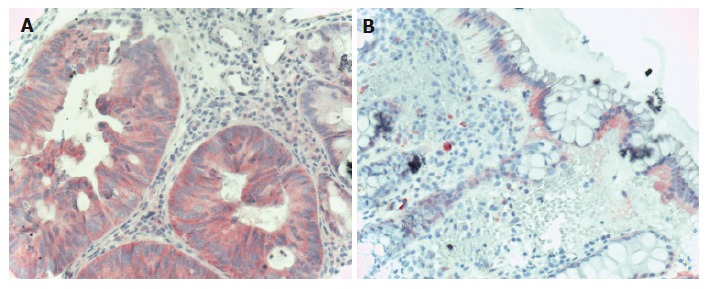
EGFR immunohistochemistry in CRC. A: Carcinomatous glands of the colon showing diffuse cytoplasmatic, moderately intensive EGFR staining (Hematoxylin co-staining × 200); B: Normal colonic epithelia showing mildly, moderately intensive basolateral intracytoplasmatic EGFR staining. The lower 2/3 of the crypts do not show EGFR positivity (Hematoxylin co-staining × 100).
DISCUSSION
We compared the gene expression pattern of biopsy samples from patients with adenoma, CRC and IBD, on Human Atlas Glass 1.0 microarrays containing 1081 target sequences. Biopsies were taken from disease-involved areas of the colon and surrounding disease-free colonic mucosa from the same patient. Multiple filtering methods for reduction of array-array variability origins from the spotted microarray hybridization procedure, and multiple statistical analyses were applied for finding colorectal carcinogenesis-associated genes, which can enhance the conventional histological diagnosis.
The Atlas Glass Human 1.0 microarray platform is a spotted microarray with each probe consisting of a single “long oligo” (80 mer) spotted on a glass slide. On the Atlas Glass microarray, all of the weight of detection rests on a single oligonucleotide. If cross-hybridization with an inappropriate target occurs, then the readout for that gene is incorrect. This could result in an erroneous quantification of mRNA levels[19,20] and relatively high standard deviation values. This main disadvantage of spotted opened system microarrays was apparent in our analyses: several hybridized microarrays (12 from 34) were removed from the further analysis after the quality testing, because of the problems mentioned above. Hence, comparison results from analysis of different microarray types, besides the facts mentioned above, requires due foresight, because of the different signal detection techniques, various sample amounts, sample types, sample processing and storage, and different experimental controls. The results are strongly affected by choice of biopsy samples or surgically resected tissue as a starting material, heterogeneous tissue samples or homogeneous cells such as laser microdissected cells. Our concept was that biopsies taken from patients during endoscopical examination with minimal intervention, are the most suitable samples for identifying early diagnostic target molecules.
CRC cases are characterized by upregulated genes in the DNA replication, cell cycle, extracellular matrix remodeling, transcription regulation, oncogenesis and growth factor related cell proliferation cell function groups; and downregulated genes in the DNA repair, tumor suppression and apoptosis cell function groups, while adenoma cases showed altered gene expression data in apoptosis, growth factors, receptors and their signal transduction, oncogenesis and tumor suppression functional groups. Despite the differences in samples and microarray types, there are genes, which were found differently expressed in our study that were also found commonly with other research groups. This fact emphasizes the importance of these CRC-related genes. Similarly to our findings, overexpression of several oncogenes (v-jun[20], BRCA2[8], growth factor genes (VEGF[22], HGF[23], TGFβ[5,9,24,25]), DNA replication and repair genes (TOP2A[8,26], tankyrase[9]), signal transduction gene (ephrin B2[27]), cell cycle genes (cell growth regulatory with zinc finger domain[8]) and transcription factors (several interferon regulatory factors[8]) was previously found by others using microarray analysis in gastrointestinal cancers.
Over- and underexpression of several apoptosis-, proliferation- and cell survival-related genes were observed in our microarray analysis (Table 1). These cell processes and their balance play a critical role in carcinogenesis and tumor progression. Functional analysis and visualization of biological association networks, especially pathways involved in apoptosis and cell survival were done using Pathway Assist 2.53 software. Figure 4 shows a graphical overview illustrating the role of the most important upregulated apoptosis-related genes. We found differently expressed genes in our study that are involved in or affected by not only one, but four different apoptotic pathways which are known in mammalian cells. These genes include TNFRSF10c (tumor necrosis factor receptor superfamily, member 10c) so called TRAILR3, TRAF6 (TNF receptor-associated factor 6), TNFRSF6 (Fas) and caspase 4 (CASP4) genes, downregulated apoptotic BIK (Bcl-2 interacting killer) and PP2A (protein phosphatase 2A). Two other genes that showed increased mRNA level in CRC, and were verified by real-time RT-PCR and/or immunohistochemistry, DNA topoisomerase I (TOP1) and EGFR (epidermal growth factor receptor), have a possible role in cell processes leading to CRC. Overexpression of EGFR correlates with CRC progression and metastatic potential[28]. EGFR activation can promote proliferation and maintain survival via MAPK (Ras/JNK), Ras/ERK and JAK/STAT signalling pathways. Amplification of receptor signaling by means of overexpression may promote tumor growth and resistance to apoptosis. Human TOP1 associated with intensive cell division and DNA replication occurring in the malignant cells.
Figure 4.
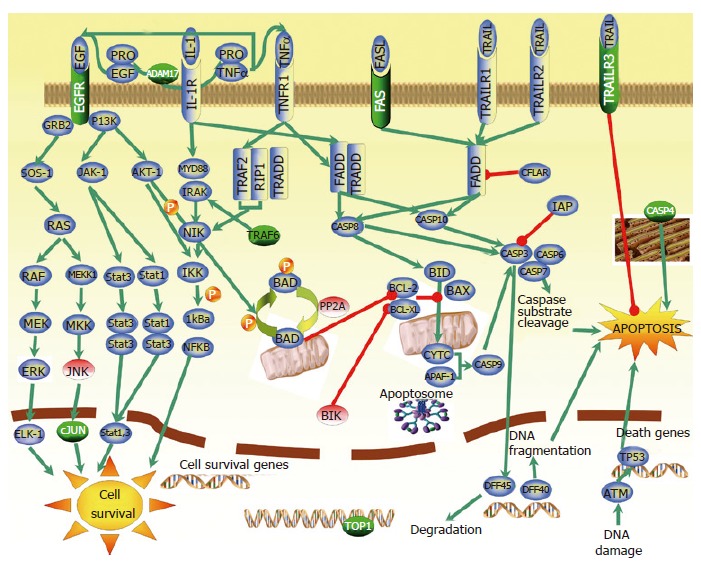
The main apoptotic and cell survival pathways identified in CRC gene expression study. Genes marked with green were found to be overexpressed, while genes marked with red were found to be underexpressed in our microarray analysis. Genes in blue are previously described as apoptotic and cell survival pathways-related genes. Green arrows refer to the positive regulation, while reds mean negative regulation (inhibition).
We also aimed to find genes that can enhance the molecular classification of CRC and its precancerous stages. Several genes from different functional groups were found to be overexpressed in both adenoma and CRC samples compared to adjacent normal mucosa, and the degree of upregulation of these genes follows the normal-adenoma-CRC sequence. Genes found to be important for this classification include cell survival promoting molecules (TRAF6, Grb10, ELF2, TACE), DNA synthesis and cell cycle involved molecules (RanGAP1, replication protein A1) and CFTR chloride channel. CFTR (cystic fibrosis transmembrane conductance regulator) is the most important chloride channel in the luminal membrane of the colon. A central role has been suggested for CFTR in coordinating electrolyte transport by changing absorption into secretion in colon carcinogenesis[29]. Recent studies demonstrate an enhanced cAMP-activated Cl- secretion in the hyperproliferative colonic mucosa that is caused by elevated CFTR expression[30].
IBD cases are featured by the gene expression changes of immune regulation, transport and growth factor-related cell proliferation in our study. In correlation with our findings, elevated chemokine (small inducible cytokine A4, interleukin-1 receptor antagonist) and matrix metalloprotease (MMP-3, MMP-12) mRNA levels were detected in IBD compared to normal mucosa by microarray analysis[16]. Small inducible protein A4 is highly expressed in IBD according to the degree of inflammation[31]. IL-1 receptor antagonists inhibit the activity of IL1 and modulate a variety of IL1-related immune and inflammatory responses. Programmed expression of MMPs is involved in tissue remodeling during inflammation, moreover MMP-12 may play a role in macrophage movement and epithelial cell shedding[32]. Several other growth factors and chemo attractants were found overexpressed in our microarray analyses. Secretion of GM-CSF2 (macrophage-granulocyte colony stimulating factor 2) is increased in mucosal lesions in IBD. B cell growth factor 1 released by T cells after antigen stimulation, and supports the clonal proliferation of B cells. CXCL13 B cell chemoattractant is mostly produced by the monocyte/macrophage lineage in UC[33].
Discrimination of colonic diseases according to the gene expression markers
Discriminant analysis shows the genes which can help us to classify an unknown sample into one of the groups of observed diseases, considering their expression levels. According to the expression changes of the following four genes the three colonic disease groups can be significantly distinguished: HSF1, calgranulin A, TNFRSF10c (TRAILR3) and bystin-like.
HSF1 is a heat-shock transcription factor, that was found to be upregulated in CRC compared to normal mucosa in both our and other research groups’ gene expression studies[34]. In adenoma and IBD cases it showed lower expression level compared to the CRC samples. Induction of HSF1 gene expression could activate the HSF1 heat shock stress signal pathway in sporadic CRC. The heat shock stress signaling pathway is highly involved in carcinogenesis since heat shock proteins are responsible for maintaining the conformation, stability and function of key oncogenic client proteins involved in signal transduction pathways leading to proliferation, cell cycle progression and apoptosis, as well as other features of the malignant phenotype such as invasion, angiogenesis and metastasis[35-37].
Calgranulin A (S100A8) is a calcium-binding protein that showed higher expression in the IBD sample group, than in CRC and adenoma cases. S100 proteins are involved in the regulation of a number of cellular processes such as cell cycle progression, differentiation and immune response[38].
Significantly increased TRAILR3 level was found in CRC samples, but not in IBD and adenoma cases. This receptor inhibits the TRAIL-induced apoptosis via binding TRAIL ligand which in this case cannot interact with the pro-apoptotic, death domain containing other TRAIL receptors[39,40].
Interestingly, significant bystin-like 7-fold mRNA overexpression was found in IBD samples in our microarray analysis. Suzuki et al[41] identified a cytoplasmic protein, named bystin, that directly binds trophinin and tastin cell adhesion molecules that are involved in the process of the embryo implantation. A role for bystin-like protein in inflammation has not been described.
The factor analysis gives information about the functional gene groups which can differentiate the observed diseases according to their different expression levels. Five of the seven genes (HSF1, TRAILR3, stem cell growth factor, interleukin-7 receptor and pleiotrophin) with the most considerable explorative variance value (belong to Factor 1) were mentioned previously in the scientific literature as cell proliferation and cancer related genes (HSF1[34-37], TRAILR3[39,40], stem cell growth factor[42], interleukin-7 receptor[43] and pleiotrophin[44,45]).
Twenty two colonic biopsy samples were clustered in correlation with the conventional histopathological diagnoses. Excluding one case, all adenoma cases belong to a significantly distinct cluster. The cluster of severely inflammed IBD cases contained one colorectal adenocarcinoma case, as it showed a similar gene expression pattern. This fact can refer that colorectal cancer can develop on the basis of chronic inflammation and some types of adenomas.
In summary, we can say that the overall mid-size glass arrays are suitable for identification of disease-specific genes which are considered as gene expression markers. Detection of the mRNA expression levels of marker gene panels gives an opportunity for classification of colonic samples, even in the case of small biopsy specimens. The limited starting RNA amount arising from the small sample size makes microarray analysis difficult. The standardization of the opened manual microarray systems is more difficult; however, automatization of this technique would further improve the efficacy, reproducibility and quality of microarray analysis.
ACKNOWLEDGMENTS
We thank C Lofton-Day, PhD and V Galamb PhD for critical reading of this manuscript.
Footnotes
Supported by Epigenomics Inc
S- Editor Wang GP L- Editor Alpini GD E- Editor Lu W
References
- 1.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galamb O, Sipos F, Fischer K, Tulassay Z, Molnar B. The results of the expression array studies correlate and enhance the known genetic basis of gastric and colorectal cancer. Cytometry B Clin Cytom. 2005;68:1–17. doi: 10.1002/cyto.b.20069. [DOI] [PubMed] [Google Scholar]
- 3.Langmann T, Moehle C, Mauerer R, Scharl M, Liebisch G, Zahn A, Stremmel W, Schmitz G. Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26–40. doi: 10.1053/j.gastro.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Okahara S, Arimura Y, Yabana T, Kobayashi K, Gotoh A, Motoya S, Imamura A, Endo T, Imai K. Inflammatory gene signature in ulcerative colitis with cDNA macroarray analysis. Aliment Pharmacol Ther. 2005;21:1091–1097. doi: 10.1111/j.1365-2036.2005.02443.x. [DOI] [PubMed] [Google Scholar]
- 5.Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, Nita ME, Takagi T, Nakamura Y, Tsunoda T. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544–3549. [PubMed] [Google Scholar]
- 6.Lin YM, Furukawa Y, Tsunoda T, Yue CT, Yang KC, Nakamura Y. Molecular diagnosis of colorectal tumors by expression profiles of 50 genes expressed differentially in adenomas and carcinomas. Oncogene. 2002;21:4120–4128. doi: 10.1038/sj.onc.1205518. [DOI] [PubMed] [Google Scholar]
- 7.Backert S, Gelos M, Kobalz U, Hanski ML, Böhm C, Mann B, Lövin N, Gratchev A, Mansmann U, Moyer MP, et al. Differential gene expression in colon carcinoma cells and tissues detected with a cDNA array. Int J Cancer. 1999;82:868–874. doi: 10.1002/(sici)1097-0215(19990909)82:6<868::aid-ijc16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 8.Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, Fleming J, Tavana D, Frenkel E, Becerra C. Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res. 2003;9:931–946. [PubMed] [Google Scholar]
- 9.Kwon HC, Kim SH, Roh MS, Kim JS, Lee HS, Choi HJ, Jeong JS, Kim HJ, Hwang TH. Gene expression profiling in lymph node-positive and lymph node-negative colorectal cancer. Dis Colon Rectum. 2004;47:141–152. doi: 10.1007/s10350-003-0032-7. [DOI] [PubMed] [Google Scholar]
- 10.Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 11.Mori Y, Yin J, Sato F, Sterian A, Simms LA, Selaru FM, Schulmann K, Xu Y, Olaru A, Wang S, et al. Identification of genes uniquely involved in frequent microsatellite instability colon carcinogenesis by expression profiling combined with epigenetic scanning. Cancer Res. 2004;64:2434–2438. doi: 10.1158/0008-5472.can-03-3508. [DOI] [PubMed] [Google Scholar]
- 12.Birkenkamp-Demtroder K, Christensen LL, Olesen SH, Frederiksen CM, Laiho P, Aaltonen LA, Laurberg S, Sørensen FB, Hagemann R, ØRntoft TF. Gene expression in colorectal cancer. Cancer Res. 2002;62:4352–4363. [PubMed] [Google Scholar]
- 13.Frederiksen CM, Knudsen S, Laurberg S, Ørntoft TF. Classification of Dukes' B and C colorectal cancers using expression arrays. J Cancer Res Clin Oncol. 2003;129:263–271. doi: 10.1007/s00432-003-0434-x. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal D, Chen T, Irby R, Quackenbush J, Chambers AF, Szabo M, Cantor A, Coppola D, Yeatman TJ. Osteopontin identified as colon cancer tumor progression marker. C R Biol. 2003;326:1041–1043. doi: 10.1016/j.crvi.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 16.Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445–456. doi: 10.1093/hmg/10.5.445. [DOI] [PubMed] [Google Scholar]
- 17.Sun XR, Wang J, Garner D, Palcic B. Detection of cervical cancer and high grade neoplastic lesions by a combination of liquid-based sampling preparation and DNA measurements using automated image cytometry. Cell Oncol. 2005;27:33–41. doi: 10.1155/2005/981612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molnar B, Szentirmay Z, Bodo M, Sugar J, Feher J. Application of multivariate, fuzzy set and neural network analysis in quantitative cytological examinations. Anal Cell Pathol. 1993;5:161–175. [PubMed] [Google Scholar]
- 19.Rogojina AT, Orr WE, Song BK, Geisert EE Jr. Comparing the use of Affymetrix to spotted oligonucleotide microarrays using two retinal pigment epithelium cell lines. Mol Vis. 2003;9:482–496. [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 21.Mori M, Mimori K, Yoshikawa Y, Shibuta K, Utsunomiya T, Sadanaga N, Tanaka F, Matsuyama A, Inoue H, Sugimachi K. Analysis of the gene-expression profile regarding the progression of human gastric carcinoma. Surgery. 2002;131:S39–S47. doi: 10.1067/msy.2002.119292. [DOI] [PubMed] [Google Scholar]
- 22.Okuno K, Yasutomi M, Nishimura N, Arakawa T, Shiomi M, Hida J, Ueda K, Minami K. Gene expression analysis in colorectal cancer using practical DNA array filter. Dis Colon Rectum. 2001;44:295–299. doi: 10.1007/BF02234309. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012–7017. [PubMed] [Google Scholar]
- 24.Birkenkamp-Demtroder K, Olesen SH, Sørensen FB, Laurberg S, Laiho P, Aaltonen LA, Orntoft TF. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut. 2005;54:374–384. doi: 10.1136/gut.2003.036848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandres E, Catalan V, Sola I, Honorato B, Cubedo E, Cordeu L, Andion E, Escalada A, Zarate R, Salgado E, et al. Dysregulation of apoptosis is a major mechanism in the lymph node involvement in colorectal carcinoma. Oncol Rep. 2004;12:287–292. [PubMed] [Google Scholar]
- 26.El-Rifai W, Frierson HF Jr, Harper JC, Powell SM, Knuutila S. Expression profiling of gastric adenocarcinoma using cDNA array. Int J Cancer. 2001;92:832–838. doi: 10.1002/ijc.1264. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radinsky R, Risin S, Fan D, Dong Z, Bielenberg D, Bucana CD, Fidler IJ. Level and function of epidermal growth factor receptor predict the metastatic potential of human colon carcinoma cells. Clin Cancer Res. 1995;1:19–31. [PubMed] [Google Scholar]
- 29.Kunzelmann K, Mall M. Electrolyte transport in the mammalian colon: mechanisms and implications for disease. Physiol Rev. 2002;82:245–289. doi: 10.1152/physrev.00026.2001. [DOI] [PubMed] [Google Scholar]
- 30.Umar S, Scott J, Sellin JH, Dubinsky WP, Morris AP. Murine colonic mucosa hyperproliferation. I. Elevated CFTR expression and enhanced cAMP-dependent Cl(-) secretion. Am J Physiol Gastrointest Liver Physiol. 2000;278:G753–G764. doi: 10.1152/ajpgi.2000.278.5.G753. [DOI] [PubMed] [Google Scholar]
- 31.Banks C, Bateman A, Payne R, Johnson P, Sheron N. Chemokine expression in IBD. Mucosal chemokine expression is unselectively increased in both ulcerative colitis and Crohn's disease. J Pathol. 2003;199:28–35. doi: 10.1002/path.1245. [DOI] [PubMed] [Google Scholar]
- 32.Vaalamo M, Karjalainen-Lindsberg ML, Puolakkainen P, Kere J, Saarialho-Kere U. Distinct expression profiles of stromelysin-2 (MMP-10), collagenase-3 (MMP-13), macrophage metalloelastase (MMP-12), and tissue inhibitor of metalloproteinases-3 (TIMP-3) in intestinal ulcerations. Am J Pathol. 1998;152:1005–1014. [PMC free article] [PubMed] [Google Scholar]
- 33.Carlsen HS, Baekkevold ES, Morton HC, Haraldsen G, Brandtzaeg P. Monocyte-like and mature macrophages produce CXCL13 (B cell-attracting chemokine 1) in inflammatory lesions with lymphoid neogenesis. Blood. 2004;104:3021–3027. doi: 10.1182/blood-2004-02-0701. [DOI] [PubMed] [Google Scholar]
- 34.Cen H, Zheng S, Fang YM, Tang XP, Dong Q. Induction of HSF1 expression is associated with sporadic colorectal cancer. World J Gastroenterol. 2004;10:3122–3126. doi: 10.3748/wjg.v10.i21.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochel HJ, Gademann G. Heat-shock protein 90: potential involvement in the pathogenesis of malignancy and pharmacological intervention. Onkologie. 2002;25:466–473. doi: 10.1159/000067442. [DOI] [PubMed] [Google Scholar]
- 36.Blagosklonny MV. Hsp-90-associated oncoproteins: multiple targets of geldanamycin and its analogs. Leukemia. 2002;16:455–462. doi: 10.1038/sj.leu.2402415. [DOI] [PubMed] [Google Scholar]
- 37.Witkin SS. Heat shock protein expression and immunity: relevance to gynecologic oncology. Eur J Gynaecol Oncol. 2001;22:249–256. [PubMed] [Google Scholar]
- 38.Roth J, Vogl T, Sorg C, Sunderkötter C. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 2003;24:155–158. doi: 10.1016/s1471-4906(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz de Almodóvar C, Ruiz-Ruiz C, Rodríguez A, Ortiz-Ferrón G, Redondo JM, López-Rivas A. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) decoy receptor TRAIL-R3 is up-regulated by p53 in breast tumor cells through a mechanism involving an intronic p53-binding site. J Biol Chem. 2004;279:4093–4101. doi: 10.1074/jbc.M311243200. [DOI] [PubMed] [Google Scholar]
- 40.Shivapurkar N, Toyooka S, Toyooka KO, Reddy J, Miyajima K, Suzuki M, Shigematsu H, Takahashi T, Parikh G, Pass HI, et al. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer. 2004;109:786–792. doi: 10.1002/ijc.20041. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki N, Zara J, Sato T, Ong E, Bakhiet N, Oshima RG, Watson KL, Fukuda MN. A cytoplasmic protein, bystin, interacts with trophinin, tastin, and cytokeratin and may be involved in trophinin-mediated cell adhesion between trophoblast and endometrial epithelial cells. Proc Natl Acad Sci USA. 1998;95:5027–5032. doi: 10.1073/pnas.95.9.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiraoka A, Yano Ki K, Kagami N, Takeshige K, Mio H, Anazawa H, Sugimoto S. Stem cell growth factor: in situ hybridization analysis on the gene expression, molecular characterization and in vitro proliferative activity of a recombinant preparation on primitive hematopoietic progenitor cells. Hematol J. 2001;2:307–315. doi: 10.1038/sj.thj.6200118. [DOI] [PubMed] [Google Scholar]
- 43.Cosenza L, Gorgun G, Urbano A, Foss F. Interleukin-7 receptor expression and activation in nonhaematopoietic neoplastic cell lines. Cell Signal. 2002;14:317–325. doi: 10.1016/s0898-6568(01)00245-5. [DOI] [PubMed] [Google Scholar]
- 44.Klomp HJ, Zernial O, Flachmann S, Wellstein A, Juhl H. Significance of the expression of the growth factor pleiotrophin in pancreatic cancer patients. Clin Cancer Res. 2002;8:823–827. [PubMed] [Google Scholar]
- 45.Souttou B, Juhl H, Hackenbruck J, Röckseisen M, Klomp HJ, Raulais D, Vigny M, Wellstein A. Relationship between serum concentrations of the growth factor pleiotrophin and pleiotrophin-positive tumors. J Natl Cancer Inst. 1998;90:1468–1473. doi: 10.1093/jnci/90.19.1468. [DOI] [PubMed] [Google Scholar]


