Abstract
AIM: To study the blood coagulation response after partial hepatectomy (PH) at transcriptional level.
METHODS: After PH of rats, the associated genes with blood coagulation were obtained through reference to the databases, and the gene expression changes in rat regenerating liver were analyzed by the Rat Genome 230 2.0 array.
RESULTS: It was found that 107 genes were associated with liver regeneration. The initially and totally expressing gene numbers occurring in initiation phase of liver regeneration (0.5-4 h after PH), G0/G1 transition (4-6 h after PH), cell proliferation (6-66 h after PH), cell differentiation and structure-function reconstruction (66-168 h after PH) were 44, 11, 58, 7 and 44, 33, 100, 71 respectively, showing that the associated genes were mainly triggered in the forepart and prophase, and worked at different phases. According to their expression similarity, these genes were classified into 5 groups: only up-, predominantly up-, only down-, predominantly down-, up- and down-regulation, involving 44, 8, 36, 13 and 6 genes, respectively, and the total times of their up- and down-regulation expression were 342 and 253, respectively, demonstrating that the number of the up-regulated genes was more than that of the down- regulated genes. Their time relevance was classified into 15 groups, showing that the cellular physiological and biochemical activities were staggered during liver regeneration. According to gene expression patterns, they were classified into 29 types, suggesting that their protein activities were diverse and complex during liver regeneration.
CONCLUSION: The blood coagulation response is enhanced mainly in the forepart, prophase and anaphase of liver regeneration, in which the response in the forepart, prophase of liver regeneration can prevent the bleeding caused by partial hepatectomy, whereas that in the anaphase contributes to the structure-function reorganization of regenerating liver. In the process, 107 genes associated with liver regeneration play an important role.
Keywords: Partial hepatectomy, Rat Genome 230 2.0 array, Blood coagulation response, Genes associated with liver regeneration
INTRODUCTION
The liver is the main site where coagulation factors are synthesized[1]. Tissue damage is often companied with angiorrhexis, bleeding and blood coagulation. Blood coagulation is a complex hemostatic process in which zymogens convert into coagulation factors, and promote blood coagulation at the site of wound[2]. This process is classified into extrinsic pathway and intrinsic pathway[3]. A proportion of coagulation factors of the former originate from blood, while that of the latter all from blood. Blood coagulation abnormity can lead to hereditary prothrombin deficiency, vitamin K deficiency, haemophilia and hepatic coagulation defects[4,5].
Partial hepatectomy (PH)[6] causes blood vessel injury and bleeding. Meanwhile, the remnant hepatocytes activated by it enter into cell cycle to compensate for the lost liver tissue, which process is called liver regeneration (LR)[7,8]. Generally, based on the physiological activities of the cells, the regeneration process is classified into 4 phases: the initiation (0.5-4 h after PH), the transition from G0 to G1 (4-6 h after PH), the cell proliferation (6-66 h after PH), the cell differentiation and structure-function reorganization (66-168 h after PH)[9]. According to time course, it is also divided into 4 phases including forepart (0.5-4 h after PH), prophase (6-12 h after PH), metaphase (16-66 h after PH), and anaphase (72-168 h after PH)[10], which are regulated by many factors including blood coagulation response[11]. To study the relationship between the blood coagulation response after PH and liver regeneration at transcriptional level[10,12], we examined the expression changes of genes in regenerating liver after partial hepatectomy by Rat Genome 230 2.0 array[13] containing 174 blood coagulation-associated genes. One hundred and seven genes were identified which were associated with liver regeneration[14], and we primarily studied the expression changes, patterns and functions of these genes.
MATERIALS AND METHODS
Regenerating liver preparation
Healthy SD rats weighing 200-250 g were obtained from the Animal Center of Henan Normal University. The rats were separated into groups at random and each group included 6 rats (male:female = 1:1). PH was performed according to Higgins and Anderson[6]: the left and middle lobes of the liver were removed. Rats were sacrificed by cervical vertebra dislocation at 0.5, 1, 2, 4, 6, 8, 12, 24, 36, 54, 66, 72, 120, 144 and 168 h after PH and the regenerating livers were examined at corresponding time points. The livers were rinsed three times in PBS at 4°C. Then 100-200 mg liver tissues from middle parts of the right lobe, six samples of each group were gathered and mixed together to 1-2 g (0.1-0.2 g × 6) of total liver tissue, then stored at -80°C. The sham-operation (SO) groups were treated the same with partial hepatectomy ones except that the liver lobes were not removed. The guidelines of animal protection of China were followed strictly.
RNA isolation and purification
Total RNA was isolated from the frozen livers according to the manual of Trizol kit (Invitrogen)[15] and then purified based on the guide of RNeasy mini kit (Qiagen)[16]. In brief, total liver tissues frozen at -80°C were homogenized liquid nitrogen, and homogenates were split in TRIzol reagent, followed by chloroform extraction and isopentyl alcohol precipitation. The total RNA integrity was confirmed by agarose electrophoresis and checked by the ratio of 28S to 18S. Total RNA concentration and purity were estimated by optical density measurements at 260/280 nm[17].
cDNA and cRNA synthesis and purification
As template, 0.4 ng total RNA was used for cDNA synthesis. cDNA purification was proceeded based on the methods established by Affymetrix[18]. cRNA labeled with biotin was synthesized using cDNA as the template and then purified[18]. Measurement of cDNA, cRNA concentration and purity were the same as above.
cRNA fragmentation and microarray detection
Fifteen microliter (1 μg/μL) cRNA incubated with 5 × fragmentation buffer at 94°C for 35 min was digested into 35-200 bp fragments. The hybridization buffer was added to the prehybridized Rat Genome 230 2.0 microarray produced by Affymetrix, then hybridization was carried out for 16 h at 45°C on a rotary mixer at 60 rotation/min. The microarray was washed and stained by GeneChip fluidics station 450 (Affymetrix Inc., USA). The chips were scanned by GeneChip Scan 3000 (Affymetrix Inc., USA), and the signal values of gene expression were observed[13].
Microarray data analysis
Signal values were quantified and normalized with GCOS1.2 software. Quantified signal intensities were obtained by deducting foreground signal values. Signal intensities were replaced by 200 when they were < 200. When experiment/control (Ri) was between 0.1 and 10, Ri was taken as natural logarithms to generate lnRi, and the normalize coefficient factor (ND) was taken by averaged Ri. The modified signal values were generated by ND multiplying control, and were replaced by 200 when it was < 200[13].
Normalization of microarray data
To minimize error from the microarray analysis, each analysis was performed three times. Results whose total ratio was maximal (Rm) and that whose average value of three housekeeping genes (β-actin, hexokinase and glyseraldehyde-3-phosphate dehydrogenase) approached 1.0 (Rh) were taken as a reference. The modified data were generated using a correction factor (Rm/Rh) multiplying the ratio of every gene in Rh at each time point. To remove spurious gene expression changes resulting from errors in the microarray analysis, the gene expression profiles at 0-4 h, 6-12 h and 12-24 h after PH were reorganized with NAP software (normalization analysis program) according to the cell cycle progression of the regenerating hepatocytes. Data statistics and cluster analysis were done using GeneMath, GeneSpring, and Microsoft Excel software[13,19,20].
Identification of genes associated with liver regeneration
Firstly, the nomenclature of blood coagulation was adopted from the GENEONTOLOGY database (www.geneontology.org), and input into blood coagulation at NCBI (www.ncbi.nlm.nih.gov) and RGD (rgd.mcw.edu) to identify the rat, mouse and human genes associated with the blood coagulation. According to maps of biological pathways embodied by GENMAPP (www.genmapp.org), KEGG (www. genome.jp/kegg/pathway.html#amino) and BIOCARTA (www.biocarta.com/genes/index.asp), the genes associated with blood coagulation were collated. The results of the analysis were codified, and compared with the results from human and mouse studies in order to identify human and mouse genes which are different from those of rat. In comparison of these genes with the analysis output of the Rat Genome 230 2.0 array, those genes which showed a greater than twofold change in expression level, regarded as meaningful[14] rat homologous genes, were referred to as rat specific genes associated with blood coagulation response under evaluation. Genes, which displayed reproducible results on three independent analyses with the chip and which showed a greater than twofold change in expression level at least at one time point during liver regeneration with significant difference (0.01 ≤ P < 0.05) or extremely significant difference (P ≤ 0.01) between PH and SO, were referred to as associated with liver regeneration.
RESULTS
Expression changes associated with blood coagulation response during liver regeneration
According to the databases at NCBI, GENMAPP, KEGG, BIOCARTA and RGD, 357 genes were involved in blood coagulation, in which, 174 genes were contained in the Rat Genome 230 2.0 array. Among them, the expression of 107 genes displayed meaningful changes at least at one time point after PH, showed significant or extremely significant differences in expression between PH and SO, and displayed reproducible results at three detections with Rat Genome 230 2.0 array, suggesting that the genes were associated with LR. Fold changes in up-regulation range were more than 2 to 46 fold compared with control, and down-regulation more than 2 to 10 fold (Table 1). The analysis indicated that 44 genes were up-regulated, 36 genes down-, and 27 genes up/down- during liver regeneration. The total up- and down-regulation times were 342 and 253, respectively (Figure 1A). At the initiation phase (0.5-4 h after PH), 24 genes displayed up-regulation, 14 genes down, and 2 genes up/down; at the transition phase from G0 to G1 (4-6 h after PH), 24 genes up, and 8 genes down; at the cell proliferation phase (6-66 h after PH), 45 genes up, 42 genes down, and 11 genes up/down; at cell differentiation and structure-function reorganization phase (66-168 h after PH), 38 genes up, and 27 genes down, and 5 genes up/down (Figure 1B).
Table 1.
Expression abundance of 107 blood coagulation response-associated genes during rat liver regeneration
| Gene | Associated | Fold | Gene | Associated | Fold | Gene | Associated | Fold | Gene | Associated | Fold |
| Abbr. | with | difference | Abbr. | with | difference | Abbr. | with | difference | Abbr. | with | difference |
| C2 | 1 | 2.1 | Anxa8 | 1, 2 | 0.4 | Fbn1 | 1, 2 | 2.8 | Rab27a | 1, 2 | 3.4 |
| Cd59 | 1 | 0.5 | Anxa9 | 1, 2 | 4.6 | Fgb | 1, 2 | 4.3 | Scube1 | 1, 2 | 3.2 |
| Coch | 1 | 0.2, 2.3 | App | 1, 2 | 6.4 | Fgl2 | 1, 2 | 3.1 | Tbxa2r | 1, 2 | 0.2, 2.0 |
| Daf1 | 1 | 0.2 | B4galt1 | 1, 2 | 3.1 | Fli1 | 1, 2 | 4.9 | Tm4sf3 | 1, 2 | 3.6 |
| Hrg | 1 | 0.5, 2.8 | Bdkrb2 | 1, 2 | 0.4 | G7c | 1, 2 | 3.9 | 3Tnf | 1, 2 | 3.2 |
| Il1b | 1 | 0.4 | C1qA | 1, 2 | 0.3 | Ggcx | 1, 2 | 0.3, 2.6 | Trove2 | 1, 2 | 0.4 |
| 3Il6 | 1 | 0.3, 6.1 | C1qr1 | 1, 2 | 5.5 | Gna12 | 1, 2 | 2.5 | 3A2m | 2 | 0.4, 46.2 |
| Lman1 | 1 | 2.0 | C3 | 1, 2 | 0.2 | Gnaq | 1, 2 | 2.5 | Adora2a | 2 | 0.5, 2.0 |
| Masp1 | 1 | 3.0 | C3ar1 | 1, 2 | 2.3 | 3Hgf | 1, 2 | 0.4 | C8g | 2 | 0.4, 2.0 |
| Mbl2 | 1 | 0.2 | C4a | 1, 2 | 0.5 | 3Hnf4a | 1, 2 | 0.1, 4.5 | Cfh | 2 | 2.5 |
| Nfe2 | 1 | 0.1 | C4bpa | 1, 2 | 2.0 | Hs6st2 | 1, 2 | 0.1, 2.6 | Crp | 2 | 0.5, 2.0 |
| Ptgdr | 1 | 0.3, 2.0 | C5r1 | 1, 2 | 0.4, 2.6 | Itgb3 | 1, 2 | 0.2 | Cspg2 | 2 | 0.4, 6.8 |
| Ptger2 | 1 | 0.5 | Cd36 | 1, 2 | 0.1 | Klkb1 | 1, 2 | 0.4 | Ctrl | 2 | 0.2, 4.6 |
| Ptgs1 | 1 | 3.4 | Cfi | 1, 2 | 6.4 | Kng1 | 1, 2 | 2.1 | Dcbld2 | 2 | 0.5, 4.3 |
| Ptgs2 | 1 | 0.1, 2.1 | Clca2 | 1, 2 | 0.5 | Kptn | 1, 2 | 0.2 | Gc | 2 | 3.4 |
| Prss1 | 1 | 0.1 | Clca3 | 1, 2 | 0.2 | Mmrn1 | 1, 2 | 0.1 | Lrp1 | 2 | 0.1, 2.0 |
| Prss2 | 1 | 0.4, 39.4 | Clca4 | 1, 2 | 0.3, 5.7 | P2rx1 | 1, 2 | 9.7 | Phyh | 2 | 2.3 |
| Serpind1 | 1 | 0.1 | Cr2 | 1, 2 | 6.0 | P2ry12 | 1, 2 | 0.1 | Proc | 2 | 0.3, 2.0 |
| Tf | 1 | 2.7 | Crry | 1, 2 | 2.4 | P2y12 | 1, 2 | 0.2 | Procr | 2 | 6.5 |
| Tfpi | 1 | 4.4 | Edn1 | 1, 2 | 0.4, 2.6 | Pabpc4 | 1, 2 | 0.3, 2.1 | Pros1 | 2 | 2.1 |
| Tfpi2 | 1 | 0.4 | Efemp2 | 1, 2 | 0.5, 2.4 | Plat | 1, 2 | 0.4, 4.9 | Serpina5 | 2 | 0.1, 7.8 |
| Tp53 | 1 | 2.9 | Entpd2 | 1, 2 | 0.4 | Plau | 1, 2 | 0.4, 3.0 | Serpinb9 | 2 | 5.3 |
| Anxa2 | 1, 2 | 4.5 | F10 | 1, 2 | 0.5 | Plaur | 1, 2 | 13.9 | Serpine1 | 2 | 16.7 |
| Anxa3 | 1, 2 | 3.9 | F2 | 1, 2 | 0.3 | Plg | 1, 2 | 2.1 | Serpinf2 | 2 | 0.2, 2.0 |
| Anxa4 | 1, 2 | 2.0 | F2rl2 | 1, 2 | 0.2 | Plscr1 | 1, 2 | 7.5 | Serpini2 | 2 | 0.4, 7.0 |
| Anxa5 | 1, 2 | 2.3 | F3 | 1, 2 | 0.1, 2.0 | Ppbp | 1, 2 | 0.1, 2.1 | 3Thbd | 2 | 9.6 |
| Anxa7 | 1, 2 | 6.8 | F5 | 1, 2 | 0.5 | Rab11fip1 | 1, 2 | 0.3, 8.6 |
1: Extrinsic blood coagulation pathway; 2: Intrinsic blood coagulation pathway;
Reported genes associated with liver regeneration.
Figure 1.
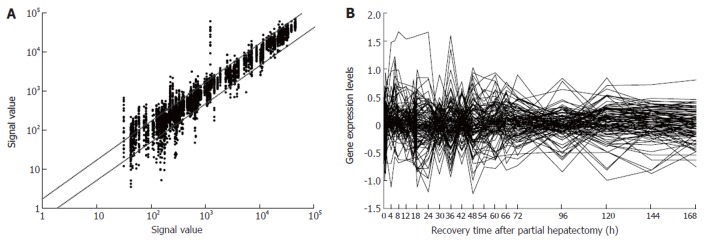
Expression frequency, abundance and changes of 107 blood coagulation response-associated genes during rat liver regeneration. Detection data of Rat Genome 230 2.0 array were analyzed and graphed with Microsoft Excel. A: Gene expression frequency. The dots above bias represent the genes up-regulated more than two fold, and total times of up-regulation were 342; those below bias down-regulated more than two fold, and times of down-regulation were 253; and the ones between biases no-sense alteration; B: Gene expression abundance and changes. Seventy-six genes were 2-46 fold up-regulated, and 62 genes 2-10 fold down- regulated.
Initial expression time points associated with blood coagulation response during liver regeneration
At each time point of liver regeneration, the numbers of initial up-, down-regulated and total up-, down-regulated genes are as follows in sequence: both 9 and 6 at 0.5 h; 6, 10 and 12, 14 at 1 h; 8, 0 and 16, 3 at 2 h; 4, 1 and 17, 4 at 4 h; 4, 2 and 21, 7 at 6 h; 1, 0 and 13, 5 at 8 h; 0, 5 and 12, 10 at 12 h; 7, 7 and 15, 13 at 16 h; 5, 8 and 17, 24 at 18 h; 3, 0 and 18, 16 at 24 h; 2, 3 and 11, 11 at 30 h; 0, 4 and 23, 18 at 36 h; 1, 1 and 9, 9 at 42 h; 1, 2 and 22, 24 at 48 h; 0, 0 and 12, 13 at 54 h; 0, 0 and 22, 11 at 60 h; 2, 0 and 21, 9 at 66 h; 0, 1 and 15, 11 at 72 h; 0, 0 and 11, 10 at 96 h; 4, 0 and 13, 9 at 120 h; 0, 0 and 15, 9 at 144 h; 0, 0 and 12, 13 at 168 h (Figure 2). Generally, gene expression changes occurred during the whole liver regeneration, with the up- and down-regulation times of 342 and 253, respectively. The initially up-regulated genes were predominantly expressed in the forepart, and the down-regulated genes in the prophase and metaphase, whereas there was little initial expression in the anaphase.
Figure 2.
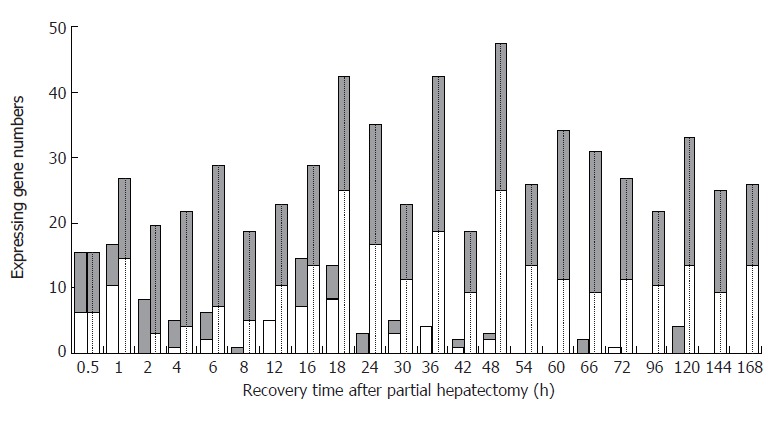
The initial and total expression profiles of 107 blood coagulation response-associated genes at each time point of liver regeneration. Grey bars: Up-regulated gene; White bars: Down-regulated. Blank bars represent initial expressing genes, in which up-regulated genes were predominant in the forepart, and the down-regulated genes in the prophase and metaphase, whereas there was little initial expression in the anaphase. Dotted bars represent the total expressing genes, in which some genes were up-regulated, and the others down regulated during the whole LR.
Expression similarity and time relevance associated with blood coagulation response during liver regeneration
Totally 107 genes could be characterized based on their similarity in expression as following: only up-, predominantly up-, only down-, predominantly down-, up-/down-regulated, involving 44, 8, 36, 13 and 6 genes, respectively (Figure 3). According to time relevance, they were classified into 15 groups, including 0.5 h, 1 h, 2 h, 4 and 6 h, 8 and 12 h, 16 h, 18 and 24 h, 30 and 42 h, 36 h, 48 h, 54 and 60 h, 66 and 72 h, 96 h, 120 h, 144 and 168 h, and the up- and down-regulation times were 9 and 6, 12 and 14, 16 and 3, 38 and 11, 25 and 15, 15 and 13, 35 and 40, 20 and 20, 23 and 18, 22 and 24, 34 and 24, 35 and 20, 11 and 10, 19 and 13, 27 and 22, respectively (Figure 3). The up-regulated expression genes were chiefly associated with promoting blood coagulation, and the down-regulated expression genes mostly associated with inhibiting blood coagulation.
Figure 3.
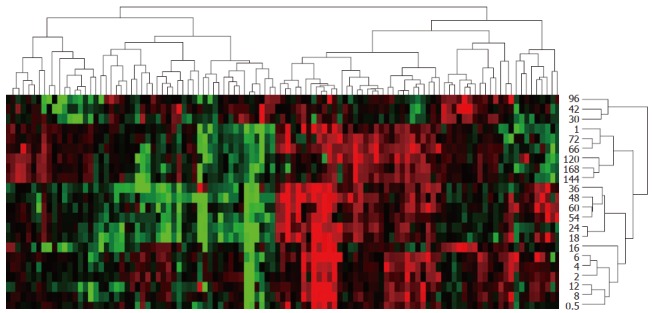
Expression similarity and time relevance clusters of 107 blood coagulation response-associated genes during liver regeneration. Detection data of Rat Genome 230 2.0 array were analyzed by H-clustering. Red represents up-regulated genes chiefly associated with promoting blood coagulation; Green represents down-regulated ones mainly associated with inhibiting blood coagulation; Black: No-sense in expression change. The upper and right trees respectively show expression similarity and time series clusters, by which the above genes were classified into 5 and 15 groups separately.
Expression patterns of the genes associated with blood coagulation response during liver regeneration
The 107 genes were categorized into 29 patterns, according to the changes in their expression. (1) up-regulation at one time point, i.e. 6, 16, 30, 66, 120 h after partial hepatectomy (Figure 4A), 8 genes involved; (2) up at two time points, i.e. 6 and 42 h, 30 and 42 h (Figure 4B), 2 genes involved; (3) up at three time points (Figure 4B), 2 genes involved; (4) up at multiple time points (Figure 4C), 5 genes involved; (5) up at one phase, i.e. 1-48, 4-6, 120-168 h (Figure 4D), 3 genes involved; (6) up at one time point/phase, i.e. 0.5 and 4-6 h, 24 and 66-72 h, 42 and 120-168 h, 66 and 120-168 h (Figure 4E), 4 genes involved; (7) up at one time point/two phases (Figure 4F), 2 genes involved; (8) up at one time point/three phases (Figure 4F), 1 gene involved; (9) up at two time points/one phase (Figure 4G), 5 genes involved; (10) up at two time points/phases (Figure 4H), 2 genes involved; (11) up at two time points/three phases (Figure 4H), 3 genes involved; (12) up at three time points/one phase (Figure 4I), 3 genes involved; (13) up at three time points/two phases (Figure 4I), 1 gene involved; (14) up at multiple time points/phases (Figure 4J), 3 genes involved; (15) down at one time point, i.e. 16, 30, 36, 42, 48, 72 h (Figure 4K), 8 genes involved; (16) down at two time points, i.e. 0.5 and 48 h, 1 and 72 h, 16 and 30 h, 16 and 96 h, 18 and 48 h, 18 and 54 h, 30 and 48 h, 30 and 96 h, 36 and 48 h (Figure 4L), 10 genes involved; (17) down at multiple time points (Figure 4M), 4 genes involved; (18) down at one phase, i.e. 6-12 h (Figure 4N), 1 gene involved; (19) down at two phases, i.e. 18-24 and 48-54 h (Figure 4N), 1 gene involved; (20) down at one time point/phase, i.e. 1 and 96-120 h, 18 and 120-144 h, 36 and 12-24 h, 96 and 16-24 h (Figure 4O), 4 genes involved; (21) down at one time point/two phases (Figure 4O), 1 gene involved; (22) down at one time point/three phases (Figure 4O), 1 gene involved; (23) down at two time points/one phase (Figure 4P), 3 genes involved; (24) down at two time points/phases (Figure 4P), 1 gene involved; (25) down at three time points/two phases (Figure 4Q), 1 gene involved; (26) down at multiple time points/phases (Figure 4Q), 1 gene involved; (27) predominantly up (Figure 4R), 8 genes involved; (28) predominantly down (Figure 4S), 13 genes involved; (29) similarly up/down (Figure 4T), 6 genes involved.
Figure 4.
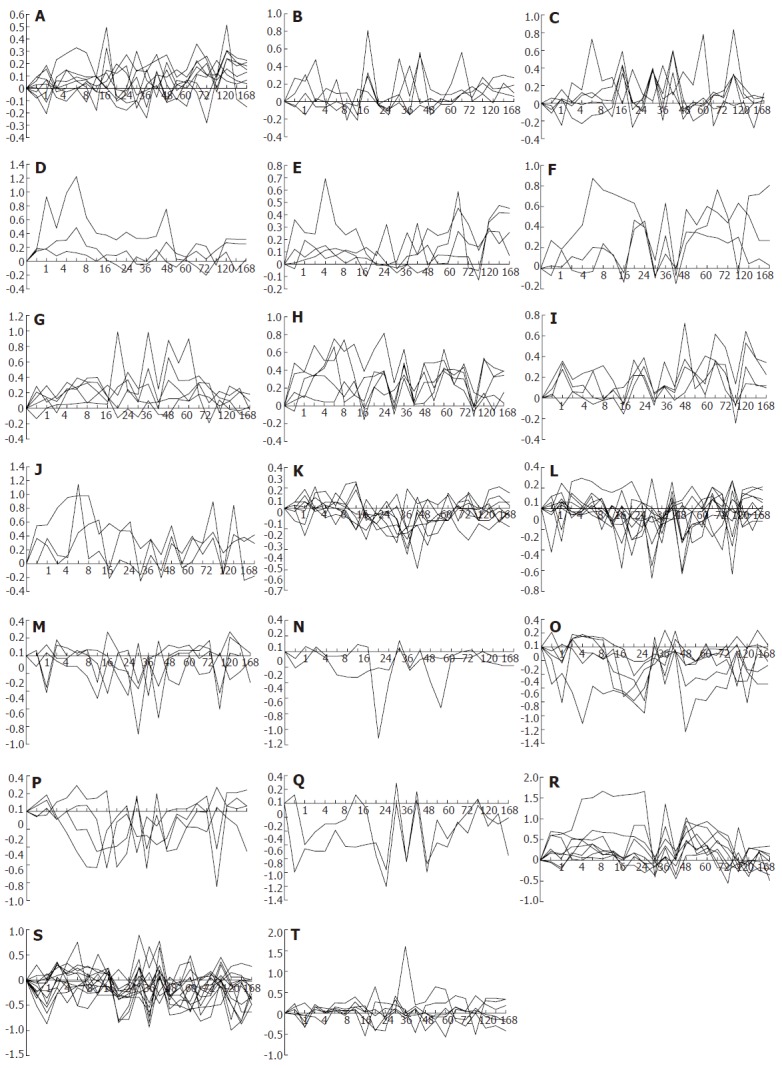
Twenty-nine gene expression patterns of 107 blood coagulation response-associated genes during liver regeneration. Expression patterns were obtained by the analysis of detection data of Rat Genome 230 2.0 array with Microsoft Excel. A-J: 44 up-regulated genes; K-Q: 36 down-regulated genes; R-T: 27 up/down-regulated genes. X-axis represents recovery time after partial hepatectomy (h); Y-axis shows logarithm ratio of the signal values of genes at each time point to control.
DISCUSSION
Blood coagulation plays an important physiological role. PH induces blood coagulation, which results from direct or indirect interaction between various proteins. Tissue factor (TF), associated with extrinsic blood coagulation, initiates extrinsic blood coagulation by acting on coagulation factor 7 (F7)[21]. Interleukin 6 (IL6) accelerates the transformation from fibrinogen to fibrin[22]. Twelve proteins including protease serine 2 (PRSS2) inhibit blood coagulation by suppressing the activities of TF and coagulation factors[23-25]. Seven proteins including annexin A 7 (ANXA7) accelerate the transformation from plasminogen to plasmin by inhibiting the activities of factor 10 (F10) and thrombin, repress blood coagulation, and induce migration and tube repair of blood vessel endothelial cells as well[26-28]. The meaningful expression profiles of the proteins encoding the above genes were the same or similar at some points while different at others, indicating that they may co-regulate extrinsic blood coagulation. Among them, il6 was up-regulated at 2-8, 18, 48, 60 and 96 h, and reached a peak at 96 h, which was 6.1 times higher than the control. This is generally in line with the result reported by Takatori et al[29]. tf was up-regulated at 16, 30 and 96 h during liver regeneration, and reached a peak at 16 h, which was 2.7 times higher than the control. prss2 was up-regulated mainly at 18, 36 and 54-60 h, and reached a peak at 36 h, which was 39.4 times higher than the control. anxa7 was up-regulated at 16, 30, 42 and 96 h, and reached a peak at 96 h, which was 6.8 times higher than the control. It suggests that these genes play a key role in blood coagulation during liver regeneration.
Moreover, six proteins including phospholipid scramblase 1 (PLSCR1), associated with intrinsic blood coagulation, promote blood coagulation by activating coagulation factors[30-33]. Amyloid beta precursor protein (APP) and beta-1,4-galactosyltransferase 1 (B4GALT1) accelerate blood coagulation[34,35]. Nine proteins including alpha-2-macroglobulin (A2M) inhibit blood coagulation by reducing thrombin and collagenase[36-39]. Five proteins including serine peptidase inhibitor clade E member 1 (SERPINE1) depress fibrinolysis by inhibiting the activities of plasma serine protease protein C and plasminogen activator[40,41]. Twelve proteins including plasminogen activator tissue (PLAT) inhibit blood coagulation by converting plasminogen to plasmin through cleaving the Arg-Val bond[42-44]. Thrombomodulin (THBD) restrains blood coagulation[45]. The meaningful expression profiles of the genes encoding the proteins mentioned above were the same -ness or similarity at some points while difference at others, suggesting that they may co-regulate intrinsic blood coagulation. Among them, thbd showed up-regulation during almost the whole LR, and had the highest abundance of 9.6 times higher than control at 6 h, which is consistent with the result reported by Takatori et al[46]. a2m was up-regulated at 0.5-24, 36 and 48-54 h, and reached a peak at 8 h, which was 46.2 times higher than the control. This is generally in conformity with the result reported by Scotte et al[47]. plscr1 was up-regulated at 2-24, 36 and 48-120 h during liver regeneration, and reached a peak at 6 h, which was 2.7 times higher than the control. app was up-regulated at multiple phases after PH, and reached a peak at 168 h, which was 3.1 times higher than the control. serpine1 was up-regulated at 1-48 h, and reached a peak at 6 h, which was 16.7 times higher than the control. plat was up-regulated at multiple phases after PH, and reached a peak at 66 h, which was 4.9 times higher than the control. It implies that these genes play a crucial role in blood coagulation during liver regeneration.
In summary, the expression changes of the genes associated with blood coagulation during liver regeneration have been investigated by high-throughput gene expression analysis and in long time range (0.5 h-7 d after PH) and at multiple time points (totally 23). It is preliminarily proved that PH can cause various physiological responses including blood coagulation, and that Rat Genome 230 2.0 array is a useful tool for analysis of the blood coagulation responses at gene transcriptional level. However, the processes of DNA→mRNA→protein are influenced by many factors including protein interactions. Therefore, our results need to be further analyzed using techniques, such as Northern blotting, protein chip, RNA interference, protein-interaction.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30270673
S- Editor Liu Y L- Editor Zhu LH E- Editor Bi L
References
- 1.Østerud B, Bjørklid E. Sources of tissue factor. Semin Thromb Hemost. 2006;32:11–23. doi: 10.1055/s-2006-933336. [DOI] [PubMed] [Google Scholar]
- 2.Machado FR, Silva E. Coagulation and sepsis. Endocr Metab Immune Disord Drug Targets. 2006;6:175–182. doi: 10.2174/187153006777442369. [DOI] [PubMed] [Google Scholar]
- 3.Celikel R, McClintock RA, Roberts JR, Mendolicchio GL, Ware J, Varughese KI, Ruggeri ZM. Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301:218–221. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]
- 4.Khrenov AV, Ananyeva NM, Saenko EL. Role of the B domain in proteolytic inactivation of activated coagulation factor VIII by activated protein C and activated factor X. Blood Coagul Fibrinolysis. 2006;17:379–388. doi: 10.1097/01.mbc.0000233368.95733.3c. [DOI] [PubMed] [Google Scholar]
- 5.Wu SY, Wang ZY, Dong NZ, Zhang W, Bai X, Ruan CG. [A novel genetic defect in a Chinese family with inherited coagulation factor XIII deficiency] Zhonghua Xueyexue Zazhi. 2006;27:145–149. [PubMed] [Google Scholar]
- 6.Higgins GM, Anderson RM. Experimental pathology of the liver I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 7.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 8.Michalopoulos GK, DeFrances M. Liver regeneration. Adv Biochem Eng Biotechnol. 2005;93:101–134. doi: 10.1007/b99968. [DOI] [PubMed] [Google Scholar]
- 9.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 10.Xu CS, Chang CF, Yuan JY, Li WQ, Han HP, Yang KJ, Zhao LF, Li YC, Zhang HY, Rahman S, et al. Expressed genes in regenerating rat liver after partial hepatectomy. World J Gastroenterol. 2005;11:2932–2940. doi: 10.3748/wjg.v11.i19.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada K, Ueshima S, Imano M, Kataoka K, Matsuo O. The regulation of liver regeneration by the plasmin/alpha 2-antiplasmin system. J Hepatol. 2004;40:110–116. doi: 10.1016/j.jhep.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Dransfeld O, Gehrmann T, Köhrer K, Kircheis G, Holneicher C, Häussinger D, Wettstein M. Oligonucleotide microarray analysis of differential transporter regulation in the regenerating rat liver. Liver Int. 2005;25:1243–1258. doi: 10.1111/j.1478-3231.2005.01158.x. [DOI] [PubMed] [Google Scholar]
- 13.Knepp JH, Geahr MA, Forman MS, Valsamakis A. Comparison of automated and manual nucleic acid extraction methods for detection of enterovirus RNA. J Clin Microbiol. 2003;41:3532–3536. doi: 10.1128/JCM.41.8.3532-3536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nuyts S, Van Mellaert L, Lambin P, Anné J. Efficient isolation of total RNA from Clostridium without DNA contamination. J Microbiol Methods. 2001;44:235–238. doi: 10.1016/s0167-7012(01)00219-6. [DOI] [PubMed] [Google Scholar]
- 15.Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Roden J, Shapiro BE, Wold BJ, Bhatia S, Forman SJ, Bhatia R. Reproducibility, fidelity, and discriminant validity of mRNA amplification for microarray analysis from primary hematopoietic cells. J Mol Diagn. 2005;7:48–56. doi: 10.1016/S1525-1578(10)60008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hood L. Leroy Hood expounds the principles, practice and future of systems biology. Drug Discov Today. 2003;8:436–438. doi: 10.1016/s1359-6446(03)02710-7. [DOI] [PubMed] [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner T. Cluster analysis and promoter modelling as bioinformatics tools for the identification of target genes from expression array data. Pharmacogenomics. 2001;2:25–36. doi: 10.1517/14622416.2.1.25. [DOI] [PubMed] [Google Scholar]
- 20.Yue H, Eastman PS, Wang BB, Minor J, Doctolero MH, Nuttall RL, Stack R, Becker JW, Montgomery JR, Vainer M, et al. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 2001;29:E41–E41. doi: 10.1093/nar/29.8.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez K, McVey JH. Tissue factor initiated blood coagulation. Front Biosci. 2006;11:1349–1359. doi: 10.2741/1888. [DOI] [PubMed] [Google Scholar]
- 22.Mannila MN, Eriksson P, Ericsson CG, Hamsten A, Silveira A. Epistatic and pleiotropic effects of polymorphisms in the fibrinogen and coagulation factor XIII genes on plasma fibrinogen concentration, fibrin gel structure and risk of myocardial infarction. Thromb Haemost. 2006;95:420–427. doi: 10.1160/TH05-11-0777. [DOI] [PubMed] [Google Scholar]
- 23.Shan J, Baguinon M, Zheng L, Krishnamoorthi R. Expression, refolding, and activation of the catalytic domain of human blood coagulation factor XII. Protein Expr Purif. 2003;27:143–149. doi: 10.1016/s1046-5928(02)00608-3. [DOI] [PubMed] [Google Scholar]
- 24.Shrader WD, Kolesnikov A, Burgess-Henry J, Rai R, Hendrix J, Hu H, Torkelson S, Ton T, Young WB, Katz BA, et al. Factor VIIa inhibitors: gaining selectivity within the trypsin family. Bioorg Med Chem Lett. 2006;16:1596–1600. doi: 10.1016/j.bmcl.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 25.de Pont AC, Moons AH, de Jonge E, Meijers JC, Vlasuk GP, Rote WE, Büller HR, van der Poll T, Levi M. Recombinant nematode anticoagulant protein c2, an inhibitor of tissue factor/factor VIIa, attenuates coagulation and the interleukin-10 response in human endotoxemia. J Thromb Haemost. 2004;2:65–70. doi: 10.1111/j.1538-7836.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 26.Park JE, Lee DH, Lee JA, Park SG, Kim NS, Park BC, Cho S. Annexin A3 is a potential angiogenic mediator. Biochem Biophys Res Commun. 2005;337:1283–1287. doi: 10.1016/j.bbrc.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Peterson EA, Sutherland MR, Nesheim ME, Pryzdial EL. Thrombin induces endothelial cell-surface exposure of the plasminogen receptor annexin 2. J Cell Sci. 2003;116:2399–2408. doi: 10.1242/jcs.00434. [DOI] [PubMed] [Google Scholar]
- 28.Battistelli S, Vittoria A, Cappelli R, Stefanoni M, Roviello F. Protein S in cancer patients with non-metastatic solid tumours. Eur J Surg Oncol. 2005;31:798–802. doi: 10.1016/j.ejso.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Fulop AK, Pocsik E, Brozik M, Karabelyos C, Kiss A, Novak I, Szalai C, Dobozy O, Falus A. Hepatic regeneration induces transient acute phase reaction: systemic elevation of acute phase reactants and soluble cytokine receptors. Cell Biol Int. 2001;25:585–592. doi: 10.1006/cbir.2000.0715. [DOI] [PubMed] [Google Scholar]
- 30.Sims PJ, Wiedmer T. Unraveling the mysteries of phospholipid scrambling. Thromb Haemost. 2001;86:266–275. [PubMed] [Google Scholar]
- 31.Katsanis N, Venable S, Smith JR, Lupski JR. Isolation of a paralog of the Doyne honeycomb retinal dystrophy gene from the multiple retinopathy critical region on 11q13. Hum Genet. 2000;106:66–72. doi: 10.1007/s004390051011. [DOI] [PubMed] [Google Scholar]
- 32.Louvain-Quintard VB, Bianchini EP, Calmel-Tareau C, Tagzirt M, Le Bonniec BF. Thrombin-activable factor X re-establishes an intrinsic amplification in tenase-deficient plasmas. J Biol Chem. 2005;280:41352–41359. doi: 10.1074/jbc.M507846200. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi M, Matsushita T, Mackman N, Ito M, Adachi T, Katsumi A, Yamamoto K, Takeshita K, Kojima T, Saito H, et al. Fatal thrombosis of antithrombin-deficient mice is rescued differently in the heart and liver by intercrossing with low tissue factor mice. J Thromb Haemost. 2006;4:177–185. doi: 10.1111/j.1538-7836.2005.01679.x. [DOI] [PubMed] [Google Scholar]
- 34.Dhainaut JF, Marin N, Mignon A, Vinsonneau C. Hepatic response to sepsis: interaction between coagulation and inflammatory processes. Crit Care Med. 2001;29:S42–S47. doi: 10.1097/00003246-200107001-00016. [DOI] [PubMed] [Google Scholar]
- 35.Hansske B, Thiel C, Lübke T, Hasilik M, Höning S, Peters V, Heidemann PH, Hoffmann GF, Berger EG, von Figura K, et al. Deficiency of UDP-galactose: N-acetylglucosamine beta-1,4-galactosyltransferase I causes the congenital disorder of glycosylation type IId. J Clin Invest. 2002;109:725–733. doi: 10.1172/JCI14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paredes N, Xu L, Berry LR, Chan AK. The effects of chemotherapeutic agents on the regulation of thrombin on cell surfaces. Br J Haematol. 2003;120:315–324. doi: 10.1046/j.1365-2141.2003.03971.x. [DOI] [PubMed] [Google Scholar]
- 37.Varani K, Portaluppi F, Gessi S, Merighi S, Ongini E, Belardinelli L, Borea PA. Dose and time effects of caffeine intake on human platelet adenosine A(2A) receptors : functional and biochemical aspects. Circulation. 2000;102:285–289. doi: 10.1161/01.cir.102.3.285. [DOI] [PubMed] [Google Scholar]
- 38.Rohlena J, Kolkman JA, Boertjes RC, Mertens K, Lenting PJ. Residues Phe342-Asn346 of activated coagulation factor IX contribute to the interaction with low density lipoprotein receptor-related protein. J Biol Chem. 2003;278:9394–9401. doi: 10.1074/jbc.M209097200. [DOI] [PubMed] [Google Scholar]
- 39.Mosnier LO, Griffin JH. Protein C anticoagulant activity in relation to anti-inflammatory and anti-apoptotic activities. Front Biosci. 2006;11:2381–2399. doi: 10.2741/1977. [DOI] [PubMed] [Google Scholar]
- 40.Sabovic M, Salobir B, Preloznik Zupan I, Bratina P, Bojec V, Buturovic Ponikvar J. The influence of the haemodialysis procedure on platelets, coagulation and fibrinolysis. Pathophysiol Haemost Thromb. 2005;34:274–278. doi: 10.1159/000093107. [DOI] [PubMed] [Google Scholar]
- 41.Nagai N, Habuchi H, Esko JD, Kimata K. Stem domains of heparan sulfate 6-O-sulfotransferase are required for Golgi localization, oligomer formation and enzyme activity. J Cell Sci. 2004;117:3331–3341. doi: 10.1242/jcs.01191. [DOI] [PubMed] [Google Scholar]
- 42.Ji X, Meng R, Zhou J, Ling F, Jia J. Dynamic change of coagulation and anticoagulation markers of patients with acute cerebral infarction during intravenous urokinase thrombolysis. Neurol Res. 2006;28:46–49. doi: 10.1179/016164106X91861. [DOI] [PubMed] [Google Scholar]
- 43.Oszajca K, Szemraj J, Bartkowiak J. [The influence of nitric oxide on the regulation of plasminogen activator inhibitor type 1 and tissue-type plasminogen activator expression] Postepy Biochem. 2005;51:407–413. [PubMed] [Google Scholar]
- 44.Shariat-Madar Z, Mahdi F, Warnock M, Homeister JW, Srikanth S, Krijanovski Y, Murphey LJ, Jaffa AA, Schmaier AH. Bradykinin B2 receptor knockout mice are protected from thrombosis by increased nitric oxide and prostacyclin. Blood. 2006;108:192–199. doi: 10.1182/blood-2006-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita T, Sekiguchi A, Iwasaki YK, Sagara K, Hatano S, Iinuma H, Aizawa T, Fu LT. Thrombomodulin and tissue factor pathway inhibitor in endocardium of rapidly paced rat atria. Circulation. 2003;108:2450–2452. doi: 10.1161/01.CIR.0000102969.09658.F2. [DOI] [PubMed] [Google Scholar]
- 46.Takatori M, Iwabuchi S, Ro S, Murayama M, Maeyama S, Uchikoshi T, Nakano M, Ishii H. Increased serum levels and sinusoidal expression of thrombomodulin in acute liver damage. Thromb Res. 1999;93:113–120. doi: 10.1016/s0049-3848(98)00167-4. [DOI] [PubMed] [Google Scholar]
- 47.Scotté M, Masson S, Lyoumi S, Hiron M, Ténière P, Lebreton JP, Daveau M. Cytokine gene expression in liver following minor or major hepatectomy in rat. Cytokine. 1997;9:859–867. doi: 10.1006/cyto.1997.0273. [DOI] [PubMed] [Google Scholar]


